Abstract
Activities of rhizosphere microbes are key to the functioning of terrestrial ecosystems. It is commonly believed that bacteria are the major consumers of root exudates and that the role of fungi in the rhizosphere is mostly limited to plant-associated taxa, such as mycorrhizal fungi, pathogens and endophytes, whereas less is known about the role of saprotrophs. In order to test the hypothesis that the role of saprotrophic fungi in rhizosphere processes increases with increased time after abandonment from agriculture, we determined the composition of fungi that are active in the rhizosphere along a chronosequence of ex-arable fields in the Netherlands. Intact soil cores were collected from nine fields that represent three stages of land abandonment and pulse labeled with 13CO2. The fungal contribution to metabolization of plant-derived carbon was evaluated using phospholipid analysis combined with stable isotope probing (SIP), whereas fungal diversity was analyzed using DNA-SIP combined with 454-sequencing. We show that in recently abandoned fields most of the root-derived 13C was taken up by bacteria but that in long-term abandoned fields most of the root-derived 13C was found in fungal biomass. Furthermore, the composition of the active functional fungal community changed from one composed of fast-growing and pathogenic fungal species to one consisting of beneficial and slower-growing fungal species, which may have essential consequences for the carbon flow through the soil food web and consequently nutrient cycling and plant succession.
Introduction
Studying chronosequences has increased our understanding of plant community succession, an important theme in ecology (Tilman, 1988). Most chronosequence studies have focused on plants, but they also offer great opportunities to study soil microbial community responses to primary or secondary vegetation succession and coinciding changes in soil characteristics (Kuramae et al., 2010; Hell et al., 2013; Dini-Andreote et al., 2014). One such series of sites in the Netherlands represents a secondary succession chronosequence and consists of ex-agricultural fields abandoned at different moments in time. All sites are located in the Veluwe, a region in the center of the Netherlands on the same parent soil material originating from glacial sand deposits. Since the Middle Ages, species-rich, seminatural grasslands, heathlands and even drifts sands have developed on this parent material as a consequence of agricultural practices. After the introduction of artificial fertilizers in the beginning of the former century and through intensification of agricultural practice, the area covered by species-rich grasslands and heathlands declined dramatically. In order to restore these seminatural ecosystems, agricultural soils have been taken out of production at different moments in time. This has resulted in a chronosequence of fields, in the present study covering 7–30 years of land abandonment, which has served as a model system to study the relationship between plant secondary succession and soil abiotic and biotic factors (Kardol et al., 2006; Van der Wal et al., 2006; Holtkamp et al., 2008; Van de Voorde et al., 2011).
It is increasingly acknowledged that soil microbes are strongly involved in plant community succession (Bardgett and van der Putten, 2014). Microbes not only control soil carbon and nutrient cycling affecting plant community performance indirectly (Wardle et al., 2004) but can also have direct effects on plants via mutualistic and pathogenic interactions (Buee et al., 2009). Fungi are assumed to be more important in natural ecosystems than in intensively managed systems that are largely dominated by bacteria (de Boer et al., 2005; de Vries et al., 2006). Thus systems in transition such as this chronosequence of abandoned arable fields can give valuable information on the shifts in microbial communities and consequently on their contribution to soil ecosystem functioning.
In initial studies, it has been assumed that saprotrophic fungal biomass would increase over time of secondary succession due to changes in substrate quality and that at the same time associations between (arbuscular) mycorrhizal fungi and plants would become more important due to depletion of easily available nutrients (Holtkamp et al., 2008). These belowground interactions have been proposed to contribute to the observed replacement in plant species composition and cover of fast-growing early successional species to slow-growing later successional plants (Kardol et al., 2006). However, when the fungal biomass and fungal:bacterial biomass ratio were investigated, an initial increase during the first 2 years after abandonment was seen without any further effect on fungal abundance or fungal:bacterial biomass ratio for the next 30 years (Van der Wal et al., 2006). Yet, an increase in carbon and nitrogen mineralization rates was observed (Holtkamp et al., 2011; Tardy et al., 2015), indicating an increase in soil conditions for microbial decomposer activity without enhancing plant productivity (Holtkamp et al., 2011). When the identities of the archaeal, bacterial and fungal communities along this chronosequence were assessed, the microbial communities appeared to differ between soils abandoned for <10 years and for >25 years (Thomson et al., 2015). However, this only partly explained changes in mineralization rates and could not be related to the changes in plant community productivity and composition.
The aim of the current study was to relate the secondary succession on these ex-arable fields to the active fungal community in the rhizosphere using 13CO2 pulse labeling of plants and subsequent isotope analysis of both fungal lipids and DNA in intact soil cores collected along this so-called ‘Veluwe chronosequence’. We focused on the active rhizosphere fungi regulated mainly by the plant community (Strickland et al., 2015) and used a pulse labeling approach to separate the fungi that are actively assimilating recently photosynthesized, plant-derived carbon from the inactive fungi or fungi fulfilling other ecological roles. Our main interest was in saprotrophs using root exudates thus priming soil organic matter decomposition and affecting mineralization rates, plant pathogens and arbuscular mycorrhizal fungi (AMF). The internal transcribed spacer (ITS) region of fungi was sequenced using 454-sequencing, and the fungi were divided into functional trait-based categories based on knowledge of the ecology of species (Tedersoo et al., 2014, Nguyen et al., 2016).
The research questions of the current study were
how does the diversity, functional composition and species interactions of the active rhizosphere fungal community change during secondary succession?
what are the main (biotic and abiotic) factors driving these changes?
Our hypothesis is that the fungal community will follow secondary succession and its functionality will change in time since agriculture from one dominated with fast-growing species common in agricultural soils to a community more often found in soils of species-rich grasslands. We further hypothesize that the change in fungal community composition is due to changes in plant community, soil abiotic parameters, autogenic succession and/or result from interactions with other soil organisms.
Materials and methods
Experimental set-up and sampling
In July 2012, intact soil cores were collected from nine grassland sites that were taken out of agricultural production 7–30 years prior to sampling. All fields are located in the south Veluwe region, in the center of the Netherlands (Supplementary Table S1). In three separate sampling locations per field, 4 soil cores (12 cm diameter, at least 20 cm deep) were collected with native vegetation growing on it. We selected sites that were dominated by grasses, Agrostis capillaris, Holcus lanatus and Plantago lanceolata to decrease variation due to differences in plant cover. More details on the sampling and sample handling are presented in Morriën et al. (2017) and in Supplementary Materials and Methods.
Nine cores per field site were labeled with 99.99 atom-% 13CO2 (Cambridge Isotope Laboratories, Andover, MA, USA) in an artificially lit air-tight growth chamber for a total of 8 h. Three other cores per field site were placed in a similar chamber and kept under identical conditions but with a 12CO2 atmosphere, representing the control treatment. Samples were collected at 24 h and 7 and 14 days after the start of labeling. Samples from rhizosphere were collected from the upper, root-filled part of the cores by brushing the roots. More details on 13C labeling and sampling are presented in Supplementary Materials and Methods. Soil chemistry and analysis of soil biota was analyzed and data are given by Morriën et al. (2017). Here these data are used as background information for correlation analysis.
Phospholipid analysis combined with stable isotope probing (PLFA and NLFA) analysis
PLFAs were extracted from freeze-dried soil and concentrations and δ13C values were measured on a Finnigan Delta-S gas chromatograph–isotope ratio mass spectrometer as described in Boschker (2004). For more details, see Supplementary Materials and Methods.
Molecular and bioinformatic analysis
DNA was extracted from rhizosphere soil using the MoBIO PowerSoil Kit according to the manufacturer’s instructions. The quantity of DNA was inspected using Quant-iT PicoGreen dsDNA Assay Kit (Thermo Life Sciences, Waltham, MA, USA) using λ-DNA as standard. 13C-enriched (‘heavy’) DNA was separated from non-labeled (‘light’) DNA by density-gradient centrifugation and analyzed as described in Neufeld et al. (2007). Fungal PCR was performed for combined ‘heavy’ and ‘light’ fractions for each sample using primers ITS4 and ITS9 (Ihrmark et al., 2012) and subsequent 454-sequencing was performed. Resulted sequences were submitted to ENA under project number PRJEB15250. For more details on molecular and bioinformatics analysis, see Supplementary Materials and Methods.
The fungi were divided into functional groups when possible. This approach is similar to the one taken earlier by Tedersoo et al. (2014) and refined by Nguyen et al. (2016). The functional groups used in this study were: AMF, ericoid mycorrhizae, coprotrophic fungi, endophytes and potential plant pathogens, molds, nematophagous fungi, saprotrophic fungi, wood parasites and decomposers, yeasts, entomopathogens, animal pathogens, lichens, ectomycorrhizal fungi, and mycoparasites. The groups not expected to have functions in rhizosphere that also had little reads/operational taxonomic units (OTUs) assigned to them (including entomopathogens, animal pathogens, lichens, ectomycorrhizal fungi and mycoparasites) were later grouped into a group ‘other’. The types that could not be identified into functional groups were categorized as unclassified Chytridiomycota, unclassified Mucoromycota, unclassified Ascomycota, unclassified Basidiomycota and unclassified fungi. Species could belong to multiple functional groups, especially yeasts and molds (lifestyles) were also members of other groups (that is, saprotrophs and pathogens). Both data on OTUs’ presence–absence and OUTs’ percentage of total reads were used in the analysis. For each analysis, the data used are highlighted.
The relative labeling of each OTU was determined as the ratio of the abundance of that OTU in the heavy 13C fraction to that of the abundance in the light 12C fraction.
Statistical analysis
The difference in the amount of reads per sample was standardized by using the percentage of total OTUs in a sample as a measure for the relative abundance of each OTU (McMurdie and Holmes, 2014). Absolute values were used for diversity and rarefaction calculations. The effect of treatment (recent, mid- and long-term abandonment) on fungal community composition at the level of phylum, class, order and OTU was estimated using analysis of variance (ANOVA) combined with Tukey’s pairwise comparisons as the post hoc test. When the distribution of data was not in accordance with assumptions of ANOVA (that is, due to complete absence of a group in one or more treatments) Kruskal–Wallis test was used in combination with Mann–Whitney as a post hoc test. The effect of abandonment time was evaluated using Pearson’s correlation coefficient.
The OTUs explaining most differences between treatments were analyzed using SIMPER in program PAST (Hammer et al., 2001) and their statistical significance was confirmed with ANOVA. Cluster analysis (using WARD linkages) was performed to analyze the similarity between ‘heavy’ and ‘ light’ fractions from the same sample. NMDS using both presence–absence data and the percentage of data was applied to visualize the effects of time since labeling, the fraction (that is, presence in ‘heavy’ or ‘light’ fraction) and the agricultural abandonment time on community composition. Differences in beta-diversity between abandonment categories were evaluated using analysis of similarity (ANOSIM) with Jaccard as a distance measure in PAST.
In order to assess α-diversity, both Simpson and Shannon H indices were calculated using the non-transformed raw data and resulting values were subjected to ANOVA to compare treatment effects. Rarefaction curves of the observed richness were calculated in PAST using 1000-fold resampling without replacement.
Variance partitioning was performed in CANOCO (Ter Braak and Smilauer, 1998) using both forward selection of the variables and canonical correspondence analysis (CCA) with data derived from Morrien et al. (2017).
In order to explore which species are most connected to others in different abandonment categories, co-occurrence networks were used (Barberán et al., 2012). Data on only labeled (that is, ‘heavy’ fractions) OTUs were used and time points were combined to get a better view on the system making the total number of samples used for correlations 3 (replicates) × 3 (time points sampled) × 3 (soils in the land abandonment category)=27. Only species present in >3 samples per abandonment category and with relative abundance of >1% were used in analysis. Significant correlations (Pearson’s rho >0.5, P<0.05, adjusted using the false discovery rate method in R) were used to plot the networks in Cytoscape (Shannon et al., 2003).
Data from labeling status of fungal feeding soil biota was taken from another data set (Morrien, et al. 2017) and correlated with the fungal data in R using Pearson’s correlation coefficient.
Results
The total number of good quality reads was 1.8 × 106, which were divided into 3562 OTUs. The average number of reads per sample was 10 123 and the average number of OTUs per sample was 412. There were no significant differences in the total number of reads or numbers of identified OTUs between heavy and light fractions, time points (incubation) or time of abandonment. The rarefaction curves of all samples are presented in Supplementary Figure S1.
Of the total raw reads, 80.5% was certainly identified using ITSx (Bengtsson-Palme et al., 2013) to be ITS sequences from fungi. The remaining reads were categorized mainly as plant derived (14%) and Alveolates (superphylum of protists) (4%). These sequences were removed from the dataset and are not presented or discussed here. Approximately 10% of the reads (and 20% of the OTUs) assigned as fungi could not be assigned to finer taxonomic level than ‘unknown fungi’. The fungal communities across the samples in these abandoned ex-arable ecosystems consisted predominantly of Sordariomycetes (making up 25.1% of all reads and 19.5% of all OTUs) and of Dothideomycetes (21.7% and 12.0%, respectively). According to the functional classification, the majority of reads (70.1%) and OTUs (52.7%) were classified as ‘saprotrophic’.
There were 18 OTUs with average abundance of >1% across the samples and fractions and 141 OTUs with an average abundance of >0.1%. These 141 OTUs are referred to as the dominant OTUs in the data set. The most commonly found OTU with an average abundance of 7.12% was a Pleosporales sp. (OTU 5302), which was also present in all samples.
Fungal contribution to the carbon flow
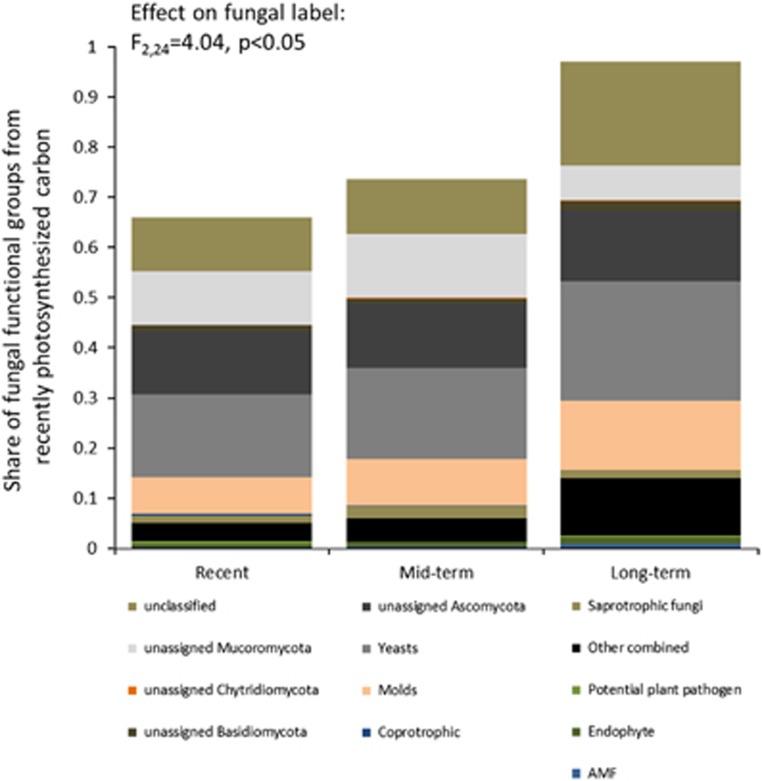
Neither the amount of fungi as based on the PLFA signature of 18:2ω6c nor the fungi-to-bacteria ratio (F:B ratio) changed during the secondary succession following land abandonment (F2,24=1.19, P=0.32). However, when the excess 13C in fungal and bacterial PLFAs (calculated compared with PLFAs in 12C labeled control samples) was used to calculate the ‘active F:B ratio’, we observed that fungi were receiving relatively more carbon from the plants in the long-term abandoned fields than in recent and mid-term abandoned fields (Figure 1). Furthermore, the ‘active F:B ratio’ was positively correlated with time since abandonment from agriculture (R2=0.48, P<0.05).
Figure 1.
The share of functional groups of fungi incorporating recently photosynthesized carbon by vegetation on soil cores of three land abandonment categories. The total amount of carbon from plants to fungi is calculated as an average of three soils for each of the three time categories using the excess 13C labeling information of the PLFA marker 18:2ω6c and set to 1 for the long-term soils. For assignment of ITS sequences to functional groups, see text.
Active vs total fungi in the rhizosphere
Heavy and light fractions (that is, fungal communities enriched in 13C and 12C) were similar to each other in the general composition of the fungi when all samples were included in the analysis (Supplementary Figure S2A, ANOSIM for 13C vs 12C R<0.20, P>0.20). Furthermore, the Ward linkage analysis revealed that in 10 of the 18 soil samples tested at the first time point, heavy and light fractions from the same sample clustered together. However, the light fractions were significantly more diverse (measured by Shannon-H index) than the heavy fractions when all the samples were combined (t=2.6, P<0.05), which was mainly explained by observed differences between the fractions at the first sampling point 24 h after labeling (Supplementary Figure S2B).
Active rhizosphere communities in soils and between treatments
Land abandonment time category had no significant effect on the α-diversities of the fungi in the heavy fraction at any time point after labeling (ANOVA: F=0.74, P=0.48) (Supplementary Figure S3). A significant effect of the abandonment time category on fungal community structure in the ‘heavy’ fraction was found in samples that were taken 1 day after labeling (ANOSIM: R=0.42, P=0.024, Supplementary Figure S4). No effect of time since abandonment was detected for the two other sampling time points or in the ‘light’ fractions. For this reason, a more detailed analysis was made for fungi assimilating recently photosynthesized carbon (that is, in the 13C labeled ‘heavy’ fraction in the first time point).
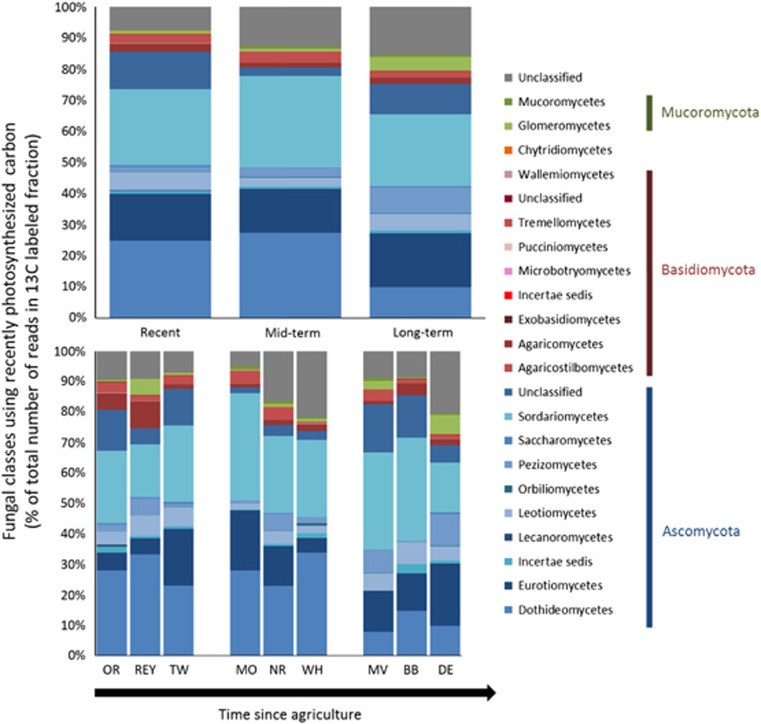
At the level of classes, using the proportional data, some significant directional changes in the composition of fungi using recently photosynthesized carbon were detected despite the large variation between soils (Figure 2). The classes most affected by the time since land abandonment were: Dothideomycetes (F2,27=13.2, P<0.01, R2=0.46, P<0.05) and Agaricomycetes (F2,27=4.5, P=0.06, R2=0.40, P=0.07), which decreased in abundance with increasing abandonment time, Leotiomycetes (F2,27=5.4, P<0.05), which showed the highest abundance in mid-term abandoned fields, and Lecanoromycetes (H=7.6, P=0.06), which were completely absent in mid- and long-term abandoned fields (Supplementary Figure S5). When looking at a more detailed level, the orders affected by the treatment were Capnodiales (F2,27=12.5, P<0.01), Pleosporales (F2,27=12.0, P<0.01) and unclassified Dothideomycetes (F2,27=5.5, P<0.05).
Figure 2.
Effect of the time since abandonment from agriculture on fungal classes in the samples calculated from the percentage of data. For significant differences between treatments, see Supplementary Figure S5.
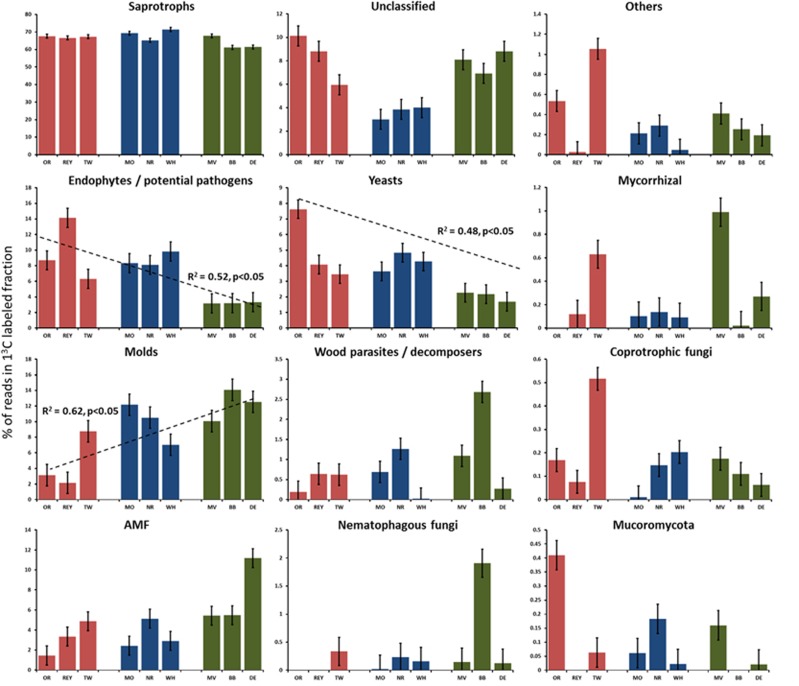
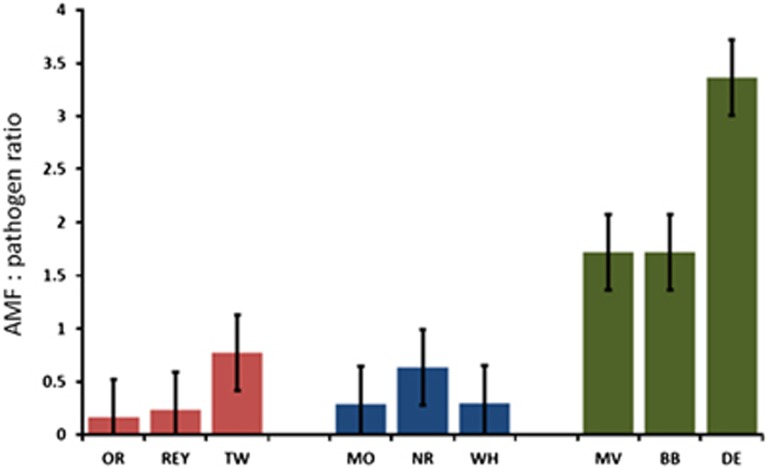
When OTUs were divided into functional categories, significant effects of time since land abandonment were detected: ‘potential plant pathogens and endophytes’ and ‘yeasts’ were proportionally more abundant in recently and mid-term abandoned fields than in long-term abandoned fields (F2,18=5.5, P<0.05 and F2,18=11.88, P<0.05, respectively) and also decreased in proportion as a function of time since abandonment (R2=0.52 and R2= 0.48, for both P<0.05; Figure 3). Saprotrophic fungi were most abundant in the mid-term abandoned fields (F2,18=11.60, P<0.01), whereas the proportion of ‘molds’ was the highest in long-term abandoned fields and increased with time since abandonment (F2,18=5.68, P<0.05, R2=0.62, P<0.05; Figure 3). Furthermore, there was a trend that the proportion of AMF of total fungi receiving carbon from the plants increased with time since abandonment (F2,18=2.64, P=0.15). When a ratio between AMF to potential pathogens was calculated, a significant effect of the time abandonment category (F2,18=9.97, P=0.01) and time since abandonment (R2=0.51, P<0.05) was found (Figure 4).
Figure 3.
Fungal functional groups using recently photosynthesized carbon from plants affected by the time since agricultural abandonment. The recently abandoned fields are represented by red bars, mid-term abandoned fields by blue and long-term abandoned fields by green. The bars represent averages and error bars represent s.e. The significant trends in time are shown in the figure; for significance between treatments, see text. The groups are ordered based on their abundances in samples starting from the most abundant group. The full colour version of this figure is available at ISME Journal online.
Figure 4.
The AMF/pathogen ratio changing in time since agricultural abandonment. The recently abandoned fields are marked as red bars, mid-term abandoned fields as blue and long-term abandoned fields with green. The bars represent averages and error bars represent s.e. The full colour version of this figure is available at ISME Journal online.
Some of the soils had a significantly larger proportion of certain functional groups of fungi: for example, long-term abandoned field BB had significantly more OTUs categorized as wood decomposers or parasites and nematophagous fungi than other fields (Figure 3) probably due to its vicinity to the forest. These differences were not consistent over abandonment time but rather related to unique attributes to the field probably related to the abiotic or biotic factors in that specific field. Likewise, each soil had several OTUs that were unique: they were present in at least two soil samples from that site but absent in other sites. One of the sites abandoned for 30 years (BB) had the most unique OTUs (64) while a recently abandoned site REY had no unique OTUs at all (Supplementary Figure S6).
There were 816 OTUs present in at least two sites for each abandonment category. Fifty-eight OTUs were found in at least two long-term abandoned fields but were not found in earlier abandoned fields. Of these, only two OTUs were consistently found in all long-term abandoned fields but not in any other field (Supplementary Table S4). The recently abandoned fields had 33 OTUs that were not present in older abandoned fields and only 4 of them were present in all 3 recently abandoned field soils (Supplementary Table S4). When the number of shared OTUs between fields was related to the number of years since abandonment, a significant negative correlation was found (R2=0.25, P<0.01), indicating that there is a directional succession in the fungal community composition.
The contribution of individual OTUs to the observed differences between the abandonment time categories was evaluated using SIMPER and the significance in the presence of OTUs between treatments was confirmed using ANOVA. The top 20 most abundant OTUs explaining 32.4% of the observed differences between abandonment time categories are shown in Table 1. Owing to large variation between soils, only 4 out of the 20 OTUs selected by SIMPER were significantly different between abandonment categories when evaluated with ANOVA (Table 1). These OTUs were mostly categorized as ascomycetes with unknown function.
Table 1. SIMPER and ANOVA analysis on the effects of time since agricultural abandonment on rhizosphere fungi actively assimilating root exudates.
|
Treatment |
Treatment effect |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OTU number | Function | Phyla | Class | Order | Taxon | Variation explained (%) | Recent | Mid-term | Long term | F (H) | P | |
| Otu05302 | Unc. Ascomycota | Ascomycota | Dothideomycetes | Pleosporales | Pleosporales sp. | 5.58 | 3.42 (±1.30)* | 14.17 (±3.26) | 0.66 (±0.19) | 16.69 | 0.004 | |
| Otu05176 | Unclassified | Unclassified | Unclassified | Unclassified | Unclassified | 2.91 | 0.95 (±0.42) | 8.11 (±3.16) | 0.37 (±0.14) | 6.91 | 0.028 | |
| Otu04821 | Unc. Ascomycota | Ascomycota | Dothideomycetes | Pleosporales | Pleosporales sp. | 2.10 | 4.52 (±0.57)* | 0.78 (±0.07) | 2.73 (±2.75)* | 2.70 | 0.146 | |
| Otu05253 | Unc. Ascomycota | Ascomycota | Dothideomycetes | Pleosporales | Dendryphion sp. | 1.96 | 0.64 (±0.16) | 0.14 (±0.04) | 0.34 (±0.14) | 2.98 | 0.126 | |
| Otu05238 | Unc. Ascomycota | Ascomycota | Sordariomycetes | Hypocreales | Trichoderma sp. | 1.95 | 1.38 (±0.22) | 2.93 (±1.02) | 1.94 (±0.32) | 1.17 | 0.372 | |
| Otu05163 | Unc. Ascomycota | Ascomycota | Sordariomycetes | Hypocreales | Fusarium sp. | 1.81 | 7.12 (±1.59) | 7.17 (±0.57) | 7.73 (±3.03) | 0.05 | 0.954 | |
| Otu05264 | Unc. Ascomycota | Ascomycota | Pezizomycetes | Unclassified | Pezizomycetes sp. | 1.63 | 1.88 (±0.67) | 3.71 (±1.09) | 3.73 (±2.86) | 0.22 | 0.811 | |
| Otu05193 | Mold | Ascomycota | Eurotiomycetes | Eurotiales | Trichocomaceae sp. | 1.59 | 2.52 (±1.75)* | 2.85 (±1.20)* | 4.57 (±1.79)* | 0.35 | 0.718 | |
| Otu05099 | Endophyte | Ascomycota | Dothideomycetes | Pleosporales | Drechslera sp. BAFC3419 | 1.54 | 4.41 (±3.44) | 1.68 (±1.31) | 1.19 (±0.52) | 0.49 | 0.638 | |
| Otu05240 | Animal pathogen | Ascomycota | Eurotiomycetes | Chaetothyriales | Exophiala equina | 1.34 | 3.84 (±1.35) | 2.86 (±0.62) | 6.26 (±1.02) | 2.10 | 0.200 | |
| Otu04695 | Mold | Ascomycota | Eurotiomycetes | Eurotiales | Aspergillus sp. | 1.31 | 0.38 (±1.58) | 1.81 (±1.40) | 2.35 (±1.53) | 1.53 | 0.290 | |
| Otu05105 | Unclassified | Unclassified | Unclassified | Unclassified | Unclassified | 1.24 | 0.79 (±0.23) | 1.13 (±0.48) | 3.78 (±1.66) | 2.52 | 0.150 | |
| Otu05177 | Unc. Ascomycota | Ascomycota | Unclassified | Unclassified | Unclassified | 1.11 | 1.37 (±0.39) | 0.37 (±0.11) | 3.57 (±2.12)* | 5.6 (H) | 0.062 | |
| Otu05254 | Unc. Ascomycota | Ascomycota | Dothideomycetes | Pleosporales | Unclassified | 1.07 | 2.38 (±1.43) | 0.29 (±0.07) | 2.41 (±1.78) | 0.90 | 0.449 | |
| Otu05212 | Unc. Ascomycota | Ascomycota | Unclassified | Unclassified | Unclassified | 1.05 | 0.62 (±0.15) | 2.32 (±0.57) | 2.17 (±1.83) | 0.53 | 0.611 | |
| Otu05235 | Yeast | Basidiomycota | Tremellomycetes | Trichosporonales | Trichosporon sp. | 1.01 | 0.97 (±0.36)* | 1.93 (±0.85) | 0.32 (±0.23) | 15.12 | 0.005 | |
| Otu05120 | Unc. Ascomycota | Ascomycota | Sordariomycetes | Unclassified | Unclassified | 0.81 | 2.49 (±2.16) | 0.09 (±0.09) | 0.63 (±0.55) | 0.6 (H) | 0.723 | |
| Otu05027 | Unc. Ascomycota | Ascomycota | Dothideomycetes | Capnodiales | Davidiellaceae sp. | 0.80 | 3.21 (±0.60) | 0.61 (±0.25) | 1.14 (±0.34) | 10.40 | 0.008 | |
| Otu05243 | Saprotrophs | Ascomycota | Sordariomycetes | Chaetosphaeriales | Chloridium sp. GHJ3 | 0.73 | 0.91 (±0.28) | 2.70 (±0.43) | 1.64 (±0.91) | 0.98 | 0.427 | |
| Otu05269 | Mold | Ascomycota | Eurotiomycetes | Eurotiales | Penicillium sp. | 0.60 | 0.14 (±0.08) | 2.38 (±1.26) | 0.18 (±0.10) | 5.42 (H) | 0.071 | |
Abbreviation: ANOVA, analysis of variance. Asterisk behind the abundance in a treatment indicates that the same OTU was identified as one of the most connected OTUs in that treatment using network approaches. Values in bold denote significant differences between treatments.
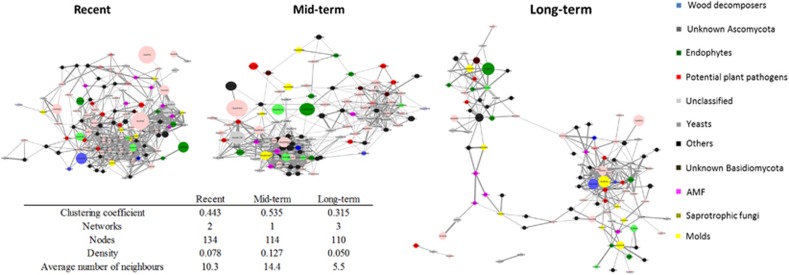
To reveal the connectedness of the species and possibly find more indicator species of land abandonment time, co-occurrence networks were constructed for the different abandonment stages (Figure 5). Network topology revealed that density of networks was the highest at mid-term abandoned fields (0.13) and the lowest at long-term abandoned fields (0.05). Species in the networks from mid-term fields also had the highest number of direct neighbors (average of 14 per OTU). Interestingly, we found a split of network into two subclusters (‘hubs’) starting in mid-term abandoned fields and continuing to long-term fields (Figure 5) where the two ‘hubs’ were completely separated.
Figure 5.
Development of network topology abandonment time. Only significant (P<0.05) positive correlations are visualized. The node size is proportional to the percentage of abundance of the organism and edge darkness is scaled to the strength of the correlation (Pearson’s rho). OTUs presented in the figure are grouped to functional groups (see text) and colored based on function. Network parameters such as clustering coefficient, number of nodes, density and number of neighbors are presented in the figure. The full colour version of this figure is available at ISME Journal online.
We looked in more detail to the OTUs with most neighbors in each of the three land abandonment categories in order to identify potentially important species in the systems. In recent fields, the most connected OTUs were two unclassified Ascomycota, OTU4843 and OTU5273, both with 24 neighbors. In mid-term abandoned fields, the most connected OTU was identified as Coniochaetales sp. with 36 neighbors. In long-term fields, the OTU with most neighbors was Lecythophora sp. with 23 neighbors (Supplementary Table S5). Furthermore, some OTUs identified as indicators by SIMPER analysis were among the top connected OTUs in respective fields (Table 1). Notably, OTU5177, unclassified ascomycete, was more abundant and also among the top connected species in the long-term fields making it a good candidate for further analyses as a keystone OTU.
Abiotic factors affecting rhizosphere fungi
Variance partitioning was performed in order to find out which abiotic and biotic factors affect the fungi in the ‘heavy’ fraction the most. Factors tested were: soil pH, N-mineralization, soil C, total N, total P, and C:N ratio. Biotic factors included were: δ13C in root, fungal PLFA and fungal feeding groups, and dominant plant species in the core. Abiotic variables explained 23.7% of the total variation in CCA (F=1.2, P=0.012) but the measured biotic variables failed to explain significant variation alone or in combination. Of the abiotic variables, pH and organic carbon in the soils were factors significantly explaining most of the active rhizosphere fungal composition (Table 2). Time since abandonment in years (explaining 3.6% of the variation, F=1.3, P=0.049), abandonment time category (4.0% of the variation, F=1.4, P=0.016) and soil (4.3% of the variation, F=1.5, P=0.012) were found to effect OTU composition when constrained and unconstrained CCAs were compared (Table 2). The sampling time (1 day, 1 week or 2 weeks) did not significantly affect the overall OTU composition in the heavy fraction.
Table 2. Variance partitioning of the treatment effect, abiotic and biotic variables influencing rhizosphere fungal communities.
| Treatment variables | % Variation explained | F | P |
|---|---|---|---|
| Soil | 4.3 | 1.5 | 0.012 |
| Treatmenta | 4.0 | 1.4 | 0.016 |
| Time since abandonment | 3.6 | 1.3 | 0.049 |
| Sampling time | 1.1 | 1.0 | 0.424 |
| Abiotic parameters | 23.7 | 1.2 | 0.012 |
| N-mineralization, mg kg−1 week−1 | 3.3 | 1.1 | 0.234 |
| pH | 5.0 | 1.7 | 0.006 |
| Total N | 2.4 | 0.8 | 0.804 |
| C/N | 3.4 | 1.2 | 0.134 |
| Organic matter | 3.8 | 1.3 | 0.016 |
| Phosphorous | 3.6 | 1.2 | 0.056 |
| Biotic parameters | 28.9 | 1.1 | 0.346 |
| Plant species in the core | 3.4 | 1.2 | 0.134 |
| Roots C | 4.2 | 1.4 | 0.082 |
| Fungal C | 2.6 | 0.9 | 0.604 |
| Protists | 1.9 | 0.6 | 0.834 |
| Fungivorous nematodes | 5.6 | 1.2 | 0.228 |
| Fungivorous collembola | 2.1 | 0.6 | 0.955 |
| Fungivorous prostigmatic mites | 3.7 | 0.5 | 0.838 |
| Fungivorous astigmatic mites | 5.0 | 0.8 | 0.766 |
| Fungivorous cryptostigmatic mites | 4.2 | 1.0 | 0.402 |
Treatment refers to categories ‘recent’, ‘mid-term’ and ‘long term’.
Fungal feeding organisms
Data on the labeling status of morphologically identified soil animals collected 1 week after labeling were correlated with fungal orders and functional groups. This revealed that relative abundance in 13C fraction of groups such as ericoid mycorrhizal fungi, molds, wood decomposers, parasites and nematophagous fungi was related to 13C carbon detected in fungal feeding cryptostigmatic mites 1 week later (Supplementary Table S7). Labeling of fungivorous nematodes with 13C was correlated with labeling of coprophilic, entomopathogenic and potential plant pathogenic fungi. Labeling of fungivorous prostigmatic mites was related to general saprotrophic fungi (Supplementary Table S7) and amount of 13C in protist PLFA was positively correlated with the percentage of Saccharomycetes (yeasts) and Chytridiomycetes in the heavy fractions of the samples (Supplementary Table S6).
Discussion
Fungal α- and β-diversity was determined for a total of 9 ex-arable fields that were abandoned 7–30 years prior to sampling. As expected, we observed that a large proportion of the variation in the diversity of fungal communities could be explained by abiotic factors such as pH and organic matter content and a lesser part by abandonment time. High explanatory values based on abiotic variables is more rule than exception for soil fungal community composition (Tedersoo et al., 2014; Thomson et al., 2015). Nevertheless, the current set-up enabled us to separate the effects of abandonment time from the abiotic effects. This approach demonstrated a shift in the active community of fungi consuming recently fixed (13C labeled) carbon with progressing time since land abandonment. We showed that fungi become a more important sink of recently photosynthesized carbon during secondary succession, in spite of no increase of the fungal:bacterial ratio or fungal biomass (Van der Wal et al., 2006; Morriën et al., 2017). Earlier it has been proposed that shifts in flow of carbon and relative contribution of fungi compared with bacteria has direct consequences for the food-web architecture and influence through microbial community function on ecosystem functions, such as carbon and nitrogen mineralization (Holtkamp et al., 2011; Malik et al., 2016; Morrien, et al., 2017). Recently, it has been proposed that fungi in many systems might be more important consumers of labile organic compounds, such as root exudates, than thought (Hannula et al., 2012; de Vries and Caruso, 2016; Morrien, et al., 2017). However, the identity and functionality of these rhizosphere fungi was unknown. Our identification of these fungi reveals that the active fungal community using these labeled compounds is highly diverse and largely influenced by soil and plant.
The division of fungi into functional guilds based on phylogeny and literature is becoming a common practice in fungal ecology (Nguyen et al., 2016). We acknowledge that such division into functional guilds is rather artificial as in different circumstances fungi can have different roles, for example, along the continuum pathogen–endophyte–saprotroph (Arnold, 2007; Aguilar-Triguerosa et al., 2015). However, when performed conservatively and based on available literature, this classification will give a more complete picture on the function of soil fungal communities. Following this approach, it appeared that abandonment time had the strongest effect on communities of potential endophytes and plant pathogens and yeasts (decrease with abandonment time), AMF and molds (increase with abandonment time) and saprotrophic fungi (hump-shaped pattern) (Figure 3). Furthermore, there was an increase in the AMF:pathogen ratio with ongoing time since abandonment (Figure 4). Our study is the first to demonstrate pathogen enrichment in early and AMF enrichment in late-secondary succession stage soils, as has been proposed based on variations in plant–soil feedback from negative to positive using the same chronosequence (Kardol et al., 2006). Furthermore, we found a significant negative relationship between agricultural abandonment time and relative abundance of yeasts, indicating that there is a shift from fast-growing, single-cell fungi to more resilient, hyphal-growing fungal community using root exudates. Yeasts are more common in relatively disturbed systems growing on labile carbon sources such as root exudates but contribute probably very little to the maintenance of the soil structure and nutrient cycling as compared with hyphal-growing fungi that are usually better at producing extracellular enzymes (Mestre et al., 2011; Hannula et al., 2012; Treseder and Lennon, 2015).
Besides observed changes in the proportion of functional groups and classes with abandonment time, changes at Order, Genus level and even OTU level were detected to be significantly affected by time since agriculture. Furthermore, highly connected species, specific to certain abandonment stages (so called keystone OTUs; for synthesis, see van der Heijden and Hartmann, 2016) could be identified and may be of importance for speeding up nature restoration (Wubs et al., 2016). A hypothesis presented recently is that plant performance in a given ecosystem depends on the presence of keystone species that make the system more stable by protecting against invasion (Banerjee et al., 2015; Agler et al., 2016; van der Heijden and Hartmann, 2016; Wubs et al., 2016). Here the fungal taxa we found to be differentially present and highly connected during secondary succession belong mainly to Ascomycota and more specifically to orders Pleosporales and Eurotiales. Also a species of yeast from the basidiomycete class Tremellomycetes was indicated as a significant keystone species by network analysis. Further work is needed to experimentally test these species responses to plant root exudates and their potential in use for speeding up nature restoration (Wubs et al., 2016).
We explored the co-occurrence networks (Barberán et al., 2012) in order to assess how the connectedness of fungi is influenced by time since abandonment. These analyses revealed that the highest network densities of fungi using plant-derived carbon in terms of species connectedness were not found in later successional stages but in the mid-term abandoned fields. In long-term abandoned fields, the network configuration was different than in recently abandoned fields: there was a clear split of the network into two ‘hubs’ instead of one tightly clustered hub. This increase in modularity and in the number of ‘ hubs’ has been recently proposed to result in increased benefits to the plant and soil health through recruitment of species and prevention of pathogen invasion through highly connected hubs (Agler et al., 2016; van der Heijden and Hartmann, 2016). If modularity increases over time, this is indicative that community development follows a non-random succession pattern (autogenic succession), which is driven by biotic interactions (Frankland, 1998). In our system, the second ‘hub’ started to form in the mid-aged fields indicating that it took some time after agriculture for the system to gain its modularity by recruiting new taxa and creating new connections. This coincided with increase in AMF receiving carbon from the plant and decrease in potential pathogenic fungi.
Last, we explored the effects of changes in fungal community on fungal-feeding organisms. We could show, based on correlations of the labeling data in time, that, in recently abandoned fields, protists and fungivorous nematodes both seem to be using yeasts, usually fast-growing and stress-tolerant fungi (Treseder and Lennon, 2015) as a primary source of carbon (Figure 6). This can be due to the shorter generation times and relatively small body size of the animals and fungi compared with, for example, mites (Yeates et al., 1993) and filamentous fungi (Treseder and Lennon, 2015). In later successional fields, the fungal feeders receiving most of the carbon were cryptostigmatic mites. In microcosms, it has been shown that fungal feeders do have preferences for the fungal species and groups they feed on (Crowther et al., 2012). Here, we provide preliminary evidence that preference of fungal feeders to certain fungal taxa also affects the communities in soils.
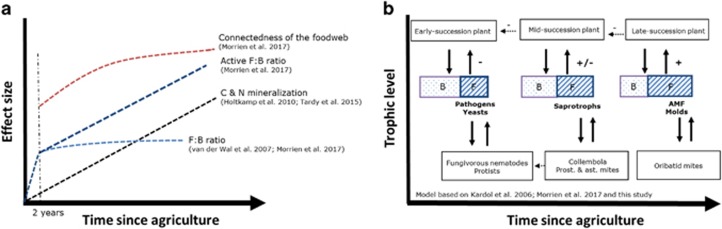
Figure 6.
Trends in mineralization rates, network topology and F:B ratio (a) and changes in soil food web and fungal functional groups (b) as a function of time since agriculture. B, bacterial channel; F, fungal channel.
In conclusion, we show that 30 years of land abandonment result in shifts of active plant-associated communities from bacterial-dominated to fungal-dominated communities. The composition of the active functional fungal community changed from one composed of fast-growing and pathogenic fungal species to one consisting of beneficial and slower-growing fungal species, which may have essential consequences for the carbon flow through the soil food web and consequently nutrient cycling and plant succession (Figure 6).
Acknowledgments
This study was carried out as part of EcoFINDERs research project (EU-FP7-264465). EM was supported by NWO-ALW-Veni 863.15.021 grant.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Agler MT, Ruhe J, Kroll S, Morhenn C, Kim ST et al. (2016). Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol 14: e1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Triguerosa CA, Hempel S, Powell JR, Anderson IC, Antonovics J, Bergmann J et al. (2015). Branching out: towards a trait-based understanding of fungal ecology. Fungal Biol Rev 29: 34–41. [Google Scholar]
- Arnold AE. (2007). Understanding the diversity of foliar endophytic fungi: progress, challenges, and frontiers. Fungal Biol Rev 21: 51–60. [Google Scholar]
- Barberán I, Scott T, Bates ST, Casamayor EO, Fierer N et al. (2012). Using network analysis to explore co-occurrence patterns in soil microbial communities. ISMEJ J 6: 343–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardgett RD, van der Putten WH. (2014). Belowground biodiversity and ecosystem functioning. Nature 515: 505–511. [DOI] [PubMed] [Google Scholar]
- Bengtsson-Palme J, Ryberg M, Hartmann M, Branco S, Wang Z, Godhe A et al. (2013). Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol Evol 4: 914–919. [Google Scholar]
- Banerjee S, Kirkby CA, Schmutter D, Bissett A, Kirkegaard JA, Richardson AE. (2015). Network analysis reveals functional redundancy and keystone taxa amongst bacterial and fungal communities during organic matter decomposition in an arable soil. Soil Biol Biochem 97: 188–198. [Google Scholar]
- de Boer W, Forlman LB, Summerbell RC, Boddy L. (2005). Living in a fungal world: impact of fungi on soil bacterial nice development. FEMS Microbiol Rev 29: 795–811. [DOI] [PubMed] [Google Scholar]
- Boschker HTS. (2004). Linking microbial community structure and functioning: stable isotope (13C) labeling in combination with PLFA analysis. In: Kowalchuk GA, FJ de Bruijn, Head IM, Akkermans AD, and van Elsas JD (eds). Moliecular Microbial Ecology Manual. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 1673-1688.
- Buee M, de Boer W, Martin F, van Overbeek l, Jurkevich E. (2009). The rhizosphere zoo: an overview of plant-associated communities of microorganisms, including phages, bacteria, archaea, and fungi, and of some of their structuring factors. Plant Soil 321: 189–212. [Google Scholar]
- Crowther TW, Boddy L, Hefin Jones T. (2012). Functional and ecological consequences of saprotrophic fungus-grazer interactions. ISME J 6: 1992–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dini-Andreote F, de Cassia Pereira e Silva M, Triado-Margarit X, Casamayor EO, van Elsas JD, Salles JF. (2014). Dynamics of bacterial community succession in a salt marsh chronosequence: evidences for temporal niche partitioning. ISME J 8: 1989–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankland JC. (1998). Fungal succession - unravelling the unpredictable. Mycol Res 102: 1–15. [Google Scholar]
- Hammer Ø, Harper DAT, Ryan PD. (2001). PAST: paleontological statistics software package for education and data analysis. Paleontol Electron 4: 9. [Google Scholar]
- Hannula SE, Boschker HTS, de Boer W, van Veen JA. (2012). 13C pulse-labeling assessment of the community structure of active fungi in the rhizosphere of a genetically starch-modified potato (Solanum tuberosum cultivar and its parental isoline. New Phytol 194: 784–799. [DOI] [PubMed] [Google Scholar]
- Hell K, Edwards A, Zarsky J, Podmirseg SM, Girdwood S, Pachebat JA et al. (2013). The dynamic bacterial communities of a melting High Arctic glacier snowpack. ISMEJ J 7: 1813–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtkamp R, Kardol P, van der Wal A, Dekker SC, van der Putten WH, de Ruiter PC. (2008). Soil food web structure during ecosystem development after land abandonment. Appl Soil Ecol 39: 23–34. [Google Scholar]
- Holtkamp R, van der Wal A, Kardol P, van der Putten WH, de Ruiter PC, Dekker SC. (2011). Modelling C and N mineralisation in soil food webs during secondary succession on ex-arable landSoil Biol Biochem 43: 251–260.
- Ihrmark K, Bödeker ITM, Cruz-Martinez K, Friberg H, Kubartova A, Schenck J et al. (2012). New primers to amplify the fungal ITS2 region – evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol Ecol 82: 666–677. [DOI] [PubMed] [Google Scholar]
- Kardol P, Martijn Bezemer T, van der Putten WH. (2006). Temporal variation in plant–soil feedback controls succession. Ecol Lett 9: 1080–1088. [DOI] [PubMed] [Google Scholar]
- Kuramae EE, Gamper HA, Yergeau E, Piceno YM, Brodie EL, Desantis TZ et al. (2010). Microbial secondary succession in a chronosequence of chalk grasslands. ISME J 4: 711–715. [DOI] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S. (2014). Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 10: e1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik AA, Chowdhury S, Schlager V, Oliver A, Puissant J, Vazquez PGM et al. (2016). Soil fungal:bacterial ratios are linked to altered carbon cycling. Front Microbiol 7: 1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestre CM, Rosa CA, Safar SVB, Libkind D, Fontenla SB. (2011). Yeast communities associated with the bulk-soil, rhizosphere and ectomycorrhizosphere of a Nothofagus pumilio forest in Northwestern Patagonia, Argentina. FEMS Microbiol Ecol 78: 531–541. [DOI] [PubMed] [Google Scholar]
- Morriën E, Hannula SE, Snoek B, Helmsing NR, Zweers H, de Hollander M et al. (2017). Linking belowground and aboveground succession in terrestrial ecosystems. Nat Commun 8: 14349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld JD, Vohra J, Dumont MG, Lueders T, Manefield M, Friedrich MW et al. (2007). DNA stable-isotope probing. Nat Protoc 2: 860–866. [DOI] [PubMed] [Google Scholar]
- Nguyen NH, Song S, Bates ST, Branco S, Tedersoo L, Menke J et al. (2016). FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol 20: 241–248. [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland MS, McCulley RL, Nelson JA, Bradford MA. (2015). Compositional differences in simulated root exudates elicit a limited functional and compositional response in soil microbial communities. Front Microbiol 6: 817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardy V, Spor A, Mathieu O, Lévèque J, Terrat S, Plassart P et al. (2015). Shifts in microbial diversity through land use intensity as drivers of carbon mineralization in soil. Soil Biol Biochem 90: 204–213. [Google Scholar]
- Tedersoo L, Bahram M, Põlme S, Kõljalg U, Yorou NS, Wijesundera R et al. (2014). Global diversity and geography of soil fungi. Science 346: 1078. [DOI] [PubMed] [Google Scholar]
- Ter Braak C, Smilauer P. (1998) CANOCO Reference Manual and User's Guide to Canoco for Windows: Software for Canonical Community Ordination (Version 5.0). Centre for Biometry: Wageningen, The Netherlands. [Google Scholar]
- Thomson BC, Tisserant E, Plassart P, Uroz S, Griffiths RI, Hannula SE et al. (2015). Soil conditions and land use intensification effects on soil microbial communities across a range of European field sites. Soil Biol Biochem 88: 403–413. [Google Scholar]
- Tilman D. (1988) Plant Strategies and the Dynamics and Structure of Plant Communities. Princeton University Press: Princeton, NJ, USA. [Google Scholar]
- Treseder KK, Lennon JT. (2015). Fungal traits that drive ecosystem dynamics on land. Microbiol Mol Biol Rev 79: 243–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Voorde TFJ, Van der Putten WH, Bezemer TM. (2011). Intra- and interspecific plant-soil interactions, soil legacies and priority effects during old-field succession. J Ecol 99: 945–953. [Google Scholar]
- van der Heijden MGA, Hartmann M. (2016). Networking in the plant microbiome. PLoS Biol 14: e1002378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Wal A, Van Veen JA, Smant W, Boschker HTS, Bloem J, Kardol P et al. (2006). Fungal biomass development in a chronosequence of land abandonment. Soil Biol Biochem 38: 51–60. [Google Scholar]
- de Vries FT, Hoffland E, van Eekeren N, Brussaard L, Bloem J. (2006). Fungal/bacterial ratios in grasslands with contrasting nitrogen management. Soil Biol Biochem 38: 2092–2103. [Google Scholar]
- de Vries FT, Caruso T. (2016). Eating from the same plate? Revisiting the role of labile carbon inputs in the soil food web. Soil Biol Biochem 102: 4–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardle DA, Bardgett RD, Klironomos JN, Setälä H, van der Putten WH, Wall DH. (2004). Ecological linkages between aboveground and belowground biota. Science 304: 1629–1633. [DOI] [PubMed] [Google Scholar]
- Wubs ERJ, van der Putten WH, Bosch M, Martijn Bezemer TM. (2016). Soil inoculation steers restoration of terrestrial ecosystems. Nat Plants 2: 16107. [DOI] [PubMed] [Google Scholar]
- Yeates GW, Bongers T, De Goede RGM, Freckman DW, Georgieva SS. (1993). Feeding habits in soil nematode families and genera—an outline for soil ecologists. J Nematol 25: 215–331. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.