Abstract
In permafrost (perennially frozen ground) microbes survive oligotrophic conditions, sub-zero temperatures, low water availability and high salinity over millennia. Viable life exists in permafrost tens of thousands of years old but we know little about the metabolic and physiological adaptations to the challenges presented by life in frozen ground over geologic time. In this study we asked whether increasing age and the associated stressors drive adaptive changes in community composition and function. We conducted deep metagenomic and 16 S rRNA gene sequencing across a Pleistocene permafrost chronosequence from 19 000 to 33 000 years before present (kyr). We found that age markedly affected community composition and reduced diversity. Reconstruction of paleovegetation from metagenomic sequence suggests vegetation differences in the paleo record are not responsible for shifts in community composition and function. Rather, we observed shifts consistent with long-term survival strategies in extreme cryogenic environments. These include increased reliance on scavenging detrital biomass, horizontal gene transfer, chemotaxis, dormancy, environmental sensing and stress response. Our results identify traits that may enable survival in ancient cryoenvironments with no influx of energy or new materials.
Introduction
Despite frozen conditions and the absence of energy and nutrient inputs, permafrost hosts a diversity of microbial life (Gilichinsky et al., 2008; Mackelprang et al., 2011; Jansson and Tas, 2014; Hultman et al., 2015). Several decades of research demonstrate unambiguously that microbial metabolism occurs in permafrost and that many community members not only perform basal metabolic functions but also grow and divide (Rivkina et al., 2000; Bakermans et al., 2003; Waldrop et al., 2010; Tuorto et al., 2014; Hultman et al., 2015). Therefore, it is likely that microbial community structure and function reflect adaptations to the permafrost biophysical environment in addition to the dead or dormant cells encased in ice at the time of permafrost formation.
Microbial communities must contend with suite of stressors associated with living in permafrost. Subzero temperatures stabilize nucleic acid secondary structure and reduce membrane fluidity and the structural flexibility of proteins. Communities are exposed to background terrestrial γ radiation from soil minerals at a level of ~2 mGy per year (Gilichinsky, 2002; Fairén et al., 2010). The concentration of reactive oxygen species, which damage DNA, RNA, proteins and lipids could also increase at low temperatures (De Maayer et al., 2014). Liquid water exists in permafrost down to ~−10 °C due to high solute concentrations (Gilichinsky, 2002). Thus, to overcome temperature effects live cells are likely patchily distributed and contained within thin brine channels or pockets of the soil matrix (Gilichinsky et al., 2003; Rivkina et al., 2004).
In the Arctic and subarctic, permafrost persists from the late Pleistocene. Stressors placed on the community likely accumulate over time, necessitating counteractive adaptations for long-term survival in the harsh environmental conditions. However, we know little about the ecological strategies utilized by microbial communities in response to the challenges presented by spending millennia in permafrost. Much of our knowledge about adaptation to permafrost is derived from isolates (Rodrigues et al., 2008; Bakermans et al., 2009; Ayala-Del-Río et al., 2010; Dieser et al., 2013; Mykytczuk et al., 2013; Goordial et al., 2015), which may not be representative of whole communities and do not address adaptations that occur over long periods. Further, studies that target community-level function are typically from Holocene (<11.7 kyr) permafrost or focus on other research questions (Mackelprang et al., 2011; Tas et al., 2014; Hultman et al., 2015; Rivkina et al., 2016). Here we asked whether we could observe functional and compositional changes consistent with physiological and biochemical adaptations enabling survival in permafrost over geologic time or if paleoclimate and -vegetation present at the time of permafrost formation are responsible for differences in microbial attributes. We present the first ‘omics’-based study investigating the influence of permafrost age on microbial community structure and function across a chronosequence of Pleistocene-aged permafrost. These data have ramifications for the search for life on other planets—by understanding adaptations to stresses associated with frozen conditions over geologic time we may gain insight into survival strategies life (if it exists) might use on other cryogenic bodies.
Materials and methods
Field sampling
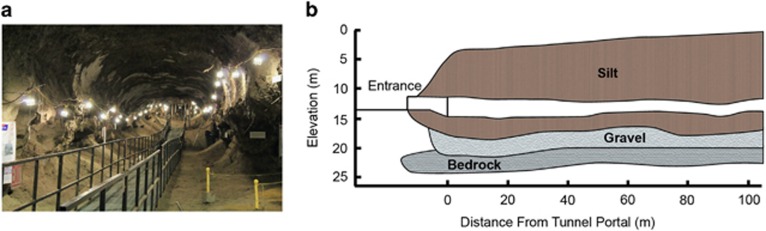
Permafrost was sampled from the United States Army Cold Regions and Research and Engineering Laboratory permafrost tunnel in Fox, Alaska (64.951°N, −147.621°W) in 2012 (Figure 1a) (Bjella et al., 2008). The tunnel penetrates a hillside providing access to increasingly ancient late Pleistocene-aged permafrost (Figure 1b). It is well suited for a chronosequence approach because while age differs, other permafrost characteristics are similar across the age gradient, minimizing potential confounding factors (Supplementary Methods).
Figure 1.
(a) Photograph of the CRREL permafrost tunnel taken ~20 m from the tunnel portal. (b) Generalized cross-section of the tunnel showing lithology. Samples were collected at 20 (19 kyr), 54 (27 kyr) and 81 m (33 kyr) from the tunnel portal. Figure adapted from Bjella et al. (2008) and Hamilton et al. (1988).
We sampled ice-cemented silt deposits along the tunnel corresponding to 19 (20 m from the tunnel portal), 27 (54 m) and 33 kyr (81 m) before present. We took four replicate cores for each permafrost age for physical, chemical and microbial analyses. To minimize contamination, we scraped away the dry loess material from the interior wall that had sublimated over time. Beneath this sublimated material (~5 cm) was the frozen permafrost wall. We used a round 10 cm diameter × 5 cm deep keyhole saw attached to a power drill to take cores from the wall. Three approximately 5 cm depths into the wall were taken (Supplementary Figure 1).
Radiocarbon dating
Calendar ages for the three samples were estimated based on radiocarbon age dates of soil CO2 and soil organic matter, and were placed within previously published dates from the tunnel (Supplementary Methods). CO2 was collected after thawing the permafrost for 48 h at 5 °C in a sealed mason jar purged with zero air, followed by purification on vacuum line. Graphite targets were prepared and 14C analysis was performed by accelerator mass spectrometry at the Keck accelerator mass spectrometry facility at University of California Irvinie (for CO2) or at the United States Geological Survey in Reston and Lawrence Livermore National Labs (for soil organic matter). Calendar ages were estimated using the Calib radiocarbon calibration program and one sigma ranges (Stuiver and Reimer, 1993). Owing to reburial and mixing, the age dates of permafrost carbon is not the same as the age of permafrost itself (Lachniet et al., 2012; Vasil'chuk and Vasil'chuk, 2014), but provides an understanding of the of the relative differences in age among permafrost units. For our youngest sample location, nearest the tunnel entrance, radiocarbon age dates for CO2 and soil organic matter (SOM) were between 16 and 22 cal kyr before present (BP) and there was agreement between the 14C of CO2 and SOM. This is slightly older than the 12 kyr-old debris fan slightly (<1 m) nearer the entrance to the tunnel (Hamilton et al., 1988). We call this earliest sample 19 kyr BP. At the other end of the tunnel, the oldest samples we collected were between 32 and 33 cal kyr BP for CO2 and 37 and 39 cal kyr BP for the SOM. We took the average of the calendar ages from CO2, and called this zone 33 cal kyr BP. The intermediate-aged samples were the most difficult to bracket in terms of age because CO2 and SOM did not agree with one another. The SOM dated to between 37 and 38 cal kyr BP, similar to the oldest site, but the CO2 in the permafrost was substantially younger and dated to between 26 and 27 cal kyr BP. These samples were located near an ice wedge that had previously been dated to 25 cal kyr BP (Katayama et al., 2007) but the wedge ice represents the time of formation of the ice feature into previously deposited permafrost soils. We argue that the younger age of the evolved CO2 represents a pool of more recently deposited C within this permafrost soil and therefore a closer approximation to the age of permafrost in this location. We thus call this intermediate-aged site 27 cal kyr BP. Further detailed information about dating, sediment composition and cryostructure is provided in the Supplementary Material.
Soil chemical analysis
Once in the laboratory frozen cores were disaggregated with a hammer while they remained in plastic bags. Frozen core material was then subsampled into a preweighed, ashed weighing dish and thawed at room temperature. Once thawed, samples were gently mixed and 10 ml of the supernatant was sampled with a syringe and passed through a 0.2 μm Supor polyethersulfone filter (Pall Corporation, Port Washington, NY, USA) pre-rinsed with ultrapure water and a small volume of sample. Filtrate was diluted (typically 10 ×) and analyzed using an Aurora 1030W Total Organic Carbon Analyzer (OI Analytical, College Station, TX, USA) for dissolved organic carbon concentrations. Remaining material was oven-dried and weighed to determine gravimetric ice content, taking into account the volume removed for dissolved organic carbon analysis. A second subsample of the frozen core material was thawed for pH analysis on an Accumet pH meter with AccuTupH Rugged Bulb (ThermoFisher Scientific, Waltham, MA, USA), after diluting 1:1 (v/v). Another sample was run through a Carlo Erba elemental analyzer (ThermoFisher Scientific) for total soil carbon and nitrogen analysis. %C and %N were measured on dried ground samples on a Micromass Optima continuous flow mass spectrometer with a Carlo Erba elemental analyzer on the front end. We used values for δ13C and δ15N from directly adjacent cores sampled in 2007 from the same tunnel locations (n=4 per age category). These data were generated using the same protocols as for %C and %N. We note that there were no differences in %C or %N between sampling years. We tested for significant differences in ice content, dissolved organic carbon and pH, δ13C, δ15N, %C, %N and C/N among the different age categories using analysis of variance followed by Tukey–Kramer post hoc tests.
Direct counts of live and dead cells
We separated cells from the soil matrix based on previously published protocols (Amalfitano and Fazi, 2008; Poté et al., 2010; Morono et al., 2013). A unit of 1.5 g of soil was disrupted in 2 ml of TTSP (0.05% Tween 80 and 50 mm tetrasodium phyrophosphate) using 5 mm glass beads and vortexing for 2 min. Large particles and debris were removed by centrifugation at 750 g for 7 min at 4 °C. A volume of 1000 μl of the supernatant was layered over 600 μl of 1.3 g ml−1 Nycodenz density buffer and centrifuged at 14 000 g for 30 min at 4 °C. The cell-containing upper and middle phases were transferred to a new tube and cells were pelleted by centrifugation at 10 000 g for 15 min at 4 °C. Cells were resuspended in 1 ml of 0.85% NaCl.
Dual staining of live (intact cell membrane) and dead (compromised cell membrane) cells was performed using the LIVE/DEAD BacLight kit (Molecular Probes, Eugene, OR, USA). Briefly, the cell suspension was diluted in 0.85% NaCl. A volume of 3 μl of a 1:1 mixture of 3.34 mm SYTO 9 and 20 mm propidium iodide was added to each sample. Samples were vacuum filtered onto a 0.2 μm black polycarbonate membrane. Live (green-fluorescing) and dead (red-fluorescing) cells were visualized (Kepner and Pratt, 1994) at a single focal plane using a Zeiss Axio Imager M2 fluorescence microscope coupled to an Apotome 2.0 System (Zeiss, Oberkochen, Germany). Fifteen fields of view were counted per sample.
DNA extraction, PCR and 16S rRNA gene sequencing
Before DNA extraction, cores were sprayed with 0.5 μm Fluoresbrite yellow green latex microbeads (Polyscience, Warrington, PA, USA). To remove contamination on core exteriors, the entire surface was removed using autoclaved knives and chisels. Interior sections were viewed under a fluorescent microscope to verify that there was no contamination from the surface.
For each sample, a single DNA extraction was performed directly from 0.5 g of soil using the FastDNA Spin Kit for Soil (MP Biomedicals, Santa Ana, CA, USA) according to the manufacturer’s protocol. Additional purification was performed using the PowerClean DNA Clean-Up Kit (MoBio Laboratories, Carlsbad, CA, USA). Amplification of the variable region four (V4) of bacterial and archaeal 16S rRNA genes was performed using the golay barcoded Illumina (San Diego, CA, USA) adapted primer set 515 F/806 R according to the methods recommended by the Earth Microbiome Project protocol version 4.13 (Caporaso et al., 2010) and sequenced on an Illumina MiSeq instrument. See Supplementary Methods for further details.
Library construction and metagenomic sequencing
Illumina paired-end libraries were constructed by linker-mediated emulsion PCR following established protocols (Blow et al., 2008; Mackelprang et al., 2011; Mason et al., 2012; Hultman et al., 2015). In all, 2 × 100 paired-end shotgun sequencing was performed on an Illumina HiSeq 2000 instrument (Illumina). The sequence quality was high, with a median quality score above 32 at all positions across the reads from all samples. Details are given in Supplementary Methods.
16S rRNA gene amplicon analysis
Raw 16S rRNA gene sequences were demultiplexed using the python script prep_fastq_for_uparse_paired.py version 0.0.1 (https://github.com/leffj/helper-code-for-uparse) (Smets et al., 2016). The UPARSE pipeline was used to merge paired-end reads, conduct quality filtering and dereplication, remove singleton sequences, create a de novo database and map merged reads back to the database (Edgar, 2013). To merge reads, we set the minimum length overlap to 20 and allowed a maximum of 1 mismatch in the overlap region. The minimum length of a merged read was 200 bp and we used a fastq truncation quality of 3. Clustering was done at a 97% similarity threshold and the merged raw reads were mapped to the de novo database at 97% similarity.
QIIME 1.9.1 (Caporaso et al., 2010) was used for downstream analysis. Low-frequency operational taxonomic units (OTUs), defined as those observed 10 or fewer times were removed from analysis. Taxonomy was assigned using the RDP classifier and the Greengenes database (Wang et al., 2007; McDonald et al., 2012). Contaminates commonly found in DNA extraction and amplification kits were also removed from further analysis (Salter et al., 2014) (Supplementary Methods). Total sequences per sample after low-frequency and contamination filtering ranged from 46 253 to 165 084. The data were rarefied to 46 000 and downstream analyses were conducted on rarefied samples.
Alpha and beta diversity metrics were calculated and visualized using the phyloseq package in R (McMurdie and Holmes, 2013). We calculated the following alpha diversity metrics: Shannon index; Chao1; and Fisher. Beta diversity was computed using the unweighted UniFrac metric. Relationships between samples were visualized using principal coordinate analysis. Permutational multivariate analysis of variance was used to test correlations between community composition and soil physiochemical characteristics (Supplementary Table 1). Analyses were performed in R using the adonis function in the vegan package (Oksanen et al., 2015).
Metagenomic analysis
Raw fastq sequence files were filter trimmed using Trimmomatic to remove adapter sequences (Bolger et al., 2014). Reads were quality-filtered using fastq_quality_filter from the FASTX toolkit at default settings. Functional annotation was performed by comparison of quality-filtered reads to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa et al., 2017) using the UBLAST algorithm within the USEARCH program with an acceleration value of 0.4 and an E-value cutoff of 1e-6 (Edgar, 2010). Reads were assigned the KEGG Orthology (KO) number of the top hit. Ordinations of KO count data were performed using nonmetric multidimensional scaling based on the Bray–Curtis dissimilarity metric as implemented in the phyloseq package (McMurdie and Holmes, 2013). As with the 16S rRNA gene amplicon data, permutational multivariate analysis of variance was used to test for correlation between functional composition and environmental parameters. To perform differential abundance analysis, a negative binomial generalized linear model was applied to KO count data using the DESeq2 package (Love et al., 2014). Genes were considered significantly different between age classes if the adjusted P-value was <0.05.
To infer the composition of past vegetation we identified plant sequences by comparing quality-filtered reads to the National Center for Biotechnology Information (NCBI) plant protein RefSeq database (downloaded August 2016) using DIAMOND v0.8.17.79 (Buchfink et al., 2015). A total of 5 million randomly selected reads per sample were used to reduce computational burden. Matching reads were placed on the NCBI taxonomic scaffold and taxonomic composition of past vegetation was inferred using MEGAN Community Edition v6.4.20 (Huson et al., 2016) with default parameters. Reads assigned to the members of the Streptophyta lineage were used for downstream analysis (Supplementary Table 2). Principal coordinate analysis was conducted within MEGAN from Bray–Curtis distances. A Mantel test (999 permutations) and Procrustes randomization test were used to test for correlation between plant taxonomy and functional gene composition as implemented in the R vegan package (Oksanen et al., 2015).
Pathway analysis
Because genes operate within the context of pathways and networks, we analyzed differences in functional potential between age categories at pathway-level resolution rather than through a gene-by-gene approach. However, pathway-centric analysis of metagenomic data is not straightforward (De Filippo et al., 2012). Common methods for identifying differentially abundant pathways rely on the aggregation of gene abundances (Kristiansson et al., 2009; White et al., 2009; Parks and Beiko, 2010). Pathways are often organized at a coarse level of resolution and may contain several sub-pathways or modules (Kanehisa et al., 2017). For example, the KEGG methane metabolism pathway encompasses both methanogenesis and methanotrophy. Aggregation of gene abundances across entire pathways may mask significant differences in sub-pathways. To circumvent this hurdle, we developed a strategy that identifies pathways in which differentially abundant genes are concentrated.
The strategy for pathway-centric analysis is illustrated by the following example. Given a pair of age categories (for example, age A and age B), to identify pathways enriched in age B, we placed genes significantly more abundant in age B compared to age A onto the KEGG pathway map (Kanehisa et al., 2017). For each pathway, we counted the number of differentially abundant genes contained within. We obtained a null distribution of the number of differentially abundant genes expected in each pathway by randomly assigning P-values to each gene from the observed set of P-values and performed 10 000 permutations. P-values obtained from the permutation test were corrected using the false discovery rate (Benjamini and Hochberg, 1995).
Another difficulty with pathway analysis is the ‘multiple mapping problem’ that arises when an enzyme catalyzes reaction steps across multiple pathways (Hanson et al., 2014). Pathways may be mistakenly identified as differing between age categories if the majority of genes driving the differentiation are highly promiscuous. For this reason, we manually inspected each pathway identified as enriched in an age category and removed it from downstream analysis if the genes differing significantly between age groups primarily mapped to multiple pathways.
To identify pathways that increased in abundance along the chronosequence, pathways were required to be significantly enriched in 33 kyr samples compared with 19 kyr samples. Pathways with the strongest evidence for age-driven differences were those that showed significant enrichment from 19 to 27 kyr, 27 to 33 kyr and 19 to 33 kyr. Comparisons between 19–27 and 27–33 kyr were not required to be significant, but pathway abundance in 27 kyr samples could not be less than in 19 kyr samples or greater than in 33 kyr samples (Table 1). Likewise, genes were considered to increase in abundance across the chronosequence if they were significantly greater in 33 kyr samples compared with 19 kyr samples (P<0.05) and followed a trend of increasing abundance between 19–27 and 27–33 kyr.
Table 1. KEGG pathways increasing in abundance along a permafrost Pleistocene chronosequence.
| Pathway | 19 vs 33 kyr | 19 vs 27 kyr | 27 vs 33 kyr |
|---|---|---|---|
| Two-component system | *** | ** | *** |
| Valine, leucine and isoleucine degradation | *** | ** | * |
| Bacterial secretion system | *** | *** | *** |
| ABC transporters | *** | NS | *** |
| Glyoxylate and dicarboxylate metabolism | *** | *** | *** |
| Bacterial chemotaxis | *** | NS | *** |
| Lipopolysaccharide biosynthesis | *** | NS | *** |
| Phosphotransferase system | *** | *** | NS |
| Toluene degradation | *** | NS | *** |
| Cysteine and methionine metabolism | *** | ** | NS |
| Butanoate metabolism | ** | *** | NS |
| Peptiodglycan biosynthesis | ** | NS | *** |
| Fatty acid biosynthesis | *** | NS | NS |
| Glutathione metabolism | *** | NS | NS |
| Xylene degradation | ** | NS | NS |
| Dioxin degradation | * | NS | NS |
| Geraniol degradation | ** | NS | NS |
| Galactose metabolism | * | NS | NS |
Abbreviations: ABC, ATP-binding cassette; KEGG, Kyoto Encyclopedia of Genes and Genomes; NS, not significant.
***P<0.005.
**P=0.01–0.005.
*P=0.05–0.01.
Data indicate whether the pathway was significantly more abundant in the older of the two age categories.
Results
16S rRNA gene-based community analysis
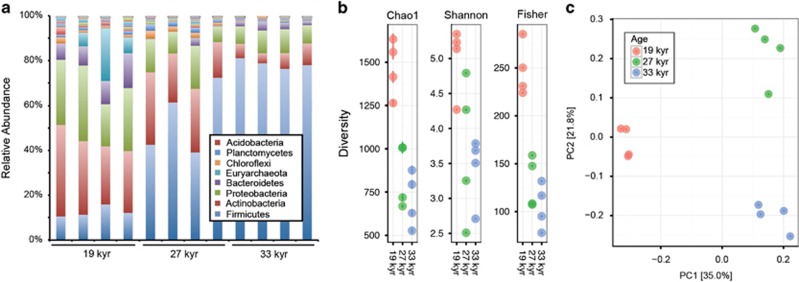
16S rRNA gene sequencing of the bacterial and archaeal communities from 19, 27 and 33 kyr permafrost (n=4 per age category; 12 samples total) from the Cold Regions and Research and Engineering Laboratory permafrost tunnel yielded 1 222 638 sequences representing 2331 OTUs. Microbial community composition and diversity changed with permafrost age. Firmicutes OTUs, primarily from the spore-forming clostridia and bacilli classes, increased from an average relative abundance of 13% in the youngest age category to 79% in the oldest samples. The average relative abundance of the three other most dominant phyla (actinobacteria, proteobacteria and bacteroidetes) declined. Actinobacteria declined from 32 to 7%, proteobacteria declined from 27 to 9% and bacteroidetes declined from 11 to 3% (Figure 2a and Supplementary Figure 2). Methanogen OTUs were most abundant in 19 kyr permafrost (averaging 10%) and declined to nearly undetectable levels in the older samples (Supplementary Table 3). One OTU from the genus Methanobacterium dominated all methanogen sequences. Alpha diversity decreased from 19 to 27 kyr and 19 to 33 kyr (P<0.05; Figure 2b) but was not significantly different in 27 kyr samples compared with 33 kyr samples.
Figure 2.
16S rRNA gene sequencing reveals age-related differences in microbial community composition and diversity. (a) Relative abundance of bacterial and archaeal phyla in 19, 27 and 33 kyr samples. (b) Alpha diversity measurements compared across age categories. (c) Principal coordinates analysis of unweighted UniFrac distances colored by age.
Principal coordinate analysis of UniFrac distances revealed distinct clustering by age (Figure 2c). When considering age along with soil chemical characteristics (Supplementary Table 1), permutational multivariate analysis of variance analysis revealed that age explained 40% of the variation in beta diversity (F=3.8, P=0.001) followed by ice content (14%, F=2.7, P=0.011) and dissolved organic carbon (11%, F=2.2, P=0.036) (Oksanen et al., 2015). Tests for differences in soil chemistry among age categories were not significant (Supplementary Table 1, analysis of variance, P>0.05 for all tests). Procrustes randomization and Mantel tests for correlation between plant taxonomic profiles and community composition were not significant.
Direct cell counts
Dual staining of live and dead cells revealed that 18–22% had an intact membrane and were thus classified as live. The 33 kyr permafrost had the lowest live/dead ratio (0.28) though differences between age categories were not significant (Supplementary Figure 3A). Similarly, the total cell count was lowest in 33 kyr samples (9.32 × 106 cells per g dry weight). We counted the most cells in 27 kyr samples (1.37 × 107 cells per g dry weight), which was significantly different from 33 kyr samples (Supplementary Figure 3B). Other pairwise comparisons were not significant.
Metagenomic sequencing and pathway analysis
Shotgun metagenomic sequencing resulted in 264 Gb of sequence for an average of 22 Gb per sample. Ordination of gene count data revealed three clearly distinct age-based clusters, giving a picture of functional genes similar to the taxonomic structure obtained through 16S rRNA gene sequencing (Supplementary Figure 4A). Analysis of microbial community function using permutational multivariate analysis of variance analysis demonstrated that age was a significant factor (F=3.7, P=0.039) driving functional composition, explaining 36% of the variation in diversity. No other chemical measurements (Supplementary Table 1) were significant. Plant community structure and microbial functional gene relative abundances were not significantly correlated (Procrustes randomization and Mantel tests). Ordination analysis based on paleovegetation taxonomy did not show three distinct age-based clusters. Instead, we observed co-clustering of 33 and 19 kyr samples—27 kyr samples were distinct from both (Supplementary Figure 4B).
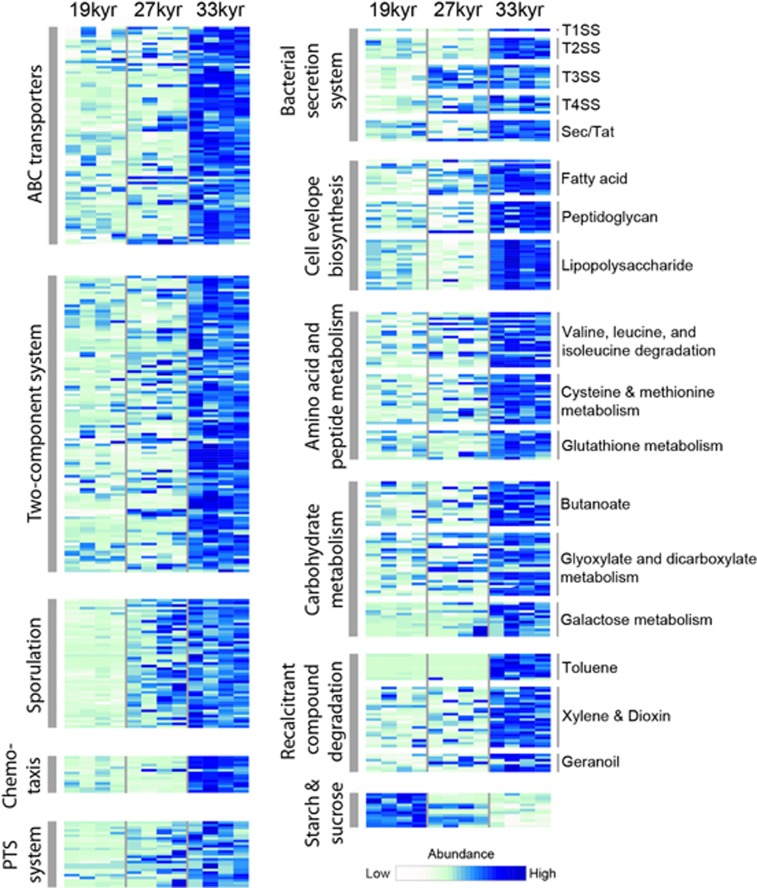
We compared the functional potential of microbial communities between age categories using a pathway-based approach. We identified 18 KEGG pathways that became increasingly abundant in older permafrost (Table 1; Figure 3; Supplementary Table 4). Pathways enriched in older permafrost compared with younger samples were involved in synthesis of cell envelope components, amino-acid and peptide metabolism, carbohydrate metabolism, environmental sensing and response, chemotaxis, membrane transport and the degradation of recalcitrant biomass (Figure 3). Only one pathway, starch and sucrose metabolism, decreased significantly in abundance over time. Genes involved in dormancy were more abundant in older permafrost soils, consistent with the increase in Firmicutes abundance (Figure 3; Supplementary Figure 5). The methane metabolism pathway was significantly more abundant in 19 kyr samples compared with 27 and 33 kyr samples. However, it was slightly but significantly more abundant in 33 kyr permafrost compared with 27 kyr permafrost, corresponding to 16 S rRNA amplicon data (Supplementary Figure 6).
Figure 3.
Heatmap of KEGG pathways increasing or decreasing in abundance along a permafrost chronosequence. In the case of all pathways except for starch and sucrose metabolism, genes that are significantly more abundant in 33 kyr permafrost compared with 19 kyr permafrost are shown (P<0.05). For the starch and sucrose metabolism pathway, genes more abundant in 19 kyr permafrost compared with 33 kyr permafrost are displayed (P<0.05). Abundances are scaled by row.
Environmental sensing and chemotaxis
The ability to respond to environmental conditions (including stressors expected in ancient cryoenvironments such as low temperature, high osmolarity and resource limitation) using the two-component regulatory system (Aguilar et al., 2001; Hyyryläinen et al., 2007; Raivio, 2014) increased along the permafrost chronosequence (Table 1). Specifically, two-component sensor system genes that increased in abundance were involved in temperature sensing, protein misfolding, H+ ion regulation, salt stress, osmolarity, oxygen limitation and cell envelope stress. We also found that genes involved in nutrient and resource sensing—ions and trace metals, nitrogen, acetoacetate, malate and glucose—increased in abundance with age (Figures 3 and Figures 4a; P<0.05).
Figure 4.
Visualization of KEGG pathways more abundant in increasingly ancient permafrost for the (a) two-component system, (b) chemotaxis, (c) ABC transporters and (d) the bacterial secretion system. Secretion system diagram based on Costa et al (2015). Selected genes that increase in abundance with increasing permafrost age are displayed in bold black text or (in the chemotaxis pathway) bold white text (P<0.05). ABC, ATP-binding cassette.
Chemotaxis, which is controlled by the two-component regulatory system, is the process by which bacteria sense and move toward a chemical stimulus. In our samples, the bacterial chemotaxis pathway was significantly enriched in older permafrost compared to younger samples (Figure 3; Table 1). Nearly every gene encoding components of the chemotaxis apparatus increased in abundance with age (P<0.05; Figure 4b).
Membrane transport and cell envelope synthesis
In 33 kyr permafrost the bacterial secretion system pathway increased in abundance compared with younger permafrost (Table 1). We mapped increasingly abundant genes to four of the six classes of secretion machinery and the two membrane-spanning systems (Figures 3 and Figures 4d). Of particular note is the Type IV secretion system, which encodes conjugation machinery and mediates the transfer of DNA between closely related species. The other secretion systems with genes more abundant in ancient permafrost are the Type I secretion system (secretes products into the extracellular milieu), Type IV secretion system (functions primarily in nutrient acquisition), Type III secretion system (involved in interactions with eukaryotic cells), and the sec and tat systems (works with Type II secretion system to excrete hydrolytic enzymes).
The abundance of pathways involving biosynthesis of three cell envelope components—fatty acids, lipopolysaccharides (LPS) and peptidoglycan—increased with age (Figure 3; Supplementary Figure 7; Table 1). Fatty acid chains in phospho- and glycolipids form the membrane and are altered to increase membrane fluidity in response to cold (Phadtare, 2004; Chattopadhyay, 2006). Peptidoglycans are aminosugar polymers that form the mesh-like cell wall, which can be mechanically damaged by ice and faces high osmotic pressure at subzero temperatures (Rodrigues et al., 2008; Mykytczuk et al., 2013; De Maayer et al., 2014). The LPS layer is a major component of the Gram-negative cell outer membrane and forms a selectively permeable protective barrier around the cell (Gao et al., 2006). Despite the 16S rRNA-based observations that there are fewer Gram-negative bacteria in 33 kyr permafrost than in 19 and 27 kyr permafrost, the LPS synthesis pathway was more abundant in 33 kyr samples (Table 1). Most genes in the pathway were more abundant (P<0.05) including a gene (lpxD) known to be involved in temperature-regulated remodeling (Li et al., 2012) (Supplementary Figure 7B).
The ATP-binding cassette transporter pathway also increased in abundance along the chronosequence (Table 1). Substrate specificity of increasingly abundant importer genes include those involved in transfer of amino acids, peptides, osmoprotectants, stress compounds and trace metals. Exporter genes include those involved in the transfer of LPS layer and cell wall components (P>0.05; Figures 3 and Figures 4c).
Carbon and nitrogen metabolism
Two amino-acid metabolism pathways became more abundant in ancient permafrost (Table 1; Figure 3): cysteine and methionine metabolism (Supplementary Figure 8B) and valine, leucine and isoleucine degradation (Supplementary Figure 8A). Genes in the methionine (which is energetically expensive to produce) sub-pathway (Berger et al., 2003; Sekowska et al., 2004) increased in abundance in older permafrost (Supplementary Figure 8B). The methionine salvage sub-pathway regulates polyamine synthesis, producing ‘stress molecules’ (Rhee et al., 2007) such as cadaverine, putrescine and spermine; genes involved in the synthesis of these polyamines were more abundant in the older samples (Supplementary Figure 9).
We found evidence for an increased capacity for the degradation of aromatic hydrocarbons in older permafrost compared with younger samples as indicated by an enrichment of the toluene, xylene and dioxin degradation pathways (Table 1; Figure 3; Supplementary Figure 10). The genes that increased in abundance were contained largely within anaerobic catabolism sub-pathways. Anaerobic catabolism of aromatic compounds is divided into two categories, peripheral pathways that funnel an array compounds into a small number of intermediates and central pathways that channel intermediates to the central metabolism of the cell (Carmona et al., 2009). Genes that increased in abundance were found in peripheral pathways that convert toluene, benzoate and 4-hydroxybenzoate to benzoyl-CoA (Supplementary Figure 10), the starting point for the major central pathway.
Three carbohydrate metabolism pathways were enriched in 33 kyr permafrost compared with 19 and 27 kyr samples—butanoate metabolism, galactose metabolism, and glyoxylate and dicarboxylate metabolism (Table 1; Figure 3). In particular, genes involved in butyrate and butanol fermentation, galactitol degradation, and the capacity to grow on the amino sugars d-galactosamine and N-acetyl-d-galactosamine were more abundant in older permafrost (Supplementary Figure 11).
Discussion
Our results demonstrated that microbial communities adapt to life in permafrost through geologic time. In addition, we identified genes and pathways important for survival in ancient permafrost. An alternative hypothesis is that the community and functional genes reflect the climate and vegetation at the time the permafrost formed. However, this alternative hypothesis cannot explain the differences in community structure and function between the 19 and 33 kyr samples. A paleoecological study of interior Alaska showed that the periods around 19 and 33 kyr were climatically similar to each other, but the period between 20 and 33 kyr was colder and drier, possibly with more herb tundra coverage than today (Muhs et al., 2001; Willerslev et al., 2014). We directly evaluated the composition of past plant communities, which was possible because high sequence coverage enabled us to recover DNA from detrital plant material preserved in permafrost (Willerslev et al., 2007; Parducci et al., 2015; 2012). Sequence-based taxonomic reconstruction and analysis demonstrated that paleovegetation and microbial community structure and function were uncorrelated. Vegetation from 19 and 33 kyr samples were similar to each other but differed from the 27 kyr samples, which corresponds to climatic data. We suggest the effect of being frozen in ice for millenia is the critical determining factor of microbial composition and functioning. Once entrained in permafrost, the climate the microbes responded to is that of subzero temperatures and not surface conditions with episodic seasonal warming and cooling. This is further supported by the observation that many genes increasing in abundance with age are consistent with adaptation by microbial communities in a frozen environment.
Despite increasing interest in permafrost microbial communities due to their importance in the climate change equation (Mackelprang et al., 2011, 2016; Graham et al., 2012; Jansson and Tas, 2014; Hultman et al., 2015), it is not known whether organisms in permafrost—particularly ancient permafrost—are adapted to an environment with no influx of new carbon and energy sources. Here we show that metabolic processes enriched in ancient permafrost suggest decreasing availability of simple labile substrates over time. Similar to deep sea-floor sediments, this cold, anoxic, energy-limited environment became depleted in sugar metabolism and enriched in amino-acid and peptide import, degradation and salvage genes, pointing to recycling and use of detrital biomass as a C and N source over geologic timescales (Lomstein et al., 2012; Lloyd et al., 2013; Orsi et al., 2013). In addition, older permafrost had a higher abundance of aromatic hydrocarbon degradation genes. Because our samples are uncontaminated and aromatic hydrocarbon degradation genes are promiscuous, we suggest that the abundance of xylene, dioxin and toluene degradation pathways indicate an increased capacity for degrading aromatic-rich humic compounds. We reconcile these data with previous results showing that 36 kyr permafrost C from a new bore of the Cold Regions and Research and Engineering Laboratory Permafrost Tunnel is highly labile (Drake et al., 2015) by suggesting that finite resources are available within the brine-filled pore spaces and that the rest is sequestered away from microbial metabolism by frozen conditions. This observation is supported by previous studies. For example, Mackelprang et al. (2011) found that nitrogen fixation genes were abundant in permafrost despite the presence of biologically available nitrogen. After short-term thaw, nitrogen fixation genes decreased and denitrification genes increased (Mackelprang et al., 2011). These data have implications for predicting greenhouse gas emissions from Pleistocene permafrost that thaws due to climate warming. After thaw, temperature no longer protects organic matter making it susceptible to microbial degradation. Labile permafrost carbon may be protected by freezing conditions—even over geologic time—and thus particularly vulnerable to degradation during thaw.
Methane—a highly potent greenhouse gas—from permafrost is also important in the context of global warming. Permafrost thaw may allow accumulated biogenically generated methane to escape into the atmosphere and may cause the ground to be inundated with water—providing favorable anaerobic conditions in which methanogens reside. We identified methanogen OTUs and methanogenesis genes in all age categories, consistent with previous work that found biogenically produced methane in the tunnel-wall sediment and in tunnel air (Kvenvolden et al., 1994). Methanogen relative abundance was highest in the youngest age category—similar to observations from other Pleistocene permafrost soils (Bischoff et al., 2013; Rivkina et al., 2016).
In ancient permafrost, long generation times may lead to increased spreading of adaptive traits through horizontal gene transfer as suggested by the increasing abundance of type 4 secretion system genes. In mesophilic environments transfer rates are low compared with asexual reproduction (Niehus et al., 2015). Because generation time in permafrost is long, horizontal gene transfer may be more effective for rapidly acquiring beneficial traits. In other cryogenic environments cold-adapted communities rely on the horizontal gene transfer to spread adaptive traits, including those involved in cold protection, metabolism, nutrient acquisition, fatty acid biosynthesis, motility and ultraviolet resistance (Ma et al., 2006; Collins and Deming, 2013; DeMaere et al., 2013; Dziewit and Bartosik, 2014; Feng et al., 2014). Cells in permafrost that rapidly acquire new capabilities may increase their phenotypic flexibility and thereby improve their fitness.
The potential for chemotaxis was higher in 33 kyr permafrost than in younger samples. This is inconsistent with the idea that it is limited by viscosity and high energy requirements (Rodrigues et al., 2008; De Maayer et al., 2014) but is supported by mounting evidence that chemotaxis occurs at subzero temperatures in permafrost and other environments (Graumann et al., 1996; Junge et al., 2003; Bresolin et al., 2006; Gao et al., 2006; Asakura et al., 2007; Mykytczuk et al., 2013; Hultman et al., 2015). Movement in this constrained environment toward higher concentrations of beneficial compounds may become increasingly necessary as availability decreases.
Cells in cold environments must maintain membrane fluidity to survive. Indeed, one of the most significant impacts of cold temperature is increased rigidity and transition to a gel-like phase (D'Amico et al., 2006; De Maayer et al., 2014). For this reason, we did not expect to observe a significant increase in the abundance of membrane synthesis genes in our older samples. We supposed that microbial communities surviving in permafrost for 19 kyr (our youngest age category) would have already optimized strategies for maintaining a functional membrane. We speculate that the observed increase in abundance is due to greater copy numbers of synthesis genes in the genomes from older samples. This is supported by previous findings that the genomes of psychrophilic bacteria Colwellia psychrerythraea, Planococcus halocryophilus and Psychrobacter arcticus all contain multiple copies of fatty acid biosynthesis genes (Methé et al., 2005; Ayala-Del-Río et al., 2010; Mykytczuk et al., 2013). Redundancy may enhance the ability to express synthesis genes when needed or allow cells to encode proteins with different specificities or activities.
We found that the abundance of some stress-related genes increased in older permafrost. Here we highlight polyamine synthesis genes (spermidine, putricine and cadaverine). These stress-related molecules maintain RNA secondary structure (Igarashi and Kashiwagi, 2000; Takahashi and Kakehi, 2010) and are implicated in response to cold (Zhu et al., 2015), low pH (Carper et al., 1991; Schneider et al., 2013; Zhu et al., 2015), oxidative stress (Tkachenko et al., 2001), membrane integrity and biofilm formation (Carper et al., 1991; Schneider et al., 2013; Zhu et al., 2015). The high abundance of these genes may also be responsible for the putrefied smell of Pleistocene permafrost.
Though we found an increasing abundance of endospore-formers with age, data from other studies demonstrate that not all spore-formers are dormant. Hultman et al., (2015) found that endospore-forming Firmicutes were more active in permafrost than expected based on abundance. Indeed, because DNA is prone to degradation, metabolic activity may be superior to dormancy as a long-term survival strategy (Willerslev et al., 2004; Dieser et al., 2013). Live/dead staining revealed that live cells persist in all age categories. Total counts and live/dead ratios were consistent with prior observations (Vorobyova et al., 1997; Vishnivetskaya et al., 2006; 2000; Hansen et al., 2007). However, we did not find clear trends between cell counts and permafrost age (Supplementary Figure 3). The data suggest that total cell counts and the live/dead ratio may be the lowest in the oldest-aged samples but highest in the middle-aged ones, though we emphasize that most comparisons were not statistically significant. If in future investigations this trend continues it would suggest that age is not the primary driver of cell counts or the percent of live cells, at least when comparing late Pleistocene samples. This is consistent with other observations that direct cell counts did not vary significantly across age categories (Vishnivetskaya et al., 2000, 2006).
These data represent a snapshot of late Pleistocene permafrost microbial communities in an environment that tightly controls for confounding factors. With these foundational data, expanding age ranges and incorporating other chronosequences of soils matched for parent material and other factors will be important for building a model of how life survives through geologic timescales.
Acknowledgments
This work was supported by the National Aeronautics and Space Administration (NASA) (grant numbers NNX15AM12G and NNH15AB58I), the United States Geological Survey (USGS) Climate and Land Use Research & Development Program, and the California State University Program for Education and Research in Biotechnology (CSUPERB) New Investigator Grant Program. TAD acknowledges the US Army Engineer Research and Development Center’s Basic Research (6.1) Program. CHC acknowledges support from the USGS National Research Program. We thank J Duah and R Hasan for assistance with sample processing and K Manies for assistance with age dating. RM acknowledges support from NASA #NNX15AM12G, CSUPERB New Investigator Grant Program and MPW acknowledges support from NASA #NNH15AB58I, USGS Climate and Land Use Research Development Program. Metagenomic and 16S rRNA gene sequence data have been deposited in NCBI's sequence read archive under accession numbers SRP093781 and SRP106346, respectively.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Aguilar PS, Hernandez-Arriaga AM, Cybulski LE, Erazo AC, de Mendoza D (2001). Molecular basis of thermosensing: a two-component signal transduction thermometer inBacillus subtilis. EMBO J 20: 1681–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amalfitano S, Fazi S (2008). Recovery and quantification of bacterial cells associated with streambed sediments. J Microbiol Methods 75: 237–243. [DOI] [PubMed] [Google Scholar]
- Asakura H, Ishiwa A, Arakawa E, Makino SI, Okada Y, Yamamoto S et al (2007). Gene expression profile of Vibrio cholerae in the cold stress‐induced viable but non‐culturable state. Environ Microbiol 9: 869–879. [DOI] [PubMed] [Google Scholar]
- Ayala-Del-Río HL, Chain PS, Grzymski JJ, Ponder MA, Ivanova N, Bergholz PW et al (2010). The genome sequence of Psychrobacter arcticus 273-4, a psychroactive Siberian permafrost bacterium, reveals mechanisms for adaptation to low-temperature growth. Appl Environ Microbiol 76: 2304–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakermans C, Bergholz PW, Ayala-del-Río H, Tiedje J. (2009). Genomic insights into cold adaptation of permafrost bacteria. InMargesin Red Soil Biology: Permafrost Soils, vol. 16. Springer: Berlin, Heidelberg, pp 159–168. [Google Scholar]
- Bakermans C, Tsapin AI, Souza-Egipsy V, Gilichinsky DA, Nealson KH (2003). Reproduction and metabolism at -10°C of bacteria isolated from Siberian permafrost. Environ Microbiol 5: 321–326. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing on JSTOR. J R Stat Soc Series B Stat Methodol 57: 289–300. [Google Scholar]
- Berger BJ, English S, Chan G, Knodel MH (2003). Methionine regeneration and aminotransferases in Bacillus subtilisBacillus cereus, and Bacillus anthracis. J Bacteriol 185: 2418–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff J, Mangelsdorf K, Gattinger A, Schloter M, Kurchatova AN, Herzschuh U et al (2013). Response of methanogenic archaea to Late Pleistocene and Holocene climate changes in the Siberian Arctic. Global Biogeochem Cycles 27: 305–317. [Google Scholar]
- Bjella K, Tantillo T, Weale J, Lever J (2008)Evaluation of the CRREL Permafrost Tunnel US Army Corps of Engineers Engineer Research and Development Center ERDC/CRREL TR-08-11 Available from http://www.dtic.mil/dtic/tr/fulltext/u2/a485745.pdf accessed May 2013.
- Blow MJ, Zhang T, Woyke T, Speller CF, Krivoshapkin A, Yang DY et al (2008). Identification of ancient remains through genomic sequencing. Genome Res 18: 1347–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresolin G, Neuhaus K, Scherer S, Fuchs TM (2006). Transcriptional analysis of long-term adaptation of Yersinia enterocolitica to low-temperature growth. J Bacteriol 188: 2945–2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH (2015). Fast and sensitive protein alignment using DIAMOND. Nat Methods 12: 59–60. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona M, Zamarro MT, Blázquez B, Durante-Rodríguez G, Juárez JF, Valderrama JA et al (2009). Anaerobic catabolism of aromatic compounds: a genetic and genomic view. Microbiol Mol Biol Rev 73: 71–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carper SW, Willis DG, Manning KA, Gerner EW (1991). Spermidine acetylation in response to a variety of stresses in Escherichia coli. J Biol Chem 266: 12439–12441. [PubMed] [Google Scholar]
- Chattopadhyay MK (2006). Mechanism of bacterial adaptation to low temperature. J Biosci 31: 157–165. [DOI] [PubMed] [Google Scholar]
- Collins RE, Deming JW (2013). An inter-order horizontal gene transfer event enables the catabolism of compatible solutes by Colwellia psychrerythraea 34H. Extremophiles 17: 601–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa TRD, Felisberto-Rodrigues C, Meir A, Prevost MS, Redzej A, Trokter M et al (2015). Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat Rev Microbiol 13: 343–359. [DOI] [PubMed] [Google Scholar]
- D'Amico S, Collins T, Marx J-C, Feller G, Gerday C (2006). Psychrophilic microorganisms: challenges for life. EMBO Rep 7: 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo C, Ramazzotti M, Fontana P, Cavalieri D (2012). Bioinformatic approaches for functional annotation and pathway inference in metagenomics data. Brief Bioinform 13: 696–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maayer P, Anderson D, Cary C, Cowan DA (2014). Some like it cold: understanding the survival strategies of psychrophiles. EMBO Rep 15: 508–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMaere MZ, Williams TJ, Allen MA, Brown MV, Gibson JAE, Rich J et al (2013). High level of intergenera gene exchange shapes the evolution of haloarchaea in an isolated Antarctic lake. Proc Natl Acad Sci USA 110: 16939–16944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieser M, Battista JR, Christner BC (2013). DNA double-strand break repair at -15ºC. Appl Environ Microbiol 79: 7662–7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake TW, Wickland KP, Spencer RGM, McKnight DM, Striegl RG (2015). Ancient low-molecular-weight organic acids in permafrost fuel rapid carbon dioxide production upon thaw. Proc Natl Acad Sci USA 112: 13946–13951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziewit L, Bartosik D (2014). Plasmids of psychrophilic and psychrotolerant bacteria and their role in adaptation to cold environments. Front Microbiol 5: 596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Edgar RC (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10: 996–998. [DOI] [PubMed] [Google Scholar]
- Fairén AG, Davila AF, Lim D, Bramall N, Bonaccorsi R, Zavaleta J et al (2010). Astrobiology through the ages of Mars: the study of terrestrial analogues to understand the habitability of Mars. Astrobiology 10: 821–843. [DOI] [PubMed] [Google Scholar]
- Feng S, Powell SM, Wilson R, Bowman JP (2014). Extensive gene acquisition in the extremely psychrophilic bacterial species Psychroflexus torquis and the link to sea-ice ecosystem specialism. Genome Biol Evol 6: 133–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Yang ZK, Wu L, Thompson DK, Zhou J (2006). Global transcriptome analysis of the cold shock response of Shewanella oneidensis MR-1 and mutational analysis of its classical cold shock proteins. J Bacteriol 188: 4560–4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilichinsky D, Rivkina E, Shcherbakova V, Laurinavichuis K, Tiedje J (2003). Supercooled water brines within permafrost—an unknown ecological niche for microorganisms: a model for astrobiology. Astrobiology 3: 331–341. [DOI] [PubMed] [Google Scholar]
- Gilichinsky D, Vishnivetskaya T, Petrova M, Spirina E, Mamykin V, Rivkina E (2008)Bacteria in the permafrost. In: Margesin R, Schinner F, Marx J-C, Gerday C (eds). Psychrophiles: From Biodiversity to Biotechnology. Springer, Berlin, Heidelberg, pp 83–102. [Google Scholar]
- Gilichinsky DA (2002)Permafrost model of extraterrestrial habitatInHorneck G, Baumstark-Khan Ceds Astrobiology. Springer, Berlin, Heidelberg, pp 125–142. [Google Scholar]
- Goordial J, Raymond-Bouchard I, Zolotarov Y, de Bethencourt L, Ronholm J, Shapiro N et al (2015). Cold adaptive traits revealed by comparative genomic analysis of the eurypsychrophile Rhodococcus sp. JG3 isolated from high elevation McMurdo Dry Valley permafrost, Antarctica. FEMS Microbiol Ecol 92: fiv154. [DOI] [PubMed] [Google Scholar]
- Graham DE, Wallenstein MD, Vishnivetskaya TA, Waldrop MP, Phelps TJ, Pfiffner SM et al (2012). Microbes in thawing permafrost: the unknown variable in the climate change equation. ISME J 6: 709–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann P, Schröder K, Schmid R, Marahiel MA (1996). Cold shock stress-induced proteins in Bacillus subtilis. J Bacteriol 178: 4611–4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton TD, Craig JL, Sellman PV (1988). The Fox permafrost tunnel: a late Quaternary geologic record in central Alaska. Geol Soc Am Bull 100: 948–969. [Google Scholar]
- Hansen AA, Herbert RA, Mikkelsen K, Jensen LL, Kristoffersen T, Tiedje JM et al (2007). Viability, diversity and composition of the bacterial community in a high Arctic permafrost soil from Spitsbergen, Northern Norway. Environ Microbiol 9: 2870–2884. [DOI] [PubMed] [Google Scholar]
- Hanson NW, Konwar KM, Hawley AK, Altman T, Karp PD, Hallam SJ (2014). Metabolic pathways for the whole community. BMC Genomics 15: 619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultman J, Waldrop MP, Mackelprang R, David MM, McFarland J, Blazewicz SJ et al (2015). Multi-omics of permafrost, active layer and thermokarst bog soil microbiomes. Nature 521: 208–212. [DOI] [PubMed] [Google Scholar]
- Huson DH, Beier S, Flade I, Górska A, El-Hadidi M, Mitra S et al (2016). MEGAN community edition—interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput Biol 12: e1004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyyryläinen H-L, Pietiäinen M, Lundén T, Ekman A, Gardemeister M, Murtomäki-Repo S et al (2007). The density of negative charge in the cell wall influences two-component signal transduction in Bacillus subtilis. Microbiology 153: 2126–2136. [DOI] [PubMed] [Google Scholar]
- Igarashi K, Kashiwagi K (2000). Polyamines: mysterious modulators of cellular functions. Biochem Biophys Res Commun 271: 559–564. [DOI] [PubMed] [Google Scholar]
- Jansson JK, Tas N (2014). The microbial ecology of permafrost. Nature Rev Microbiol 12: 414–425. [DOI] [PubMed] [Google Scholar]
- Junge K, Eicken H, Deming JW (2003). Motility of Colwellia psychrerythraea strain 34H at subzero temperatures. Appl Environ Microbiol 69: 4282–4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K (2017). KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucelic Acids Res 45: D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama T, Tanaka M, Moriizumi J, Nakamura T, Brouchkov A, Douglas TA et al (2007). Phylogenetic analysis of bacteria preserved in a permafrost ice wedge for 25,000 years. Appl Environ Microbiol 73: 2360–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepner RL, Pratt JR (1994). Use of fluorochromes for direct enumeration of total bacteria in environmental samples: past and present. Microbiol Rev 58: 603–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansson E, Hugenholtz P, Dalevi D (2009). ShotgunFunctionalizeR: an R-package for functional comparison of metagenomes. Bioinformatics 25: 2737–2738. [DOI] [PubMed] [Google Scholar]
- Kvenvolden KA, Lorenson TD, Barber V (1994)Methane in the Fox permafrost tunnel near Fairbanks, Alaska. In: Till AB, Moore TE (eds). Geologic Studies in Alaska by the U.S. Geological Survey, US Geological Survey Bulletin 2107. US Government Printing Office: Washington DC, USA, pp 31–37. [Google Scholar]
- Lachniet MS, Lawson DE, Sloat AR (2012). Revised 14C dating of ice wedge growth in interior Alaska (USA) to MIS 2 reveals cold paleoclimate and carbon recycling in ancient permafrost terrain. Quater Res 78: 217–225. [Google Scholar]
- Li Y, Powell DA, Shaffer SA, Rasko DA, Pelletier MR, Leszyk JD et al (2012). LPS remodeling is an evolved survival strategy for bacteria. Proc Natl Acad Sci USA 109: 8716–8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd KG, Schreiber L, Petersen DG, Kjeldsen KU, Lever MA, Steen AD et al (2013). Predominant archaea in marine sediments degrade detrital proteins. Nature 496: 215–218. [DOI] [PubMed] [Google Scholar]
- Lomstein BA, Langerhuus AT, D'Hondt S, Jørgensen BB, Spivack AJ (2012). Endospore abundance, microbial growth and necromass turnover in deep sub-seafloor sediment. Nature 484: 101–104. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Wang L, Shao Z (2006). Pseudomonas, the dominant polycyclic aromatic hydrocarbon‐degrading bacteria isolated from Antarctic soils and the role of large plasmids in horizontal gene transfer. Environ Microbiol 8: 455–465. [DOI] [PubMed] [Google Scholar]
- Mackelprang R, Saleska SR, Jacobsen CS, Jansson JK, Tas N (2016). Permafrost meta-omics and climate change. Annu Rev Earth Planet Sci 44: 439–462. [Google Scholar]
- Mackelprang R, Waldrop MP, DeAngelis KM, David MM, Chavarria KL, Blazewicz SJ et al (2011). Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature 480: 368–371. [DOI] [PubMed] [Google Scholar]
- Mason OU, Hazen TC, Borglin S, Chain PSG, Dubinsky EA, Fortney JL et al (2012). Metagenome, metatranscriptome and single-cell sequencing reveal microbial response to Deepwater Horizon oil spill. ISME J 6: 1715–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A et al (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6: 610–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8: e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methé BA, Nelson KE, Deming JW, Momen B, Melamud E, Zhang X et al (2005). The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analyses. Proc Natl Acad Sci USA 102: 10913–10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morono Y, Terada T, Kallmeyer J, Inagaki F (2013). An improved cell separation technique for marine subsurface sediments: applications for high-throughput analysis using flow cytometry and cell sorting. Environ Microbiol 15: 2841–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhs DR, Ager TA, Begét JE (2001). Vegetation and paleoclimate of the last interglacial period, central Alaska. Quat Sci Rev 20: 41–61. [Google Scholar]
- Mykytczuk NCS, Foote SJ, Omelon CR, Southam G, Greer CW, Whyte LG (2013). Bacterial growth at −15 °C; molecular insights from the permafrost bacterium Planococcus halocryophilus Or1. ISME J 7: 1211–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehus R, Mitri S, Fletcher AG, Foster KR (2015). Migration and horizontal gene transfer divide microbial genomes into multiple niches. Nat Commun 6: 8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P (2015). Vegan: community ecology package. R Package Vegan, Vers 2.2-1 Available at https://cran.r-project.org/web/packages/vegan/index.html.
- Orsi WD, Edgcomb VP, Christman GD, Biddle JF (2013). Gene expression in the deep biosphere. Nature 499: 205–208. [DOI] [PubMed] [Google Scholar]
- Parducci L, Jørgensen T, Tollefsrud MM, Elverland E, Alm T, Fontana SL et al (2012). Glacial survival of boreal trees in northern Scandinavia. Science 335: 1083–1086. [DOI] [PubMed] [Google Scholar]
- Parducci L, Väliranta M, Salonen JS, Ronkainen T, Matetovici I, Fontana SL et al (2015). Proxy comparison in ancient peat sediments: pollen, macrofossil and plant DNA. Philos Trans R Soc Lond Biol 370: 20130382–20130382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Beiko RG (2010). Identifying biologically relevant differences between metagenomic communities. Bioinformatics 26: 715–721. [DOI] [PubMed] [Google Scholar]
- Phadtare S (2004). Recent developments in bacterial cold-shock response. Curr Issues Mol Biol 6: 125–136. [PubMed] [Google Scholar]
- Poté J, Bravo AG, Mavingui P, Ariztegui D, Wildi W (2010). Evaluation of quantitative recovery of bacterial cells and DNA from different lake sediments by Nycodenz density gradient centrifugation. Ecol Indic 10: 234–240. [Google Scholar]
- Raivio TL (2014). Everything old is new again: an update on current research on the Cpx envelope stress response. Biochim Biophys Acta 1843: 1529–1541. [DOI] [PubMed] [Google Scholar]
- Rhee HJ, Kim EJ, Lee JK (2007). Physiological polyamines: simple primordial stress molecules. J Cell Mol Med 11: 685–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivkina E, Laurinavichius K, McGrath J, Tiedje J, Shcherbakova V, Gilichinsky D (2004). Microbial life in permafrost. Adv Space Res 33: 1215–1221. [DOI] [PubMed] [Google Scholar]
- Rivkina E, Petrovskaya L, Vishnivetskaya T, Krivushin K, Shmakova L, Tutukina M et al (2016). Metagenomic analyses of the late Pleistocene permafrost—additional tools for reconstruction of environmental conditions. Biogeosciences 13: 2207–2219. [Google Scholar]
- Rivkina EM, Friedmann EI, McKay CP, Gilichinsky DA (2000). Metabolic activity of permafrost bacteria below the freezing point. Appl Environ Microbiol 66: 3230–3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues DF, Ivanova N, He Z, Huebner M, Zhou J, Tiedje JM (2008). Architecture of thermal adaptation in an Exiguobacterium sibiricum strain isolated from 3 million year old permafrost: a genome and transcriptome approach. BMC Genomics 9: 547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF et al (2014). Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 12: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider BL, Hernandez VJ, Reitzer L (2013). Putrescine catabolism is a metabolic response to several stresses in Escherichia coli. Mol Microbiol 88: 537–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekowska A, Dénervaud V, Ashida H, Michoud K, Haas D, Yokota A et al (2004). Bacterial variations on the methionine salvage pathway. BMC Microbiol 4: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smets W, Leff JW, Bradford MA, McCulley RL, Lebeer S, Fierer N (2016). A method for simultaneous measurement of soil bacterial abundances and community composition via 16 S rRNA gene sequencing. Soil Biology and Biochemistry 96: 145–151. [Google Scholar]
- Stuiver M, Reimer PJ (1993). Extended 14 C data base and revised CALIB 3.0 14 C age calibration program. Radiocarbon 35: 215–230. [Google Scholar]
- Takahashi T, Kakehi J-I (2010). Polyamines: ubiquitous polycations with unique roles in growth and stress responses. Ann Bot 105: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tas N, Prestat E, McFarland JW, Wickland KP, Knight R, Berhe AA et al (2014). Impact of fire on active layer and permafrost microbial communities and metagenomes in an upland Alaskan boreal forest. ISME J 8: 1904–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkachenko AG, Pshenichnov MR, Nesterova LY (2001). Putrescine as a factor protecting Escherichia coliagainst oxidative stress. Microbiology 70: 422–428. [Google Scholar]
- Tuorto SJ, Darias P, McGuinness LR, Panikov N, ZHANG T, Häggblom MM et al (2014). Bacterial genome replication at subzero temperatures in permafrost. ISME J 8: 139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasil'chuk YK, Vasil'chuk AC (2014). Strategy of valid 14 C dates choice in syngenetic permafrost. Cryosphere 8: 5589–5621. [Google Scholar]
- Vishnivetskaya T, Kathariou S, McGrath J, Gilichinsky D, Tiedje JM (2000). Low-temperature recovery strategies for the isolation of bacteria from ancient permafrost sediments. Extremophiles 4: 165–173. [DOI] [PubMed] [Google Scholar]
- Vishnivetskaya TA, Petrova MA, Urbance J, Ponder M, Moyer CL, Gilichinsky DA et al (2006). Bacterial community in ancient Siberian permafrost as characterized by culture and culture-independent methods. Astrobiology 6: 400–414. [DOI] [PubMed] [Google Scholar]
- Vorobyova E, Soina V, Gorlenko M, Minkovskaya N, Zalinova N, Mamukelashvili A et al (1997). The deep cold biosphere: facts and hypothesis. FEMS Microbiol Rev 20: 277–290. [Google Scholar]
- Waldrop MP, Wickland KP, White R III, Berhe AA, Harden JW, Romanovsky VE (2010). Molecular investigations into a globally important carbon pool: permafrost-protected carbon in Alaskan soils. Global Change Biol 16: 2543–2554. [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JR, Nagarajan N, Pop M (2009). Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol 5: e1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willerslev E, Cappellini E, Boomsma W, Nielsen R, Hebsgaard MB, Brand TB et al (2007). Ancient biomolecules from deep ice cores reveal a forested southern Greenland. Science 317: 111–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willerslev E, Davison J, Moora M, Zobel M, Coissac E, Edwards ME et al (2014). Fifty thousand years of Arctic vegetation and megafaunal diet. Nature 506: 47–51. [DOI] [PubMed] [Google Scholar]
- Willerslev E, Hansen AJ, Rønn R, Brand TB, Barnes I, Wiuf C et al (2004). Long-term persistence of bacterial DNA. Curr Biol 14: R9–10. [DOI] [PubMed] [Google Scholar]
- Zhu X, Li Q, Yin C, Fang X, Xu X (2015). Role of spermidine in overwintering of cyanobacteria. J Bacteriol 197: 2325–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.