Abstract
Acquisition of additional photosynthetic pigments enables photosynthetic organisms to survive in particular niches. To reveal the ecological significance of chlorophyll (Chl) f, we investigated the distribution of Chl and cyanobacteria within two microbial mats. In a 7-mm-thick microbial mat beneath the running water of the Nakabusa hot spring, Japan, Chl f was only distributed 4.0–6.5 mm below the surface, where the intensity of far-red light (FR) was higher than that of photosynthetically active radiation (PAR). In the same mat, two ecotypes of Synechococcus and two ecotypes of Chl f-producing Leptolyngbya were detected in the upper and deeper layers, respectively. Only the Leptolyngbya strains could grow when FR was the sole light source. These results suggest that the deeper layer of the microbial mat was a habitat for Chl f-producing cyanobacteria, and Chl f enabled them to survive in a habitat with little PAR.
Introduction
Light quality is one of the most important drivers in niche differentiation of phototrophs, and the spectrum of available light determines the distribution and community composition of photosynthetic organisms in natural environments. These niche differentiations are generally caused by variations in pigment composition and/or photosynthetic apparatus, which have diverged to adapt to various colors of light. Close relationships between the absorption spectra of photosynthetic organisms and the surrounding light environments and niche differentiation have been reported in many previous studies, such as those observed in Synechococcus with different types of phycobilisomes (Wood et al., 1998; Stomp et al., 2007), planktonic bacteria with green and blue types of proteorhodopsins in ocean water columns (Béjà et al., 2001; Sabehi et al., 2007), and cyanobacteria containing chlorophyll (Chl) a and b (Prochloron) and Chl d (Acaryochloris) coexisting with colonial ascidians (Kühl et al., 2005).
Chl f is a newly discovered red-shifted Chl with an absorption peak in the far-red light (FR) region (Chen et al., 2010). The first reported Chl f-containing organism was a filamentous cyanobacterium, Halomicronema hongdechloris, isolated from stromatolites in Australia (Chen et al., 2012). To date, this strain has been used to study the extinction coefficient (Li et al., 2012), biochemical structure (Willows et al., 2013) and biophysical characteristics (Li et al., 2013; Tomo et al., 2014; Akimoto et al., 2015) of Chl f. Several other cyanobacteria producing Chl f have also been found from various environments. A unicellular cyanobacterium strain, KC1, was isolated from Lake Biwa in Japan (Akutsu et al., 2011; Miyashita et al., 2014), and a phylogenetically related species was found in biofilms at Jenolan Caves in Australia (Behrendt et al., 2015). The Leptolyngbya sp. strain JSC-1 from a hot spring in Yellowstone National Park in the United States (Gan et al., 2014) and Chlorogloeopsis fritschii from a rice paddy in India (Airs et al., 2014) were reported to contain both Chl d and Chl f.
Some previous studies have reported that Chl f in various cyanobacterial species was reversibly induced and decreased in laboratory experiments (Chen et al., 2012; Airs et al., 2014; Gan et al., 2014; Miyashita et al., 2014). In all newly discovered cyanobacterial strains, Chl f was produced when their cells were grown under FR as the sole light source and decreased to an undetectable level under light conditions of white or another color. Five previously identified cyanobacterial strains were also reported to accumulate Chl f, as well as to synthesize a small amount of Chl d and FR shifted phycobiliproteins when they were cultured under FR (Gan et al., 2015). The mechanism of this reversible switch in pigmentation was referred to as FaRLiP (far red light photoacclimation). Some key genes of this FaRLiP response and the Chl f-synthesis enzyme have previously been identified (Zhao et al., 2015; Ho et al., 2016). The available data suggest that the distribution of Chl f in natural environments must be restricted to habitats in which FR dominates, such as soils, under the plant canopy, microbial mats and aquatic blooms (Gan and Bryant, 2015). Indeed, the presence of Chl f in nature has recently been confirmed in cyanobacterial biofilms located in a cave environment and beachrock in Australia (Behrendt et al., 2015; Trampe and Kühl, 2016).
The light spectra in natural and artificial microbial mats have been measured (Pierson et al., 1990; Boomer et al., 2000; Kühl and Fenchel, 2000; Ferris et al., 2003) and found that photosynthetically active radiation (PAR) decreased rapidly within a few millimeters through its absorption by cyanobacteria and/or anoxygenic photosynthetic bacteria living in the mats. Light with wavelengths longer than 700 nm remained, even in deeper layers of the mats. Based on these observations, we expected Chl f-producing cyanobacteria to be distributed in microbial mats and Chl f to be produced in deeper layers of the mat, where organisms use FR for photosynthesis. The Nakabusa hot spring in Japan is famous for its thick, colorful microbial mats that contain cyanobacteria (Nakagawa and Fukui, 2002; Kubo et al., 2011). The present study was conducted to reveal the niche of Chl f-producing cyanobacteria and the ecological role of Chl f in microbial mats from this hot spring. We investigated the spectra and strength of penetrated light, the vertical distribution of Chl, and the community structure of cyanobacteria at each depth of the mat. After isolating the cyanobacteria from the mat, we also evaluated the presence of Chl f and growth characteristics of those isolates under white light (WL) and FR.
Materials and methods
Sampling
We collected microbial mat samples from the Nakabusa hot spring, Nagano, Japan (36.394367 N, 137.747183E), on 14 October 2012. At this site, dense colorful microbial mats are formed on the wall of the sand control dam, on which hot spring water falls continuously. Two pieces of green-colored mats Nb3 and Nb4 (ca 5 × 10 cm) were cut from the wall with a knife and put in different plastic containers. The distance between these two mat samples was ~10 cm, and their in situ temperatures were 49.9 °C and 46.8 °C, respectively. The samples were transported to the laboratory under dark, ambient temperature conditions and stored in a refrigerator at 4 °C for 2 days until the culturing experiment was carried out, after which they were frozen at −20 °C for further experiments.
Measurement of light spectra in microbial mats
The downward spectral transmittances as a function of depth in the microbial mats were measured using a USB4000 spectrophotometer (Ocean Optics, Inc., Dunedin, FL, USA) with a fiber optic tip (Pierson et al., 1990). Solar spectral irradiance was measured at the sampling site with USB4000 at 10:00, 17 June 2012. The downward spectral irradiances in the microbial mats were calculated from these data. The light environments in the microbial mats at different times of day and under different weather conditions were also estimated. For those calculations, we used the solar radiation spectra data collected at Gifu (~140 km away from Nakabusa) by the New Energy and Industrial Technology Development Organization (NEDO) for 1 week before sampling the mats.
Pigment composition analyses
The frozen microbial mats were cut into a rectangular parallelepiped, whose base was 1 cm square, and embedded in 2% agarose gel. The gel blocks were then bored 19 mm in diameter, frozen again at −20 °C and sliced horizontally into sections 0.5 mm thick from the surface down to 6.5 mm (Nb3) and 4.0 mm (Nb4) using a simple microtome (Artec Co. Ltd, Osaka, Japan). Pigments were extracted twice from each freeze-dried section with 100 μl of methanol for 15 min by sonication in the dark. Next, a total of 200 μl of extract was centrifuged for 5 min at 16 000 g, after which its supernatant was analyzed by reverse phase high performance liquid chromatography using a Spherisorb 5 μm ODS2 column (4.6 × 250 mm, Waters, Milford, MA, USA) at 40 °C and a methanol–water–acetone gradient program (2.5 ml min−1, 0–10 min linear gradient 90–100% methanol, 10–20 min 100% methanol, 20–22 min linear gradient methanol to acetone, 22–23 min 100% acetone). The absorption spectra of eluted pigments (350–800 nm) were obtained by using a Waters 2996 PDA detector (Waters, Milford, MA, USA). Chls a and f and BChl a were eluted at retention times of 9.8, 8.3 and 7.3 min, respectively. The specific absorption coefficients of 78.36 (665.5 nm), 78.36 (707 nm) and 60 (771 nm) L g−1 cm−1 in methanol were used to calculate the relative amounts of Chl a, Chl f and BChl a, respectively (Namsaraev, 2009; Li et al., 2012).
Microscopy
Cross-sectioned images of the microbial mat Nb3 were acquired using a BX51 microscope equipped with a U-MWU2 mirror unit and a DP72 digital camera (OLYMPUS, Tokyo, Japan). The range of excitation wavelengths was 330–385 nm and fluorescence at >420 nm could be observed. Cells of the newly isolated cyanobacteria were observed using an ECLIPSE 80i microscope and a DS-5Mc digital camera (Nikon, Tokyo, Japan).
PCR-DGGE
DNAs were extracted from each horizontally sliced and pigment-extracted section of mats using the method described by Ohkubo and Miyashita (2012). PCR and denaturant gradient gel electrophoresis (DGGE) were performed with the Cyanobacteria-specific primer set CYA353F-CYA781R as described in Ohkubo et al. (2006), except for the concentrations of polyacrylamide (9%) and denaturant (30–70%) in gel, and the electrophoresis conditions (60 °C, 100 V, 13 h). The gel was stained with SYBR Gold Nucleic Acid Gel Stain (Life Technologies Japan, Tokyo, Japan) and photographed on a Visi-Blue transilluminator (LLC., Upland, CA, USA) with a CCD camera AE-9000 N E-Graph (ATTO CORPORATION, Tokyo, Japan).
Isolation of cyanobacteria
A piece of mat Nb3 was homogenized in a microtube containing BG11 medium using sterile pestles. Aliquots of homogenates were inoculated to liquid BG11 medium in two 24-well plastic culture plates that were placed in 22 °C or 35 °C under FR LEDs as a sole light source. After 3 months, cyanobacteria that had increased under these conditions were isolated under microscopy were transferred to new media with sterile pipette tips and cultured under FR or WL.
Sequencing and phylogenetic analyses
Sequencing of DGGE bands was performed as described in Ohkubo and Miyashita (2012). DNA extraction from cyanobacterial strains was conducted as described above. Two primer sets, CYA359F-1510 R and 9 F-CYA781R (Nübel et al., 1997; Ahmed et al., 2007), were used for PCR amplification of 16S rDNA fragments from strains Nb3F1 and Nb3U1. In the case of strain Nb3F2, a Bacteria-universal primer set, 341 F-907 R (Nakagawa and Fukui, 2002) was used. Direct sequencing was performed with the primers 9 F, 341 F, CYA359F, 515 F, 536 R, CYA781R, 907 F, 907 R, 936 R and 1510 R (Nübel et al., 1997; Ahmed et al., 2007; Nakagawa and Fukui, 2002). The sequences obtained were compared with GenBank sequences by NCBI BLAST searches (Altschul et al., 1990) and deposited in the DDBJ database under accession numbers LC192821–LC192830, LC270317. These sequences and 68 known cyanobacterial sequences were aligned using PyNAST (Caporaso et al., 2010), and the phylogenetic relationship was analyzed using the maximum likelihood method in the RAxML program (Stamatakis, 2014) with the GTRGAMMA model and a rapid bootstrap procedure (500 replicates). The constructed tree was visualized using FigTree ver. 1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
Growth experiment
Cyanobacterial strains Nb3U1, Nb3F1 and Nb3F2 were pre-cultured in BG11 medium (pH 8.0) with aeration under continuous irradiation (110 μmol photon m−2 s−1) provided by a WL LED (Color Changing LED Light Bulb, Lighting EVER, Las Vegas, NV, USA). After 20–30 days, pre-cultured cells were harvested and homogenized using plastic pestles. The same volumes of homogenates were then inoculated into glass tubes containing 50 ml of BG11 and cultivated for 30–35 days with aeration. All three strains were incubated under 110 μmol photon m−2 s−1 provided by WL LED and FR LED (MR60L-5LED, Aster, Toyohashi, Japan), and Nb3F1 and Nb3F2 were incubated in the dark, too. Each experiment was triplicated. The emission spectra of LEDs are shown in Supplementary Figure S1. Strains Nb3U1 and Nb3F1 were cultured at 45 °C, whereas Nb3F2 was cultured at 25 °C. Cell growth was monitored by measuring the OD780 with a V-650 spectrophotometer (JASCO, Tokyo, Japan).
Results
Penetrating light spectra and distribution of Chls within the microbial mats
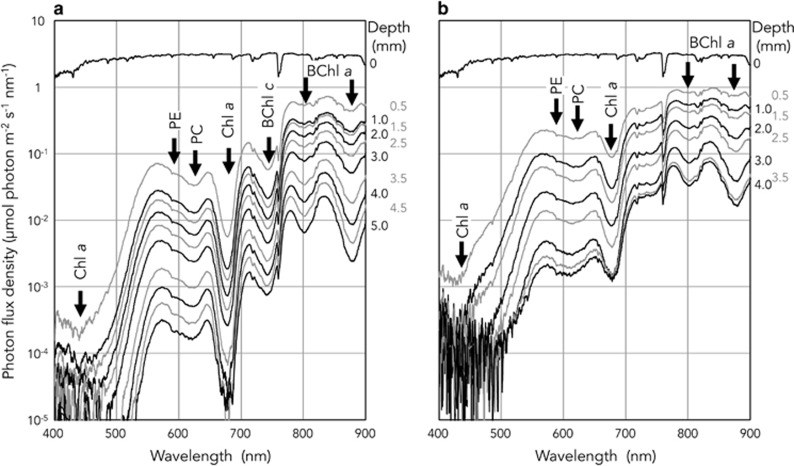
The spectral compositions and the photon flux density of the downward irradiance changed with depth within the microbial mats (Figure 1). The on-site solar radiation was 1.4–3.3 μmol photon m−2 s−1 nm−1 over the wavelength range of 400–900 nm. There were some attenuation peaks in the spectra within the microbial mats. Those peaks could be assigned to light absorption by photosynthetic pigments such as Chl a (440 and 678 nm), phycobiliproteins (588 and 615 nm), BChl a (802 and 880 nm) and BChl c (742 nm) based on previous reports (Boomer et al., 2000; Ferris et al., 2003). The results showed that the PAR (400–700 nm) was depleted within both microbial mats, and the depletion was faster than that of FR (700–750 nm), indicating that FR of certain strengths could penetrate deeper layers (Figure 1). Although PAR (813.4 μmol photon m−2 s−1) was 5.4 times higher than that of FR (149.3 μmol photon m–2 s–1) at the surface, the photon flux density of FR exceeded that of PAR at a depth of 3.5 mm in Nb3 (PAR; 0.185, FR; 0.223 μmol photon m−2 s−1) and 2.5 mm in Nb4 (PAR; 2.808, FR; 1.767 μmol photon m−2 s−1). Although the solar radiation spectra changed depending on the time and the weather for one week before sampling of mats, the calculated light spectra within the mats did not differ significantly (Supplementary Figure S2).
Figure 1.
Downward irradiance spectra with depth in microbial mats Nb3 (a) and Nb4 (b) from Nakabusa hot spring. Arrows indicate major attenuation peaks from absorption of photosynthetic pigments (Chl a, 440 and 678 nm; phycoerythrin (PE), 588 nm; phycocyanin (PC), 615 nm; BChl c, 742 nm; BChl a, 802 and 880 nm).
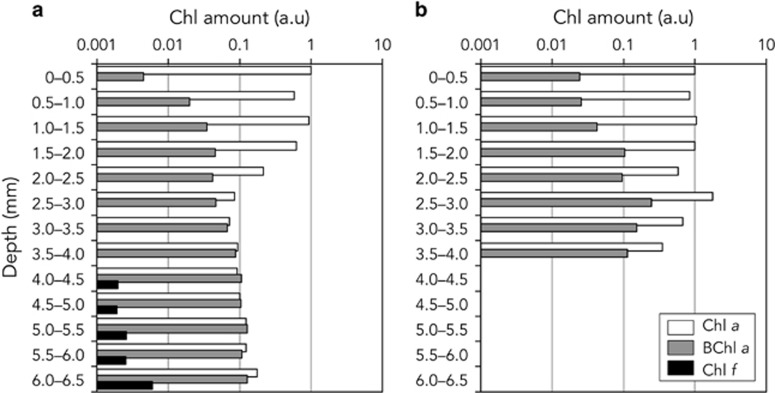
Chl compositions in each horizontal serial section (0.5 mm thickness) of both microbial mats are shown in Figure 2. The amounts of Chl a, BChl a and Chl f in each section were expressed as relative values with respect to the amount of Chl a in the surface (0–0.5 mm) section of each mat. In mat Nb3, the Chl a content was similar from the surface to 2 mm in depth. The content in sections deeper than 2.5 mm were less than one-tenth that of the surface. BChl a increased in proportion to depth, with the relative amount of BChl a in the 2.5–3.0 mm section being 10 times higher than that in the 0–0.5 mm section. Chl f was only detected from five sections deeper than 4.0 mm. The amounts of Chl f detected were 1.9–3.4% of those of Chl a detected at each depth. In the mat Nb4, Chl a content varied in the range of 35.2–176% from the surface to bottom. BChl a gradually increased from the surface to deep layers. Chl f were below the detection limits in all sections of the mat.
Figure 2.
Relative amounts of chlorophyll in each 0.5 mm thick horizontal section of microbial mats Nb3 (a) and Nb4 (b). This graph shows relative amounts of chlorophyll a (white), bacteriochlorophyll a (gray) and chlorophyll f (black).
Vertical distribution of cyanobacteria within the microbial mats
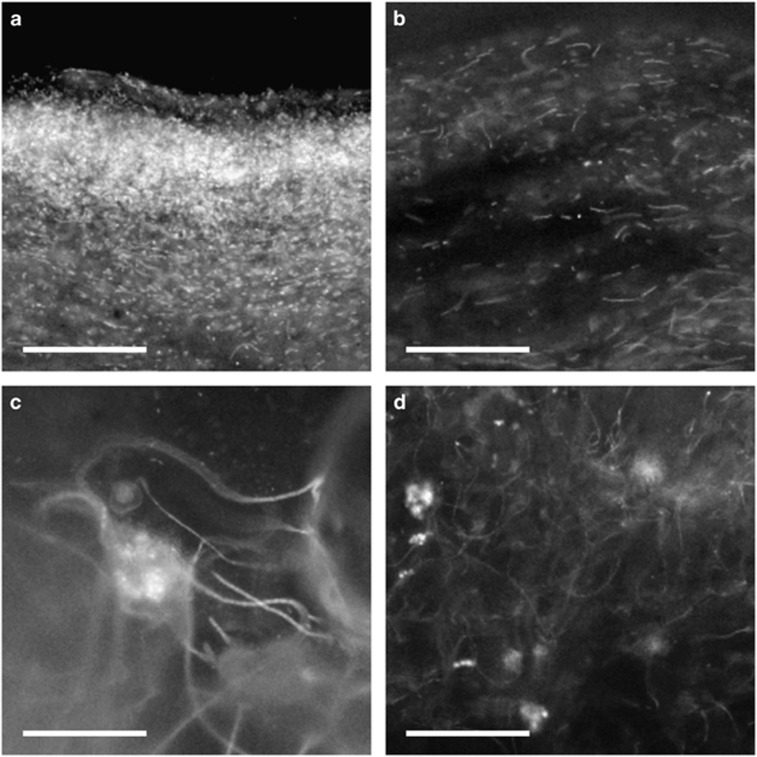
The vertical distribution of cyanobacteria in microbial mat Nb3 was observed by both microscopy and PCR-DGGE analysis. The mat appeared to consist of four layers, a bright-green surface layer, a glossy white or gelatinous brown layer, a black-green layer and a white or brown layer that was similar to the second layer. Upon microscopic observation, strong red autofluorescence was only observed at the surface of the mat (Supplementary Figure S3). Cells observed in this layer were unicellular (0.8–3 μm in diameter) and rod shaped or elongated (0.8–1 μm in diameter and 5–30 μm in length) (Figures 3a and b). The cells were scattered within the surface layer of the mat. Below the greenish surface layer, the green color and the red autofluorescence weakened with increasing depth (Supplementary Figure S3). Although the clearly visible green color was not observed in the mat at depths of >1.0 mm upon light microscopy, weak red fluorescence was observed at depths of 4.0–7.0 mm under fluorescence microscopy. Cells in these layers were long, thin filaments 1–3 μm in diameter and >100 μm in length that were loosely tangled together (Figures 3c and d).
Figure 3.
Images from vertical sections of microbial mat Nb3 shown under epifluorescence microscopy at depths of 0–0.2 mm (a), 0.4–0.7 mm (b), 4.8–5.0 nm (c) and 6.4–6.7 mm (d). Scale bars indicate 100 μm.
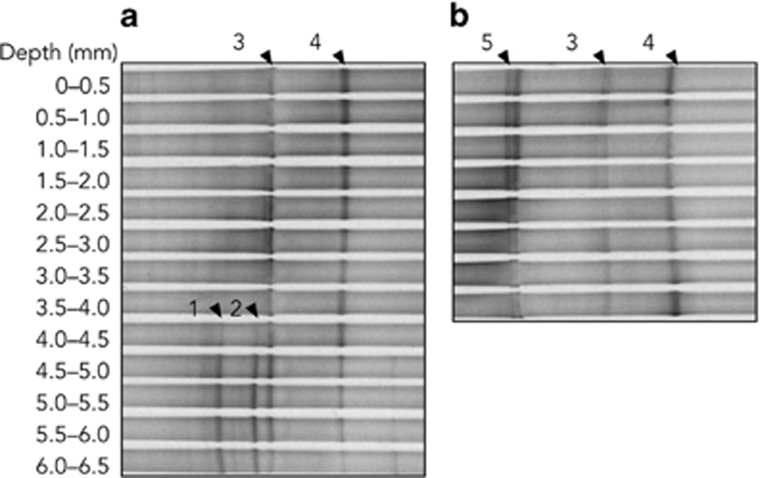
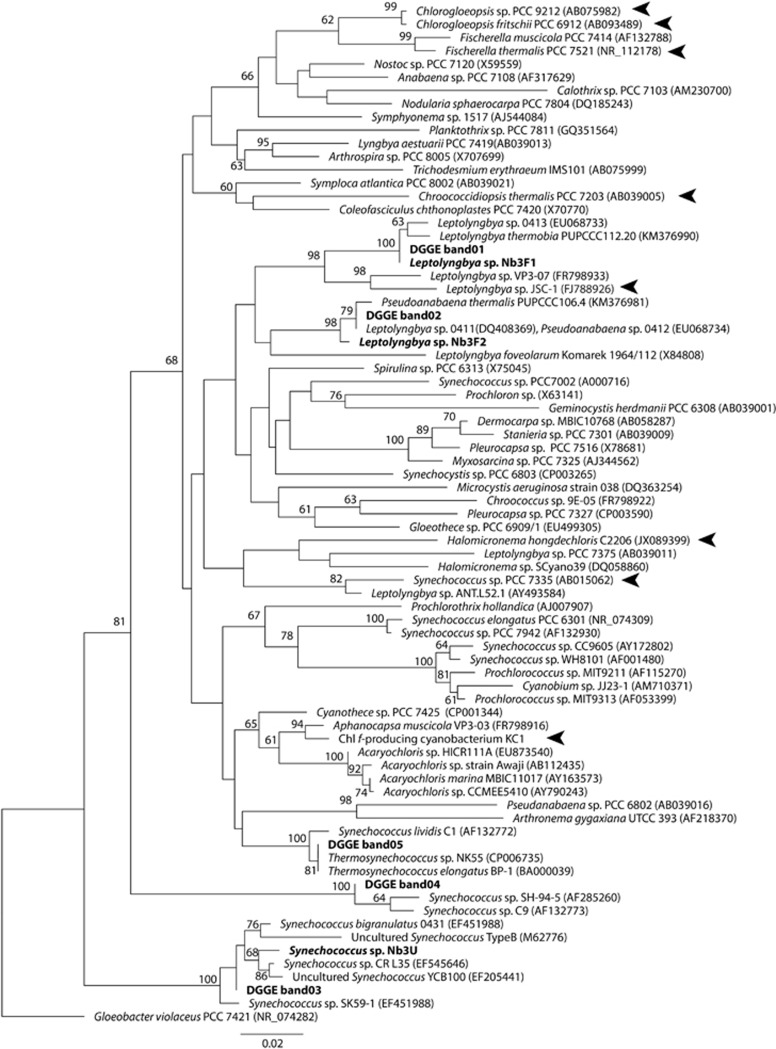
The PCR-DGGE profile showed that mat Nb3 contained four dominant cyanobacteria (Figure 4a). Two cyanobacteria (band3 and band4) were predominant at depths of 0–4.0 mm, whereas two others (band1 and band2) were detected in deeper layers of the mat. The total intensity of band1 and band2 accounted for 17% of all bands in the 4.0–4.5 mm section, then increased to 70% with increasing depth. Four cyanobacteria detected by PCR-DGGE were phylogenetically distinct from one another. Cyanobacteria from band1 and band2 were independently clustered with filamentous cyanobacteria in the phylogenetic tree (Figure 5). Band1 was closely related to Leptolyngbya sp. 0413, whereas band2 was clustered with Leptolyngbya sp. 0412 and Pseudanabaena sp. 0411. They were isolated from Russian hot springs (Sorokovikova et al., 2008). Band3 was clustered within the Synechococcus T1 clade containing strain CR_L35 and the uncultured clone YCB100, which were originated from hot springs in Costa Rica and Tibet, respectively (Lau et al., 2009; Ward et al., 2012). Band4 was also closely related to thermophilic Synechococcus spp. strains SH-94-5 and C9, which were both isolated from hot springs in North America (Ferris et al., 1996; Miller and Castenholz, 2000). These results show that at least two species of unicellular Synechococcus (band3 and band4) were predominant cyanobacteria in the surface to middle layers of mat Nb3 (0–4.0 mm), and that two species of filamentous Leptolyngbya (band1 and band2) were present in the deep layer of the same mat (> 4.0 mm).
Figure 4.
Vertical distribution of cyanobacterial phylotypes detected by PCR-DGGE within microbial mats Nb3 (a) and Nb4 (b). DNA was extracted from each 0.5 mm thick horizontal section of the microbial mat shown in Figure 2, after pigment extraction.
Figure 5.
Phylogenetic relationship of sequences obtained from PCR-DGGE bands in Figure 4 and strains newly isolated in this study. Bold text indicates the bands and strains isolated from the microbial mat, and arrowheads indicate the sequences of reported Chl f-producing cyanobacteria. Numbers at each node show the bootstrap values represented as a percentage. Nodes without any values received <60% support.
Three DGGE bands were detected from mat Nb4 (Figure 4b). Two bands were identical to band3 and band4 in mat Nb3, whereas band5 was detected from all depths. Although two bands were present at the position of band5, the DNA sequences of both bands were identical. Band5 showed an identical sequence to some unicellular cyanobacterial strains containing the thermophilic cyanobacteria, Thermosynechococcus elongatus BP-1 and Thermosynechococcus sp. NK55 (Figure 5), which were isolated from Beppu and Nakabusa hot springs in Japan, respectively (Nakamura et al., 2002; Stolyar et al., 2014). Conversely, neither of the Leptolyngbya spp. (band1 and band2) were detected from the mat. Therefore, the dominant cyanobacteria in mat Nb4 were three unicellular thermophilic cyanobacteria, which were all distributed from the surface to the bottom of the mat.
Characteristics of isolated cyanobacteria
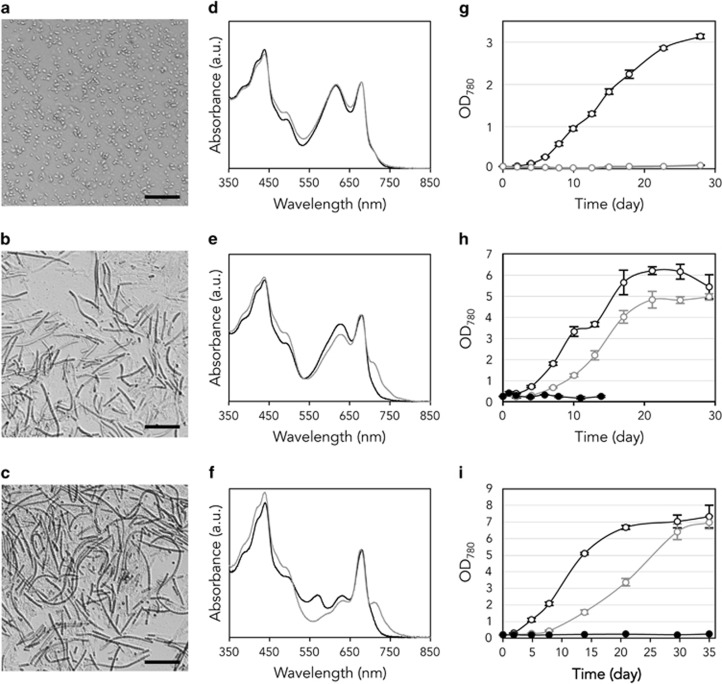
To reveal which cyanobacteria detected by PCR-DGGE produced Chl f in mat Nb3, we cultured and isolated cyanobacteria from the mat. Analysis of the cultured organisms revealed three strains, Nb3U1, Nb3F1 and Nb3F2. Strain Nb3U1 was a blue-green (under both WL and FR LEDs) ellipsoidal or rod-shaped unicellular cyanobacterium 1.5–1.9 μm in diameter and 3.0–5.7 μm in length (Figure 6a). Strain Nb3F1 was a blue-green (under WL) or yellow green (under FR) thin filamentous cyanobacterium 1.7–2.0 μm in width (Figure 6b). Strain Nb3F2 was filamentous, 1.6–1.8 μm in width and blackish green and green under WL and FR, respectively (Figure 6c). Trichomes of both filamentous strains were not constricted at the cross walls nor coiled into aggregates or biofilms in flasks. Phylogenetic analysis showed that the 16S rDNA sequence of strain Nb3U1 was closely related to that of DGGE band3 (99.2%, 381/384). The 16S rDNA sequence of strains Nb3F1 and Nb3F2 revealed identical or similar sequences to that of DGGE band1 (100%, 386/386) and band2 (97.2%, 344/354), respectively. Based on these observations, these three strains were identified at the genus level as Synechococcus sp. Nb3U1, Leptolyngbya sp. Nb3F1, and Leptolyngbya sp. Nb3F2.
Figure 6.
(a–c) Microscopic observation of Synechococcus sp. strain Nb3U1 (a), Leptolyngbya spp. strain Nb3F1 (b), and Nb3F2 (c). Scale bars indicate 30 μm. (d–f) Cell absorption spectra of strains Nb3U1 (d), Nb3F1 (e) and Nb3F2 (f) grown under WL (black lines) and FR (gray lines) LEDs. (g–i) Growth curves of strains Nb3U1 (g), Nb3F1 (h) and Nb3F2 (i) under conditions of WL (black open circles), FR (gray open circles) and dark (black filled circles).
When the three isolated strains were cultured under FR as the sole light source, Chl f accumulated in the cells of Leptolyngbya spp. strains Nb3F1 and Nb3F2, but not in Synechococcus sp. Nb3U1. The absorption spectra of strain Nb3U1 cells grown under WL and FR were almost identical (Figure 6d). However, there was a distinct shoulder at ~715–750 nm in the absorption spectra of strains Nb3F1 and Nb3F2 cells grown under the FR (Figures 6e and f). Absorptions at 450–500 nm were also enhanced, but they decreased under FR at 550–650 nm, and the peak at 568.5 nm in Nb3F2 disappeared completely. High performance liquid chromatography analyses also showed that strain Nb3F1 accumulated Chl f when the cells were grown under FR, whereas no peak in Chl f was detected from strain Nb3U1 (Supplementary Figure S4).
The growth characteristics of these cyanobacteria were compared under WL and FR. Synechococcus sp. strain Nb3U1 grew under WL, but showed almost no growth under FR (Figure 6g). The maximum specific growth rate (μmax) was 0.40±0.01 per day, and cell growth reached a plateau in ~1 month under WL. Conversely, Leptolyngbya spp. grew under both colors of light, and did not grow in the dark (Figures 6h and i). The lag phase of cell growth under FR was 3–5 days longer when compared with that of cells grown under WL. The growth rate under FR was slower than that under WL. The μmax of Leptolyngbya sp. strain Nb3F1 under WL and FR was 0.30±0.05 and 0.26±0.02 per day, respectively. Cell densities plateaued in ~20 days under both light conditions, and the OD780 in the stationary phase under FR was ~80% of that under WL. In the case of Leptolyngbya sp. strain Nb3F2, the μmax under FR (0.21±0.02 per day) was ~50% of that under WL (0.42±0.05 per day). However, the density of cells grown under FR rose to approximately the same level as that of cells grown under WL about 10 days later (Figure 6i).
Discussion
We investigated the distribution of Chl f within microbial mats in a stream flowing from the hot spring at Nakabusa, Japan. In a 7-mm-thick microbial mat (Nb3), we only detected Chl f from 4.0–6.5 mm below the surface (Figure 2), where the photon flux density of FR was higher than that of PAR (Figure 1). Chl f may be produced to use the FR that can reach to deeper layers of the mat. Our findings confirm an exception that Chl f is distributed in microbial mats as described in a previous study (Gan and Bryant, 2015).
The vertical distribution of cyanobacterial species and Chl relative to light quality at each depth showed that the habitat of Chl f-producing cyanobacteria is an environment dominated by FR. Microbial mat Nb3 contained four dominant species of cyanobacteria, two Leptolyngbya species (DGGE band1 and band2) and two Synechococcus species (band3 and band4). Of these, Synechococcus was mainly distributed in the surface and middle layers (0–4.0 mm below the surface), whereas Leptolyngbya was in the deep layers (4.0–6.5 mm), corresponding to the presence of Chl f (Figure 4). These distribution patterns indicate that Chl f in the mat originated from both Leptolyngbya species, which supports our analysis of the pigment composition in the isolated cyanobacterial strains. Two strains of Leptolyngbya spp. (Nb3F1 and Nb3F2) accumulated Chl f under FR, whereas Synechococcus sp. Nb3U1 did not (Figures 6d–f). Not all strains were identical to the cyanobacteria detected by PCR-DGGE based on their 16S rDNA sequences. However, strains Nb3F1, Nb3F2 and Nb3U1 are thought to be equivalent to band1, band2 and band3, respectively, based on the species level. Therefore, the physiological characteristics are thought to be similar between each isolated strain and cyanobacterium within the mat. These results indicate that the spatial segregation between unicellular and filamentous cyanobacteria within the mat was caused by differences in the light wavelength they could use for photosynthesis. In fact, both strains of Leptolyngbya grew under FR, whereas strain Nb3U1 could not grow under the same conditions (Figures 6g–i). Previous studies have suggested that two Synechococcus strains have the capacity to grow at deep layers in microbial mats of Mushroom Spring in Yellowstone National Park (Nowack et al., 2015; Olsen et al., 2015). These Synechococcus strains have genes involved in a special form of allophycocyanin and IsiA-like protein, which can probably absorb FR. It is unclear whether the cyanobacteria living in mat Nb3 have these genes, but strain Nb3U1 has no capacity to grow under FR-dominated environments. The growth rate of strain Nb3U1 was similar to or greater than that of filamentous strains under WL, indicating that Synechococcus cells might be able to grow much faster than Leptolyngbya spp. by using PAR. In contrast, in deep layers in which FR is dominant, Synechococcus could be out-competed by Leptolyngbya. It is possible that Synechococcus were detected from the deep layer by PCR-DGGE because their small unicellular cells could be easily carried by water flow within the mat. We concluded that Chl f caused niche differentiation in two types of cyanobacteria within microbial mat Nb3, and that the deep layer was a niche for Chl f-producing cyanobacteria.
Even though the distance between the two mats analyzed was only about 10 cm, Chl f was not detected from mat Nb4 (Figure 2). DGGE bands of Leptolyngbya (band3 and band4) were also absent from mat Nb4 (Figure 4). These results correspond to the above suggestion that Chl f in mat Nb3 originated from these Leptolyngbya species. However, it is still unknown whether Thermosynechococcus sp. in mat Nb4 (band5) has the capacity to synthesize Chl f; therefore, this should be investigated in future studies. The in situ light environments in mat Nb4 might also cause a lack of Chl f. Although FR was enriched in both mats, the light intensity (400–900 nm) in mat Nb4 was always higher than that at a depth of 4.0 mm in Nb3 (Figure 1). If the intensity of PAR in Nb4 is too high for a FaRLiP response, Chl f should not be synthesized, even if Chl f-producing cyanobacteria are present. Therefore, the microbial mats in hot springs do not always contain Chl f, and both the presence of Chl f-producing organisms and environmental factors in the mats would be important.
Previous studies have confirmed that anoxygenic photosynthetic bacteria are major components in the microbial mats at Nakabusa hot spring (Nakagawa and Fukui, 2002; Kubo et al., 2011; Everroad et al., 2012). In this study, we observed attenuation peaks from bacteriochlorophylls (BChls a and c) in downward irradiance spectra within mats (Figure 1). A bacteria-targeted PCR-DGGE profile also indicated the presence of anoxygenic photosynthetic bacteria (Supplementary Figure S5). Supplementary Table S1 shows that the sequence of bandS1 was closely related to the 16S rRNA gene sequences of Chloracidobacterium thermophilum strains A, E and S (96.6%, 515/533), which were recently discovered in microbial mats at hot springs in Yellowstone National Park (Bryant et al., 2007). It was also related to C. thermophilum strain B (96.1%, 512/533), the type strain of this species (Tank and Bryant, 2015a). BandS2 showed an identical sequence (534/534) to Roseiflexus castenholzii DSM 13941 (Supplementary Table S2), which was isolated from a microbial mat at Nakabusa (Hanada et al., 2002). C. thermophilum is a unicellular aerobic anoxygenic phototrophic bacterium containing BChl a and c, and a peak at 745 nm was observed in the cell absorption spectrum (Tank and Bryant, 2015b). In contrast, R. castenholzii is a filamentous anoxygenic facultative phototrophic bacterium containing BChl a, but it lacks BChl c and chromosomes, whereas it produces in vivo absorption peaks at 801 and 878 nm (Hanada et al., 2002). This bacterium is photoheterotrophic under anaerobic light conditions. Judging from the DGGE band densities and the downward irradiance spectra, Chloracidobacterium sp. was distributed in the upper layers (0–4.0 mm) and R. castenholzii was dominant in deeper layers (3.0–6.5 mm) of the mat corresponding to their oxygen requirements. Accordingly, there were four types of photosynthetic bacteria within mat Nb3: Synechococcus (Chl a), Leptolyngbya (Chl a+f), Chloracidobacterium (BChl c+a) and Roseiflexus (BChl a), which could coexist within same mat by avoiding competition for light.
We thought that cyanobacteria acquired the capacity to synthesize Chl f as a result of evolutionary adaptation to environments within microbial mats. Both strains of Leptolyngbya isolated in this study could grow photoautotrophically under FR, which is the dominant light in the deeper layers of microbial mats (Figure 6). This characteristic would have allowed Chl f-producing cyanobacteria to expand their distribution range vertically within microbial mats. The acquisition of Chl f by cyanobacteria would have occurred just once during the early stage of their evolutionary process, as at least eight species of Chl f-producing cyanobacteria were widespread in the phylogenetic tree of the Cyanobacteria phylum (Figure 5). Moreover, the deep layer of microbial mats might have been a safe habitat for cyanobacteria in the early Earth, where UV radiation was probably much greater than it is today (Garcia-Pichel, 1998; Nisbet and Sleep, 2001).
This study provides a new example of niche differentiation along the light spectrum. Within a microbial mat, Chl f was distributed in the deeper layer, where FR was dominant. Two species of Chl f-producing filamentous cyanobacteria containing Chl a and f were also distributed there and able to grow under FR. Two other species of unicellular cyanobacteria containing only Chl a were distributed in the upper layer reached by PAR, whereas two species of anoxygenic phototrophic bacteria containing only BChl a or BChl c+a also existed within the same mat. The results suggest that Chl f contributes to cyanobacterial survival in habitats with little PAR, as well as to coexistence with other photosynthetic organisms in microbial mats. However, our results only showed a temporary state of the microbial mat, as well as the physiological characteristics of cyanobacteria isolated and cultured in the laboratory. Further studies are needed to reveal how Chl f-producing cyanobacteria increase and decrease in the life cycle of microbial mats and the expression patterns of genes of FaRLiP and Chl f-synthesis (Gan et al., 2014, 2015; Ho et al., 2016).
Acknowledgments
This research was supported by MEXT KAKENHI Grant Number 21247005. We thank Katsumi Matsuura, Hiroyo Otaki and Satomi Iwata of Tokyo Metropolitan University for their help in sampling microbial mats at the Nakabusa hot spring. We also thank Yukako Hihara of Saitama University for lending a spectrophotometer to measure the absorption spectra of cyanobacteria in the growth experiments.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Ahmed I, Yokota A, Fujiwara T. (2007). A novel highly boron tolerant bacterium, Bacillus boroniphilus sp. nov., isolated from soil, that requires boron for its growth. Extremophiles 11: 217–224. [DOI] [PubMed] [Google Scholar]
- Airs RL, Temperton B, Sambles C, Farnham G, Skill SC, Llewellyn C. (2014). Chlorophyll f and chlorophyll d are produced in the cyanobacterium Chlorogloeopsis fritschii when cultured under natural light and near-infrared radiation. FEBS Lett 588: 3770–3777. [DOI] [PubMed] [Google Scholar]
- Akimoto S, Shinoda T, Chen M, Allakhverdiev SI, Tomo T. (2015). Energy transfer in the chlorophyll f-containing cyanobacterium, Halomicronema hongdechloris, analyzed by time-resolved fluorescence spectroscopies. Photosynth Res 125: 115–122. [DOI] [PubMed] [Google Scholar]
- Akutsu S, Fujinuma D, Furukawa H, Watanabe T, Ohnishi-Kameyama M, Ono H et al. (2011). Pigment analysis of a chlorophyll f-containing cyanobacterium strain KC1 isolated from Lake Biwa. Photomed Photobiol 33: 35–40. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990). Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Behrendt L, Brejnrod A, Schliep M, Sørensen SJ, Larkum AW, Kühl M. (2015). Chlorophyll f-driven photosynthesis in a cavernous cyanobacterium. ISME J 9: 2108–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béjà O, Spudich EN, Spudich JL, Leclerc M, DeLong EF. (2001). Proteorhodopsin phototrophy in the ocean. Nature 411: 786–789. [DOI] [PubMed] [Google Scholar]
- Boomer SM, Pierson BK, Austinhirst R, Castenholz RW. (2000). Characterezation of novel bacteriochlorophyll-a-containing red filaments from alkaline hot springs in Yellowstone National Park. Arch Microbiol 174: 152–161. [DOI] [PubMed] [Google Scholar]
- Bryant DA, Costas AMG, Maresca JA, Chew AGM, Klatt CG, Bateson MM et al. (2007). Candidatus Chloracidobacterium thermophilum: an aerobic phototrophic Acidobacterium. Science 317: 523–526. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Bittinger K, Bushman FD, Desantis TZ, Andersen GL, Knight R. (2010). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26: 266–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Li Y, Birch D, Willows RD. (2012). A cyanobacterium that contains chlorophyll f - A red-absorbing photopigment. FEBS Lett 586: 3249–3254. [DOI] [PubMed] [Google Scholar]
- Chen M, Schliep M, Willows RD, Cai Z-L, Neilan BA, Scheer H. (2010). A red-shifted chlorophyll. Science 329: 1318–1319. [DOI] [PubMed] [Google Scholar]
- Everroad RC, Otaki H, Matsuura K, Haruta S. (2012). Diversification of bacterial community composition along a temperature gradient at a thermal spring. Microbes Environ 27: 374–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris MJ, Kühl M, Wieland A, Ward DM. (2003). Cyanobacterial ecotypes in different optical microenvironments of a 68 °C hot spring mat community revealed by 16S-23S rRNA internal transcribed spacer region variation. Appl Environ Microbiol 69: 2893–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris MJ, Ruff-Roberts AL, Kopczynski ED, Bateson MM, Ward DM. (1996). Enrichment culture and microscopy conceal diverse thermophilic Synechococcus populations in a single hot spring microbial mat habitat. Appl Environ Microbiol 62: 1045–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan F, Bryant DA. (2015). Adaptive and acclimative responses of cyanobacteria to far-red light. Environ Microbiol 17: 3450–3465. [DOI] [PubMed] [Google Scholar]
- Gan F, Shen G, Bryant DA. (2015). Occurrence of far-red light photoacclimation (FaRLiP) in diverse cyanobacteria. Life 5: 4–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan F, Zhang S, Rockwell NC, Martin SS, Lagarias C, Bryant DA. (2014). Extensive remodeling of a cyanobacterial photosynthetic apparatus in far-red light. Science 345: 1312–1317. [DOI] [PubMed] [Google Scholar]
- Garcia-Pichel F. (1998). Solar ultraviolet and the evolutionary history of cyanobacteria. Orig Life Evol Biosph 28: 321–347. [DOI] [PubMed] [Google Scholar]
- Hanada S, Takaichi S, Matsuura K, Nakamura K. (2002). Roseiflexus castenholzii gen. nov., sp. nov., a thermophilic, filamentous, photosynthetic bacterium that lacks chlorosomes. Int J Syst Evol Microbiol 52: 187–193. [DOI] [PubMed] [Google Scholar]
- Ho M-Y, Shen G, Canniffe DP, Zhao C, Bryant DA, Flombaum P et al. (2016). Light-dependent chlorophyll f synthase is a highly divergent paralog of PsbA of photosystem II. Science 353: 213–227. [DOI] [PubMed] [Google Scholar]
- Kubo K, Knittel K, Amann R, Fukui M, Matsuura K. (2011). Sulfur-metabolizing bacterial populations in microbial mats of the Nakabusa hot spring, Japan. Syst Appl Microbiol 34: 293–302. [DOI] [PubMed] [Google Scholar]
- Kühl M, Chen M, Ralph PJ, Schreiber U, Larkum AWD. (2005). A niche for cyanobacteria containing chlorophyll d. Nature 433: 820. [DOI] [PubMed] [Google Scholar]
- Kühl M, Fenchel T. (2000). Bio-optical characteristics and the vertical distribution of photosynthetic pigments and photosynthesis in an artificial cyanobacterial mat. Microb Ecol 40: 94–103. [DOI] [PubMed] [Google Scholar]
- Lau MCY, Aitchison JC, Pointing SB. (2009). Bacterial community composition in thermophilic microbial mats from five hot springs in central Tibet. Extremophiles 13: 139–149. [DOI] [PubMed] [Google Scholar]
- Li Y, Cai ZL, Chen M. (2013). Spectroscopic properties of chlorophyll f. J Phys Chem B 117: 11309–11317. [DOI] [PubMed] [Google Scholar]
- Li Y, Scales N, Blankenship RE, Willows RD, Chen M. (2012). Extinction coefficient for red-shifted chlorophylls: chlorophyll d and chlorophyll f. Biochim Biophys Acta - Bioenerg 1817: 1292–1298. [DOI] [PubMed] [Google Scholar]
- Miller SR, Castenholz RW. (2000). Evolution of thermotolerance in hot spring cyanobacteria of the genus Synechococcus. Appl Environ Microbiol 66: 4222–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita H, Ohkubo S, Komatsu H, Sorimachi Y, Fukayama D, Fujinuma D et al. (2014). Discovery of chlorophyll d in Acaryochloris marina and chlorophyll f in a unicellular cyanobacterium, strain KC1, isolated from Lake Biwa. J Phys Chem Biophys 4: 149. [Google Scholar]
- Nakagawa T, Fukui M. (2002). Phylogenetic characterization of microbial mats and streamers from a Japanese alkaline hot spring with a thermal gradient. J Gen Appl Microbiol 48: 211–222. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Kaneko T, Sato S, Ikeuchi M, Katoh H, Sasamoto S et al. (2002). Complete genome structure of the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1. DNA Res 9: 123–130. [DOI] [PubMed] [Google Scholar]
- Namsaraev ZB. (2009). Application of extinction coefficients for quantification of chlorophylls and bacteriochlorophylls. Microbiology 78: 794–797. [Google Scholar]
- New Energy and Industrial Technology Development Organization (NEDO) Original 'Solar radiation spectrum database' edited with permission.
- Nisbet EG, Sleep NH. (2001). The habitat and nature of early life. Nature 409: 1083–1091. [DOI] [PubMed] [Google Scholar]
- Nowack S, Olsen MT, Schaible GA, Becraft ED, Shen G, Klapper I et al. (2015). The molecular dimension of microbial species: 2. Synechococcus strains representative of putative ecotypes inhabiting different depths in the Mushroom Spring microbial mat exhibit different adaptive and acclimative responses to light. Front Microbiol 6: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nübel U, Garcia-Pichel F, Muyzer G. (1997). PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl Environ Microbiol 63: 3327–3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkubo S, Miyashita H. (2012). Selective detection and phylogenetic diversity of Acaryochloris spp. that exist in association with didemnid ascidians and sponge. Microbes Environ 27: 217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkubo S, Miyashita H, Murakami A, Takeyama H, Tsuchiya T, Mimuro M. (2006). Molecular detection of epiphytic Acaryochloris spp. on marine macroalgae. Appl Environ Microbiol 72: 7912–7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen MT, Nowack S, Wood JM, Becraft ED, LaButti K, Lipzen A et al. (2015). The molecular dimension of microbial species: 3. Comparative genomics of Synechococcus strains with different light responses and in situ diel transcription patterns of associated putative ecotypes in the Mushroom Spring microbial mat. Front Microbiol 6: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson BK, Sands VM, Frederick JL. (1990). Spectral irradiance and distribution of pigments in a highly layered marine microbial mat. Appl Environ Microbiol 56: 2327–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabehi G, Kirkup BC, Rozenberg M, Stambler N, Polz MF, Béjà O. (2007). Adaptation and spectral tuning in divergent marine proteorhodopsins from the eastern Mediterranean and the Sargasso Seas. ISME J 1: 48–55. [DOI] [PubMed] [Google Scholar]
- Sorokovikova EG, Tikhonova IV, Belykh OI, Klimenkov IV, Likhoshwai EV. (2008). Identification of two cyanobacterial strains isolated from the Kotel’nikovskii hot spring of the Baikal rift. Microbiology 77: 365–372. [PubMed] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolyar S, Liu Z, Thiel V, Tomsho LP, Pinel N, Nelson WC et al. (2014). Genome sequence of the thermophilic cyanobacterium Thermosynechococcus sp. strain NK55a. Genome Announc 2: 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stomp M, Huisman J, Vörös L, Pick FR, Laamanen M, Haverkamp T et al. (2007). Colourful coexistence of red and green picocyanobacteria in lakes and seas. Ecol Lett 10: 290–298. [DOI] [PubMed] [Google Scholar]
- Tank M, Bryant DA. (2015. a). Chloracidobacterium thermophilum gen. nov., sp. nov.: an anoxygenic microaerophilic chlorophotoheterotrophic acidobacterium. Int J Syst Evol Microbiol 65: 1426–1430. [DOI] [PubMed] [Google Scholar]
- Tank M, Bryant DA. (2015. b). Nutrient requirements and growth physiology of the photoheterotrophic acidobacterium, Chloracidobacterium thermophilum. Front Microbiol 6: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomo T, Shinoda T, Chen M, Allakhverdiev SI, Akimoto S. (2014). Energy transfer processes in chlorophyll f-containing cyanobacteria using time-resolved fluorescence spectroscopy on intact cells. Biochim Biophys Acta 1837: 1484–1489. [DOI] [PubMed] [Google Scholar]
- Trampe E, Kühl M. (2016). Chlorophyll f distribution and dynamics in cyanobacterial beachrock biofilms. J Phycol 996: 990–996. [DOI] [PubMed] [Google Scholar]
- Ward DM, Castenholz RW, Miller SR. (2012) Cyanobacteria in Geothermal Habitats. In: Whitton BA (ed.). Ecology of Cyanobacteria II: Their Diversity in Space and Time. Springer Science, pp 39–63. [Google Scholar]
- Willows RD, Li Y, Scheer H, Chen M. (2013). Structure of chlorophyll f. Org Lett 15: 1588–1590. [DOI] [PubMed] [Google Scholar]
- Wood AM, Phinney DA, Yentsch CS. (1998). Water column transparency and the distribution of spectrally distinct forms of phycoerythrin-containing organisms. Mar Ecol Prog Ser 162: 25–31. [Google Scholar]
- Zhao C, Gan F, Shen G, Bryant DA. (2015). RfpA, RfpB, and RfpC are the master control elements of far-red light photoacclimation (FaRLiP). Front Microbiol 6: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.