Abstract
Small-molecule ligands targeting nucleic acids have been explored as potential therapeutic agents. Duplex groove-binding ligands have been shown to recognize DNA in a sequence-specific manner. On the other hand, quadruplex-binding ligands exhibit high selectivity between quadruplex and duplex, but show limited discrimination between different quadruplex structures. Here we propose a dual-specific approach through the simultaneous application of duplex- and quadruplex-binders. We demonstrated that a quadruplex-specific ligand and a duplex-specific ligand can simultaneously interact at two separate binding sites of a quadruplex-duplex hybrid harbouring both quadruplex and duplex structural elements. Such a dual-specific targeting strategy would combine the sequence specificity of duplex-binders and the strong binding affinity of quadruplex-binders, potentially allowing the specific targeting of unique quadruplex structures. Future research can be directed towards the development of conjugated compounds targeting specific genomic quadruplex-duplex sites, for which the linker would be highly context-dependent in terms of length and flexibility, as well as the attachment points onto both ligands.
Introduction
Small-molecule ligands with strong binding affinity to nucleic acids have been explored as potential therapeutic agents1–3. These ligands recognize DNA either in its canonical double helical form3,4, or in alternative forms such as four-stranded G-quadruplex structure5,6. There are various classes of duplex-binding ligands, including groove-binders3,4,7, intercalators8–11, cross-linking agents12, and triplex-forming oligonucleotides (TFOs)13–15. Sequence-specific recognition of duplex DNA was achieved with groove-binders and triplex-forming oligonucleotides through the explicit establishment of hydrogen-bond interactions. Such targeting approach enables selective gene silencing at the target site through down-regulation of transcription2. On the other hand, the majority of quadruplex-binding drugs investigated to date mainly recognize G-quadruplex structures through stacking onto terminal G-tetrads16–30. Current generation quadruplex-binding ligands exhibit very high binding affinity to G-quadruplex, and they show a high level of discrimination of G-quadruplex from all other DNA structural forms. However, the selectivity of these ligands between different quadruplex structures is still limited. With more than 700,000 potential G-quadruplex-forming sites within the human genome31, the challenge is to develop a quadruplex-binding drug specific to a single genomic G-quadruplex with minimal off-target binding. Here we propose a dual-specific targeting strategy based on the simultaneous application of duplex- and quadruplex-binding ligands. Using NMR spectroscopy we demonstrated that a quadruplex-specific ligand and a duplex-specific ligand can simultaneously interact at two separate binding sites of a quadruplex-duplex hybrid harbouring both quadruplex and duplex structural elements. This approach combines the sequence specificity of duplex-binders with the tight binding affinity of quadruplex-binders, and can be directed towards the selective targeting of quadruplex-duplex hybrid-forming sequences32. Chemical linkage of the two ligands could potentially lead to their synergistic recognition of the target quadruplex-duplex hybrid structure.
Results and Discussion
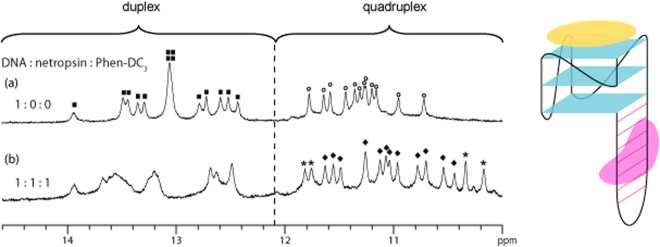
To begin with, a quadruplex-duplex hybrid33 (QD H1; Table S1 and Figure S1, Supporting Information) containing both a duplex segment harbouring six continuous A • T base pairs and a quadruplex was titrated with the duplex-binder netropsin34 (Fig. 1d), which was shown to recognize AT-rich regions, and the bisquinolinium quadruplex-binder Phen-DC3 35 (Fig. 2d), individually. 1D imino proton NMR spectrum of free QDH1 is shown in Fig. 1a: twelve major peaks (indicated with open circles) were observed at 10.6–11.8 ppm, corresponding to the formation of a three-layered G-tetrad core; fourteen major peaks (indicated with filled squares) were observed at 12.4–14 ppm, corresponding to the formation of the duplex stem. Upon adding half the molar equivalent of netropsin to QDH1, additional peaks emerged in the duplex region with a concomitant reduction in the intensity of the original duplex peaks (Fig. 1b), while the quadruplex peaks remained largely unchanged. This indicated the specific binding of netropsin onto the duplex stem, giving rise to equal populations of bound and unbound duplex stems. At 1:1 DNA-to-ligand ratio, the duplex stem was fully bound with netropsin at the AT-rich binding site, as evidenced by the disappearance of the unbound duplex peaks (Fig. 1c), whereas the quadruplex peaks showed minimal change.
Figure 1.
1D imino proton NMR spectrum of (a) free QDH1, (b) QDH1 bound with half the molar equivalent of netropsin, (c) QDH1 bound with equimolar ratio of netropsin, and (d) chemical structure of netropsin, a DNA minor groove-binder. G-quadruplex imino proton peaks are labelled with open circles, whereas duplex imino proton peaks of free QDH1 are labelled with filled squares. Schematic structure of netropsin (in pink) binding to the duplex stem is shown on the right.
Figure 2.
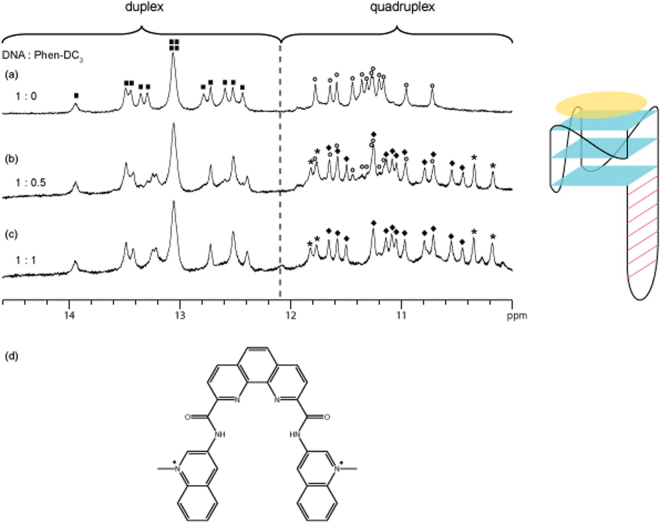
1D imino proton NMR spectrum of (a) free QDH1, (b) QDH1 bound with half the molar equivalent of Phen-DC3, and (c) QDH1 bound with equimolar ratio of Phen-DC3, and (d) chemical structure of Phen-DC3, a quadruplex-binder. Unbound and bound G-quadruplex imino proton peaks are labelled with open circles and filled diamonds, respectively, whereas duplex imino proton peaks of free QDH1 are labelled with filled squares. Peaks labelled with asterisks originate from the ligand Phen-DC3. Schematic structure of Phen-DC3 (in yellow) binding to the G-tetrad is shown on the right.
Titration of QDH1 with Phen-DC3 produced comparable results. Addition of half the molar equivalent of Phen-DC3 to QDH1 led to the emergence of additional peaks in the quadruplex region (indicated by diamonds), with a concomitant reduction in the intensity of the original quadruplex peaks (Fig. 2a, b), while the duplex peaks showed minimal change. This indicated the specific binding of Phen-DC3 onto the G-quadruplex. At 1:1 DNA-to-ligand ratio, the quadruplex was fully bound, as evidenced by the disappearance of the unbound quadruplex peaks (Fig. 2c). Upon Phen-DC3 binding, the quadruplex peaks were generally upfield-shifted, reflecting the aromatic stacking effect of Phen-DC3. Furthermore, these quadruplex peaks were sharp and showed similar linewidths as those in the free DNA, indicating the tight binding of Phen-DC3 to the quadruplex.
Our dual-specific targeting strategy involves the simultaneous application of both duplex and quadruplex ligands for enhanced binding specificity and/or affinity. We demonstrated this approach with the addition of both netropsin and Phen-DC3 to QDH1. The resulting 1D imino proton NMR spectrum displayed features corresponding to those of the respective bound segments; duplex peaks at 12.4–14 ppm showed similar distribution patterns as in QDH1:netropsin complex, while tetrad peaks at 10.4–11.7 ppm matched those of QDH1:Phen-DC3 complex (Fig. 3). These observations indicated that the binding of both netropsin and Phen-DC3 to QDH1 is compatible and non-interfering. 2D NOESY spectrum of QDH1:netropsin:Phen-DC3 complex showed signature G(H1)-C(H41)/G(H1)-C(H42) and T(H3)-A(H2) cross-peaks indicative of Watson-Crick G • C and A • T base pair formation, as well as characteristic guanine imino-H8 cross-peaks consistent with G-tetrad formation (Figure S2, Supporting Information). In addition, cross-peaks were observed between QDH1 and Phen-DC3 (boxed in red; Figure S2, Supporting Information), further supporting their interactions. A structural model of the QDH1:netropsin:Phen-DC3 complex showing the simultaneous binding of netropsin to the duplex AT-rich region and Phen-DC3 to the terminal G-tetrad of QDH1 was built (Figure S3, Supporting Information).
Figure 3.
1D imino proton NMR spectrum of (a) free QDH1 and (b) QDH1 bound with equimolar ratio of netropsin and Phen-DC3. Unbound and bound G-quadruplex imino proton peaks are labelled with open circles and filled diamonds, respectively, whereas duplex imino proton peaks of free QDH1 are labelled with filled squares. Peaks labelled with asterisks originate from the ligand Phen-DC3. Schematic structure of simultaneous netropsin (in pink) and Phen-DC3 (in yellow) binding to QDH1 is shown on the right.
CD-melting experiments were performed to investigate the binding of netropsin and Phen-DC3 to duplex and quadruplex motifs, either individually or in concert. The melting temperature (T m) of the reference duplex hairpin dx (Table S1, Supporting Information), as monitored at 267 nm, was increased by more than 19 °C in the presence of netropsin, but was not affected by the presence of Phen-DC3 (Figure S4, Supporting Information). This indicated that netropsin binds to the duplex hairpin, whereas Phen-DC3 does not. The CD spectrum of free QDH1 was similar to those of QDH1 bound with either or both of netropsin and Phen-DC3 (Figure S5, Supporting Information), suggesting that ligand binding did not alter the folding topology of QDH1. Melting of the quadruplex component of QDH1 (Figure S6, Supporting Information) was monitored at 256 nm – a wavelength at which the CD signal of the duplex was close to zero and only exhibited a small variation over the temperature range. We observed that netropsin did not affect the T m of the G-quadruplex, whereas Phen-DC3 led to an increase in T m of more than 11 °C (Figure S6, Supporting Information). In the presence of both netropsin and Phen-DC3, the T m of the quadruplex was close to that in the presence of Phen-DC3 alone, suggesting that netropsin does not negatively affect the binding of Phen-DC3 onto the quadruplex. Our data were consistent with previous observations of the high selectivity of Phen-DC3 between quadruplex and duplex35, and indicated that netropsin and Phen-DC3 do not negatively interfere with their respective binding of duplex and quadruplex motifs. It was reported that distamycin, a minor groove-binder similar to netropsin, could bind to the groove of a G-quadruplex36,37 or stack on a terminal G-tetrad38. In our case, there exists the possibility that netropsin, aside from binding to the duplex minor groove, could bind to the quadruplex yet do not significantly affect the binding of the high-affinity quadruplex-binder Phen-DC3.
To demonstrate the general applicability of this dual-specific targeting strategy, we further titrated various quadruplex-duplex hybrid constructs (Table S1, Supporting Information) with the same ligands. In all cases, we observed respective binding of both duplex and quadruplex segments by netropsin and Phen-DC3 (Figures S7 and S8, Supporting Information), as indicated by the change in chemical shift of the imino proton peaks. On the other hand, we have also performed titration of QDH1 with different quadruplex-binding ligands including BRACO-1939 and pyridostatin40 (Figures S9–S12, Supporting Information), demonstrating that these ligands too can be utilized in the current approach. Herein, we have employed just the single duplex ligand netropsin, which binds to the minor groove of AT-rich duplexes. In principle, other duplex ligands with different sequence selectivity can also be applied. For instance, the polyamide class of duplex ligands can be designed to target specific sequences of choice2,4,41.
Previously, various approaches for the two-pronged targeting of DNA structures were proposed. These include the combined use of TFO and intercalator to target duplex42, tetrad-stacking ligand and groove-binder to target G-quadruplex43, and tetrad stacking-ligand and antibody to target G-quadruplex44. In these cases, the target elements all arise from the same DNA conformation. On the other hand, the targets in our proposed strategy are duplex and quadruplex structures, two distinct structural conformations.
Duplex groove-binding ligands have been shown to exhibit sequence selectivity with a good binding affinity. In contrast, quadruplex-binding ligands have been shown to exhibit exceptional binding affinity and high selectivity for quadruplex over duplex, but limited discrimination between different quadruplex structures. Our present targeting approach would combine the advantages of the two ligand types: sequence specificity of duplex-binders and tight binding affinity of quadruplex-binders. In certain aspects, we can relate this strategy with bi-specific targeting45–47 and fragment-based drug discovery (FBDD)48–50, which has received mounting interest towards the design of protein-binding ligands. In FBDD, small fragments were screened for their weak binding to the target protein, and subsequently combined into larger lead molecules for further optimization of target affinity and potency. On the other hand, our proposal to combine the use of separate high-affinity quadruplex-binders and sequence specific duplex-binders to target specific genomic sites might address the lack of specificity for G-quadruplex targets.
Furthermore, we propose that a linker could chemically join the two ligands into a single entity, which could provide synergistic binding of the two modules to the quadruplex-duplex hybrid target. The linker should exhibit a certain extent of flexibility, and should take into consideration the structural context at the junction between the duplex and quadruplex segments33. Examples of such linkers that can be used are shown in Figure S13 (Supporting Information). Even though the notion of joining the two ligands by a chemical linker is straightforward, the synthesis of an actual construct targeting a genomic quadruplex-duplex hybrid is non-trivial; the design of the linker would be highly context-dependent (e.g. length and flexibility of the linker, as well as the attachment points onto both ligands) and might not apply between different quadruplex-duplex hybrid systems. On a separate note, the duplex-quadruplex junction can also serve as a unique recognition site by a junction binder33,51,52, which can be applied independently or in combination with the duplex- and quadruplex-binding ligands.
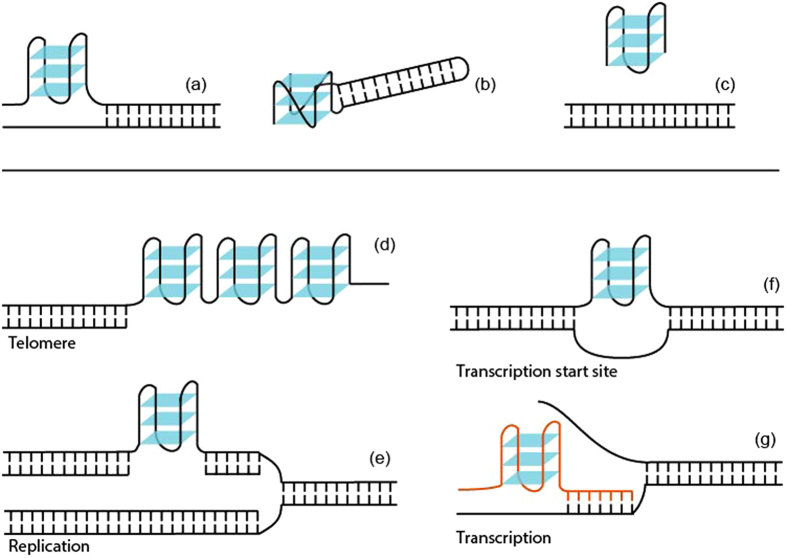
A conjoined duplex- and quadruplex-targeting ligand would bind to duplex and quadruplex elements which are spatially close. This can arise in one of three arrangements: (i) a quadruplex immediately upstream/downstream of a duplex (Fig. 4a), (ii) a quadruplex which harbours within its loop a duplex element (Fig. 4b), and (iii) a quadruplex and a duplex originating from two distinct strands (or that are sequentially far apart) (Fig. 4c). In a biological context, these can happen under various scenarios. We have identified previously that diverse quadruplex-duplex hybrid-forming sequences can be found in the human genome, many of which overlap with regulatory important regions32. These sequences, and their RNA counterparts, would be forthright targets for the dual-targeting approach. On the other hand, simultaneous existence of duplex and quadruplex structures can also occur at the end of telomere53 (Fig. 4d), during replication54 (Fig. 4e), recombination55, transcription56,57 (Fig. 4f,g), and splicing58. In this manner, sequence selectivity provided by the duplex-binding ligand would help in discrimination between the myriad G-quadruplex structures31, thereby enabling the targeting of a unique genomic site.
Figure 4.
Examples of structural (a–c) and biological (d–g) contexts in which both duplex and quadruplex elements exist in close spatial proximity. (a) A quadruplex immediately up-/downstream of a duplex. (b) A quadruplex-duplex hybrid. (c) A quadruplex and a duplex formed by distinct strands. (d) Formation of quadruplexes at the telomere end. (e) Formation of a quadruplex during replication. (f) Formation of a quadruplex at the transcription start site. (g) Formation of a DNA-RNA quadruplex-duplex hybrid during transcription. The RNA transcript is shown in red.
Conclusion
In summary, we have proposed a targeting strategy based on the simultaneous application of duplex- and quadruplex-binding ligands. The simultaneous binding of netropsin and Phen-DC3 to a quadruplex-duplex hybrid construct has been demonstrated, and the approach can be applied to different duplex and quadruplex binders. Hence the targeting approach provides a potential route for the specific targeting of unique sites in the human genome harbouring both quadruplex and duplex structural elements.
Methods
DNA sample preparation
DNA oligonucleotides were chemically synthesized on an ABI 394 DNA/RNA synthesizer using reagents from Glen Research. The oligonucleotides were de-protected using ammonium hydroxide and purified with Poly-PakTM cartridges. DNA samples were successively dialyzed against water and 20 mM KCl solution. They were subsequently lyophilized and dissolved in a buffer containing 20 mM potassium phosphate (pH 7.0) and 20 mM KCl.
Ligand preparation
Netropsin, BRACO-19, and pyridostatin were purchased from Sigma Aldrich. Phen-DC3 was a gracious gift from Marie-Paule Teulade-Fichou. Lyophilized Phen-DC3 was dissolved in dimethyl sulfoxide. All other ligands in lyophilized form were dissolved in water.
NMR spectroscopy
Strand concentration of NMR samples was typically 0.2–1.5 mM. 1D spectra were recorded on Bruker AVANCE 600-MHz spectrometer at 25 °C and processed with the software TopSpinTM.
Circular dichroism
Circular dichroism (CD) spectra were recorded at 25 °C on a Jasco-815 spectropolarimeter over the range of 220–320 nm using a 1-cm path length quartz cuvette with a reaction volume of 500 µL. The DNA concentration was typically 4 µM. For each spectrum, an average of three scans was taken, the spectrum of the buffer was subtracted, and the data were zero-corrected at 320 nm. For CD-melting experiments, heating was performed across the temperature range of 15–95 °C. The full spectrum was recorded at intervals of 1 °C.
Construction of structural model
The model of a quadruplex was built with the XPLOR-NIH program59 using constraints adapted from reported quadruplex-duplex hybrid structures33. The quadruplex model was combined with a duplex generated by LEaP60 in the PYMOL program to obtain the structural model of QDH1. Netropsin and Phen-DC3 were then aligned onto QDH1 in PyMOL based on reported duplex:netropsin (PDB code: 2LWH) and quadruplex:Phen-DC3 (PDB code: 2MGN) structures.
Electronic supplementary material
Acknowledgements
This research was supported by Singapore Ministry of Education Academic Research Fund Tier 3 (MOE2012-T3-1-001) and Tier 2 (MOE2012-T2-1-102) and Nanyang Technological University grants to A. T. Phan. We thank Marie-Paule Teulade-Fichou for generously providing the bisquinolinium compound Phen-DC3.
Author Contributions
A.T.P. conceived the idea. T.Q.N.N. performed experiments under the supervision of K.W.L. All authors designed experiments, analysed data, and co-wrote the paper.
Competing Interests
A provisional patent on the dual-specific targeting method has been filed by Nanyang Technological University with all three authors as inventors.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-10583-9
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hélène C, Toulme JJ. Specific regulation of gene expression by antisense, sense and antigene nucleic acids. Biochim. Biophys. Acta. 1990;1049:99–125. doi: 10.1016/0167-4781(90)90031-V. [DOI] [PubMed] [Google Scholar]
- 2.Gottesfeld JM, Neely L, Trauger JW, Baird EE, Dervan PB. Regulation of gene expression by small molecules. Nature. 1997;387:202–205. doi: 10.1038/387202a0. [DOI] [PubMed] [Google Scholar]
- 3.Neidle S. DNA minor-groove recognition by small molecules. Nat. Prod. Rep. 2001;18:291–309. doi: 10.1039/a705982e. [DOI] [PubMed] [Google Scholar]
- 4.Trauger JW, Baird EE, Dervan PB. Recognition of DNA by designed ligands at subnanomolar concentrations. Nature. 1996;382:559–561. doi: 10.1038/382559a0. [DOI] [PubMed] [Google Scholar]
- 5.Balasubramanian S, Hurley LH, Neidle S. Targeting G-quadruplexes in gene promoters: A novel anticancer strategy? Nat. Rev. Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neidle S. A personal history of quadruplex-small molecule targeting. Chem. Rec. 2015;15:691–710. doi: 10.1002/tcr.201500011. [DOI] [PubMed] [Google Scholar]
- 7.Spink N, Brown DG, Skelly JV, Neidle S. Sequence-dependent effects in drug-DNA interaction: the crystal structure of Hoechst 33258 bound to the d (CGCAAATTTGCG)2 duplex. Nucleic Acids Res. 1994;22:1607–1612. doi: 10.1093/nar/22.9.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lerman LS. The structure of the DNA-acridine complex. Proc. Natl. Acad. Sci. 1963;49:94–102. doi: 10.1073/pnas.49.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peek ME, et al. DNA distortion in bis-intercalated complexes. Biochemistry. 1994;33:3794–3800. doi: 10.1021/bi00179a002. [DOI] [PubMed] [Google Scholar]
- 10.Malinina L, Soler-Lopez M, Aymami J, Subirana JA. Intercalation of an acridine-peptide drug in an AA/TT base step in the crystal structure of [d(CGCGAATTCGCG)]2 with six duplexes and seven Mg2+ ions in the asymmetric unit. Biochemistry. 2002;41:9341–9348. doi: 10.1021/bi020135c. [DOI] [PubMed] [Google Scholar]
- 11.Horowitz ED, Lilavivat S, Holladay BW, Germann MW, Hud NV. Solution structure and thermodynamics of 2′,5′ RNA intercalation. J. Am. Chem. Soc. 2009;131:5831–5838. doi: 10.1021/ja810068e. [DOI] [PubMed] [Google Scholar]
- 12.Brulikova L, Hlavac J, Hradil P. DNA interstrand cross-linking agents and their chemotherapeutic potential. Curr. Med. Chem. 2012;19:364–385. doi: 10.2174/092986712803414295. [DOI] [PubMed] [Google Scholar]
- 13.Felsenfeld G, Davies DR, Rich A. Formation of a three-stranded polynucleotide molecule. J. Am. Chem. Soc. 1957;79:2023–2024. doi: 10.1021/ja01565a074. [DOI] [Google Scholar]
- 14.Moser HE, Dervan PB. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987;238:645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- 15.Beal PA, Dervan PB. Second structural motif for recognition of DNA by oligonucleotide-directed triple-helix formation. Science. 1991;251:1360–1363. doi: 10.1126/science.2003222. [DOI] [PubMed] [Google Scholar]
- 16.Monchaud D, Teulade-Fichou MP. A hitchhiker’s guide to G-quadruplex ligands. Org. Biomol. Chem. 2008;6:627–636. doi: 10.1039/B714772B. [DOI] [PubMed] [Google Scholar]
- 17.Alzeer J, Vummidi BR, Roth PJ, Luedtke NW. Guanidinium-modified phthalocyanines as high-affinity G-quadruplex fluorescent probes and transcriptional regulators. Angew. Chem. Int. Ed. 2009;48:9362–9365. doi: 10.1002/anie.200903685. [DOI] [PubMed] [Google Scholar]
- 18.Dai J, Carver M, Hurley LH, Yang D. Solution structure of a 2:1 quindoline-c-MYC G-quadruplex: insights into G-quadruplex-interactive small molecule drug design. J. Am. Chem. Soc. 2011;133:17673–17680. doi: 10.1021/ja205646q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collie GW, et al. Structural basis for telomeric G-quadruplex targeting by naphthalene diimide ligands. J. Am. Chem. Soc. 2012;134:2723–2731. doi: 10.1021/ja2102423. [DOI] [PubMed] [Google Scholar]
- 20.Nicoludis JM, et al. Optimized end-stacking provides specificity of N-methyl mesoporphyrin IX for human telomeric G-quadruplex DNA. J. Am. Chem. Soc. 2012;134:20446–20456. doi: 10.1021/ja3088746. [DOI] [PubMed] [Google Scholar]
- 21.Bessi I, et al. Spectroscopic, molecular modeling, and NMR-spectroscopic investigation of the binding mode of the natural alkaloids berberine and sanguinarine to human telomeric G-quadruplex DNA. ACS Chem. Biol. 2012;7:1109–1119. doi: 10.1021/cb300096g. [DOI] [PubMed] [Google Scholar]
- 22.Bazzicalupi C, Ferraroni M, Bilia AR, Scheggi F, Gratteri P. The crystal structure of human telomeric DNA complexed with berberine: an interesting case of stacked ligand to G-tetrad ratio higher than 1:1. Nucleic Acids Res. 2013;41:632–638. doi: 10.1093/nar/gks1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung WJ, et al. Solution structure of an intramolecular (3 + 1) human telomeric G-quadruplex bound to a telomestatin derivative. J. Am. Chem. Soc. 2013;135:13495–13501. doi: 10.1021/ja405843r. [DOI] [PubMed] [Google Scholar]
- 24.Chung WJ, Heddi B, Hamon F, Teulade-Fichou MP, Phan AT. Solution structure of a G-quadruplex bound to the bisquinolinium compound Phen-DC3. Angew. Chem. Int. Ed. 2014;53:999–1002. doi: 10.1002/anie.201308063. [DOI] [PubMed] [Google Scholar]
- 25.Trajkovski M, et al. Interactions of Pt-ttpy with G-quadruplexes originating from promoter region of the c-myc gene deciphered by NMR and gel electrophoresis analysis. Chem. Eur. J. 2015;21:7798–7807. doi: 10.1002/chem.201500347. [DOI] [PubMed] [Google Scholar]
- 26.Riva B, et al. Molecular recognition in naphthoquinone derivatives - G-quadruplex complexes by NMR. Biochim. Biophys. Acta. 2015;1850:673–680. doi: 10.1016/j.bbagen.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 27.Kotar A, et al. NMR structure of a triangulenium-based long-lived fluorescence probe bound to a G-quadruplex. Angew. Chem. Int. Ed. 2016;55:12508–12511. doi: 10.1002/anie.201606877. [DOI] [PubMed] [Google Scholar]
- 28.Pavan Kumar Y, et al. Fluorescent dansyl-guanosine conjugates that bind c-MYC promoter G-quadruplex and downregulate c-MYC expression. ChemBioChem. 2016;17:388–393. doi: 10.1002/cbic.201500631. [DOI] [PubMed] [Google Scholar]
- 29.Scaglioni L, Mondelli R, Artali R, Sirtori FR, Mazzini S. Nemorubicin and doxorubicin bind the G-quadruplex sequences of the human telomeres and of the c-MYC promoter element Pu22. Biochim Biophys Acta. 2016;1860:1129–1138. doi: 10.1016/j.bbagen.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 30.Funke A, Dickerhoff J, Weisz K. Towards the development of structure-selective G-quadruplex-binding Indolo[3,2-b]quinolines. Chem. Eur. J. 2016;22:3170–3181. doi: 10.1002/chem.201504416. [DOI] [PubMed] [Google Scholar]
- 31.Chambers VS, et al. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015;33:877–881. doi: 10.1038/nbt.3295. [DOI] [PubMed] [Google Scholar]
- 32.Lim KW, et al. Duplex stem-loop-containing quadruplex motifs in the human genome: a combined genomic and structural study. Nucleic Acids Res. 2015;43:5630–5646. doi: 10.1093/nar/gkv355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim KW, Phan AT. Structural basis of DNA quadruplex–duplex junction formation. Angew. Chem. Int. Ed. 2013;52:8566–8569. doi: 10.1002/anie.201302995. [DOI] [PubMed] [Google Scholar]
- 34.Wartell RM, Larson JE, Wells RD. Netropsin. A specific probe for A-T regions of duplex deoxyribonucleic acid. J. Biol. Chem. 1974;249:6719–6731. [PubMed] [Google Scholar]
- 35.De Cian A, Delemos E, Mergny JL, Teulade-Fichou MP, Monchaud D. Highly efficient G-quadruplex recognition by bisquinolinium compounds. J. Am. Chem. Soc. 2007;129:1856–1857. doi: 10.1021/ja067352b. [DOI] [PubMed] [Google Scholar]
- 36.Martino L, et al. Structural and thermodynamic studies of the interaction of distamycin A with the parallel quadruplex structure [d(TGGGGT)]4. J. Am. Chem. Soc. 2007;129:16048–16056. doi: 10.1021/ja075710k. [DOI] [PubMed] [Google Scholar]
- 37.Cosconati S, et al. Structural and conformational requisites in DNA quadruplex groove binding: Another piece to the puzzle. J. Am. Chem. Soc. 2010;132:6425–6433. doi: 10.1021/ja1003872. [DOI] [PubMed] [Google Scholar]
- 38.Cocco MJ, Hanakahi LA, Huber MD, Maizels N. Specific interactions of distamycin with G-quadruplex DNA. Nucleic Acids Res. 2003;31:2944–2951. doi: 10.1093/nar/gkg392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Read M, et al. Structure-based design of selective and potent G quadruplex-mediated telomerase inhibitors. Proc. Natl. Acad. Sci. USA. 2001;98:4844–4849. doi: 10.1073/pnas.081560598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodriguez R, et al. A novel small molecule that alters shelterin integrity and triggers a DNA-damage response at telomeres. J. Am. Chem. Soc. 2008;130:15758–15759. doi: 10.1021/ja805615w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawamoto Y, et al. Development of a new method for synthesis of tandem hairpin pyrrole-imidazole polyamide probes targeting human telomeres. J. Am. Chem. Soc. 2013;135:16468–16477. doi: 10.1021/ja406737n. [DOI] [PubMed] [Google Scholar]
- 42.Mergny JL, et al. Triple helix-specific ligands. Science. 1992;256:1681–1684. doi: 10.1126/science.256.5064.1681. [DOI] [PubMed] [Google Scholar]
- 43.Zhao P, et al. Novel porphyrin-daunomycin hybrids: synthesis and preferential binding to G-quadruplexes over i-motif. Spectrochim. Acta A. 2015;137:227–235. doi: 10.1016/j.saa.2014.08.123. [DOI] [PubMed] [Google Scholar]
- 44.Yangyuoru PM, et al. Dual binding of an antibody and a small molecule increases the stability of TERRA G-quadruplex. Angew. Chem. Int. Ed. 2015;127:924–927. doi: 10.1002/ange.201408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodrik-Outmezguine VS, et al. Overcoming mTOR resistance mutations with a new-generation mTOR inhibitor. Nature. 2016;534:272–276. doi: 10.1038/nature17963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waring MJ, et al. Potent and selective bivalent inhibitors of BET bromodomains. Nat. Chem. Biol. 2016;12:1097–1104. doi: 10.1038/nchembio.2210. [DOI] [PubMed] [Google Scholar]
- 47.Kontermann RE, Brinkmann U. Bispecific antibodies. Drug Discov. Today. 2015;20:838–847. doi: 10.1016/j.drudis.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 48.Murray CW, Rees DC. The rise of fragment-based drug discovery. Nat. Chem. 2009;1:187–192. doi: 10.1038/nchem.217. [DOI] [PubMed] [Google Scholar]
- 49.Scott DE, Coyne AG, Hudson SA, Abell C. Fragment-based approaches in drug discovery and chemical biology. Biochemistry. 2012;51:4990–5003. doi: 10.1021/bi3005126. [DOI] [PubMed] [Google Scholar]
- 50.Erlanson DA, Fesik SW, Hubbard RE, Jahnke W, Jhoti H. Twenty years on: the impact of fragments on drug discovery. Nat. Rev. Drug. Discov. 2016;15:605–619. doi: 10.1038/nrd.2016.109. [DOI] [PubMed] [Google Scholar]
- 51.Oleksy A, et al. Molecular recognition of a three-way DNA junction by a metallosupramolecular helicate. Angew. Chem. Int. Ed. 2006;45:1227–1231. doi: 10.1002/anie.200503822. [DOI] [PubMed] [Google Scholar]
- 52.Russo Krauss I, Ramaswamy S, Neidle S, Haider S, Parkinson GN. Structural insights into the quadruplex-duplex 3′ interface formed from a telomeric repeat: A potential molecular target. J. Am. Chem. Soc. 2016;138:1226–1233. doi: 10.1021/jacs.5b10492. [DOI] [PubMed] [Google Scholar]
- 53.Phan AT. Human telomeric G-quadruplex: Structures of DNA and RNA sequences. FEBS J. 2010;277:1107–1117. doi: 10.1111/j.1742-4658.2009.07464.x. [DOI] [PubMed] [Google Scholar]
- 54.Rhodes D, Lipps HJ. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015;43:8627–8637. doi: 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat. Struct. Mol. Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 56.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu RY, Zheng KW, Zhang JY, Hao YH, Tan Z. Formation of DNA:RNA hybrid G-quadruplex in bacterial cells and its dominance over the intramolecular DNA G-quadruplex in mediating transcription termination. Angew. Chem. Int. Ed. 2015;54:2447–2451. doi: 10.1002/anie.201408719. [DOI] [PubMed] [Google Scholar]
- 58.Weldon C, et al. Identification of G-quadruplexes in long functional RNAs using 7-deazaguanine RNA. Nat. Chem. Biol. 2017;13:18–20. doi: 10.1038/nchembio.2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/S1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 60.Case DA, et al. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.