Abstract
Membrane fusion induced by the envelope glycoprotein enables the intracellular replication of HIV-1; hence, this process constitutes a major target for antiretroviral compounds. It has been proposed that peptides having propensity to interact with membrane interfaces might exert broad antiviral activity against enveloped viruses. To test this hypothesis, in this contribution we have analyzed the antiviral effects of peptides derived from the membrane-proximal external region and the transmembrane domain of the envelope glycoprotein subunit gp41, which display different degrees of interfacial hydrophobicity. Our data support the virucidal activity of a region that combines hydrophobic-at-interface membrane-proximal external region aromatics with hydrophobic residues of the transmembrane domain, and contains the absolutely conserved 679LWYIK/R683 sequence, proposed to embody a “cholesterol recognition/interaction amino acid consensus” motif. We further sought to correlate the antiviral activity of these peptides and their effects on membranes that mimic lipid composition and biophysical properties of the viral envelope. The data revealed that peptides endowed with virucidal activity were membrane active and induced permeabilization and fusion of virus-like lipid vesicles. In addition, they modulated lipid packing and miscibility of laterally segregated liquid domains, two properties that depend on the high cholesterol content of the viral membrane. Thus, the overall experimental evidence is consistent with a pattern of HIV inhibition that involves direct alteration of the physical chemistry of the virus membrane. Furthermore, the sequence-dependent effects observed might guide the development of new virucidal peptides.
Introduction
Among the major drug classes approved for human immunodeficiency virus (HIV) antiretroviral therapy, entry inhibitors are unique at blocking the function of an extracellular target, the envelope glycoprotein (Env) (1, 2, 3). The Env subunits gp120 (surface) and gp41 (trans-membrane) are responsible for receptor/coreceptor binding and virus-cell fusion, respectively (4, 5, 6). Upon activation of the fusion process, gp41 ectodomain trimers refold into low-energy 6-helix bundles that pull cell and virus membranes into close contact. The conformational transition undergone by gp41 trimers constitutes the target for the clinically approved fusion inhibitor Enfuvirtide (7), which blocks 6-helix bundle completion and hence membrane merger (8).
Development of alternative fusion inhibitors displaying broad and sustained antiretroviral activity against HIV-1 remains a pursued goal to date (9, 10). It has been recently proposed that membrane-targeting virucidal compounds may comprise broad-spectrum inhibitors of enveloped virus entry (10, 11, 12). Arguably, compounds interfering with fusion by acting on the lipid component of the viral envelope could provide the basis for escape-proof antiviral therapies (13, 14). In addition, as opposed to host cell membranes that are subject to ongoing synthesis, degradation, and influx and efflux of their components, static viral membranes lacking repairing mechanisms cannot escape the major perturbations induced by virucide activity. In this regard, a series of works support the possibility that peptides derived from hydrophobic-at-interface envelope glycoprotein sequences may comprise new generic antivirals (15, 16, 17, 18, 19). Along this same line of thinking, in recent reviews it has been contended that these virucidal peptides could block viral entry by changing directly the physical chemistry of the viral membrane upon partitioning (20, 21).
We have recently established a synthetic virus-like (VL) mixture, which includes the main seven lipid species found in the HIV-1 membrane (22) and exhibits the same order degree and phase behavior (termed “VL-4” in our previous report (23)). Here, to get new insights into the molecular mechanisms governing antiviral activity by membrane-partitioning peptides, we first compare the inhibitory potencies of several membrane-proximal external region-transmembrane domain (MPER-TMD)-derived peptides that possess distinct interfacial hydrophobicity patterns (20, 24), and then establish their membrane-restructuring capacities using the VL mixture as a surrogate of the HIV membrane. Our experimental data support a virucide-like activity that alters the physical chemistry of the HIV lipid envelope for peptides combining the hydrophobic-at-interface C-terminal MPER sequence with the N-terminal hydrophobic section of the TMD. We speculate that these sequences may serve as platforms for further development of antiretroviral peptides.
Materials and Methods
1-palmitoyl-2-oleoylphosphatidylcholine (POPC), 1-palmitoyl-2-oleoylphosphatidylethanolamine (POPE), 1-palmitoyl-2-oleoylphosphatidylserine (POPS), cholesterol (Chol), egg sphingomyelin (SM, containing ∼86% n-palmitoyl SM), dodecanoyl dihydrosphingomyelin (DHSM), and 1-(1Z-octadecenyl)-2-oleoyl-sn-glycero-3-phosphatidylethanolamine (plasmologen PE, pl-PE) were purchased from Avanti Polar Lipids (Alabaster, AL). The 8-aminonaphtalene-1,3,6-trisulfonic acid sodium salt (ANTS), p-xylenebis(pyridinium)bromide (DPX), N-(7-nitro-benz-2-oxa-1,3-diazol-4-yl)phosphatidylethanolamine, N-(lissamine Rhodamine B sulfonyl)phosphatidylethanolamine, and 6-dodecanoyl-2-dimethylaminonaphthalene (Laurdan) were obtained from Molecular Probes (Eugene, OR). Phospholipid stock concentrations were determined by phosphate assay. The collection of peptides displayed in Fig. 1 B were synthesized in C-terminal carboxamide form by solid-phase methods using Fmoc chemistry, purified by reverse-phase high-performance liquid chromatography, and characterized by matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (purity >95%).
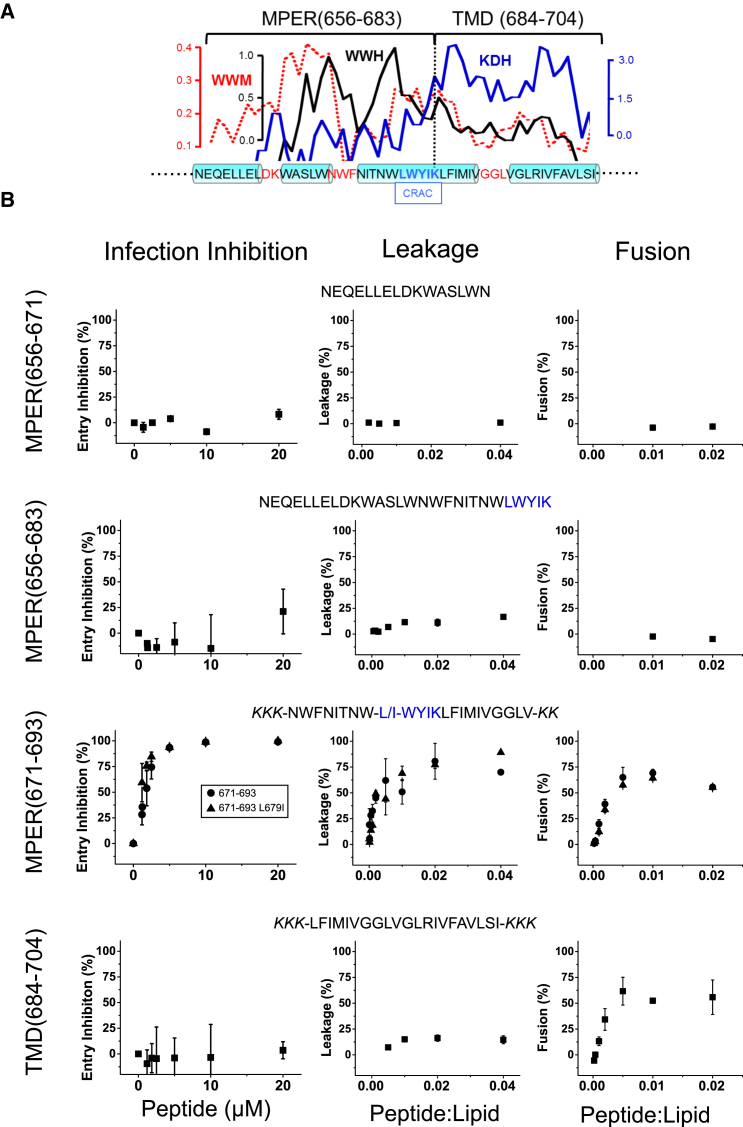
Figure 1.
Designation of hydrophobic-at-interface HIV-1 peptides and their antiviral and membrane-restructuring activities. (A) Given here is the hydrophobicity distribution within the gp41 MPER-TMD region (top) and amino acid sequence (bottom). For calculating mean WW hydrophobicity (33) and KD hydropathy (36), a window of five amino acids was used (WWH and KDH, respectively). The dotted line represents the WW moment (window of 11 amino acids) calculated for a fixed δ = 100° (helical periodicity), using the hydrophobicity-at-interface scale (59). The cylinders on the sequence highlight helical subdomains, which are connected by nonhelical joints. Position for the postulated CRAC motif is also indicated. (B) Shown here are sequences covered by the overlapping peptides used in this study and their effects on pseudovirus infectivity, vesicle permeability, and vesicle-vesicle fusion (left, center, and right panels, respectively). In cell entry inhibition assays, HIV-1 pseudoviruses were preattached to lysine-coated plates (26) and treated with increasing concentrations of the peptides as indicated in the panels. After washing, reporter TZM-bl cells were layered on top, and infectivity inferred from the number of total cells expressing GFP (Fig. S1A). To see this figure in color, go online.
Cell-entry assays were performed by measuring green fluorescent protein (GFP) gene transduction according to Bobardt et al. (25), with the modifications described by Markosyan et al. (26). In brief, HIV-1 pseudoviruses were produced by transfection of human kidney HEK293T cells with the full-length Env clone JRCSF (kindly provided by Jamie K. Scott and Naveed Gulzar, Simon Fraser University, Burnaby, British Columbia, Canada) using calcium phosphate. Cells were cotransfected with vectors pWXLP-GFP and pCMV8.91, encoding a GFP and an Env-deficient HIV-1 genome, respectively (provided by Patricia Villace, CSIC, Madrid, Spain). After 24 h, the medium was replaced with Optimem-Glutamax II (Invitrogen, Paisley, UK) without serum. Two days after transfection, the pseudovirus particles were harvested, passed through 0.45-μm-pore sterile filters (Millex-HV; Millipore NV, Brussels, Belgium) and finally concentrated by ultracentrifugation in a sucrose gradient. HIV entry was determined using TZM-bl target cells (AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, contributed by J. Kappes). HIV pseudoviruses diluted to a 10–15% tissue culture infectious dose in PBS were deposited onto poly-L-lysine-coated 96-well plates, and incubated at 4°C for 40 min. After washing, free poly-lysine was blocked for 20 min by medium addition (90% DMEM; 10% FBS) at 37°C. Several dilutions of a given peptide in PBS were subsequently applied for 90 min at 37°C. After three washing steps, 1.1 × 104 TZM-bl target cells were layered on top of immobilized virions in the presence of 30 μg/mL DEAE-dextran (Sigma-Aldrich, St. Louis, MO). Infection levels after 72 h were inferred from the number of GFP-positive cells as determined by flow cytometry using a BD FACSCalibur Flow Cytometer (Becton Dickinson Immunocytometry Systems, Mountain View, CA).
Large unilamellar vesicles (LUVs) made of the VL lipid mixture (POPC/SPM/CHOL/DHSM/POPE/pl-PE/POPS (14:12:46:5:3:13:7 mol ratio)) were produced following the extrusion method. To that end, lipid suspensions were subjected to 10 freeze-thaw cycles before extrusion 10 times through two stacked polycarbonate membranes with a nominal pore-size of 0.1 μm (Nucleopore, Pleasanton, CA). Vesicle permeability changes and vesicle-vesicle fusion were determined by the standard assays of ANTS/DPX leakage and Rho-PE/NBD-PE-based lipid mixing, respectively (27).
Giant unilamellar vesicles (GUVs) were produced by spontaneous swelling following procedures described in (28) and (29). For preparation of peptide-containing GUVs, lipids (0.125 mg of the VL lipid mixture) and peptides were mixed at the desired peptide-to-lipid molar ratio in CHCl3/CH3OH (9:1) before desiccation for 1 h to remove the organic solvent. Dried silica beads covered with lipid-peptide mixtures were collected and transferred to a 7.5 g/L sucrose buffer to induce spontaneous swelling of GUVs. Formed vesicles were transferred to the observation dish in an isosmotic 10 mM HEPES, 150 mM KCl (pH 7.4) buffer.
Images were acquired on a TCS SP5 II microscope (Leica Microsystems, Wetzlar, Germany) as described in (23) and (29). Laurdan-stained GUVs were excited at 780 nm using a ×63 water-immersion objective (numerical aperture = 1.2) and 512 × 512 pixel images were acquired at 400 Hz per scanning line. The fluorescence emission was simultaneously imaged at 435 ± 20 nm and at 500 ± 10 nm. Generalized polarization (GP) images were computed for every pixel in the image (Eq. 1), where IB is the intensity in the blue channel, and IR is the intensity in the green channel. The G factor accounts for the relative sensitivity of the two channels, calibrated with a 5 μM Laurdan solution in pure DMSO at 21°C (30):
| (1) |
To produce planar-supported phospholipid layers, phospholipid monolayers were first spread from chloroform/methanol 9:1 (v/v) solutions onto a 5 mM Tris (pH 7.4), 150 mM NaCl subphase, in a thermostated Langmuir-Blodgett trough (NIMA Technologies, Coventry, UK), and then transferred onto mica as described in (31, 32). AFM images were obtained in an Nanowizard II atomic force microscope (JPK Instruments, Berlin, Germany) as described in Huarte et al. (23).
Results
The plot in Fig. 1 A displays the distribution of Wimley-White interfacial hydrophobicity (33) along the MPER-TMD region of HIV-1 gp41. At the N-terminus, an amphipathic-at-interface helix (residues 656–671) is followed by a fully hydrophobic-at-interface stretch (residues 672–683). Supporting the biological relevance of these MPER segments, they consecutively span the broadly neutralizing epitopes 2F5 and 4E10 (27, 34, 35). These interfacial sequences are followed by the TMD (residues 684–704), which is mostly hydrophobic according to the Kyte-Doolittle hydropathy index (36). The TMD is also segmented into two subdomains due to the presence of midway polar residues (37). Interestingly, the MPER C-terminal and the TMD N-terminal subregions are part of a continuous helix (38, 39). Finally, residues 679LWYIK683 at the MPER-TMD juncture have been proposed to embody a cholesterol recognition/interaction amino acid consensus (CRAC) motif (31, 32, 40, 41, 42). This region is absolutely conserved across viral strains and isolates, and would be involved in generating the type of lipid perturbations that prime the rigid viral membrane for fusion (43, 44, 45).
The particular organization of the MPER-TMD region allows, on the one hand, testing the prediction that CRAC-mediated, Chol-dependent membrane perturbations may sustain antiviral activity of the derived peptides (31). On the other hand, one can assess the effect of combining distinct hydrophobicity patterns on the inhibitory potency of peptides (20). The sequence range covered by each of the peptides displayed in the panels of Fig. 1 B was thus selected on the basis of the presence of the CRAC motif and the hydrophobicity profiles. The list included a peptide bearing an Ile residue at the position of the absolutely conserved residue Leu-679. The resulting L679I substitution was shown to attenuate interactions of CRAC-representative peptides with Chol and reduce Env fusogenicity (45, 46).
Fig. 1 B compares the antiviral activity of the peptides with the effects exerted on VL LUVs (leakage and fusion). Antiviral activity was evaluated on virions that were first attached to poly-lysine-coated plates (26) (see Fig. S1 A), and cell infectivity subsequently quantified following procedures described in Bobardt et al. (25). The amphipathic-at-interface peptide MPER(656−671) (34) did not display significant anti-HIV activity in the 0.1−20 μM range (Fig. 1 B, left panels). Addition of interfacial hydrophobicity and the CRAC motif at the C terminus to render the peptide MPER(656−683) did not elicit activity. Thus, it appears that bearing high interfacial hydrophobicity was not sufficient to trigger virucidal activity, even if the peptide contained the CRAC sequence. In contrast, the peptide MPER(671−693), which combined the C-terminal hydrophobic-at-interface MPER stretch with the N-terminal section of the TMD, induced robust antiviral activity with a mean IC50 value of 1.67 ± 0.04 μM. Interestingly, the peptide MPER(671−693)L679I with the mutated CRAC motif displayed a comparable virucidal effect (IC50 value of 0.96 ± 0.02 μM). Finally, the TMD(684−704) sequence representing the full-length gp41 TMD, as defined by Cohen et al. (47), did not have any apparent effect on pseudovirus cell entry.
To ascertain whether the infection-blocking effect observed in the case of the peptides MPER(671−693) and MPER(671−693)L679I was due to a virucide-type of activity, and not as a consequence of the specific inhibition of HIV-1 Env-mediated entry/fusion, experiments were also conducted with virions pseudotyped with the VSV G glycoprotein (VSV-G) (Fig. S1 B). Incubation of VSV-G pseudoviruses with the peptides resulted in comparable levels of infection blocking, thereby supporting a virucide-like behavior for the inhibitory peptides, rather than an activity dependent on sequence-specific mechanisms.
To determine if the observed virucidal activity correlated with the capacity of the peptides for altering the properties of the viral membrane, we used the seven-lipid VL mixture as an envelope surrogate (23). We first assayed leakage and lipid mixing induced by the peptides on VL LUVs (Fig. 1 B, center and right panels, respectively). Significant leakage of contents and lipid mixing was observed upon VL LUV incubation with MPER(671−693) or MPER(671−693)L679I peptides. Interestingly, the TMD(684−704) peptide also induced lipid mixing, an observation consistent with data previously reported in the literature (see for a recent review (48)). Thus, VL vesicle destabilization correlated with the observed virucidal effects of the peptides MPER(671−693) or MPER(671−693)L679I.
However, peptide-induced perturbations that in the vesicular system can relax through vesicle-vesicle aggregation and fusion, must persist in the immobilized viral particles that are used in the infectivity assays (Fig. S1 A). Control experiments also indicated that lipids were not solubilized or extracted from virions treated with the inhibitory peptides (Fig. S2). Thus, we hypothesized that the local perturbations leading to LUV leakage and fusion may affect in an interrelated way additional physicochemical properties of the lipid envelope in the immobilized virions.
The HIV particle is enriched in Chol (∼50 mol %), which underpins increased membrane order and the potential for liquid phase coexistence (22, 23, 49). The lipids extracted from the infectious virus particles and the VL surrogate actually display a high lipid packing level and lateral demixing into nanodomains (23). Therefore, to identify additional effects that might correlate with virus inactivation in our cell infectivity assays, we next evaluated the changes on the Chol-dependent lipid packing and lateral organization induced by the peptides MPER(671−693) and MPER(671−693)L679I (Figs. 2 and 3).
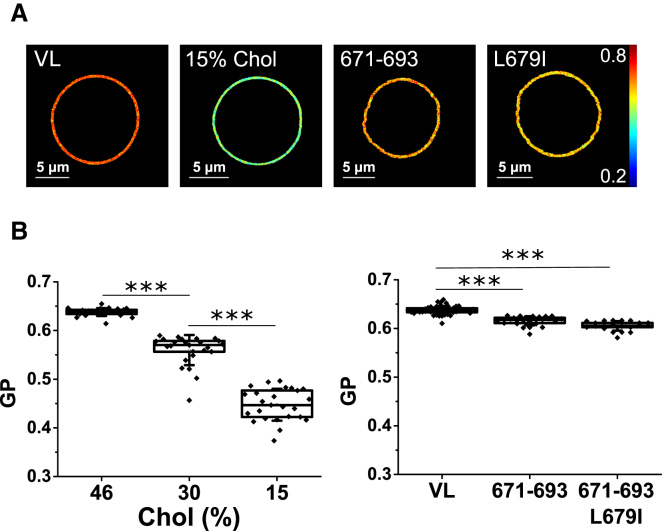
Figure 2.
Comparison of the effects induced by Chol depletion versus addition of peptides MPER(671–693) or MPER(671–693)L679I on the membrane lateral packing as determined by two-photon excitation GP images of Laurdan-labeled VL GUVs. Laurdan GP imaging of single GUVs (A) and average GP values were determined in the different samples (B). GP value distributions were determined for at least 30 single vesicles, and mean values were depicted as box-and-whisker plots (the ends of the whiskers represent SDs). The mole percents in the lipid mixtures were as follows: POPC/SPM/CHOL/DHSM/POPE/pl-PE/POPS (14:12:46:5:3:13:7) (VL), POPC/SPM/CHOL/DHSM/POPE/pl-PE/POPS (18:16:30:6:4:17:9) (VL-30 mol % Chol), and POPC/SPM/CHOL/DHSM/POPE/pl-PE/POPS (22:18:15:8:5:20:12) (VL-15 mol % Chol). Peptide concentration was 20 nM. Mann-Whitney test: ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, not significant (n.s.) ≥ 0.05. To see this figure in color, go online.
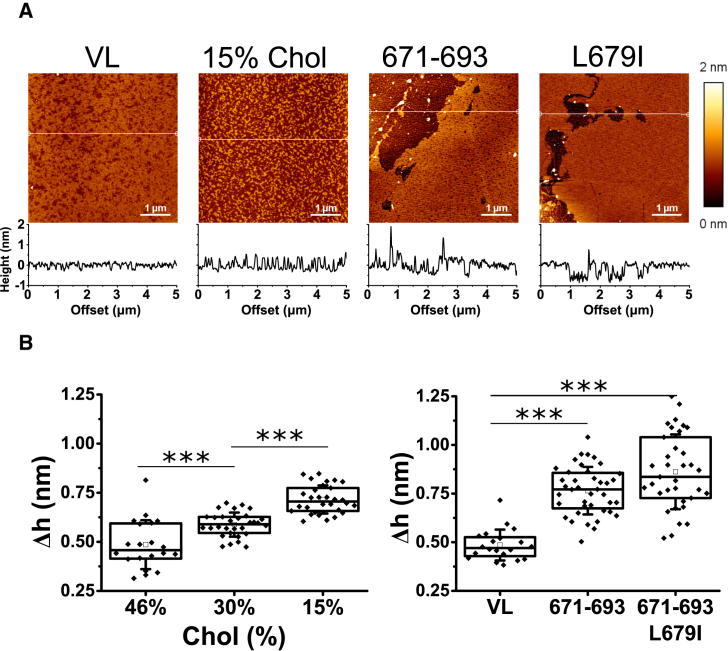
Figure 3.
Comparison of the effects induced by Chol depletion versus inclusion of peptides MPER(671–693) or MPER(671–693)L679I on the lateral organization of films made of the VL mixture. (A) AFM images were taken from VL mixture films compressed at the indicated surface pressures. Plots below images display the height profiles for the trajectories indicated by the white lines. AFM images were obtained at fixed Π = 10 mN/m. (B) Given here is the height difference between demixed domains and the surrounding lipid surface. Mann-Whitney test: ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, not significant (n.s.) ≥ 0.05. To see this figure in color, go online.
Lipid packing was first determined quantitatively by two-photon imaging of Laurdan-stained VL GUVs (Fig. 2). Laurdan’s fluorescent emission undergoes a large spectral shift due to the reorientation of polar molecules in the fluorophore vicinity during the time it spends in its excited state. When incorporated in a lipid bilayer, Laurdan’s fluorescent emission is a function of the hydration and viscosity of the membrane, reflecting its phase state.
A wavelength-ratiometric parameter called “generalized polarization” (GP; Eq. 1) quantifies Laurdan’s emission spectral shift from tightly packed (low water content, high GP values) to loosely packed (high water content, low GP values) bilayers, thus providing an indirect measurement of membrane order through packing (50, 51). Under the assumption that the 679LWYIK683 sequence can interact with, and sequester Chol (31, 46), the effect of depleting this compound from the mixtures was assessed in parallel.
GP images of the different VL GUV samples displayed a single macroscopic phase (Fig. 2 A), which was reflected in a normal, unimodal, distribution of pixel-GP values for every GUV (23). As expected, decreasing the Chol content resulted in lower mean GP values, consistent with a decrease of the VL membrane order (Fig. 2 B, left panel). A more subtle reduction of Laurdan GP values was observed for the GUVs treated with peptides MPER(671−693) and MPER(671−693)L679I (Fig. 2 B, right panel). Furthermore, the scored GP reduction was comparable for the two peptides, suggesting that preservation of the CRAC motif was not required for the fluidifying effect.
We next compared the capacity of the peptides to alter liquid-liquid miscibility of the VL membrane at the nanoscale (Fig. 3). Langmuir-Blodgett films were compressed to 10 mN/m, transferred onto mica and analyzed by AFM. We caution that VL lipid monolayers were used as a model system that allowed comparison of the potential of the different interfacial peptides to modify the Chol-dependent liquid-liquid miscibility. We further highlight that the supported monolayers under our experimental conditions are limited in the sense that they do not mimic the actual envelope and therefore can only be used as a tool to tackle specific questions of the lipid-peptide interplay. As such, the observed phenomena provided additional insights into the action mechanism of MPER-TMD-derived antiviral peptides.
The results displayed in Fig. 3 A revealed demixed nanoscopic domains in the untreated VL lipid monolayer. Upon reduction of Chol content nanodomains retained rough boundaries and convoluted morphologies. These features were consistent with the low line tension at the domain interfaces under these conditions (23). Inclusion of the peptides MPER(671−693) or MPER(671−693)L679I into the lipid mixture (peptide-to-lipid mole ratio, 1:100) resulted, in both cases, in segregation of much larger lipid platforms limited by smooth contours, which were not observed in lipid monolayers containing lower amounts of Chol.
Line tension at the rims of demixing phases intensifies as the height differences between rigid domains and surrounding membrane regions (Δh) increase (52). The high interfacial energy can be reduced by coalescence and perimeter minimization of the rigid domains. The value of the Δh parameter increased upon Chol depletion from the mixtures (Fig. 3 B, left panel). However, consistently with the smoother contours and larger domain sizes, Δh values were significantly higher in peptide-containing monolayers (Fig. 3 B, right panel). Thus, inclusion of 1 mol % of peptide induced a more marked effect on lipid miscibility than reducing the Chol content of the mixture by 30 mol %. Together with the fact that the presence of a functional CRAC was not required for inducing the effect, these data clearly indicate that the observed alterations were not the consequence of peptide-induced Chol depletion through the formation of equimolar peptide-Chol complexes.
In conclusion, the previous results suggest that the MPER(671−693) peptide does not require the establishment of direct interactions with Chol through the CRAC motif to alter lipid packing and lipid-lipid miscibility.
To gain further insights into the mechanism by which MPER(671−693) exerts its antiviral activity, we next compared its effects with those produced by the inactive MPER-TMD sequences (Fig. 4). Fig. 4 A illustrates the effects of the peptides on lipid packing as inferred from Laurdan GP determinations in single vesicles. In these experiments, peptide and lipids were mixed at defined 1:500 mol ratio before desiccation on the silica beads. The data revealed a slight decrease of the mean GP value in GUVs that contained the virucidal peptide MPER(671−693), but comparable decreases in GUVs containing the inactive peptides MPER(656−671), MPER(656−683), or TMD(684−704).
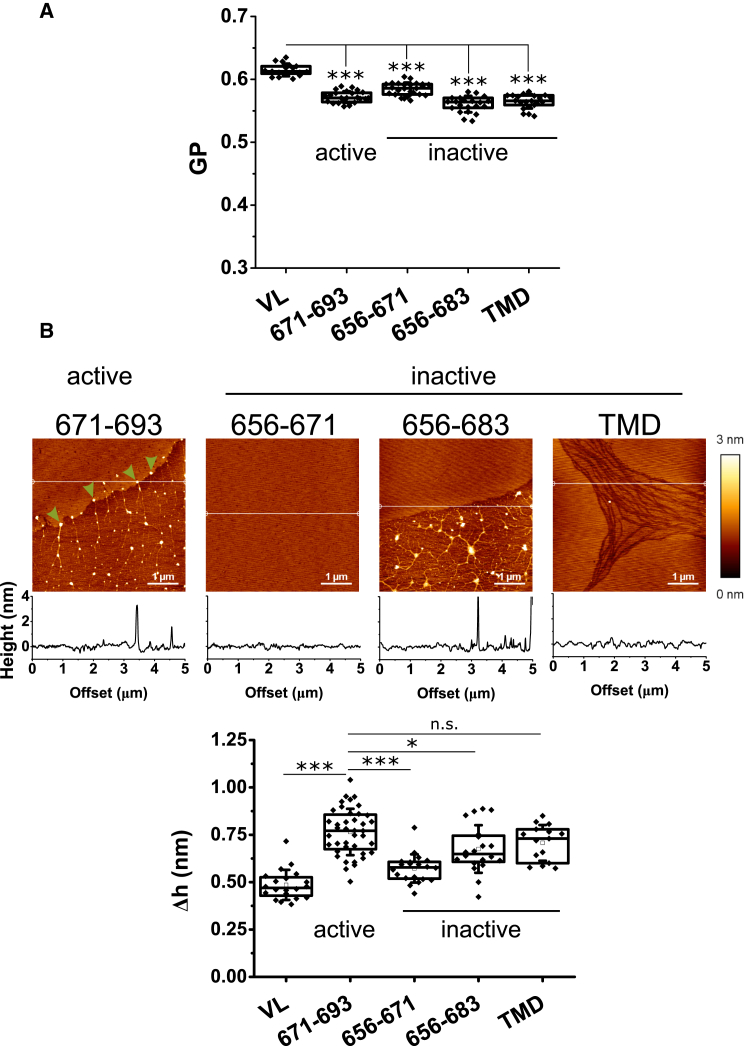
Figure 4.
Comparison of the effects induced by the different gp41 MPER-TMD-derived peptides on the packing and lateral organization of VL membranes. (A) GP values were determined for VL GUVs that contained peptide (peptide/lipid 1:500). (B) Given here is the nanoscopic structure, as determined by AFM analyses of the peptide-containing VL monolayers (top) and height difference between demixed domains and the surrounding lipid surface (bottom). Monolayers were supplemented with peptide to a peptide/lipid 1:100. Conditions otherwise as in the previous Figs. 2 and 3. Mann-Whitney test: ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, not significant (n.s.) ≥ 0.05. To see this figure in color, go online.
Differences between the active and the inactive sequences became apparent at the nanoscopic level (Fig. 4 B). Besides the segregation of large lipid platforms, AFM measurements disclosed in the samples containing MPER(671−693) an accumulation of material in the fluid areas, and also at the contacts between condensed and fluid domains (arrowheads). Lipid monolayers containing the inactive peptides displayed distinct nanostructures. No large segregated platforms nor accumulation of material could be discerned in the samples containing MPER(656−671) peptide. In contrast, segregated domains could be observed in the monolayers containing MPER(656−683) peptide. Although in these samples material was observed to accumulate in the fluid areas, no material was detected at the periphery of the rigid domains. Finally, liquid-liquid immiscibility was also observed in the samples that contained the TMD(684−704) peptide. In this case, the pattern was completely different, with the rigid areas surrounded by streams of fluid lipid. The height differences between domains displayed in the bottom panel further emphasizes that the most substantial effect was induced by the active peptide MPER(671−693).
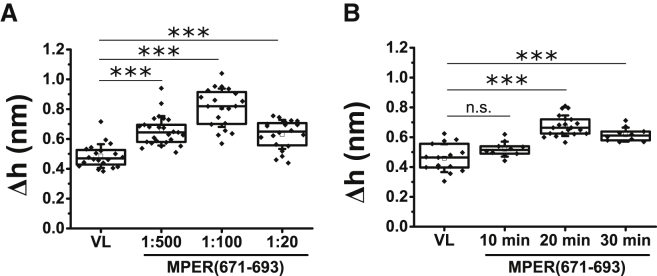
Thus, it appears that the capability to accumulate on the edges of the lipid domains and alter line tension is a distinctive property of the active MPER(671−693) sequence. Measurements of the height differences between segregated domains in VL monolayers displayed in Fig. 5 support this idea. Consistent with an intrinsic activity of the peptide, the data indicate that MPER(671−693) effect on line tension is dose- and time-dependent (Fig. 5, A and B, respectively).
Figure 5.
Height differences between demixed domains and the surrounding lipid surface induced by MPER(671–693) in VL monolayers as function of the peptide-to-lipid mole ratio (A) or time after injection into the subphase (B). Mann-Whitney test: ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, not significant (n.s.) ≥ 0.05.
Discussion
The basis for the broad antiviral activity displayed by some hydrophobic and/or amphipathic peptides is not well understood (15, 16, 17, 18, 19, 20, 21). Two factors emphasize the suitability of the HIV envelope as a targetable component for this type of peptides: 1) the HIV particle is covered with a solvent-accessible lipid bilayer mostly devoid of protein (53); and 2) preservation of the chemical composition and biophysical properties of that lipid bilayer seems to be critical for the entry function (14, 22, 23, 49, 54). In addition, the Env subunit gp41 is anchored to the HIV membrane through the MPER-TMD region (Env residues 656−704, HXB2c numbering), which encompasses interfacially active sequences enriched in aromatic residues (24). To map potential virucidal activity within the MPER-TMD region, in this work we have employed several overlapping peptides, which feature different degrees of interfacial hydrophobicity (Fig. 1). Furthermore, using the VL surrogate we have comparatively assessed the selected peptides in their capacity for altering membrane stability, as well as for modulating the Chol-dependent lipid packing and liquid-liquid miscibility. In the following, we summarize our observations and discuss them in relation to MPER virucidal activity, providing, to our knowledge, new insights into the mechanism of action of hydrophobic-at-interface antiviral peptides (20).
Identification of an MPER-TMD sequence with virucidal activity
The comparative assessment of MPER-TMD-derived peptides identified a sequence, MPER(671−693), which inhibited HIV infection with an IC50 of ∼1.5 μM (Fig. 1). The fact that a similar inhibition pattern was observed for VSV pseudoviruses (Fig. S1) supports a virucide-like activity for this peptide. Preservation of interfacial hydrophobicity, although required, was not sufficient to elicit this activity. The peptide MPER(656−683), combining within a single construct amphipathic-at-interface and fully hydrophobic-at-interface sections, was not virucidal. Antiviral capacity was not attained by the fully hydrophobic peptide TMD(684−704) either. Notably, the peptide MPER(671−693), displaying virucidal efficacy, combined hydrophobic-at-interface aromatic and hydrophobic-aliphatic residues.
The antiviral activity of the peptide MPER(671−693) and the alterations that induced on membranes also did not require an intact CRAC motif (Figs. 1 B, 2, and 3). This strongly suggests that the virucidal effect was not dependent on a direct interaction of the peptide with Chol. In fact, the observation that 1 mol % peptide induced lipid demixing and augmented the line tension more efficiently than reducing Chol content by 30 mol %, excludes the possibility that MPER(671−693) acted by depleting this compound (Fig. 3).
Correlation between virucidal activity and VL LUV destabilization
As compared to the inactive peptides, the MPER(671−693) sequence endowed with antiviral activity destabilized the seven-lipid VL membranes, causing significant LUV permeabilization (ANTS leakage) and fusion (lipid mixing) (Fig. 1 B). Thus, the virucidal peptides displayed membrane activity in lipid mixtures that resembled the composition and biophysical properties of the viral envelope.
However, the mechanism relating this membrane activity to infection inhibition is not clear. One plausible mechanism to explain the inhibitory effect of MPER(671−693) could be the general solubilization or disaggregation of the viral envelope by the peptide, either following a detergent-like mechanism, or by extracting lipids, as described for the Chol-depleting compound β-cyclodextrin (54, 55). However, control experiments under conditions emulating the infectivity assays (i.e., isolated virions immobilized onto a solid substrate; Fig. S1 A), indicated that incubation with peptides did not remove membrane lipids (Fig. S2, top). In contrast, treatment of the samples with the detergent Triton X-100 or the Chol-depleting β-cyclodextrin totally or partially depleted the solid supports of lipid, respectively (Fig. S2, bottom).
The fact that the immobilized viral particles remained isolated in the cell infectivity experimental setup (Fig. S1 A) also rules out virion-virion aggregation/fusion induced by peptides as the mechanism underlying their virucidal activity. Thus, we infer that perturbations resulting in membrane permeabilization and intervesicle lipid mixing might also affect additional biophysical properties of the membranes in the isolated virions, whose preservation are required to sustain infectivity.
Decrease in Chol-dependent lateral lipid packing
High quantities of Chol incorporate into the viral membrane during HIV assembly (22), whereas depletion of this compound inactivates the virus (54). It has been argued that the high levels of lipid packing induced by Chol are required to preserve the functionality of the HIV particle (49, 54). We observed that the virucidal peptides MPER(671−693) and MPER(671−693)L679I slightly, although significantly, decreased lipid packing of the VL mixture (Fig. 2). However, a comparable fluidifying effect was observed when inactive sequences MPER(656−683) or TMD were incorporated into VL GUVs (Fig. 4 A). Thus, a general, slight fluidification of the viral membrane does not seem to explain the virucidal activity of MPER(671−693).
Alteration of Chol-dependent lipid miscibility
Chol levels also modulate the liquid-liquid miscibility of the VL mixture (Fig. 3). We approached the effects of the MPER-TMD-derived peptides on lateral segregation using lipid monolayers that were compressed to 10 mN/m, which represents a miscibility transition pressure for the HIV lipids and ternary mixtures containing Chol (23, 56). This condition was also selected based on previous observations indicating that increasing lateral pressure to 30 mN/m induces collapse of the lipid monolayer made of the viral lipids, whereas its lateral organization, consisting of demixed nanodomains, was preserved at the lower values of 20 and 10 mN/m (23).
Thus, AFM analysis of VL lipid monolayers allowed scoring at the nanoscopic length scale the effect of the different peptides on the demixed liquid domain pattern (Figs. 3 and 4). The topographic information obtained under our experimental conditions confirmed that the active peptide MPER(671−693) can induce nanodomain clustering. However, a similar effect was observed for the inactive peptides MPER(656−683) and TMD(684−704), Thus, the capacity of the peptides to induce nanodomain clustering does not seem to explain the observed virucidal activity.
Accumulation at edges of lipid domains
The AFM analyses of lipid monolayers also revealed accumulation of material at the rim of rigid domains in VL monolayers containing MPER(671−693) virucidal peptide, a pattern not observed in the case of the inactive peptides (Fig. 4 B). In addition, the active peptide induced the highest measured domain height difference after nanodomain clustering (Figs. 4 B and 5). This observation suggests that, in comparison with the inactive peptides, MPER(671−693) can alter line tension directly by accumulating at the periphery of the rigid domains.
Conclusion
Our data provide conclusive evidence to support the virucidal activity of a peptide derived from the HIV-1 MPER-TMD region, and a mechanism of action that involves the change of the physical chemistry of the virus membrane. The comparative analysis identifies the two properties of the virucidal sequence that more robustly correlate with its inhibitory activity: the faculty of permeabilizing VL LUVs and its potential to accumulate at the periphery of rigid domains and modulate line tension. Both effects can be interrelated, because accumulation on the edges of lipid domains might help the assembly of the membrane permeating structures inducing vesicle leakage, as previously shown in the case of some pore-forming toxins (57, 58). The establishment of correlations among interfacial activity, membrane activity, and antiviral activity as a function of the sequence hydrophobicity pattern, might help future development of virucidal peptides.
Author Contributions
P.C., A.C., J.P.-G., J.R.-I., N.H., and J.L.N. conceived and designed the experiments. P.C. and I.M.-U. performed infectivity assays. I.M.-U., J.T., and B.A. performed leakage and fusion assays. P.C. and I.R.O.-A. prepared GUVs and determined GP values by two-photon excitation Laurdan microscopy. P.C. performed monolayer work. P.C., A.C., J.P.-G., J.R.-I., N.H., and J.L.N. analyzed and interpreted the data. P.C. and J.L.N. wrote the article with input from N.H., J.P.-G., and J.R.-I. and all authors reviewed it.
Acknowledgments
Technical assistance by Miguel Garcia-Porras is greatly acknowledged.
This work was supported by National Institutes of Health (NIH) (grant No. AI097051 to J.L.N. and N.H.), the Basque Government (grant No. IT838-13 to J.L.N.), the Spanish Ministry of Economy (grant No. BIO2015-67930-R to A.C. and J.P.-G.), and the Regional Government of Madrid (grant No. P2013/MIT2807 to A.C. and J.P.-G.). P.C. is a recipient of a predoctoral fellowship from the Basque Government.
Editor: Kalina Hristova.
Footnotes
Two figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30747-6.
Contributor Information
Nerea Huarte, Email: nerea.huarte@ehu.es.
José L. Nieva, Email: joseluis.nieva@ehu.es.
Supporting Material
References
- 1.Wong A. The HIV pipeline. Nat. Rev. Drug Discov. 2014;13:649–650. doi: 10.1038/nrd4364. [DOI] [PubMed] [Google Scholar]
- 2.Haqqani A.A., Tilton J.C. Entry inhibitors and their use in the treatment of HIV-1 infection. Antiviral Res. 2013;98:158–170. doi: 10.1016/j.antiviral.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 3.De Clercq E., Li G. Approved antiviral drugs over the past 50 years. Clin. Microbiol. Rev. 2016;29:695–747. doi: 10.1128/CMR.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Checkley M.A., Luttge B.G., Freed E.O. HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. J. Mol. Biol. 2011;410:582–608. doi: 10.1016/j.jmb.2011.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blumenthal R., Durell S., Viard M. HIV entry and envelope glycoprotein-mediated fusion. J. Biol. Chem. 2012;287:40841–40849. doi: 10.1074/jbc.R112.406272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melikyan G.B. Common principles and intermediates of viral protein-mediated fusion: the HIV-1 paradigm. Retrovirology. 2008;5:111. doi: 10.1186/1742-4690-5-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lalezari J.P., Henry K., Salgo M., TORO 1 Study Group Enfuvirtide, an HIV-1 fusion inhibitor, for drug-resistant HIV infection in North and South America. N. Engl. J. Med. 2003;348:2175–2185. doi: 10.1056/NEJMoa035026. [DOI] [PubMed] [Google Scholar]
- 8.Eckert D.M., Kim P.S. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 9.Flexner C., Saag M. The antiretroviral drug pipeline: prospects and implications for future treatment research. Curr. Opin. HIV AIDS. 2013;8:572–578. doi: 10.1097/COH.0000000000000011. [DOI] [PubMed] [Google Scholar]
- 10.Wisskirchen K., Lucifora J., Protzer U. New pharmacological strategies to fight enveloped viruses. Trends Pharmacol. Sci. 2014;35:470–478. doi: 10.1016/j.tips.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolf M.C., Freiberg A.N., Lee B. A broad-spectrum antiviral targeting entry of enveloped viruses. Proc. Natl. Acad. Sci. USA. 2010;107:3157–3162. doi: 10.1073/pnas.0909587107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.St Vincent M.R., Colpitts C.C., Schang L.M. Rigid amphipathic fusion inhibitors, small molecule antiviral compounds against enveloped viruses. Proc. Natl. Acad. Sci. USA. 2010;107:17339–17344. doi: 10.1073/pnas.1010026107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vigant F., Santos N.C., Lee B. Broad-spectrum antivirals against viral fusion. Nat. Rev. Microbiol. 2015;13:426–437. doi: 10.1038/nrmicro3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wojcechowskyj J.A., Doms R.W. A potent, broad-spectrum antiviral agent that targets viral membranes. Viruses. 2010;2:1106–1109. doi: 10.3390/v2051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giannecchini S., Di Fenza A., Bendinelli M. Antiviral activity and conformational features of an octapeptide derived from the membrane-proximal ectodomain of the feline immunodeficiency virus transmembrane glycoprotein. J. Virol. 2003;77:3724–3733. doi: 10.1128/JVI.77.6.3724-3733.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hrobowski Y.M., Garry R.F., Michael S.F. Peptide inhibitors of dengue virus and West Nile virus infectivity. Virol. J. 2005;2:49. doi: 10.1186/1743-422X-2-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sainz B., Jr., Mossel E.C., Garry R.F. Inhibition of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) infectivity by peptides analogous to the viral spike protein. Virus Res. 2006;120:146–155. doi: 10.1016/j.virusres.2006.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galdiero S., Falanga A., Galdiero M. Peptides containing membrane-interacting motifs inhibit herpes simplex virus type 1 infectivity. Peptides. 2008;29:1461–1471. doi: 10.1016/j.peptides.2008.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spence J.S., Melnik L.I., Garry R.F. Inhibition of arenavirus infection by a glycoprotein-derived peptide with a novel mechanism. J. Virol. 2014;88:8556–8564. doi: 10.1128/JVI.01133-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Badani H., Garry R.F., Wimley W.C. Peptide entry inhibitors of enveloped viruses: the importance of interfacial hydrophobicity. Biochim. Biophys. Acta. 2014;1838:2180–2197. doi: 10.1016/j.bbamem.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galdiero S., Falanga A., Galdiero M. gH625: a milestone in understanding the many roles of membranotropic peptides. Biochim. Biophys. Acta. 2015;1848(1 Pt A):16–25. doi: 10.1016/j.bbamem.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brügger B., Glass B., Kräusslich H.G. The HIV lipidome: a raft with an unusual composition. Proc. Natl. Acad. Sci. USA. 2006;103:2641–2646. doi: 10.1073/pnas.0511136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huarte N., Carravilla P., Nieva J.L. Functional organization of the HIV lipid envelope. Sci. Rep. 2016;6:34190. doi: 10.1038/srep34190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lorizate M., Huarte N., Nieva J.L. Interfacial pre-transmembrane domains in viral proteins promoting membrane fusion and fission. Biochim. Biophys. Acta. 2008;1778:1624–1639. doi: 10.1016/j.bbamem.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bobardt M.D., Cheng G., Gallay P.A. Hepatitis C virus NS5A anchor peptide disrupts human immunodeficiency virus. Proc. Natl. Acad. Sci. USA. 2008;105:5525–5530. doi: 10.1073/pnas.0801388105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Markosyan R.M., Cohen F.S., Melikyan G.B. Time-resolved imaging of HIV-1 Env-mediated lipid and content mixing between a single virion and cell membrane. Mol. Biol. Cell. 2005;16:5502–5513. doi: 10.1091/mbc.E05-06-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huarte N., Lorizate M., Nieva J.L. The broadly neutralizing anti-human immunodeficiency virus type 1 4E10 monoclonal antibody is better adapted to membrane-bound epitope recognition and blocking than 2F5. J. Virol. 2008;82:8986–8996. doi: 10.1128/JVI.00846-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mattila J.-P., Shnyrova A.V., Frolov V.A. A hemi-fission intermediate links two mechanistically distinct stages of membrane fission. Nature. 2015;524:109–113. doi: 10.1038/nature14509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carravilla P., Nieva J.L., Huarte N. Two-photon Laurdan studies of the ternary lipid mixture DOPC:SM:cholesterol reveal a single liquid phase at sphingomyelin:cholesterol ratios lower than 1. Langmuir. 2015;31:2808–2817. doi: 10.1021/la504251u. [DOI] [PubMed] [Google Scholar]
- 30.Gaus K., Gratton E., Jessup W. Visualizing lipid structure and raft domains in living cells with two-photon microscopy. Proc. Natl. Acad. Sci. USA. 2003;100:15554–15559. doi: 10.1073/pnas.2534386100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Epand R.M., Sayer B.G., Epand R.F. Peptide-induced formation of cholesterol-rich domains. Biochemistry. 2003;42:14677–14689. doi: 10.1021/bi035587j. [DOI] [PubMed] [Google Scholar]
- 32.Epand R.F., Sayer B.G., Epand R.M. The tryptophan-rich region of HIV gp41 and the promotion of cholesterol-rich domains. Biochemistry. 2005;44:5525–5531. doi: 10.1021/bi0500224. [DOI] [PubMed] [Google Scholar]
- 33.Wimley W.C., White S.H. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 1996;3:842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- 34.Lorizate M., de la Arada I., Nieva J.L. Structural analysis and assembly of the HIV-1 Gp41 amino-terminal fusion peptide and the pretransmembrane amphipathic-at-interface sequence. Biochemistry. 2006;45:14337–14346. doi: 10.1021/bi0612521. [DOI] [PubMed] [Google Scholar]
- 35.Apellániz B., Nieva J.L. The use of liposomes to shape epitope structure and modulate immunogenic responses of peptide vaccines against HIV MPER. Adv. Protein Chem. Struct. Biol. 2015;99:15–54. doi: 10.1016/bs.apcsb.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 36.Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 37.Ashkenazi A., Faingold O., Shai Y. HIV-1 fusion protein exerts complex immunosuppressive effects. Trends Biochem. Sci. 2013;38:345–349. doi: 10.1016/j.tibs.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 38.Apellániz B., Rujas E., Nieva J.L. The atomic structure of the HIV-1 gp41 transmembrane domain and its connection to the immunogenic membrane-proximal external region. J. Biol. Chem. 2015;290:12999–13015. doi: 10.1074/jbc.M115.644351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rujas E., Caaveiro J.M., Nieva J.L. Structural basis for broad neutralization of HIV-1 through the molecular recognition of 10E8 helical epitope at the membrane interface. Sci. Rep. 2016;6:38177. doi: 10.1038/srep38177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vincent N., Genin C., Malvoisin E. Identification of a conserved domain of the HIV-1 transmembrane protein gp41 which interacts with cholesteryl groups. Biochim. Biophys. Acta. 2002;1567:157–164. doi: 10.1016/s0005-2736(02)00611-9. [DOI] [PubMed] [Google Scholar]
- 41.Schroeder C. Cholesterol-binding viral proteins in virus entry and morphogenesis. Subcell. Biochem. 2010;51:77–108. doi: 10.1007/978-90-481-8622-8_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang S.T., Kreutzberger A.J., Tamm L.K. The role of cholesterol in membrane fusion. Chem. Phys. Lipids. 2016;199:136–143. doi: 10.1016/j.chemphyslip.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vishwanathan S.A., Hunter E. Importance of the membrane-perturbing properties of the membrane-proximal external region of human immunodeficiency virus type 1 gp41 to viral fusion. J. Virol. 2008;82:5118–5126. doi: 10.1128/JVI.00305-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen S.S., Yang P., Huang S.C. Identification of the LWYIK motif located in the human immunodeficiency virus type 1 transmembrane gp41 protein as a distinct determinant for viral infection. J. Virol. 2009;83:870–883. doi: 10.1128/JVI.01088-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vishwanathan S.A., Thomas A., Epand R.M. Hydrophobic substitutions in the first residue of the CRAC segment of the gp41 protein of HIV. Biochemistry. 2008;47:124–130. doi: 10.1021/bi7018892. [DOI] [PubMed] [Google Scholar]
- 46.Epand R.F., Thomas A., Epand R.M. Juxtamembrane protein segments that contribute to recruitment of cholesterol into domains. Biochemistry. 2006;45:6105–6114. doi: 10.1021/bi060245+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohen T., Cohen S.J., Shai Y. HIV-1 gp41 and TCRα trans-membrane domains share a motif exploited by the HIV virus to modulate T-cell proliferation. PLoS Pathog. 2010;6:e1001085. doi: 10.1371/journal.ppat.1001085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klug Y.A., Rotem E., Shai Y. Mapping out the intricate relationship of the HIV envelope protein and the membrane environment. Biochim. Biophys. Acta. 2017;1859:550–560. doi: 10.1016/j.bbamem.2016.10.012. [DOI] [PubMed] [Google Scholar]
- 49.Aloia R.C., Tian H., Jensen F.C. Lipid composition and fluidity of the human immunodeficiency virus envelope and host cell plasma membranes. Proc. Natl. Acad. Sci. USA. 1993;90:5181–5185. doi: 10.1073/pnas.90.11.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klymchenko A.S., Kreder R. Fluorescent probes for lipid rafts: from model membranes to living cells. Chem. Biol. 2014;21:97–113. doi: 10.1016/j.chembiol.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 51.Bagatolli L.A. To see or not to see: lateral organization of biological membranes and fluorescence microscopy. Biochim. Biophys. Acta. 2006;1758:1541–1556. doi: 10.1016/j.bbamem.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 52.Lee D.W., Min Y., Zasadzinski J.A. Relating domain size distribution to line tension and molecular dipole density in model cytoplasmic myelin lipid monolayers. Proc. Natl. Acad. Sci. USA. 2011;108:9425–9430. doi: 10.1073/pnas.1106368108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu P., Liu J., Roux K.H. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 54.Campbell S.M., Crowe S.M., Mak J. Virion-associated cholesterol is critical for the maintenance of HIV-1 structure and infectivity. AIDS. 2002;16:2253–2261. doi: 10.1097/00002030-200211220-00004. [DOI] [PubMed] [Google Scholar]
- 55.Guyader M., Kiyokawa E., Trono D. Role for human immunodeficiency virus type 1 membrane cholesterol in viral internalization. J. Virol. 2002;76:10356–10364. doi: 10.1128/JVI.76.20.10356-10364.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Veatch S.L., Keller S.L. Organization in lipid membranes containing cholesterol. Phys. Rev. Lett. 2002;89:268101. doi: 10.1103/PhysRevLett.89.268101. [DOI] [PubMed] [Google Scholar]
- 57.Barlic A., Gutiérrez-Aguirre I., González-Mañas J.M. Lipid phase coexistence favors membrane insertion of equinatoxin-II, a pore-forming toxin from Actinia equina. J. Biol. Chem. 2004;279:34209–34216. doi: 10.1074/jbc.M313817200. [DOI] [PubMed] [Google Scholar]
- 58.Schön P., García-Sáez A.J., Schwille P. Equinatoxin II permeabilizing activity depends on the presence of sphingomyelin and lipid phase coexistence. Biophys. J. 2008;95:691–698. doi: 10.1529/biophysj.108.129981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sáez-Cirión A., Arrondo J.L., Nieva J.L. Structural and functional roles of HIV-1 gp41 pretransmembrane sequence segmentation. Biophys. J. 2003;85:3769–3780. doi: 10.1016/S0006-3495(03)74792-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.