ABSTRACT
Hypervirulent Klebsiella pneumoniae (hvKP) is an emerging pathotype that is capable of causing tissue-invasive and organ- and life-threatening infections in healthy individuals from the community. Knowledge on the virulence factors specific to hvKP is limited. In this report, we describe a new factor (PEG344) that increases the virulence of hvKP strain hvKP1. peg-344 is present on the hvKP1 virulence plasmid, is broadly prevalent among hvKP strains, and has increased RNA abundance when grown in human ascites. An isogenic derivative of hvKP1 (hvKP1Δpeg-344) was constructed and compared with its wild-type parent strain in in vitro, ex vivo, and infection model studies. Both survival and competition experiments with outbred CD1 mice demonstrated that PEG344 was required for full virulence after pulmonary challenge but, interestingly, not after subcutaneous challenge. In silico analysis suggested that PEG344 serves as an inner membrane transporter. Compared to hvKP1, a small but significant decrease in the growth/survival of hvKP1Δpeg-344 was observed in human ascites, but resistance to the bactericidal activity of complement was similar. These data suggested that PEG344 may transport an unidentified growth factor present in ascites. The data presented are important since they expand our limited knowledge base on virulence factors unique to hvKP, which is needed to lay the groundwork for translational approaches to prevent or treat these devastating infections.
KEYWORDS: Klebsiella pneumoniae, hypervirulent, infection model, pathogenesis, reference gene, superbug, virulence factors
INTRODUCTION
In the Western world, the majority of infections caused by Klebsiella pneumoniae are due to the “classical” strains of K. pneumoniae (cKP) (1). Although historically, cKP has been described as causing community-acquired pneumonia, primarily in alcoholics, with the advent of antibiotics and their liberal usage, the epidemiology of Klebsiella pneumoniae infection has shifted dramatically. Most infections now occur in hospitals and long-term-care facilities (2). With the less-than-judicious use of antibiotics, antimicrobial resistance has also been on the rise. cKP strains have received increased notoriety due to their propensity to obtain various antimicrobial resistance mechanisms, primarily carbapenemases. As such, these infections are becoming much more difficult to treat (3–5). Recently, cKP was responsible for the first death in the United States due to a panresistant strain (6).
During the mid-1980s, reports from Taiwan described a unique clinical syndrome of community-acquired K. pneumoniae infection. These patients presented with community-acquired pyogenic liver abscesses, had no history of hepatobiliary disease, and demonstrated a propensity for metastatic spread (7). A number of clinical and phenotypic characteristics set this pathotype apart from cKP. The first characteristic was its ability to cause serious, potentially life-threatening, infections in otherwise healthy hosts from the community. Second, these strains were responsible for a number of unusual primary sites of infection for K. pneumoniae, such as meningitis, necrotizing fasciitis, endophthalmitis, and osteomyelitis (8–12). The final characteristic was the predilection for metastatic spread from the site of primary infection to virtually any organ or site (13–16). The ability to spread from the initial site of infection is more commonly seen with certain Gram-positive pathogens (e.g., Staphylococcus aureus) but is relatively uncommon for enteric Gram-negative bacilli (e.g., extraintestinal pathogenic Escherichia coli, Proteus, and cKP) in the absence of an immunocompromised host (1). This pathotype has been termed hypervirulent K. pneumoniae (hvKP) to both differentiate it from cKP and increase awareness of its unique pathogenic potential (PP) (1). While initially recognized in the Asian Pacific Rim (e.g., Taiwan, South Korea, Vietnam, and Japan), increasing numbers of hvKP infections have been reported throughout the world. More concerning is that initial isolates of hvKP were resistant to ampicillin only, but recently, increasing antimicrobial resistance has been described for these strains (17–20).
In the majority of strains, the increased virulence of hvKP1 compared to that of cKP has been associated with the presence of a virulence plasmid (140 to 220 kb), although occasionally, the plasmid appears to have integrated into the chromosome (21, 22). The plasmid contains genes that result in increased capsule synthesis (mediated by rmpA-rmpA2) and increased siderophore production (mediated by the aerobactin biosynthetic genes iucABCD). rmpA (23) and the production of the siderophore aerobactin (24, 25) have been established as important virulence factors for hvKP. Curing of this plasmid results in the loss of the hypermucoviscous phenotype conferred by increased capsule production, the ability to synthesize aerobactin, and a decrease in virulence (26, 27). It is likely that this plasmid also carries other virulence-associated genes, which, to date, have yet to be identified.
Through transcriptome sequencing (RNA-Seq) experiments, we identified a gene located on the virulence plasmid present in hvKP strain hvKP1 (annotated peg-344 [28]) with a significantly increased RNA abundance when the strain is grown in human ascites compared to defined minimal and rich laboratory media. Homology analysis suggested that the putative gene product of peg-344 is a permease of the drug/metabolite (DMT) transporter superfamily. Based on these data, we hypothesized that peg-344 encoded a novel, unrecognized virulence factor. The primary goal of this study was to determine whether PEG344 was an unrecognized virulence factor for hvKP. This was accomplished by assessing the clinical isolate hvKP1 and its isogenic derivative hvKP1Δpeg-344 (Δpeg-344) for growth and survival in vitro, ex vivo in various clinically relevant media, and in vivo in mouse infection models. The data presented extend our understanding of the pathogenesis of hvKP infection.
RESULTS
peg-344 RNA abundance is increased when hvKP1 is grown in log phase in human ascites but not rich laboratory or minimal media.
To confirm the RNA-Seq data, the RNA abundance of peg-344 in cultures grown in lysogeny broth (LB), chelated M9 minimal medium supplemented with Casamino Acids (c-M9-CA), and human ascites was assessed via quantitative PCR (qPCR) with two primer pairs. The RNA abundance of peg-344 when cultures were grown in LB was designated 1.0. Compared to LB, when cultures were grown in ascites, the RNA abundances of peg-344 for assay 1 were increased 18.18-fold ± 0.35-fold (standard error of the mean [SEM]) (mean quantification cycle [Cq] value ± SEM, 20.58 ± 0.00035) and 14.89-fold ± 0.07-fold (mean Cq ± SEM, 20.75 ± 0.00686), and those for assay 2 were increased 17.75-fold ± 0.36-fold (mean Cq ± SEM, 20.11 ± 0.0099) and 13.59-fold ± 0.22-fold (mean Cq ± SEM, 20.34 ± 0.02341) in two independent qPCR experiments.
To compare the RNA abundance of peg-344 during growth in ascites to that during growth in c-M9-CA, the RNA abundance of peg-344 in cultures grown in c-M9-CA was designated 1.0. Compared to c-M9-CA, when cultures were grown in ascites, the RNA abundances of peg-344 for assay 1 were increased 444.74-fold ± 8.44-fold (mean Cq ± SEM, 20.58 ± 0.00035) and 500.87-fold ± 2.51-fold (mean Cq ± SEM, 20.75 ± 0.00686), and those for assay 2 were increased 1,012.19-fold ± 20.38-fold (mean Cq ± SEM, 20.11 ± 0.0099) and 792.72-fold ± 12.67-fold (mean Cq ± SEM, 20.34 ± 0.02341) in two independent qPCR experiments.
Taken together, the increased RNA abundance of peg-344 in a clinically relevant human body fluid compared to rich laboratory and minimal media suggests that its gene product may contribute to the pathogenesis of hvKP strain hvKP1.
Disruption of peg-344 does not affect growth/survival of hvKP1 in LB medium, siderophore production, or capsule production.
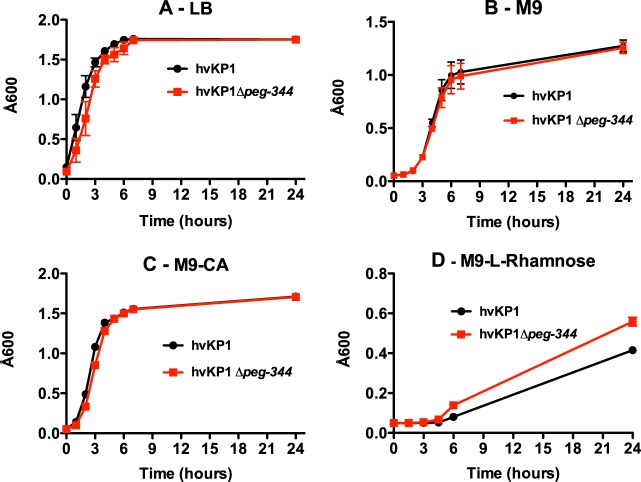
First, a growth assessment of hvKP1 and the Δpeg-344 strain was performed with LB, a rich laboratory medium, to exclude a generalized growth defect, which would affect the interpretation of subsequent experimental results. As shown in Fig. 1A, the levels of growth of hvKP1 and the Δpeg-344 strain were similar, thereby excluding a generalized growth defect. Next, since capsule and increased siderophore production are defined virulence traits of hvKP, we assessed whether these phenotypes were affected by the disruption of peg-344, albeit this was unlikely given its putative function. The Δpeg-344 strain remained string test positive, a surrogate for increased capsule production (1). A more rigorous evaluation of capsule production was performed by purifying capsular polysaccharide from hvKP1 and the Δpeg-344 strain, resolving these polysaccharides by SDS–8% polyacrylamide gel electrophoresis, and staining with alcian blue; the amounts of capsule were similar for these strains (see Fig. S1 in the supplemental material). A quantitative assay for total siderophore production was performed as described previously (25). When grown in c-M9-CA (iron chelated), hvKP1 produced 380 μg/ml of siderophores, and the Δpeg-344 strain produced 420 μg/ml. Therefore, peg-344 did not affect these defined virulence phenotypes for hvKP.
FIG 1.
Comparison of growth of hvKP1 and growth of hvKP1Δpeg-344 in LB, M9 medium, and M9-CA broth. Growth was measured by the A600 on 5 to 7 replicate cultures per strain. (A) LB (n = 4); (B) M9 medium (n = 7); (C) M9-CA (n = 5); (D) M9–l-rhamnose medium (n = 5). Data are plotted as averages, and the bars correspond to the SEM.
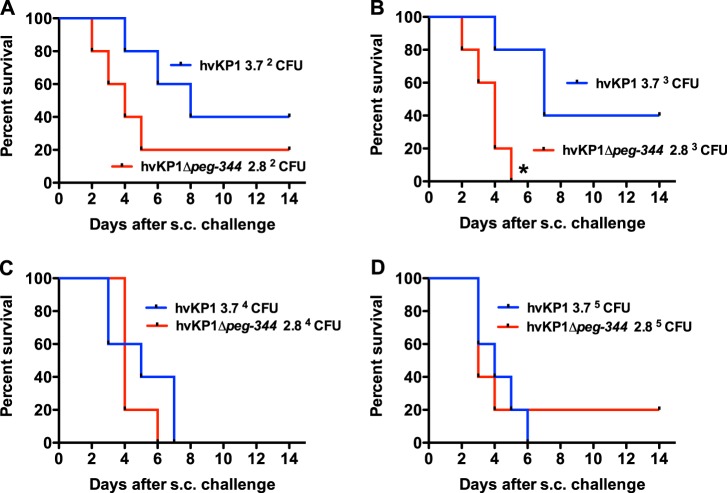
PEG344 is not required for full virulence of hvKP1 in an outbred CD1 mouse subcutaneous challenge model.
To assess the relative role of putative hvKP virulence factors, our group previously developed a subcutaneous (s.c.) challenge systemic infection model that recapitulates the dissemination that occurs with hvKP infection (29). The impact of PEG344 on the virulence of hvKP1 in vivo was assessed in this model by using survival as the endpoint. Outbred CD1 mice were injected s.c. with the Δpeg-344 strain (challenge inocula of 2.8 × 102, 2.8 × 103, 2.8 × 104, and 2.8 × 105 CFU) and compared to mice challenged with its wild-type (wt) parent strain, hvKP1 (challenge inocula of 3.7 × 102, 3.7 × 103, 3.7 × 104, and 3.7 × 105 CFU). The mortality of mice challenged with the Δpeg-344 strain was similar to that of hvKP1-challenged mice, except for animals challenged with 2.8 × 103 CFU of the Δpeg-344 strain compared to animals challenged with 3.7 × 103 CFU of hvKP1 (Fig. 2). For this inoculum, morality was increased in Δpeg-344 strain-challenged mice. Taken together, these data do not support a role for PEG344 in contributing to the virulence of hvKP1 in this infection model.
FIG 2.
Survival of outbred CD1 mice after subcutaneous challenge with hvKP1 (wild-type) and hvKP1Δpeg-344. Strains were grown overnight in LB medium and diluted in 1× PBS, and animals were challenged s.c. with the titers listed in each panel. An in extremis state or death was scored as nonsurvival for animals monitored for 14 days (n = 5 for each titer of each strain). The growth/survival of animals challenged with hvKP1Δpeg-344 was significantly decreased compared to that of animals challenged with hvKP1 at the titers listed in panel B (*, P < 0.05, as determined by a log rank [Mantel-Cox] test).
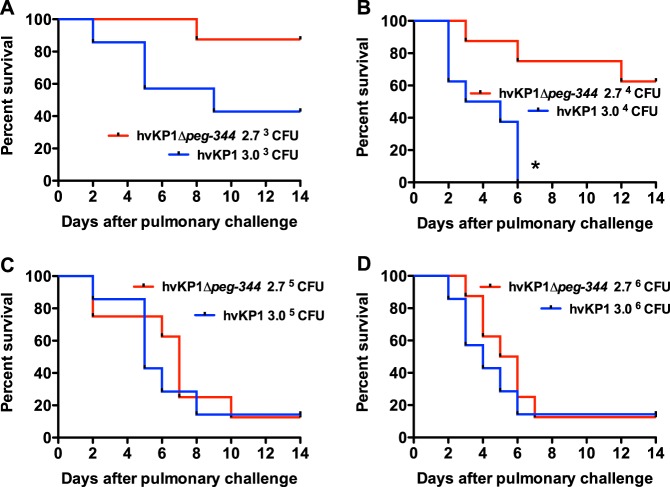
PEG344 is required for full virulence of hvKP1 in an outbred CD1 mouse pulmonary challenge model.
The lungs are a common site for both cKP and hvKP infections. Therefore, the impact of PEG344 on the virulence of hvKP1 in vivo was assessed in a mouse pulmonary infection model previously used by our group, using survival as the endpoint (24). Outbred CD1 mice underwent pulmonary challenge with the Δpeg-344 strain (challenge inocula of 2.8 × 103, 2.8 × 104, 2.8 × 105, and 2.8 × 106 CFU) and the wild-type parent strain, hvKP1 (challenge inocula of 3.0 × 103, 3.0 × 104, 3.0 × 105, and 3.0 × 106 CFU). The morality rate of mice challenged with the Δpeg-344 strain was lower and the timing of the mortality observed occurred later for all challenge inocula than with its wild-type parent strain (Fig. 3). These differences achieved statistical significance (P < 0.05) for animals challenged with 2.8 × 104 CFU of the Δpeg-344 strain compared to 3.0 × 104 CFU of hvKP1. These data were further analyzed by using the recently proposed concept of quantitating the PP of a microbe (30). Calculations of the PPs of hvKP1 and the Δpeg-344 and their PPs taking into account the element of time (PPT) were performed (Table 1). These data demonstrate that the PP of hvKP1 is greater than that of the Δpeg-344 strain with the 2 lower challenge inocula. When the element of time is incorporated into the analysis, the PPT of hvKP1 was increased relative to that of the Δpeg-344 strain for all challenge inocula. Taken together, these data support that PEG344 is required for the full virulence of hvKP1 after pulmonary challenge. Furthermore, the effect of PEG344 was site specific, contributing to the virulence of hvKP1 after pulmonary but not s.c. challenge.
FIG 3.
Survival of outbred CD1 mice after pulmonary challenge with hvKP1 (wild type) and hvKP1Δpeg-344. Strains were grown overnight in LB medium and diluted in 1× PBS, and animals underwent pulmonary challenge with the titers listed in each panel. An in extremis state or death was scored as nonsurvival for animals monitored for 14 days (n = 7 to 8 for each titer of each strain). The growth/survival of animals challenged with hvKP1Δpeg-344 was significantly decreased compared to that of animals challenged with hvKP1 at the titers listed in panel B (*, P < 0.05, as determined by a log rank [Mantel-Cox] test).
TABLE 1.
Analysis of the pathogenic potential (PP) of hvKP1 and hvKP1Δpeg-344 after pulmonary challenge with or without the element of time
| Strain | Inoculum (CFU) | Ma | PPb | PP1/PP2c | PPTd | PPT1/PPT2e |
|---|---|---|---|---|---|---|
| hvKP1 (1) | 3.0 × 103 | 0.570 | 12.4 | 2.34 | ||
| hvKP1Δpeg-344 (2) | 2.8 × 103 | 0.125 | 4.75 | 2.6 | 0.59 | 3.97 |
| hvKP1 | 3.0 × 104 | 1.000 | 3.33 | 0.833 | ||
| hvKP1Δpeg-344 | 2.8 × 104 | 0.375 | 0.85 | 3.9 | 0.122 | 6.83 |
| hvKP1 | 3.0 × 105 | 0.86 | 0.24 | 0.046 | ||
| hvKP1Δpeg-344 | 2.8 × 105 | 0.875 | 0.27 | 0.88 | 0.046 | 1.0 |
| hvKP1 | 3.0 × 106 | 0.860 | 0.024 | 0.008 | ||
| hvKP1Δpeg-344 | 2.8 × 106 | 0.875 | 0.027 | 0.88 | 0.005 | 1.6 |
M, mortality fraction.
PP was calculated as 1/inoculum × 10M × 10,000.
PP1/PP2, PP of hvKP1/PP of hvKP1Δpeg-344.
PPT, pathogenic potential incorporating the element of time (PP × 1/T, where T is the mean number of days to death).
PPT1/PPT2, PPT of hvKP1/PPT of hvKP1Δpeg-344.
hvKP1 outcompetes the Δpeg-344 strain after pulmonary but not s.c. challenge.
To further assess the role for PEG344 in contributing to the virulence of hvKP1 in vivo, competition experiments were performed with hvKP1 and the Δpeg-344 strain. Mice underwent either s.c. or pulmonary challenge with approximately equal CFU of each strain (1.2 × 104 CFU and 1.7 × 104 CFU for hvKP1 and the Δpeg-344 strain, respectively), and titers in blood and selected organs were determined at 24 and 48 h postchallenge. After s.c. challenge, titers were low at 24 h, making the interpretation of results difficult, but at 48 h, the median titers of hvKP1 and the Δpeg-344 strain were similar (Fig. 4B). In contrast, after pulmonary challenge, the median titers of hvKP1 were higher than those of the Δpeg-344 strain at every site at 24 and 48 h (Fig. 4A). Although these differences fell short of statistical significance (P = 0.06), the biological difference was unequivocal, particularly for dissemination outside the lungs at 24 h. Taken together, these data were consistent with survival data that demonstrate that PEG344 is needed for the full virulence of hvKP1 with pulmonary but not s.c. challenge.
FIG 4.
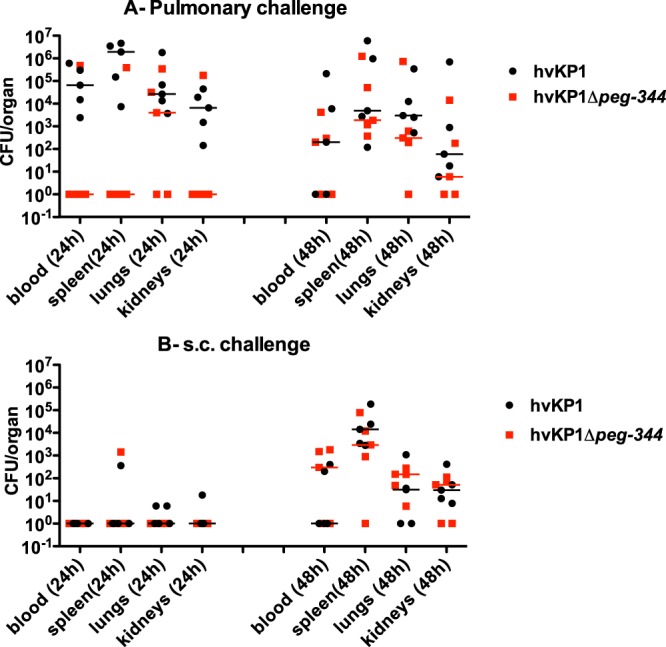
In vivo competition of hvKP1 (wt) and hvKP1Δpeg-344 in CD1 mice after subcutaneous or pulmonary challenge. Strains were grown overnight in LB medium and diluted in 1× PBS, and equal volumes of hvKP1 and hvKP1Δpeg-344 were mixed to enable codelivery via pulmonary or s.c. challenge. At 24 and 48 h, animals were euthanized, and bacterial titers of each strain in blood, lungs, kidney, and spleen were determined as described in Materials and Methods. (A) Mice underwent pulmonary challenge with 1.2 × 104 CFU of hvKP1 and 1.7 × 104 CFU of hvKP1Δpeg-344. The line indicates the median for each strain (P = 0.06, as determined by a Wilcoxon signed-rank test, for hvKP1 compared to hvKP1Δpeg-344 at 24 h for blood, lungs and spleen). (B) Mice underwent s.c. challenge with 1.2 × 104 CFU of hvKP1 and 1.7 × 104 CFU of hvKP1Δpeg-344. The line indicates the median for each strain.
The mechanism by which PEG344 contributes to the virulence of hvKP1 after intrapulmonary challenge does not appear to be due to the transport of amino acids, l-rhamnose, or rhamnolipids.
The selective role for PEG344 in contributing to the virulence of hvKP1 after pulmonary challenge was surprising. The putative function of PEG344 is as a transporter of metabolites; therefore, it was hypothesized that it could contribute to selective nutrient transport within the pulmonary compartment that could enhance the local growth and/or enhance the extrapulmonary dissemination of hvKP1. The RNA abundance of peg-344 was not increased when cells were grown in c-M9-CA compared to ascites, suggesting that it could contribute to amino acid transport. Likewise, orthologs of peg-344 have been implicated in the transport of l-rhamnose (31), suggesting that this could be a possible carbon source in vivo. To test this hypothesis, the levels of growth of the hvKP1 and Δpeg-344 strains in M9, M9-CA, and M9–l-rhamnose media were compared. However, the levels of growth of the hvKP1 and Δpeg-344 strains were similar in M9 and M9-CA media (Fig. 1B and C), and the level of growth of the Δpeg-344 strain was higher than that of hvKP1 in M9–l-rhamnose medium (Fig. 1D). Therefore, these data do not support that PEG344 contributes to amino acid transport or l-rhamnose transport.
The ability to produce and secrete rhamnolipids has been demonstrated to contribute to the pathogenesis of Pseudomonas aeruginosa (32, 33). Therefore, it was hypothesized that PEG344 could transport this product if produced by hvKP. As a first step, since hvKP1 has been completely sequenced, we performed a bioinformatics search using the MetaCyc tool (34) to search for homologs or orthologs of rhamnolipid biosynthesis genes (rhlA, rhlB, and rhlC). No genes, enzymes, or such a pathway was identified. This in silico prediction was verified by assessing hvKP1 for the production of rhamnolipids. SW agar plates, used for the detection of rhamnolipids, were made as described previously (35). hvKP1, the Δpeg-344 strain, and a strain of Pseudomonas aeruginosa (positive control) were inoculated into wells in SW plates and incubated at 37°C for 48 h, followed by incubation at 4°C for 48 h. A precipitate surrounds strains that produce and secrete rhamnolipids. This was observed for the P. aeruginosa strain but not the hvKP1 or Δpeg-344 strain (Fig. 5). Taken together, these data do not support that hvKP1 is a producer of rhamnolipids, and as a result, this is not the mechanism by which PEG344 contributes to the full virulence of hvKP1.
FIG 5.
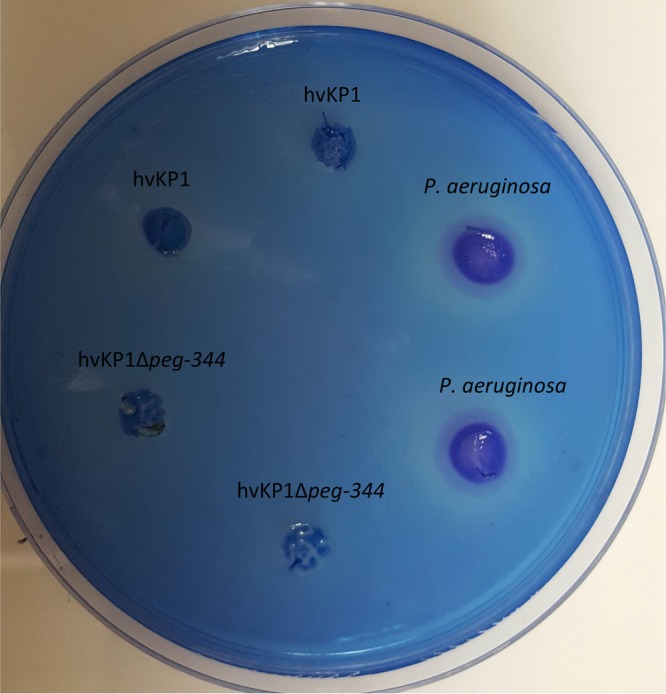
Plate assay for rhamnolipids. SW agar plates were inoculated with hvKP1, hvKP1Δpeg-344, and a clinical isolate of Pseudomonas aeruginosa. The SW plates were incubated at 37°C for 48 h, followed by 4°C for 48 h. The precipitant around the bacterial colony identifies the presence of rhamnolipid.
PEG344 may transport an undefined nutrient present in human ascites.
Next, the growth of these strains was assessed under the more clinically relevant growth conditions of 90% human ascites, an environment in which the RNA abundance of peg-344 is increased compared to that in LB or c-M9-CA. A small but statistically significant decrease in growth was observed for the Δpeg-344 strain compared to its parent strain, hvKP1 (Fig. 6A). Over the first 6 h, the complemented construct grew less well in human ascites than did hvKP1Δpeg-344. However, at 24 h, its plateau density was similar to that of its wild-type parent strain, hvKP1.
FIG 6.
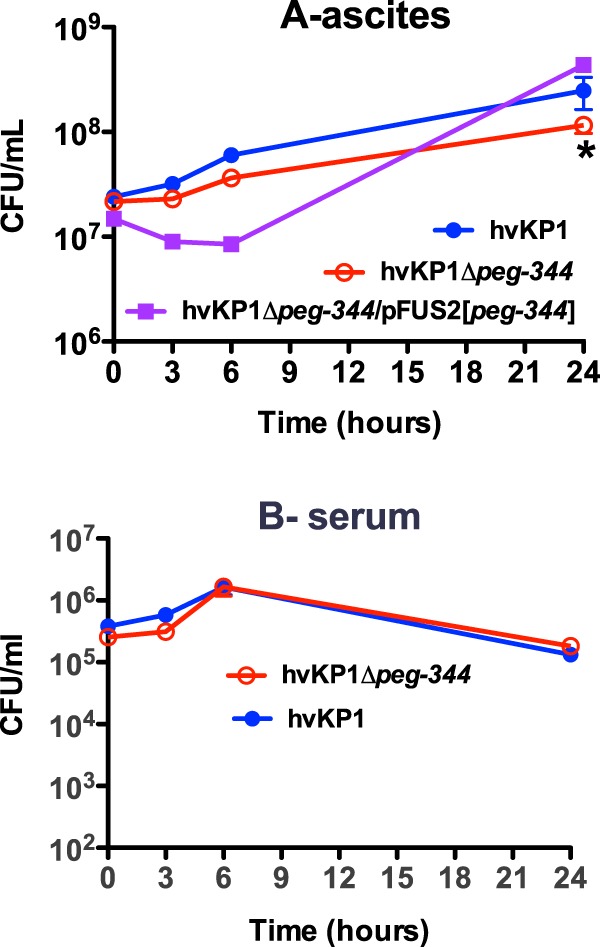
Comparison of the growth/survival rates of hvKP1, hvKP1Δpeg-344, and hvKP1Δpeg-344/pFUS2[peg-344] in 90% human ascites and 90% serum. Growth/survival in human ascites and serum was assessed by measurement of CFU at 0, 3, 6, and 24 h. (A) Ascites; (B) serum. Data are means ± SEM (n = 6 and n = 7 for each strain in ascites and serum, respectively). *, P < 0.05 (as determined by two-tailed unpaired t tests) for hvKP1 compared to hvKP1Δpeg-344 in ascites.
Human ascites is known to possess active complement. Therefore, to determine whether the observed difference in the growth/survival of the Δpeg-344 strain was due to an increased sensitivity to complement-mediated bactericidal activity, its growth/survival was compared to that of hvKP1 in 90% human serum. In serum, the growth/survival rates of the Δpeg-344 and hvKP1 strains were similar (Fig. 6B). Taken together, these data suggest that PEG344 may contribute to the transport of an undefined, non-amino-acid nutrient present in human ascites.
DISCUSSION
Understanding microbial pathogenesis is an important process that has the potential to lead to improved prevention and/or treatment of infection. hvKP is an emerging pathotype for which our knowledge of its unique traits that enhance its virulence compared to that of cKP is limited. To date, increased levels of capsule and siderophore production (predominantly aerobactin) are established virulence traits defined by genetic studies (23, 24). Both of these phenotypes are due to genes present on a virulence plasmid that appears to be a defining genetic element (36). Therefore, we hypothesized that additional virulence factors were encoded by genes present on this plasmid. An RNA-Seq approach was used to identify genes present on this plasmid with increased RNA abundances when cells were grown in human ascites compared to rich laboratory medium. This resulted in the identification of PEG344, a putative metabolite transporter. To examine the role of PEG344 in contributing to the virulence of hvKP1, an isogenic mutant derivative of hvKP1 (Δpeg-344) was generated and initially assessed in a survival model in which outbred CD1 mice developed disseminated infection after s.c. challenge. This model was previously used to establish the importance of aerobactin as a virulence factor for hvKP (24). Surprisingly, not only was Δpeg-344 not less virulent, but with certain challenge inocula, it also appeared to be more virulent (Fig. 3). Similar findings were observed previously when CD1 mice were challenged s.c. with mutant constructs that were unable to produce the siderophore yersiniabactin or salmochelin (24). We speculated that this could be due to a decreased metabolic demand for producing factors unnecessary for infection at this site and thereby perhaps enabled enhanced growth (37). Next, we compared the Δpeg-344 and hvKP1 strains in a pulmonary challenge survival model that our group has also used to establish the importance of aerobactin as a virulence factor for hvKP (24). The use of this model demonstrated that PEG344 was needed for the full virulence of hvKP1 after pulmonary challenge (Fig. 3). Competition studies that measured CFU in blood, lungs, spleen, and kidneys 24 and 48 h after pulmonary challenge not only confirmed these findings (Fig. 4A) but also suggested that PEG344 may be an important factor that enables the dissemination of hvKP1 from the pulmonary compartment. It remains unclear why PEG344 did not play a similar role after s.c. challenge. It should be noted that a limitation of this experimental design is that secreted factors could trans-complement a mutant construct and thereby obscure or diminish potential differences. However, it is unlikely the PEG344 is secreted.
In silico analysis identified the presence of two EamA superfamily domains within peg-344. This family of transporters contains inner membrane proteins involved in the transport of amino acids, drugs, metabolites, and sugar phosphates (38). In silico analysis also demonstrated that peg-344 shares homology with an l-rhamnose transporter in Salmonella enterica serovar Typhimurium and Escherichia coli (31). To gain insight into the mechanism(s) by which PEG344 enabled the increased growth/survival of hvKP1 after pulmonary challenge, a series of in vitro and ex vivo studies was performed using defined minimal medium and human ascites and serum. These data suggested that PEG344 could contribute to the transport of a non-amino-acid, non-l-rhamnose growth factor present in ascites, as evidenced by the decreased growth of the Δpeg-344 strain compared to that hvKP1 in ascites and similar growth in M9, M9-CA, and M9-rhamnose media (Fig. 1C and D and 6A). However, this statistically significant but small difference is of questionable biological significance compared to the unequivocal differences observed in vivo. Further studies are needed to identify the growth factor(s) present in ascites that could be transported by PEG344 as well as to explore alternative mechanisms.
peg-344 is located on the virulence plasmid contained in hvKP1 (28) and encodes a putative protein of 300 amino acids. It should be noted that the previously reported annotation is incorrect for hvKP in that peg-344 contains the 23 upstream amino acids in addition to the designated open reading frame (ORF) (28). A genomic analysis was recently performed on 45 liver abscess isolates of K. pneumoniae with genomic sequences in the public domain and an additional 40 K. pneumoniae strains isolated from patients with liver abscesses in China (21). The overwhelming majority of these strains were either known hvKP strains or predicted to be hvKP strains based on clinical and genomic characteristics (e.g., linkage with known hvKP plasmid-associated virulence factors such as rmpA and iucABCD). peg-344 (GenBank accession no. BAH65947.1) was present in 95.56% of the 45 strains with sequences in the public domain and in 100% of the 40 liver abscess isolates reported in that study (21), demonstrating the strong association of peg-344 with hvKP strains. For clarification, that study designated peg-344 (GenBank accession no. BAH65947.1) pagO (21). In fact, pagO is distinct but is a homolog of peg-344 (see below for details).
A study that used an oral infection model to identify virulence factors in hvKP identified a homolog of peg-344 that was designated pagO (39). pagO is present on the virulence plasmid in hvKP1 and has been designated peg-1860. peg-1860 (pagO) shares 81.6% similarity and 67.3% identity with peg-344. Oral challenge of BALB/c mice with hvKP strain CG43 in which pagO was disrupted (VA27) resulted in decreased mortality compared to that with its wild-type parent strain, CG43, albeit it was not statistically significant, perhaps due to the small number of mice (n = 5) (39). In competition experiments, 48 h after oral challenge, CG43 outcompeted VA27, as demonstrated by liver and spleen titers. However, the abilities of VA27 to colonize the small and large intestines were similar. It is intriguing that both peg-344 and peg-1860 are present on the hvKP1 virulence plasmid and that both genes appear to be involved in transport. Whether these factors prove to have independent or overlapping functions remains to be seen.
Although RNA-Seq is a powerful screening tool to identify genes with increased RNA abundances under defined conditions, it is critical to confirm findings for a gene of interest via qPCR. However, in order to accomplish this goal, it was important to establish suitable reference genes with which to compare RNA abundance levels. Surprisingly, there was a paucity of suitably identified reference genes for K. pneumoniae in the literature. We felt that a rigorous validation of a reference gene for K. pneumoniae would be particularly valuable since both cKP and hvKP are increasingly important pathogens that will hopefully become the object of an increasing number of studies. Reference gene validation studies and results are reported in Fig. S2 to S4 and Tables S1 to S3 in the supplemental material. As a first step, reference genes that had been verified in other bacterial genera were studied to determine their validity for K. pneumoniae. These genes were subsequently assessed in three different growth media (LB, c-M9-CA, and human ascites) and in three different growth phases (log, early stationary, and stationary phases). It was determined that in the different media and growth phases, the 23S rRNA gene elicited the least variation under these conditions (Fig. S4). Therefore, the 23S rRNA gene was established as a suitable reference gene with which to compare other gene expression patterns. Although this gene was previously used as a qPCR reference gene for K. pneumoniae, the lack of validation details, combined with our interest in the use of this reference gene for various growth phases and media, including ascites, which is uncommonly used, warranted a reassessment. While we evaluated RNA abundance only in log phase in this study, the reported validation will enable its use for all growth phases in subsequent studies.
In summary, this report describes the identification of PEG344 as an unrecognized factor required for the full virulence of hvKP. The data presented are important since they expand our limited knowledge base on virulence factors unique to hvKP, which is needed to lay the groundwork for translational approaches to prevent or treat these deadly infections. peg-344 is located on the hvKP1 virulence plasmid, is broadly prevalent among hvKP strains (21), and has increased RNA abundance when grown in human ascites. Interestingly, it is needed for the full virulence of hvKP1 after pulmonary challenge but not s.c. challenge. Whether this is due to increased growth or survival within the pulmonary compartment or whether PEG344 increases the ability of hvKP1 to disseminate from this site is unclear. In silico analysis suggests that PEG344 is an inner membrane transporter, but the mechanism by which PEG344 contributes to the virulence of hvKP1 requires additional study. Data are also reported that validate the 23S rRNA gene as a reference gene for qPCR of K. pneumoniae, a critical tool for future studies given that both cKP and hvKP are pathotypes of ever-increasing concern. This situation will be magnified if hvKP strains develop extensive antimicrobial resistance, as has been occurring with cKP strains.
MATERIALS AND METHODS
Strain description and construction of hvKP1Δpeg-344 (Δpeg-344).
hvKP1 (sequence type 86 [ST86]; KL1 serotype) was isolated from the blood and a liver abscess aspirate of a previously healthy 24-year-old man from Buffalo, NY, with community-acquired pyogenic liver abscess and metastatic spread to the spleen (40). The construction of the Δpeg-344 strain was performed by allelic exchange. First, hvKP1 cells were prepared for transformation via electroporation by growth in lysogeny broth (LB) medium (5 g yeast extract, 10 g tryptone, 10 g NaCl) supplemented with 0.125 mM bismuth nitrate pentahydrate and 2.5 mM sodium salicylate to suppress the expression of the capsular polysaccharide (41). Next, these cells were transformed with pKD46 that contains λRed recombinase genes under the control of the araB promoter (42), resulting in hvKP1/pKD46. Finally, hvKP1/pKD46 electroporation-competent cells were transformed with a PCR-generated linear amplicon that consisted of a kanamycin resistance gene flanked by peg-344 bp 211 to 351 and 738 to 834 plus 75 bases from the intergenic region 3′ of peg-344 and outgrown for 2 h in LB medium supplemented with 10 mM l-arabinose. The Δpeg-344 strain was selected on LB agar plates containing kanamycin, and the disruption of peg-344 was confirmed by sequence analysis. pKD46 was cured from the Δpeg-344 strain by growth at 42°C overnight. Polar effects due to the disruption of peg-344 were excluded by reverse transcription-PCR (RT-PCR) as described previously (43). The gene immediately downstream of peg-344 is iroN (peg-345), which encodes the receptor of the siderophore salmochelin. Since it is transcribed in the opposite direction of peg-344, polar effects were not anticipated. Nonetheless, the transcript for iroN was present (data not shown), thereby excluding a polar effect. Furthermore, a complemented construct (Δpeg-344/pFUS2[peg-344]) was generated for complementation experiments. A PCR-generated amplicon (forward [BamHI, sequence italicized and underlined] primer 5′-TAGGATCCAGATAAGTGAAAAAACAGCCATTAG-3′ and reverse [KpnI, sequence italicized and underlined] primer 5′-TAAGGTACCGCAGGGAAGCAAATTAGCAGA-3′) containing the peg-344 open reading frame plus the 209- and 82-nucleotide (nt) 5′ and 3′ regions, respectively, was ligated into pFUS2 (gentamicin resistant), and pFUS2[peg-344] was selected in E. coli strain XL1-Blue. pFUS2[peg-344] was then purified, and the ligated amplicon was sequenced to confirm sequence fidelity. Next, pFUS2[peg-344] was electroporated into Δpeg-344 cells grown in LB medium containing 0.125 mM bismuth nitrate pentahydrate and 2.5 mM sodium salicylate. The Δpeg-344/pFUS2[peg-344] strain required antibiotic pressure to maintain plasmid retention.
Media.
Chelated M9 minimal medium (15.62 mg of the iron chelator 2,2′-dipyridyl per 500 ml) was supplemented without (c-M9) or with (c-M9-CA) 1.5 g of Casamino Acids per 500 ml (catalog no. BP1424-100; Fisher). For M9–l-rhamnose medium, 0.5% l-rhamnose was substituted for glucose as the carbon source. SW plates used for the detection of rhamnolipids were made as described previously (35). The procedures for obtaining human ascites and serum were reviewed and approved by the Western New York Veterans Administration or the University at Buffalo-State University of New York (SUNY) Institutional Review Board. Serum was collected from healthy volunteers. Serum was used on the day of collection or stored at −80°C prior to use. Ascites was collected from deidentified patients who were undergoing therapeutic paracentesis for symptoms due to abdominal distension. These individuals were not being treated with antimicrobials. The ascites was cultured to confirm sterility, divided into aliquots, and stored at −80°C. Each batch was obtained from a different patient and was designated by the date of removal. Ascites batches 8/13/2012 and 10/18/2012 were used for RNA extraction for RNA-Seq, batch 10/18/2012 was used for qPCR, and batch 10/18/2012 was used for ex vivo growth studies. For various ex vivo growth studies, ascites or serum (90% fluid, 10% 1× phosphate-buffered saline [PBS] [pH 7.4]) was used. Media used for the growth of the Δpeg-344 strain were supplemented with 40 μg/ml of kanamycin, and media used for the growth of the Δpeg-344/pFUS2[peg-344] strain were supplemented with 40 μg/ml of kanamycin and 2.5 μg/ml of gentamicin.
RNA extraction for RNA-Seq.
Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines were utilized (44). Nuclease-free barrier tips and tubes were used (0.65-ml and 1.7-ml natural PP Eppendorf tubes [catalog no. L211151 and L211511; Laboratory Product Sales], Avant Gμard Barrier pipette tips [catalog no. L138550 and L139480; Laboratory Product Sales], and Plastibrand Bio-Cert 0.1- to 1-μl low-retention filter tips for pipetting of <1-μl volumes [catalog no. Z740033; Sigma]). The workspace and pipettes were cleaned with RNase Zap (catalog no. 9780; Ambion) to deactivate RNases. Bacteria were harvested after growth overnight in c-M9-CA, ascites, or LB medium. One milliliter of the bacterial culture was added to 2 ml of Qiagen RNAprotect Bacteria reagent (catalog no. 76506), followed by RNA extraction using the Qiagen RNeasy minikit (catalog no. 74104). The RNA was treated twice with DNase, and PCR was performed to ensure the absence of DNA. The RNA concentration was determined by using a NanoDrop 1000 spectrophotometer. The purity of the samples was measured according to the A260/A280. The Ribo-Zero Magnetic kit (bacteria, catalog no. MRZMB126; Illumina) was used to deplete rRNA from 5 μg of total RNA according to the manufacturer's protocol. The Qiagen RNeasy kit was used to clean the residual RNA. The rates of recovery were 1.8% for LB, 8.5% for c-M9-CA, 2.9% for batch 8/13/12 ascites, and 5.5% for batch 10/18/2012 ascites, which corresponded to the 2 to 8% expected recovery. The purified RNA was stored in water at −80°C.
RNA-Seq analysis.
RNA sequencing was performed at the University at Buffalo-SUNY Genomics and Bioinformatics Core Facility. Details on the methodology were reported previously (45). In brief, an Illumina TruSeq RNA sample preparation kit was used to prepare cDNA libraries from RNA samples. Ribosome-depleted RNA samples were cleaved into fragments, and the first strand was reverse transcribed to cDNA by using SuperScript II reverse transcriptase (Invitrogen) and random primers, followed by second-strand cDNA synthesis using second-strand master mix supplied with the kit. After end repair, the addition of a single “A” base, and ligation with adapters, the products were enriched and purified by PCR to create the final cDNA library according to the manufacturer's protocol. cDNA libraries were quantified by using a PicoGreen assay (Invitrogen) and a library quantification kit (Kapa Biosystems). The Fragment Analyzer High Sensitivity NGS kit (Advance Analyticals) was used to confirm the quality and size of the cDNA libraries. The cDNA libraries were then normalized, pooled, and sequenced by using an Illumina HiSeq2500 instrument according to the manufacturer's instructions.
qPCR assay. (i) Growth conditions for RNA harvest.
hvKP1 was grown overnight in 5 ml of LB at 37°C. In the morning, 1.0 ml of the culture was centrifuged, washed once with 1× PBS (pH 7.4), and resuspended in 1.0 ml of 1× PBS. Four hundred microliters was added to 30 ml of LB, ascites, or c-M9-CA. The cultures were grown to log phase in 125-ml Erlenmeyer flasks at 37°C at 250 rpm. Bacteria grown in LB were harvested at 2 h, whereas bacteria grown in ascites and c-M9-CA were harvested after 3 h of growth. To achieve similar CFU upon harvest, volumes of 2.6 ml were used for LB and ascites and a volume of 5 ml was used for c-M9-CA, yielding 9.1 × 108, 4.2 × 108, and 2.5 × 108 CFU, respectively. After RNA extraction, yields of 2,643 ng/ml (LB), 322 ng/ml (ascites), and 794 ng/ml (c-M9-CA) and A260/A280 values of 2.12 (LB), 2.05 (ascites), and 2.18 (c-M9-CA) were obtained.
(ii) RNA integrity, primer design, qPCR assay specificity, annealing temperature optimization, primer efficiency for peg-344 and reference gene candidates, and reverse transcription.
Methods for RNA integrity determination, primer design, qPCR assay specificity, annealing temperature optimization, primer efficiency for peg-344 and reference gene candidates, and reverse transcription are described in Tables S1 to S3 and Fig. S2 and S3 in the supplemental material.
(iii) qPCR protocol.
The plate setup was done under low-light conditions on ice. The reaction mixture per well consisted of 10.0 μl SsoAdvanced Universal SYBR green Supermix (catalog no. 172-5271; Bio-Rad), 1.0 μl of the primer pair (500 nM each; Integrated DNA Technologies), 4.0 μl of nuclease-free water (catalog no. 9932; Ambion), and 5 μl of diluted cDNA (5 ng total) (Hard Shell PCR plates, 96 wells, thin wall, clear well, catalog no. HSP 9601; Bio-Rad). The plate was sealed (Microseal “B” optically clear adhesive seals, catalog no. MSB1001; Bio-Rad), wrapped with aluminum foil, and briefly vortexed at a low setting, followed by a quick spin at 300 × g for 1 min. Two-step real-time PCR was performed by using the Bio-Rad CFX96 Touch instrument under the following cycling conditions: 95.0°C for 2 min (95.0°C for 15 s and 60°C for 20 s) for 40 cycles and 98.0°C for 10 s. Melt curves (65.0°C to 95.0°C, in 0.5°C increments) were included for each experiment. Before analysis of the qPCR data, the raw data were visually examined, and any outliers with a Cq difference of >0.5 cycles were discarded. Data analysis was performed by using Bio-Rad CFX Manager 3.1 qPCR analysis software (file version 3.1.1517.0823). The algorithm-based regression analysis tool was applied to determine the Cq of each sample. The Gene Study tool was employed to combine data from two qPCR experiments, each with 3 replicates per sample, into a single file for analysis. Relative normalized expression (fold change) values were used for data analysis. The value for the control LB sample was set to 1, and the 23S rRNA gene was used as the reference gene (see validation of the 23S rRNA gene as a qPCR reference gene for K. pneumoniae and Fig. S4 in the supplemental material). The software set the expression level of the control sample to 1 and normalized the relative expression levels of all target genes to the expression level of the control.
Growth in M9 medium, M9-CA, LB, ascites, and serum.
Growth in M9 medium, M9-CA, LB, ascites, and serum was performed as described previously (46). For each experiment, cultures of the hvKP1 (wt), Δpeg-344, or Δpeg-344/pFUS2[peg-344] strain were grown overnight at 37°C in LB, with the mutant strain being grown in the presence of kanamycin and the complemented strain being grown in the presence of kanamycin and gentamicin. For LB growth experiments, a fresh culture was inoculated from the cultures grown overnight to an A600 of 0.05 and grown at 37°C at 150 rpm, and the A600 was measured. For the M9 and M9-CA Å600 growth experiments, 1 ml of a culture grown overnight in LB was washed and resuspended in 1× PBS before inoculation into the respective media. The Δpeg-344 strain was grown in the presence of kanamycin. For the assessment of growth/survival in ascites, 1 ml of a culture grown overnight was washed in 1× PBS, and bacteria were appropriately diluted, added to fresh ascites, and incubated at 37°C in a shaking water bath. Aliquots were removed at 0, 3, 6, and 24 h, and CFU were enumerated by serial dilution. For the assessment of growth/survival in serum, a culture grown overnight in LB broth was diluted to an A600 of 0.10 in fresh LB medium and allowed to achieve logarithmic-phase growth at 37°C in a shaking water bath (A600 of 0.30). Bacteria were appropriately diluted in 1× PBS, and 50 μl of bacteria was added to 450 μl of human serum. Serum with bacteria was incubated at 37°C in a shaking water bath; aliquots were removed at 0, 3, 6, and 24 h; and CFU were enumerated by serial dilution.
Analysis of capsule production.
The hvKP1 and Δpeg-344 strains were grown overnight in LB medium, and an aliquot was removed to enumerate bacterial CFU. Capsule extraction, resolution by SDS–8% polyacrylamide gel electrophoresis, and analysis by alcian blue staining were performed as described previously (47). The cell pellet and supernatant volumes used for gel loading were normalized to a bacterial titer of 4.9 × 105 CFU.
Mouse subcutaneous and pulmonary challenge infection models.
Mouse subcutaneous and pulmonary challenge infection models were described previously (24, 29). Animal studies were reviewed and approved by University at Buffalo-SUNY and Veterans Administration Institutional Animal Care Committees. This study was carried out in strict accordance with recommendations of the Guide for the Care and Use of Laboratory Animals (48), and all efforts were made to minimize suffering. In brief, outbred male CD1 mice (18 to 22 g) were injected s.c. or underwent pulmonary challenge using the tongue pull technique, with various titers of the bacterial strain being assessed. For survival experiments, mice were challenged by each of the two strains being evaluated (hvKP1 and Δpeg-344). Four different challenge titers at log intervals were used per strain. For s.c. challenge, titers of hvKP1 were 3.6 × 102, 3.6 × 103, 3.6 × 104, and 3.6 × 105 CFU, and those of the Δpeg-344 strain were 2.8 × 102, 2.8 × 103, 2.8 × 104, and 2.8 × 105 CFU; for intrapulmonary challenge, titers of hvKP1 were 3.0 × 103, 3.0 × 104, 3.0 × 105, and 3.0 × 106 CFU, and those of the Δpeg-344 strain were 2.8 × 103, 2.8 × 104, 2.8 × 105, and 2.8 × 106 CFU. Animals were monitored for 14 days, with an in extremis state or death being used as the study endpoint.
For competition experiments, animals underwent s.c. or pulmonary challenge with a mixture of 1.2 × 104 CFU and 1.7 × 104 CFU of the hvKP1 and Δpeg-344 strains, respectively. Blood and organs were harvested at 24 and 48 h post-bacterial challenge. First, blood was obtained from the left ventricle of anesthetized animals for culture. Next, the right atrium was incised, and 5 ml of 1× PBS was injected through the left ventricle so that the entire blood volume was flushed from the vasculature; bacterial counts reflected solely organ-specific CFU. Next, lungs, kidneys, and spleen were harvested; washed; and homogenized for the enumeration of bacterial CFU. Undiluted aliquots and serial dilutions were plated onto LB medium, on which both the hvKP1 and Δpeg-344 strain could grow, and on LB medium with kanamycin (40 μg/ml), on which only the Δpeg-344 strain could grow. The CFU per milliliter of the Δpeg-344 strain were calculated from the LB-kanamycin plates, and the hvKP1 CFU per milliliter were calculated from the LB plates after subtraction of Δpeg-344 strain CFU. However, if the CFU per milliliter for each strain were within a log of each other, colonies from the LB plates were gridded onto LB and LB-kanamycin plates to determine the percentage of cells of each strain, which enabled a more accurate determination of each strain's titer.
Analyses of PP and PP taking into account the element of time.
Analyses of PP and PP taking into account the element of time were performed as described previously (30), except that the PP and PPT values were multiplied by 10,000. Time was calculated as the mean number of days required for death; survivors were excluded.
Statistical analyses.
Data are presented as means ± SEM. P values of <0.05/n (where n is the number of comparisons) are considered statistically significant based on the Bonferroni correction for multiple comparisons. To normalize ex vivo growth/survival data, log10-transformed values were utilized, the area under each curve was calculated, and these areas were compared by using two-tailed unpaired t tests (Fig. 1 and 6) (Prism 4 for Macintosh; GraphPad Software Inc.). A log rank (Mantel-Cox) test was used for the analysis of the Kaplan-Meier plots (Fig. 2 and 3) (Prism 4 for Macintosh; GraphPad Software Inc.). The Wilcoxon signed-rank test was used for the analysis of the in vivo competition data (Fig. 4) (Prism 4 for Macintosh; GraphPad Software Inc.).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by National Institutes of Health grants 1R21AI088318-01A1 and R21 AI123558-01 (T.A.R.) and by a VA Merit Review from the Department of Veterans Affairs (T.A.R.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00093-17.
REFERENCES
- 1.Shon AS, Bajwa RP, Russo TA. 2013. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence 4:107–118. doi: 10.4161/viru.22718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuehn BM. 2013. “Nightmare” bacteria on the rise in US hospitals, long-term care facilities. JAMA 309:1573–1574. doi: 10.1001/jama.2013.2922. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2013. Vital signs: carbapenem-resistant enterobacteriaceae. MMWR Morb Mortal Wkly Rep 62:165–170. [PMC free article] [PubMed] [Google Scholar]
- 4.Moellering RC., Jr 2010. NDM-1—a cause for worldwide concern. N Engl J Med 363:2377–2379. doi: 10.1056/NEJMp1011715. [DOI] [PubMed] [Google Scholar]
- 5.Snitkin ES, Zelazny AM, Thomas PJ, Stock F, Henderson DK, Palmore TN, Segre JA. 2012. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med 4:148ra116. doi: 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L, Todd R, Kiehlbauch J, Walters M, Kallen A. 2017. Notes from the field: pan-resistant New Delhi metallo-beta-lactamase-producing Klebsiella pneumoniae—Washoe County, Nevada, 2016. MMWR Morb Mortal Wkly Rep 66:33. doi: 10.15585/mmwr.mm6601a7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu YC, Cheng DL, Lin CL. 1986. Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch Intern Med 146:1913–1916. doi: 10.1001/archinte.1986.00360220057011. [DOI] [PubMed] [Google Scholar]
- 8.Melot B, Brisse S, Breurec S, Passet V, Malpote E, Lamaury I, Thiery G, Hoen B. 2016. Community-acquired meningitis caused by a CG86 hypervirulent Klebsiella pneumoniae strain: first case report in the Caribbean. BMC Infect Dis 16:736. doi: 10.1186/s12879-016-2065-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng NC, Yu YC, Tai HC, Hsueh PR, Chang SC, Lai SY, Yi WC, Fang CT. 2012. Recent trend of necrotizing fasciitis in Taiwan: focus on monomicrobial Klebsiella pneumoniae necrotizing fasciitis. Clin Infect Dis 55:930–939. doi: 10.1093/cid/cis565. [DOI] [PubMed] [Google Scholar]
- 10.Jung J, Park KH, Park SY, Song EH, Lee EJ, Choi SH, Choo EJ, Kwak YG, Sung H, Kim SH, Lee SO, Kim MN, Kim YS, Woo JH, Choi SH. 2015. Comparison of the clinical characteristics and outcomes of Klebsiella pneumoniae and Streptococcus pneumoniae meningitis. Diagn Microbiol Infect Dis 82:87–91. doi: 10.1016/j.diagmicrobio.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Prokesch BC, TeKippe M, Kim J, Raj P, TeKippe EM, Greenberg DE. 2016. Primary osteomyelitis caused by hypervirulent Klebsiella pneumoniae. Lancet Infect Dis 16:e190–e195. doi: 10.1016/S1473-3099(16)30021-4. [DOI] [PubMed] [Google Scholar]
- 12.Kashani AH, Eliott D. 2013. The emergence of Klebsiella pneumoniae endogenous endophthalmitis in the USA: basic and clinical advances. J Ophthalmic Inflamm Infect 3:28. doi: 10.1186/1869-5760-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng DL, Liu YC, Yen MY, Liu CY, Wang RS. 1991. Septic metastatic lesions of pyogenic liver abscess. Their association with Klebsiella pneumoniae bacteremia in diabetic patients. Arch Intern Med 151:1557–1559. doi: 10.1001/archinte.1991.00400080059010. [DOI] [PubMed] [Google Scholar]
- 14.Ko WC, Paterson DL, Sagnimeni AJ, Hansen DS, Von Gottberg A, Mohapatra S, Casellas JM, Goossens H, Mulazimoglu L, Trenholme G, Klugman KP, McCormack JG, Yu VL. 2002. Community-acquired Klebsiella pneumoniae bacteremia: global differences in clinical patterns. Emerg Infect Dis 8:160–166. doi: 10.3201/eid0802.010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shon AS, Russo TA. 2012. Hypervirulent Klebsiella pneumoniae: the next superbug? Future Microbiol 7:669–671. doi: 10.2217/fmb.12.43. [DOI] [PubMed] [Google Scholar]
- 16.Wang JH, Liu YC, Lee SS, Yen MY, Chen YS, Wang JH, Wann SR, Lin HH. 1998. Primary liver abscess due to Klebsiella pneumoniae in Taiwan. Clin Infect Dis 26:1434–1438. doi: 10.1086/516369. [DOI] [PubMed] [Google Scholar]
- 17.Li W, Sun G, Yu Y, Li N, Chen M, Jin R, Jiao Y, Wu H. 2013. Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin Infect Dis 58:225–232. doi: 10.1093/cid/cit675. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Li XY, Wan LG, Jiang WY, Yang JH, Li FQ. 2014. Virulence and transferability of resistance determinants in a novel Klebsiella pneumoniae sequence type 1137 in China. Microb Drug Resist 20:150–155. doi: 10.1089/mdr.2013.0107. [DOI] [PubMed] [Google Scholar]
- 19.Siu LK, Huang DB, Chiang T. 2014. Plasmid transferability of KPC into a virulent K2 serotype Klebsiella pneumoniae. BMC Infect Dis 14:176. doi: 10.1186/1471-2334-14-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei DD, Wan LG, Deng Q, Liu Y. 2016. Emergence of KPC-producing Klebsiella pneumoniae hypervirulent clone of capsular serotype K1 that belongs to sequence type 11 in Mainland China. Diagn Microbiol Infect Dis 85:192–194. doi: 10.1016/j.diagmicrobio.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 21.Ye M, Tu J, Jiang J, Bi Y, You W, Zhang Y, Ren J, Zhu T, Cao Z, Yu Z, Shao C, Shen Z, Ding B, Yuan J, Zhao X, Guo Q, Xu X, Huang J, Wang M. 2016. Clinical and genomic analysis of liver abscess-causing Klebsiella pneumoniae identifies new liver abscess-associated virulence genes. Front Cell Infect Microbiol 6:165. doi: 10.3389/fcimb.2016.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen YT, Chang HY, Lai YC, Pan CC, Tsai SF, Peng HL. 2004. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene 337:189–198. doi: 10.1016/j.gene.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 23.Cheng HY, Chen YS, Wu CY, Chang HY, Lai YC, Peng HL. 2010. RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43. J Bacteriol 192:3144–3158. doi: 10.1128/JB.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russo TA, Olson R, MacDonald U, Beanan J, Davidson BA. 2015. Aerobactin, but not yersiniabactin, salmochelin, or enterobactin, enables the growth/survival of hypervirulent (hypermucoviscous) Klebsiella pneumoniae ex vivo and in vivo. Infect Immun 83:3325–3333. doi: 10.1128/IAI.00430-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russo TA, Olson R, MacDonald U, Metzger D, Maltese LM, Drake EJ, Gulick AM. 2014. Aerobactin mediates virulence and accounts for increased siderophore production under iron-limiting conditions by hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Infect Immun 82:2356–2367. doi: 10.1128/IAI.01667-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nassif X, Fournier JM, Arondel J, Sansonetti PJ. 1989. Mucoid phenotype of Klebsiella pneumoniae is a plasmid-encoded virulence factor. Infect Immun 57:546–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nassif X, Sansonetti PJ. 1986. Correlation of the virulence of Klebsiella pneumoniae K1 and K2 with the presence of a plasmid encoding aerobactin. Infect Immun 54:603–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russo TA, Gill SR. 2013. Draft genome sequence of the hypervirulent Klebsiella pneumoniae strain hvKP1, isolated in Buffalo, New York. Genome Announc 1:e00065-13. doi: 10.1128/genomeA.00065-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russo TA, Shon AS, Beanan JM, Olson R, MacDonald U, Pomakov AO, Visitacion MP. 2011. Hypervirulent K. pneumoniae secretes more and more active iron-acquisition molecules than “classical” K. pneumoniae thereby enhancing its virulence. PLoS One 6:e26734. doi: 10.1371/journal.pone.0026734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Casadevall A. 2017. The pathogenic potential of a microbe. mSphere 2(1):e00015-17. doi: 10.1128/mSphere.00015-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tate CG, Muiry JA, Henderson PJ. 1992. Mapping, cloning, expression, and sequencing of the rhaT gene, which encodes a novel l-rhamnose-H+ transport protein in Salmonella typhimurium and Escherichia coli. J Biol Chem 267:6923–6932. [PubMed] [Google Scholar]
- 32.Laabei M, Jamieson WD, Lewis SE, Diggle SP, Jenkins AT. 2014. A new assay for rhamnolipid detection—important virulence factors of Pseudomonas aeruginosa. Appl Microbiol Biotechnol 98:7199–7209. doi: 10.1007/s00253-014-5904-3. [DOI] [PubMed] [Google Scholar]
- 33.Abdel-Mawgoud AM, Lepine F, Deziel E. 2010. Rhamnolipids: diversity of structures, microbial origins and roles. Appl Microbiol Biotechnol 86:1323–1336. doi: 10.1007/s00253-010-2498-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caspi R, Billington R, Ferrer L, Foerster H, Fulcher CA, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Mueller LA, Ong Q, Paley S, Subhraveti P, Weaver DS, Karp PD. 2016. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 44:D471–D480. doi: 10.1093/nar/gkv1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinzon NM, Ju LK. 2009. Improved detection of rhamnolipid production using agar plates containing methylene blue and cetyl trimethylammonium bromide. Biotechnol Lett 31:1583–1588. doi: 10.1007/s10529-009-0049-7. [DOI] [PubMed] [Google Scholar]
- 36.Bialek-Davenet S, Criscuolo A, Ailloud F, Passet V, Jones L, Delannoy-Vieillard AS, Garin B, Le Hello S, Arlet G, Nicolas-Chanoine MH, Decre D, Brisse S. 2014. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis 20:1812–1820. doi: 10.3201/eid2011.140206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lv H, Hung CS, Henderson JP. 2014. Metabolomic analysis of siderophore cheater mutants reveals metabolic costs of expression in uropathogenic Escherichia coli. J Proteome Res 13:1397–1404. doi: 10.1021/pr4009749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vastermark A, Almen MS, Simmen MW, Fredriksson R, Schioth HB. 2011. Functional specialization in nucleotide sugar transporters occurred through differentiation of the gene cluster EamA (DUF6) before the radiation of Viridiplantae. BMC Evol Biol 11:123. doi: 10.1186/1471-2148-11-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tu YC, Lu MC, Chiang MK, Huang SP, Peng HL, Chang HY, Jan MS, Lai YC. 2009. Genetic requirements for Klebsiella pneumoniae-induced liver abscess in an oral infection model. Infect Immun 77:2657–2671. doi: 10.1128/IAI.01523-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pomakova DK, Hsiao CB, Beanan JM, Olson R, MacDonald U, Keynan Y, Russo TA. 2011. Clinical and phenotypic differences between classic and hypervirulent Klebsiella pneumoniae: an emerging and under-recognized pathogenic variant. Eur J Clin Microbiol Infect Dis 31:981–989. doi: 10.1007/s10096-011-1396-6. [DOI] [PubMed] [Google Scholar]
- 41.Domenico P, Marx JL, Schoch PE, Cunha BA. 1992. Rapid plasmid DNA isolation from mucoid gram-negative bacteria. J Clin Microbiol 30:2859–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Umland TC, Schultz LW, MacDonald U, Beanan JM, Olson R, Russo TA. 2012. In vivo-validated essential genes identified in Acinetobacter baumannii by using human ascites overlap poorly with essential genes detected on laboratory media. mBio 3:e00113-12. doi: 10.1128/mBio.00113-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 45.Tsompana M, Valiyaparambil S, Bard J, Marzullo B, Nowak N, Buck MJ. 2015. An automated method for efficient, accurate and reproducible construction of RNA-seq libraries. BMC Res Notes 8:124. doi: 10.1186/s13104-015-1089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Russo TA, MacDonald U, Beanan JM, Olson R, MacDonald IJ, Sauberan SL, Luke NR, Schultz LW, Umland TC. 2009. Penicillin-binding protein 7/8 contributes to the survival of Acinetobacter baumannii in vitro and in vivo. J Infect Dis 199:513–521. doi: 10.1086/596317. [DOI] [PubMed] [Google Scholar]
- 47.Russo TA, Manohar A, Beanan JM, Olson R, MacDonald U, Graham J, Umland TC. 2016. The response regulator BfmR is a potential drug target for Acinetobacter baumannii. mSphere 1(3):e00082-16. doi: 10.1128/mSphere.00082-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.