Abstract
Arbuscular mycorrhizal fungi (AMF) are widespread root symbionts that perform important ecological services, such as improving plant nutrient and water acquisition. Some AMF from the Gigasporaceae family host a population of endobacteria, Candidatus Glomeribacter gigasporarum (Cagg). The analysis of the Cagg genome identified six putative toxin–antitoxin modules (TAs), consisting of pairs of stable toxins and unstable antitoxins that affect diverse physiological functions. Sequence analysis suggested that these TA modules were acquired by horizontal transfer. Gene expression patterns of two TAs (yoeB/yefM and chpB/chpS) changed during the fungal life cycle, with the expression during the pre-symbiotic phase higher than during the symbiosis with the plant host. The heterologous expression in Escherichia coli demonstrated the functionality only for the YoeB–YefM pair. On the basis of these observations, we speculate that TA modules might help Cagg adapt to its intracellular habitat, coordinating its proliferation with the physiological state of the AMF host.
Arbuscular mycorrhizal fungi (AMF) perform key ecological services, improving nutrient acquisition and water uptake by their plant hosts, while receiving fixed carbon from the host (Smith and Read, 2010). Many fungi, particularly from basal clades, harbor bacterial endosymbionts (Bonfante and Desirò, 2017) and AMF from the Gigasporaceae family host a population of Burkholderia-related microbes (Bianciotto et al., 1996) named Candidatus Glomeribacter gigasporarum (Cagg). Cagg is vertically transmitted and currently uncultivable; although not essential for Gigaspora survival, Cagg can enhance fungal fitness (Bianciotto et al., 2003; Lumini et al., 2007; Salvioli et al., 2016). Analysis of the 1.72-Mb Cagg genome revealed strong nutritional dependence on the fungal host (Ghignone et al., 2012). We wondered whether the Cagg genome possesses genetic determinants involved in its endocellular lifestyle and environmental sensing. Our previous study identified potentially secreted proteins, which could act as effectors (Ghignone et al., 2012); the present study identified genes encoding toxin–antitoxin (TA) systems.
Bacterial TA systems (TAs) are typically encoded in operons located on plasmids or on the bacterial chromosome. They can be classified into five types (I–V) with the antitoxin acting as a protein (types II, IV and V) or as a noncoding RNA (types I and III) (Schuster and Bertram, 2013). The well-studied type II TAs include a stable toxin protein and an unstable antitoxin that counteracts the effect of the toxin (Leplae et al., 2011). Depending on the TA superfamily, the type II toxins either interfere with DNA replication (for example, CcdB) or with the translation of mRNA (for example, MazE and RelE; Mruk and Kobayashi, 2014). It was originally proposed that obligate host-associated bacteria lost their TAs as a consequence of the reduction in genome size experienced during evolution (Pandey and Gerdes, 2005). For example, Mycobacterium tuberculosis possesses 88 TAs, but the genome of the obligate intracellular pathogen Mycobacterium leprae has no TAs, supporting the idea that TAs help free-living bacteria cope with the changing environment. However, data from high-throughput sequencing have challenged this view by revealing TAs in endocellular bacteria, including pathogens and beneficial symbionts (Sevin and Barloy-Hubler, 2007). By contrast, the genomes of insect endosymbionts do not seem to carry TA operons, except Wolbachia, the secondary symbiont of Drosophila melanogaster, which encodes seven relBE-like TAs (Sevin and Barloy-Hubler, 2007).
TAs have multiple roles, including those in programmed cell death (Bayles, 2014), stress adaptation (Ramage et al., 2009) and survival in host cells (Helaine et al., 2014). For example, persistence of Salmonella within macrophages requires the presence of TAs (Helaine et al., 2014). In the symbiotic nitrogen-fixing Sinorhizobium meliloti, a mutation of the toxin from the ntrPR system improved symbiotic efficiency and mutations of the vapBC-5 pair influenced nitrogen fixation capacity and bacteroid senescence (Olah et al., 2001; Lipuma et al., 2014). These data suggest that intracellular endobacteria may exploit TAs to modulate their interactions with host cells.
To date, characterization of TAs from fungal endobacteria has been limited to descriptions of their presence in the genome. However, due to their involvement in modulation of growth under stress conditions and survival in host cells, TAs have been hypothesized to play important roles in regulating the life of fungal endobacteria (Lackner et al., 2011). Here we characterize TAs present in the Cagg genome and test the functionality of two of them.
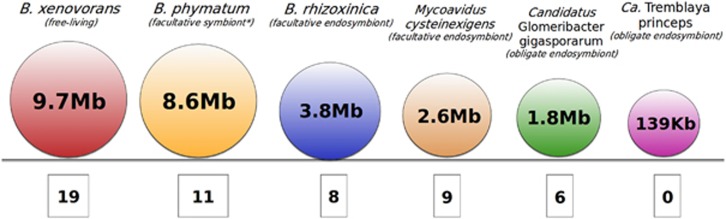
Analysis of the Cagg genome revealed six complete TAs and three orphan coding sequences (Supplementary Table 1). We also conducted a similar analysis of the genomes of Cagg relatives with different lifestyles, considering two endofungal bacteria, namely Burkholderia rhizoxinica, living inside Rhizopus microsporus (Partida-Martinez and Hertweck, 2005), and Mycoavidus cysteinexigens living inside Mortierella (Ohshima et al., 2016; Uehling et al., 2017), as well as the obligate endosymbiont of the citrus mealybug, Candidatus Tremblaya princeps (López-Madrigal et al., 2011). The genomes of B. rhizoxinica, M. cysteinexigens and Cagg encode TAs, albeit fewer than in their free-living relatives (Figure 1). By contrast, no TA could be confidently predicted for Ca. Tremblaya princeps, probably due to its extremely reduced genome (only 139 kb). Comparison of Cagg TAs with those from its close relative B. rhizoxinica revealed a low sequence identity and a lack of collinearity between the two species (Supplementary Table 2). In most cases, the sequences showed more similarity to phylogenetically distant bacteria than to each other (Supplementary Table 1), as also supported by phylogenetic analyses (Supplementary Figure 1). These results are consistent with the acquisition of TAs through lateral gene transfer, as extensively demonstrated for other bacteria (Makarova et al., 2009), including endocellular ones as Rickettsia (Audoly et al., 2011).
Figure 1.
The number of TAs found in the genome of the Cagg endosymbiont and its closest relatives. The approximate genome size is given in circles and the number of TAs is given below the circles. Genomes were scanned for TAs using the RASTA and TA finder online tools. *B. phymatum is a free-living microbe that can nodulate legume roots (Vandamme et al., 2002).
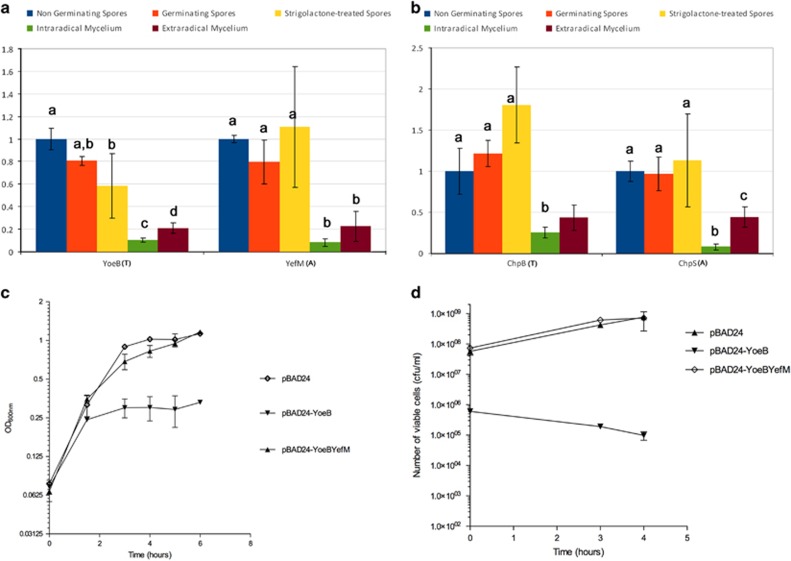
To test whether the Cagg TAs play a functional role, we selected two of them, a yoeB–yefM-like TA pair (CAGGBEG34_v5_20154- CAGGBEG34_v5_20012) and a chpB-chpS-like pair (CAGGBEG34_v5_60038- CAGGBEG34_v5_60039) from the complete TAs. These modules are likely to have a chromosomal and plasmidic localization, respectively (see Supplementary Information) and they both belong to the type II TAs (RelE and MazF superfamilies, respectively). Measurement of the expression of these TAs at different stages in the life cycle of G. margarita, from the pre-symbiotic stages (non-germinating, germinating, strigolactone-treated spores) to the formation of the symbiotic mycelium (see Supplementary Figure 2 for a depiction of the fungal life cycle), showed that TA gene expression changed throughout the fungal life cycle (Figures 2a and b), with more expression during the fungal pre-symbiotic stages. G. margarita is a biotroph, requiring the plant environment to complete its life cycle. The pre-symbiotic stage, when fungal growth mostly depends on its endogenous nutrient supplies, represents a critical step for the fungus as it explores the surrounding soil to find and associate with a plant root. Under these conditions, excessive proliferation of the endobacterium, which acts as an energy sink, might be deleterious for the survival of the fungal/bacterial system. Such stressful conditions might trigger the activation of TA transcription, in contrast to the symbiotic stage, when there is a balanced plant-fungal nutrient exchange. Thus, it is possible that Cagg TAs are involved in the bacterial response to the stress experienced during the fungal pre-symbiotic stages and/or in the survival inside spores, by the induction of a state comparable to the TA-induced persistence state described in Salmonella (Helaine et al., 2014). Interestingly, the expression of the Cagg cell division gene ftsZ inversely mirrors the expression of the tested TAs, suggesting that TA activity negatively correlates with bacterial growth (Anca et al., 2009).
Figure 2.
Relative expression of the bacterial gene pairs yoeB/yefM (a) and chpB/chpS (b) obtained by RT-qPCR according to the 2−ΔΔct method. Means with different superscripts differ significantly (Kruskal–Wallis, P<0.05). The biological material includes the fungal host G. margarita, which contains the endobacterium Cagg. The tested conditions were the following: non-germinating spores (control); germinating spores; strigolactone-treated spores; symbiotic intra-radical mycelium colonizing Lotus japonicus roots; extra-radical mycelium, developing at the root surface. Strigolactones are phytohormones that stimulate AMF hyphal growth and branching (Akiyama et al., 2005). Compared with the pre-symbiotic stages (spores), the expression of the TA operons decreased when the fungus formed the symbiotic mycelium (extra- and intra-radical). Growth (c) and viability (d) of E. coli expressing the YoeB toxin from plasmid pBAD24-YoeB and the YoeB and YefM proteins from plasmid pBAD24-YoeBYefM. The expression was induced by the addition of 1% arabinose to the bacterial cultures. Control cells carried the empty vector pBAD24. Samples taken at 0, 3 and 4 h after induction were plated and living cell numbers were calculated from three independent experiments. Bars represent standard deviations.
To test whether the Cagg TAs encode functional toxins and antitoxins, we heterologously expressed the proteins from an inducible promoter, as Cagg currently cannot be cultivated in vitro. The expression of the YoeB protein strongly affected the growth of E. coli cells, producing a decrease in numbers of living cells as early as 3 h after toxin induction. By contrast, the expression of YoeB toxin together with its cognate antitoxin YefM did not affect E. coli growth (Figures 2c and d, Supplementary Figure 3). These results demonstrate that YoeB affects E. coli cell viability, while YefM prevents its toxic effect, and that this system acts as a TA module. The activity of the ChpB toxin was also analyzed but ChpB showed no effect on E. coli growth and viability in our experimental conditions (data not shown).
TAs from the RelE and MazF superfamilies help bacteria cope with nutritional stresses (Gerdes et al., 2005). Recent evidence indicates that Mycobacterium tuberculosis Rel loci also react to other stresses, such as oxidative and nitrogen-limiting conditions (Korch et al., 2015). For this reason, we next tested whether an oxidative stress can induce the YoeB–YefM TA gene expression. Cagg has been shown to promote antioxidative responses in its fungal host (Vannini et al., 2016); therefore, we challenged germinating spores with H2O2. This stress induced the upregulation of TA gene expression, with statistically significant upregulation of the yefM antitoxin gene (Supplementary Figure 4).
The observations described above demonstrate the functionality of a TA module from an endobacterium that lives inside a fungus that lives inside a plant. TA gene expression is regulated throughout the fungal life cycle, and we speculate that it can respond to external stimuli by modulating bacterial cell division. In conclusion, our findings suggest that TAs might represent one of the genetic determinants that coordinate the Cagg population dynamics with the life cycle of its fungal host.
Acknowledgments
Research was funded by the local project, 60% to PB. Exchanges between Torino and Nice University were supported by the Italian-French PRES Agreement. The authors wish to express their thanks to Jennifer Mach for the language revision. They also thank Stefano Ghignone, Mara Novero and Veronica Volpe, as well as the RASTA software developers.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Akiyama K, Matsuzaki K, Hayashi H. (2005). Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827. [DOI] [PubMed] [Google Scholar]
- Anca IA, Lumini E, Ghignone S, Salvioli A, Bianciotto V, Bonfante P. (2009). The ftsZ gene of the endocellular bacterium 'Candidatus Glomeribacter gigasporarum' is preferentially expressed during the symbiotic phases of its host mycorrhizal fungus. Mol Plant Microbe Interact 22: 302–310. [DOI] [PubMed] [Google Scholar]
- Audoly G, Vincentelli R, Edouard S, Georgiades K, Mediannikov O, Gimenez G et al. (2011). Effect of rickettsial toxin VapC on its eukaryotic host. PLoS One 6: e26528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayles KW. (2014). Bacterial programmed cell death: making sense of a paradox. Nat Rev Microbiol 12: 63–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianciotto V, Bandi C, Minerdi D, Sironi M, Tichy HV, Bonfante P. (1996). An obligately endosymbiotic mycorrhizal fungus itself harbors obligately intracellular bacteria. Appl Environ Microbiol 62: 3005–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianciotto V, Lumini E, Bonfante P, Vandamme P. (2003). ‘Candidatus Glomeribacter gigasporarum’ gen. nov., sp. nov., an endosymbiont of arbuscular mycorrhizal fungi. Int Syst Evol Micr 53: 121–124. [DOI] [PubMed] [Google Scholar]
- Bonfante P, Desirò A. (2017). Who lives in a fungus? The diversity, origins and functions of fungal endobacteria living in the Mucoromycota. ISME J e-pub ahead of print 7 April 2017 doi:10.1038/ismej.2017.21. [DOI] [PMC free article] [PubMed]
- Gerdes K, Christensen SK, Løbner-Olesen A. (2005). Prokaryotic toxin–antitoxin stress response loci. Nat Rev Microbiol 3: 371–382. [DOI] [PubMed] [Google Scholar]
- Ghignone S, Salvioli A, Anca I, Lumini E, Ortu G, Petiti L et al. (2012). The genome of the obligate endobacterium of an AM fungus reveals an interphylum network of nutritional interactions. ISME J 6: 136–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helaine S, Cheverton AM, Watson KG, Faure LM, Matthews SA, Holden DW. (2014). Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 343: 204–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korch SB, Malhotra V, Contreras H, Clark-Curtiss JE. (2015). The Mycobacterium tuberculosis relBE toxin:antitoxin genes are stress-responsive modules that regulate growth through translation inhibition. J Microbiol 53: 783–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner G, Moebius N, Partida-Martinez LP, Boland S, Hertweck C. (2011). Evolution of an endofungal lifestyle: deductions from the Burkholderia rhizoxinica genome. BMC Genomics 12: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leplae R, Geeraerts D, Hallez R, Guglielmini J, Drèze P, Van Melderen L. (2011). Diversity of bacterial type II toxin-antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Res 39: 5513–5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipuma J, Cinege G, Bodogai M, Olah B, Kiers A, Endre G et al. (2014). A vapBC-type toxin-antitoxin module of Sinorhizobium meliloti influences symbiotic efficiency and nodule senescence of Medicago sativa. Environ Microbiol 16: 3714–3729. [DOI] [PubMed] [Google Scholar]
- López-Madrigal S, Latorre A, Porcar M, Moya A, Gil R. (2011). Complete genome sequence of ‘Candidatus Tremblaya princeps’ strain PCVAL, an intriguing translational machine below the living-cell status. J Bacteriol 193: 5587–5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumini E, Bianciotto V, Jargeat P, Novero M, Salvioli A, Faccio A et al. (2007). Presymbiotic growth and sporal morphology are affected in the arbuscular mycorrhizal fungus Gigaspora margarita cured of its endobacteria. Cell Microbiol 9: 1716–1729. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Koonin EV. (2009). Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol Direct 4: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mruk I, Kobayashi I. (2014). To be or not to be: regulation of restriction-modification systems and other toxin-antitoxin systems. Nucleic Acids Res 42: 70–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima S, Sato Y, Fujimura R, Takashima Y, Hamada M, Nishizawa T et al. (2016). Mycoavidus cysteinexigens gen. nov., sp. nov., an endohyphal bacterium isolated from a soil isolate of the fungus Mortierella elongata. Int J Syst Evol Microbiol 66: 2052–2057. [DOI] [PubMed] [Google Scholar]
- Olah B, Kiss E, Gyorgypal Z, Borzi J, Cinege G, Csanadi G et al. (2001). Mutation in the ntrR gene, a member of the vap gene family, increases the symbiotic efficiency of Sinorhizobium meliloti. Mol Plant Microbe Interact 14: 887–894. [DOI] [PubMed] [Google Scholar]
- Pandey DP, Gerdes K. (2005). Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res 33: 966–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partida-Martinez LP, Hertweck C. (2005). Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature 437: 884–888. [DOI] [PubMed] [Google Scholar]
- Ramage HR, Connolly LE, Cox JS. (2009). Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet 5: e1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvioli A, Ghignone S, Novero M, Navazio L, Venice F, Bagnaresi P et al. (2016). Symbiosis with an endobacterium increases the fitness of a mycorrhizal fungus, raising its bioenergetic potential. ISME J 10: 130–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster CF, Bertram R. (2013). Toxin-antitoxin systems are ubiquitous and versatile modulators of prokaryotic cell fate. FEMS Microbiol Lett 340: 73–85. [DOI] [PubMed] [Google Scholar]
- Sevin EW, Barloy-Hubler F. (2007). RASTA-bacteria: a web-based tool for identifying toxin-antitoxin loci in prokaryotes. Genome Biol 8: R155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SE, Read DJ. (2010) Mycorrhizal Symbiosis. Academic Press: London. [Google Scholar]
- Uehling J, Gryganskyi A, Hameed K, Tschaplinski T, Misztal PK, Wu S et al. (2017). Comparative genomics of Mortierella elongata and its bacterial endosymbiont Mycoavidus cysteinexigens. Environ Microbiol 2017; e-pub ahead of print 11 January 2017; doi:10.1111/1462-2920.13669. [DOI] [PubMed]
- Vandamme P, Goris J, Chen WM, de Vos P, Willems A. (2002). Burkholderia tuberum sp. nov. and Burkholderia phymatum sp. nov., nodulate the roots of tropical legumes. Syst Appl Microbiol 25: 507–512. [DOI] [PubMed] [Google Scholar]
- Vannini C, Carpentieri A, Salvioli A, Novero M, Marsoni M, Testa L et al. (2016). An interdomain network: the endobacterium of a mycorrhizal fungus promotes antioxidative responses in both fungal and plant hosts. New Phytol 211: 265–275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.