Abstract
Aedes aegypti is one of the species most favored by changes in the environment caused by urbanization. Its abundance increases rapidly in the face of such changes, increasing the risk of disease transmission. Previous studies have shown that mosquito species that have adapted to anthropogenic environmental changes benefit from urbanization and undergo population expansion. In light of this, we used microsatellite markers to explore how urbanization processes may be modulating Ae. aegypti populations collected from three areas with different levels of urbanization in the city of São Paulo, Brazil. Specimens were collected at eleven sites in three areas with different degrees of urbanization in the city of São Paulo: conserved, intermediate and urbanized. Ten microsatellite loci were used to characterize the populations from these areas genetically. Our findings suggest that as urbanized areas grow and the human population density in these areas increases, Ae. aegypti populations undergo a major population expansion, which can probably be attributed to the species’ adaptability to anthropogenic environmental changes. Our findings reveal a robust association between, on the one hand, urbanization processes and densification of the human population and, on the other, Ae. aegypti population structure patterns and population expansion. This indicates that this species benefits from anthropogenic effects, which are intensified by migration of the human population from rural to urban areas, increasing the risk of epidemics and disease transmission to an ever-increasing number of people.
Background
Urbanization continues to increase as more and more people migrate from rural to urban areas. Indeed, there are now more people living in cities than in rural areas. This phenomenon is responsible for the extinction of species that are not adapted to the urban environment and is especially important for the epidemiology of vector-borne diseases, as there is a clear association between reduced species richness in urban areas and an increased incidence of these diseases [1,2]. This decrease in species richness can in turn be a result of human changes to the environment and the consequent increase in abundance of the few mosquito species that are adapted to the urbanized environment, very often with epidemiological consequences [2–4]. Mosquito surveys performed in urban parks in the metropolitan region of São Paulo, Brazil, indicated high abundance of species with epidemiological relevance, including the dengue vector, Aedes aegypti [5–7].
Dengue is a tropical disease caused by a flavivirus transmitted by Aedes mosquitoes. It endangers over 2 billion people of the world's population, causing approximately 390 million infections a year [8]. Ae. aegypti [9], which can be found in tropical and subtropical regions, is the main vector of dengue and also one of the mosquitoes that is best adapted to the urban environment; it can complete its entire life cycle within a human domicile, laying eggs in artificial breeding sites and blood feeding on human hosts [10].
For these reasons, Ae. aegypti is one of the most favored species when a rural area is urbanized. In this situation there is a rapid increase in its abundance and a consequent increase in the risk of dengue transmission [11–13]. The dynamics of dengue epidemics are fueled by several factors that are usually present in developing countries, such as unplanned urbanization, chaotic population growth and ineffective public health systems and vector surveillance [2,12,14,15].
Also, Ae. aegypti is considered the primary vector for the Zika and chikungunya viruses, as well as several other arboviruses, and is responsible for numerous outbreaks of insect-borne diseases worldwide [16,17]. Identifying the genetic structure of urban populations of Ae. aegypti, especially on a microgeographic scale, can lead to a better understanding of how human modifications to the physical environment are modulating population structure in this mosquito and the implications of this modulation for disease transmission and vector control.
Microsatellites are highly polymorphic neutral markers often used in genetic population studies [18]. Recently, their use has provided valuable information on microgeographic population structures of insects in urbanized areas [19–21]. Using DNA microsatellite loci, Multini et al. [19] found that Aedes fluviatilis populations in São Paulo, Brazil, had undergone population expansion primarily as a result of urban growth, highlighting the epidemiological significance of the association between urbanization processes and population structuring in vector mosquitoes.
In light of the high abundance of Ae. aegypti in the city of São Paulo and the major role played by this species in the transmission of the dengue, Zika and chikungunya viruses, this study used microsatellite markers to explore how urbanization processes can modulate populations of Ae. aegypti collected in three areas with different levels of urbanization in the city of São Paulo.
Methods
Specimen collection
Ae. aegypti mosquitoes were collected from eleven sites, no more than 30 km apart, in three areas with different levels of urbanization in the city of São Paulo, Brazil.
Conserved areas (CON): five municipal parks consisting of large green areas (with more than 97% of vegetal cover) open to the public from 5:00 am to 8:00 pm. No biological or chemical measures are used to control mosquitoes in the parks, which are home to wild birds, reptiles and mammals. Intermediate areas (INT): four collection sites on the University of São Paulo Armando de Salles Oliveira campus (with 80% of vegetal cover). The campus extends over 3,648,944.40 m2, of which some 800,000 m2 has been built on. Over 100,000 people travel through, visit or work or study on the campus every day. Urbanized areas (URB): two collection sites on the University of São Paulo health sciences campus, which is in a highly urbanized, densely populated area extending over 83,050.82 m2 (with less than 3% of vegetation cover). Much of this area (79.923,72 m2) has been built on (Table 1).
Table 1. Aedes aegypti collection sites and collection data.
| Collection Site | Code | Coordinates | N* | Collection Year |
|---|---|---|---|---|
| Anhanguera Park | CON-1 | 23°24′54”S, 46°47′60” W | 13 | 2013 |
| Eucalipto Park | CON-2 | 23°36’54“S, 46°45’18”W | 30 | 2012 |
| Independência Park | CON-3 | 26°35’60”S, 46°36’18”W | 26 | 2015 |
| Piqueri Park | CON-4 | 23°31’30“S, 46°35’30”W | 30 | 2013 |
| Previdência Park | CON-5 | 23°35’60“S, 46° 43’30”W | 30 | 2015 |
| University of São Paulo Student Accommodations | INT-1 | 23°33’18”S, 46°43’30”W | 30 | 2014 |
| School of Communication and Arts | INT-2 | 23°33’18“S, 46°43’30”W | 29 | 2014 |
| Physics Institute | INT-3 | 23°33’54“S, 46°44’60” W | 30 | 2014 |
| Veterinary School | INT-4 | 23°33’54”S, 46°44’60”W | 29 | 2014 |
| School of Public Health | URB-1 | 23°33’18”S, 46°40’30”W | 30 | 2015 |
| Medical School | URB-2 | 23°33’18”S, 46°40’30”W | 30 | 2015 |
*Number of females used
Mosquito collections were performed monthly from 2012 to 2015. Adult mosquitoes were collected outdoors with portable battery-powered aspirators [22], and immature mosquitoes were collected with dippers and randomly selected for further analysis to avoid testing siblings. Immature specimens were kept under laboratory conditions and fed with fish food (Tetra BettaMin) until they developed into adults. Specimens were identified with the aid of taxonomic keys [23] and stored at -20°C until DNA was extracted.
The study was approved by the University of São Paulo Research Ethics Committee, and collection permits were provided by the City of São Paulo Department of the Environment and Green Areas.
DNA extraction and polymerase chain reaction
DNA extractions were performed with the DNEasy Blood and Tissue Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. Polymerase chain reactions (PCRs) were carried out individually with ten microsatellite primers originally designed by Chambers et al. [24] and Slotman et al. [25] (Table 2) using the SuperMix PCR kit (Invitrogen, Carlsbad, CA, USA).
Table 2. Microsatellite loci amplified in Aedes aegypti.
| Locus | Sequences 5’-3’ | Repetitive motif | T (°C) | Size range (bp) | References |
|---|---|---|---|---|---|
| A10 | F: AATCGTGACGCGTCTTTTG | CT | 60 | 232–242*(232–239) | Chambers et al. [24] |
| R: TAACTGCATCGAGGGAAACC | |||||
| B07 | F: CAAACAACGAACTGCTCACG | GA | 60 | 100–272*(157–183) | Chambers et al. [24] |
| R: TCGCAATTTCAACAGGTAGG | |||||
| AT1 | F: CGTCGACGTTATCTCCTTGTT | AT | 55 | 134–170*156–174) | Slotman et al. [25] |
| R: GGACCGGAAAGACACAGACA | |||||
| AG7 | F: CGTGCGAGTGAATGAGAGAC | GA | 55 | 112–190*(153–185) | Slotman et al. [25] |
| R: CATCCTCTCATCAGCTTCTAATAAA | |||||
| AG2 | F: TCCCCTTTCAAACCTAATGG | AG | 55 | 96–152*(115–178) | Slotman et al. [25] |
| R: TTTGCCCTCGTATGCTCTCT | |||||
| AG5 | F: TGATCTTGAGAAGGCATCCA | AG | 55 | 140–164*(170–180) | Slotman et al. [25] |
| R: CGTTATCCTTTCATCACTTGTTTG | |||||
| AC7 | F: TCGGCAAATTACCACAAACA | CA | 55 | 106–130*(129–143) | Slotman et al. [25] |
| R: CATTGGACTCGCTATAACACACA | |||||
| AC5 | F: TGGATTGTTCTTAACAAACACGAT | CA | 55 | 104–156*(149–163) | Slotman et al. [25] |
| R: CGATCTCACTACGGGTTTCG | |||||
| AC1 | F: TCCGGTGGGTTAAGGATAGA | CA | 55 | 140–196*(193–209) | Slotman et al. [25] |
| R: ACTTCACGCTCCAGCAATCT | |||||
| AG1 | F: AATCCCCACACAAACACACC | AG | 55 | 90–106*(113–129) | Slotman et al. [25] |
| R: GGCCGTGGTGTTACTCTCTC |
T = annealing temperature, bp = base pairs
*Values found in the Aedes aegypti tested. In parentheses, size range found in the original study.
Ten microsatellite primers were labeled with a fluorescent dye (FAM, HEX or NED) (Thermo Fisher Scientific, Waltham, MA, USA), and the PCRs were carried out according to the manufacture’s protocol using an E6331000025 Eppendorf Thermocycler (Mastercycler Nexus Gradient, Eppendorf, Hamburg, Germany). PCR products were visualized on a 1% agarose gel stained with GelRed™ (Biotium, Hayward, CA, USA).
PCR products were diluted 1:7 by mixing 3 μL of each product labeled with a different dye with 21 μL of Ultra-Pure Water (Applied Biosystems, Foster City, CA, USA) to a final volume of 30 μL. Next, 2 μL of the diluted PCR products were mixed with 8.925 μL of Hi-Di formamide (Applied Biosystems, Foster City, CA, USA) and 0.075 μL of the size standard GeneScan-500 ROX (Applied Biosystems, Foster City, CA, USA) to a final volume of 11 μL. The samples were submitted to the University of São Paulo Center for Human Genome Studies and size-sorted in an ABI 3730 automatic sequencer (Applied Biosystems, Foster City, CA, USA). Fragments were sized with GeneMarker (v1.85 SoftGenetics, State College, PA, USA).
Genetic analysis
Allele frequency, observed heterozygosity (HO) and expected heterozygosity (HE), deviations from the Hardy-Weinberg equilibrium, inbreeding coefficient (FIS) and pairwise FST with significance values (after 10,000 permutations) were calculated with Arlequin (v3.5) [26]. Linkage disequilibrium (using Bonferroni-corrected P-values) and number of migrants (Nm) were calculated using Genepop (v4.2 http://genepop.curtin.edu.au/) [27]. Allelic richness and private allelic richness were calculated using HP-Rare (v1.0) [28].
The probability of null alleles, genetic heterogeneity (FST) and Cavalli-Sforza and Edwards chord distance (taking into account the null allele bias) were calculated per locus per population with FreeNa [29]. A Mantel test was used to compare both FST values using Past (v2.17c) [30].
A dendrogram based on the Cavalli-Sforza and Edwards chord distance was constructed using Statistica (v7.0) [31]. A linear correlation analyses between FST/(1-FST) and geographic distance (km) was performed using Past (v2.17c).
Bayesian analysis was performed with Structure (v2.3.3) [32], and the number of clusters (K) representing the best fit for the data was calculated with Structure Harvester (Web v0.6.94) [33]. Bottleneck (v1.2.2) was used to test for heterozygosity deficiency using the Stepwise Mutation Model (SMM) [34–36], a signature of population expansion (HE<HEQ), or heterozygosity excess, a signature of a bottleneck event (HE>HEQ), where HE = the expected heterozygosity based on allele frequencies and HEQ = the expected heterozygosity based on observed alleles.
Results
Marker assessment
Linkage disequilibrium for the 495 possible tests, per locus per population, was found only between loci A10 and AC7, AG2 and AC5, AT1 and B07, AT1 and AG2 and AT1 and AG7 after Bonferroni correction (P = 0.0001). These results are not considered significant because there were no repetitions of pairs of loci combinations across the populations tested.
The estimate for the probability of null alleles was high (>0.2) for locus AC1 in five populations, AC5 in three, AC7 in two, AG1 in seven, AG2 in two, AG5 in one, AG7 in one, AT1 in two and B07 in one. Locus A10 had a low probability of null alleles (<0.2) (S1 Table). Allelic richness ranged from 4.8 (CON-1) to 6.38 (CON-4), and private allelic richness from 0.13 (CON-1) to 0.43 (CON-5) (S2 Table). Hardy-Weinberg equilibrium tests were performed for each locus and population. HE was higher than HO in 103 of 110 possible tests, and average FIS was 0.35 (S3 Table).
Genetic differentiation
The results for FST indicated significant genetic structure between populations. Values ranged from 0.00629 to 0.11169, and only 7% of the values (4 out of 55) were not significant. When the potential bias due to null alleles was taken into account, the results ranged from 0.002133 to 0.093649. There were no statistically significant differences between the corrected and uncorrected FST values (r = 0.986, P <0.0001) (Table 3).
Table 3. Pairwise FST* estimates for Aedes aegypti populations.
| CON-1 | CON-2 | CON-3 | CON-4 | CON-5 | INT-1 | INT-2 | INT-3 | INT-4 | URB-1 | URB-2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CON-1 | - | 0.03702 | 0.01002 | 0.02572 | 0.02288 | 0.04623 | 0.03654 | 0.05234 | 0.03347 | 0.03411 | 0.08131 |
| CON-2 | 0.05607 | - | 0.02190 | 0.02702 | 0.03122 | 0.04340 | 0.06356 | 0.09364 | 0.03765 | 0.05332 | 0.07913 |
| CON-3 | 0.02546 | 0.02873 | - | 0.00505 | 0.00837 | 0.02641 | 0.01798 | 0.04967 | 0.01815 | 0.02624 | 0.06727 |
| CON-4 | 0.03912 | 0.03013 | 0.00629 | - | 0.00213 | 0.02540 | 0.01845 | 0.03103 | 0.01027 | 0.03539 | 0.05266 |
| CON-5 | 0.04003 | 0.03813 | 0.01320 | 0.00663 | - | 0.02117 | 0.01910 | 0.03939 | 0.01902 | 0.02437 | 0.03992 |
| INT-1 | 0.06617 | 0.05217 | 0.03149 | 0.02723 | 0.02787 | - | 0.03339 | 0.06738 | 0.05008 | 0.05172 | 0.07275 |
| INT-2 | 0.05498 | 0.08431 | 0.02893 | 0.02648 | 0.02762 | 0.04627 | - | 0.04515 | 0.02662 | 0.03921 | 0.06233 |
| INT-3 | 0.07058 | 0.11169 | 0.05802 | 0.04074 | 0.05214 | 0.08097 | 0.05280 | - | 0.05777 | 0.06322 | 0.08800 |
| INT-4 | 0.04804 | 0.05050 | 0.02227 | 0.01768 | 0.02562 | 0.06004 | 0.03408 | 0.06863 | - | 0.03269 | 0.05180 |
| URB-1 | 0.04879 | 0.07165 | 0.03582 | 0.04371 | 0.03371 | 0.06853 | 0.05312 | 0.07553 | 0.03861 | - | 0.06039 |
| URB-2 | 0.10940 | 0.10668 | 0.09181 | 0.07164 | 0.05299 | 0.09418 | 0.07653 | 0.11107 | 0.06932 | 0.07741 | - |
*Below the diagonal: FST values without correction for null alleles. Significant values are in bold.
Above the diagonal: FreeNA corrected FST values.
Number of migrants between the populations was 3.94766 per population per generation based on 30 specimens per population, showing a low degree of allelic similarity. There was no correlation between genetic and geographical distances (r = 0,18; r2 = 0,03; P = 0,18) between populations.
Genetic distance
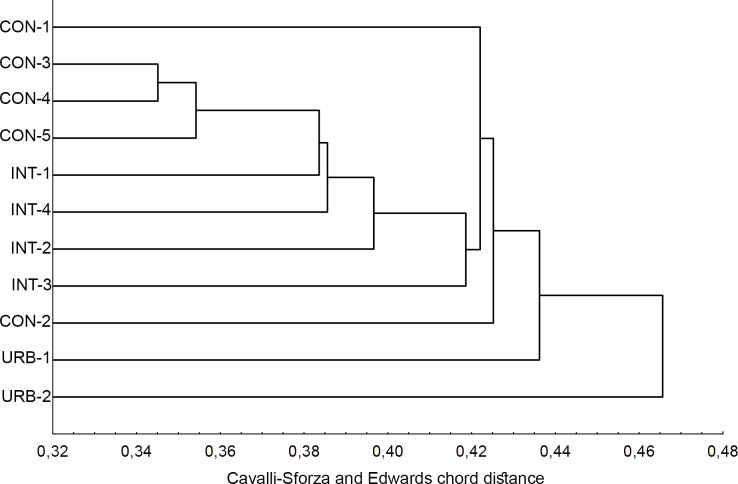
The dendrogram based on Cavalli-Sforza and Edwards chord distance was consistent with the level of urbanization in the areas where the specimens were collected. All eleven populations from the three urban areas grouped separately with no overlapping between populations from different areas. The two populations from highly urbanized areas, URB-1 and URB-2, were the most distinct, followed by two from the conserved area (CON-2 and CON-1, in that order). The CON-3, CON-4 and CON-5 populations segregated close to each other, as did the INT-1, INT-2, INT-3 and INT-4 populations (Fig 1).
Fig 1. Genetic-distance dendrogram for Aedes aegypti based on Cavalli-Sforza and Edwards chord distance.
Bayesian cluster analysis
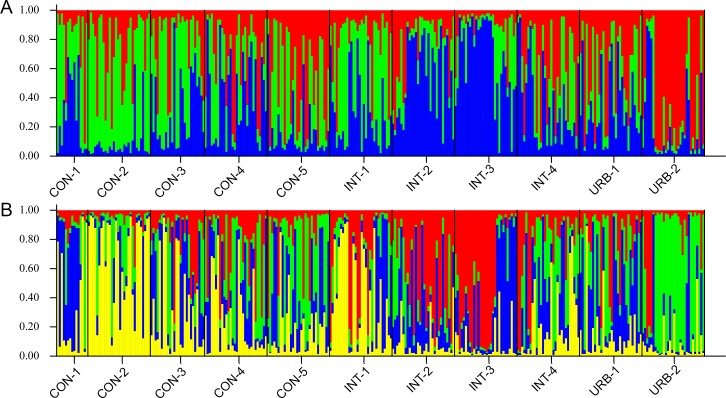
The method described by Evanno et al. [33] was used with the results of the Bayesian analysis to identify the K value that best represents the number of genetic groups in the populations tested. The K value found in this way was 4 (S1 Fig). Two subsequent analyses with different K values were performed. K = 3 indicated that the segregation of the populations depended on the area where the specimens were collected. For the CON populations, the color green predominated; for the INT populations, blue; and for the URB populations, red (Fig 2A). K = 4 showed that the URB-2 population belongs to a different genetic group than the remaining populations; this group is represented by the color yellow. The CON and INT populations formed a distinct genetic cluster in which green and red, respectively, predominated (Fig 2B).
Fig 2. Bayesian analysis of population structure for all Aedes aegypti populations showing the subdivision of individuals for K = 3 (A) and K = 4 (B).
Each of the 311 individuals from eleven populations is represented by a vertical line divided into different colored segments. The length of each segment represents the probability of the individual belonging to the genetic cluster represented by that color.
Population expansion analysis
Tests to assess heterozygosity deficiency using the SMM showed that there was a significant degree of deficiency in 5 of the 11 populations tested (P<0.05). Although not observed in any of the CON populations, heterozygosity deficiency was detected in all the INT and URB populations (except for INT-4), indicating a recent population expansion (Table 4) in the more urbanized areas. In a further analysis in which all 310 specimens were considered a metapopulation, all 10 loci displayed heterozygosity deficiency (HE<HEQ) with a highly significant P-value (0.00015). This result supports the hypothesis that the Ae. aegypti populations in this study have suffered a major recent population expansion.
Table 4. Tests to identify heterozygosity deficiency in Aedes aegypti.
| CON-1 | CON-2 | CON-3 | CON-4 | CON-5 | INT-1 | INT-2 | INT-3 | INT-4 | URB-1 | URB-2 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SMM | HE < HEQ | 4 | 5 | 5 | 7 | 6 | 8 | 8 | 8 | 6 | 8 | 8 |
| HE > HEQ | 6 | 5 | 5 | 3 | 4 | 2 | 2 | 2 | 4 | 2 | 2 | |
| P (HE < HEQ) | 0.61024 | 0.36864 | 0.39316 | 0.06188 | 0.17758 | 0.01291 | 0.01421 | 0.01439 | 0.18442 | 0.01456 | 0.01583 |
Number of loci with heterozygosity excess (HE) and number of loci with expected heterozygosity excess based on the number of observed alleles (HEQ) under the SMM. Significant P-values (<0.05) for heterozygosity deficit in bold.
Discussion
The global distribution of Ae. aegypti is intimately connected with man-made changes to the physical environment. While urbanization modifies the ecosystem to create an environment that is more suitable for one particular species, Homo sapiens [1], the mosquito Ae. aegypti also benefits from these changes. This situation, allied to the lack of sanitation and sewage treatment and inadequate epidemiologic surveillance common in developing countries, results in ideal conditions for a major increase in the abundance of this species, increasing the risk of disease transmission.
Our findings suggest that the Ae. aegypti populations in this study have undergone a major population expansion, probably as a result of increased urbanization and human population density as well as the species’ adaptability to anthropogenic changes in the physical environment. When urbanized areas expand and human population density increases, the availability of human hosts and breeding sites increases, while larval competition and the number of natural predators decrease. This agrees with the findings of Multini et al. [19] for Ae. fluviatilis, Donnelly et al. [36] for Anopheles arabiensis and Anopheles gambiae, Michel [37] for Anopheles funestus and Mirabello & Conn [38] for Anopheles darlingi.
The Ae. aegypti populations from more preserved ecosystems (CON) did not show signs of population expansion, unlike the INT and URB populations, corroborating the hypothesis that the urban environment benefits this species and that it is highly adapted to human changes to the landscape.
Bayesian analysis showed that structuring has been occurring in Ae. aegypti populations and is correlated with the level of urbanization, supported by the lack of correlation between genetic and geographic distances. This can be explained by the fact that this species is well adapted to the urban environment and therefore does not have to actively seek new areas in order to find hosts or breeding sites as these are widely available in cities regardless of the level of urbanization and are plentiful even in the more conserved environments, such as urban parks.
A possible hypothesis to explain this situation is that after Ae. aegypti was reintroduced in Brazil in the 1970’s [39], the growth of the city favored its expansion, preventing the genetic structuring of its populations. However, once urbanization in a given area was complete and these populations reached their peak abundance, genetic structuring restarted, resulting in the population structures observed here. This hypothesis is corroborated by the low Nm between populations despite their geographic proximity as revealed by the dendrogram in Fig 1 and the Bayesian analysis.
Further, the domiciliation of Ae. aegypti, which resulted from anthropogenic impacts on its evolution may be closely associated with microevolutionary processes [40], such as the structuring found in this study. The results for allelic and private allelic richness for Ae. aegypti were consistent with the results of previous studies of this species [20,39,41], indicating that the loci used here revealed the genuine allelic richness for the populations in this study.
Our finding of urbanization-dependent structuring is of great significance for the dynamics of disease transmission, particularly because there appears to be low Nm between the populations collected in areas with different levels of urbanization in this study. Furthermore, with increasing urbanization the demographic expansion of Ae. aegypti populations tends to intensify since this species is favored by urbanization. As a result, there is an increase in diseases transmission indirectly caused by human effects on the environment, such as the development of urban heat islands [42], and a consequent increase in the incidence of dengue [13].
The findings of this study indicate a robust association between, on the one hand, urbanization and increasing human population density and, on the other, Ae. aegypti population structure patterns and population expansion. This suggests that Ae. aegypti benefits from human changes to the environment, which increase in intensity as the human population migrates from rural to urban areas, increasing the risk of disease transmission and epidemics for an ever-increasing number of people.
Supporting information
(TIF)
(DOCX)
Allelic richness (Na) and private allelic richness (Np).
(DOCX)
Significant P-values in bold.
(DOCX)
Acknowledgments
The authors would like to thank Walter Ceretti-Junior, Paulo Roberto Urbinatti, Antônio Ralph Medeiros-Sousa and the City of São Paulo Zoonosis Control Center for helping us with field collections and Aristides Fernandes and Marcia Bicudo de Paula for identifying the specimens.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo: Grant 2013/15313-4 (www.fapesp.br) MTM; Fundação de Amparo à Pesquisa do Estado de São Paulo: Grant 2012/19117-2 (www.fapesp.br) ABBW; Fundação de Amparo à Pesquisa do Estado de São Paulo: Grant 2015/01172-5 (www.fapesp.br) RWS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.McKinney ML. Urbanization as a major cause of biotic homogenization. Biol Conserv. 2006;127: 247–260. doi: 10.1016/j.biocon.2005.09.005 [Google Scholar]
- 2.Ferraguti M, Martínez-de la Puente J, Roiz D, Ruiz S, Soriguer R, Figuerola J. Effects of landscape anthropization on mosquito community composition and abundance. Sci Rep. Nature Publishing Group; 2016;6: 29002 doi: 10.1038/srep29002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thongsripong P, Green A, Kittayapong P, Kapan D, Wilcox B, Bennett S. Mosquito vector diversity across habitats in central thailand endemic for dengue and other arthropod-borne diseases. PLoS Negl Trop Dis. 2013;7: e2507 doi: 10.1371/journal.pntd.0002507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kraemer MUG, Sinka ME, Duda KA, Mylne AQN, Shearer FM, Barker CM, et al. The global distribution of the arbovirus vectors Aedes aegypti and Ae. Albopictus. Elife. 2015;4: 1–18. doi: 10.7554/eLife.08347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Medeiros-Sousa AR, Ceretti W, Urbinatti PR, de Carvalho GC, de Paula MB, Fernandes A, et al. Mosquito fauna in municipal parks of São Paulo City, Brazil: a preliminary survey. J Am Mosq Control Assoc. 2013;29: 275–9. doi: 10.2987/12-6304R.1 [DOI] [PubMed] [Google Scholar]
- 6.Ceretti-Júnior W, Medeiros-Sousa AR, Multini LC, Urbinatti PR, Vendrami DP, Natal D, et al. Immature mosquitoes in bamboo internodes in municipal parks, City of São Paulo, Brazil. J Am Mosq Control Assoc. 2014;30: 268–274. doi: 10.2987/14-6403R.1 [DOI] [PubMed] [Google Scholar]
- 7.Ceretti-Junior W, de Oliveira Christe R, Rizzo M, Strobel RC, de Matos Junior MO, de Mello MHSH, et al. Species composition and ecological aspects of immature mosquitoes (Diptera: Culicidae) in bromeliads in urban parks in the city of Sao Paulo, Brazil. J Arthropod Borne Dis. 2016;10: 102–112. [PMC free article] [PubMed] [Google Scholar]
- 8.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, et al. The global distribution and burden of dengue. Nature. 2013;496: 504–7. doi: 10.1038/nature12060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilkerson RC, Linton Y-M, Fonseca DM, Schultz TR, Price DC, Strickman DA. Making mosquito taxonomy useful: a stable classification of tribe Aedini that balances utility with current knowledge of evolutionary relationships. PLoS One. 2015;10: e0133602 doi: 10.1371/journal.pone.0133602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.World Health Organization. Dengue and severe dengue. WHO Fact Sheet. 2012. Available: www.who.int/mediacentre/factsheets/fs117/en/index.htm
- 11.Descloux E, Mangeas M, Menkes CE, Lengaigne M, Leroy A, Tehei T, et al. Climate-based models for understanding and forecasting dengue epidemics. PLoS Negl Trop Dis. 2012;6: e1470 doi: 10.1371/journal.pntd.0001470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodrigues MDM, Marques GRAM, Serpa LLN, Arduino MDB, Voltolini JC, Barbosa GL, et al. Density of Aedes aegypti and Aedes albopictus and its association with number of residents and meteorological variables in the home environment of dengue endemic area, São Paulo, Brazil. Parasit Vectors. 2015;8: 1–9. doi: 10.1186/s13071-015-0703-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Araujo RV, Albertini MR, Costa-da-Silva AL, Suesdek L, Franceschi NCS, Bastos NM, et al. São Paulo urban heat islands have a higher incidence of dengue than other urban areas. Brazilian J Infect Dis. 2015;19: 146–155. doi: 10.1016/j.bjid.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buhaug H, Urdal H. An urbanization bomb? Population growth and social disorder in cities. Glob Environ Chang. Elsevier Ltd; 2013;23: 1–10. doi: 10.1016/j.gloenvcha.2012.10.016 [Google Scholar]
- 15.Gubler DJ. Epidemic dengue/dengue hemorrhagic fever as a public health, social and economic problem in the 21st century. Trends Microbiol. 2002;10: 100–3. [DOI] [PubMed] [Google Scholar]
- 16.Staples JE, Breiman RF, Powers AM. Chikungunya fever: an epidemiological review of a re-emerging infectious disease. Clin Infect Dis. 2009;49: 942–8. doi: 10.1086/605496 [DOI] [PubMed] [Google Scholar]
- 17.WHO. Zika situation report. 2016; 1–15. Available: http://www.who.int/emergencies/zika-virus/situation-report/4-march-2016/en/
- 18.Oliveira EJ, Pádua JG, Zucchi MI, Vencovsky R, Vieira MLC. Origin, evolution and genome distribution of microsatellites. Genet Mol Biol. 2006;29: 294–307. doi: 10.1590/S1415-47572006000200018 [Google Scholar]
- 19.Multini LC, Wilke ABB, Suesdek L, Marrelli MT. Population genetic structure of Aedes fluviatilis (Diptera: Culicidae). PLoS One. 2016;11: e0162328 doi: 10.1371/journal.pone.0162328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olanratmanee P, Kittayapong P, Chansang C, Hoffmann A a., Weeks AR, Endersby NM. Population genetic structure of Aedes (Stegomyia) aegypti (L.) at a micro-spatial scale in thailand: implications for a dengue suppression strategy. PLoS Negl Trop Dis. 2013;7 doi: 10.1371/journal.pntd.0001913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piccinali RV, Gürtler RE. Fine-scale genetic structure of Triatoma infestans in the Argentine Chaco. Infect Genet Evol. Elsevier B.V.; 2015;34: 143–52. doi: 10.1016/j.meegid.2015.05.030 [DOI] [PubMed] [Google Scholar]
- 22.Natal D, Marucci D. Aparelho de sucção tipo aspirador para captura de mosquitos. Rev Saude Publica. 1984;18: 418–420. doi: 10.1590/S0034-89101984000500013 [DOI] [PubMed] [Google Scholar]
- 23.Forattini OP. Culicidologia Médica. Vol. 2. EDUSP, Editora da universidade de São Paulo; 2002.
- 24.Chambers EW, Meece JK, McGowan J a., Lovin DD, Hemme RR, Chadee DD, et al. Microsatellite isolation and linkage group identification in the yellow fever mosquito Aedes aegypti. J Hered. 2007;98: 202–210. doi: 10.1093/jhered/esm015 [DOI] [PubMed] [Google Scholar]
- 25.Slotman MA, Kelly NB, Harrington LC, Kitthawee S, Jones JW, Scott TW, et al. Polymorphic microsatellite markers for studies of Aedes aegypti (Diptera: Culicidae), the vector of dengue and yellow fever. Mol Ecol Notes. 2007;7: 168–171. doi: 10.1111/j.1471-8286.2006.01533.x [Google Scholar]
- 26.Excoffier L, Laval G, Schneider S. Arlequin (version 3. 0): An integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1: 47–50. [PMC free article] [PubMed] [Google Scholar]
- 27.Rousset F. GENEPOP’007: A complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour. 2008;8: 103–106. doi: 10.1111/j.1471-8286.2007.01931.x [DOI] [PubMed] [Google Scholar]
- 28.Kalinowski ST. HP-RARE 1.0: A computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes. 2005;5: 187–189. doi: 10.1111/j.1471-8286.2004.00845.x [Google Scholar]
- 29.Chapuis M-P, Estoup A. Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol. 2006;24: 621–631. doi: 10.1093/molbev/msl191 [DOI] [PubMed] [Google Scholar]
- 30.Hammer Ø, Harper DATT, Ryan PD. PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4: 9. [Google Scholar]
- 31.StatSoft I. STATISTICA (data analysis software system). 2012. Available: www.statsoft.com
- 32.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol Ecol Notes. 2007;7: 574–578. doi: 10.1111/j.1471-8286.2007.01758.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol. 2005;14: 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- 34.Cornuet JM, Luikart G, Cornuet J.M, LUikartt G, Cornuet JM, Luikart G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics. 1996;144: 2001–2014. Article [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conn JE, Vineis JH, Bollback JP, Onyabe DY, Wilkerson RC, Póvoa MM. Population structure of the malaria vector Anopheles darlingi in a malaria-endemic region of eastern Amazonian Brazil. Am J Trop Med Hyg. 2006;74: 798–806. 74/5/798 [PubMed] [Google Scholar]
- 36.Donnelly MJ, Licht MC, Lehmann T. Evidence for recent population expansion in the evolutionary history of the malaria vectors Anopheles arabiensis and Anopheles gambiae. Mol Biol Evol. 2001;18: 1353–1364. doi: 10.1093/oxfordjournals.molbev.a003919 [DOI] [PubMed] [Google Scholar]
- 37.Michel AP, Grushko O, Guelbeogo WM, Lobo NF, Sagnon N, Costantini C, et al. Divergence with gene flow in Anopheles funestus from the Sudan Savanna of Burkina Faso, West Africa. Genetics. 2006;173: 1389–1395. doi: 10.1534/genetics.106.059667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mirabello L, Conn JE. Molecular population genetics of the malaria vector Anopheles darlingi in central and South America. Heredity (Edinb). 2006;97: 438–438. doi: 10.1038/sj.hdy.6800913 [DOI] [PubMed] [Google Scholar]
- 39.Monteiro FA, Shama R, Martins AJ, Gloria-Soria A, Brown JE, Powell JR. Genetic diversity of Brazilian Aedes aegypti: patterns following an eradication program. PLoS Negl Trop Dis. 2014;8: e3167 doi: 10.1371/journal.pntd.0003167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown JE, Evans BR, Zheng W, Obas V, Barrera-Martinez L, Egizi A, et al. Human impacts have shaped historical and recent evolution in Aedes aegypti, the dengue and yellow fever mosquito. Evolution. 2014;68: 514–525. doi: 10.1111/evo.12281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Louise C, Vidal PO, Suesdek L. Microevolution of Aedes aegypti. PLoS One. 2015;10: e0137851 doi: 10.1371/journal.pone.0137851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosenthal JK, Sclar ED, Kinney PL, Knowlton K, Crauderueff R, Brandt-Rauf PW. Links between the built environment, climate and population health: Interdisciplinary environmental change research in New York City. Ann Acad Med Singapore. 2007;36: 834–846. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(DOCX)
Allelic richness (Na) and private allelic richness (Np).
(DOCX)
Significant P-values in bold.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.