ABSTRACT
Integration of antibiotic and probiotic therapy has the potential to lessen the public health burden of antimicrobial-associated diseases. Clostridium difficile infection (CDI) represents an important example where the rational design of next-generation probiotics is being actively pursued to prevent disease recurrence. Because intrinsic resistance to clinically relevant antibiotics used to treat CDI (vancomycin, metronidazole, and fidaxomicin) is a desired trait in such probiotic species, we screened several bacteria and identified Lactobacillus reuteri to be a promising candidate for adjunct therapy. Human-derived L. reuteri bacteria convert glycerol to the broad-spectrum antimicrobial compound reuterin. When supplemented with glycerol, strains carrying the pocR gene locus were potent reuterin producers, with L. reuteri 17938 inhibiting C. difficile growth at a level on par with the level of growth inhibition by vancomycin. Targeted pocR mutations and complementation studies identified reuterin to be the precursor-induced antimicrobial agent. Pathophysiological relevance was demonstrated when the codelivery of L. reuteri with glycerol was effective against C. difficile colonization in complex human fecal microbial communities, whereas treatment with either glycerol or L. reuteri alone was ineffective. A global unbiased microbiome and metabolomics analysis independently confirmed that glycerol precursor delivery with L. reuteri elicited changes in the composition and function of the human microbial community that preferentially targets C. difficile outgrowth and toxicity, a finding consistent with glycerol fermentation and reuterin production. Antimicrobial resistance has thus been successfully exploited in the natural design of human microbiome evasion of C. difficile, and this method may provide a prototypic precursor-directed probiotic approach. Antibiotic resistance and substrate bioavailability may therefore represent critical new determinants of probiotic efficacy in clinical trials.
KEYWORDS: Clostridium difficile, Lactobacillus reuteri, next-generation probiotics, reuterin, antimicrobial resistance
INTRODUCTION
Rising rates of antimicrobial resistance and the discovery of dwindling numbers of new antibiotics are increasing the risk of a global infectious disease crisis that will burden the health care system. In the United States, the Centers for Disease Control and Prevention estimates that antibiotic resistance causes over 2 million illnesses and 23,000 deaths each year (1). An additional half a million individuals are hospitalized with antibiotic-associated Clostridium difficile infection (CDI), contributing a further 29,000 deaths (2) and a $5.4 billion burden to the U.S. health care system each year (3). While the majority of patients with a primary diagnosis of CDI respond to frontline antibiotic treatment (vancomycin or off-label metronidazole), up to 35% experience a CDI relapse (4, 5) and have a significantly increased risk for multiple disease recurrences (6). The limited options for the treatment of recurrent CDI include extended antibiotic regimens (7), further contributing to the cycle of reinfection. Patients who suffer several antibiotic treatment failures are often referred for fecal microbiota transplantation, an investigational therapy with unknown long-term consequences (8). The National Action Plan to Combat Antibiotic-Resistant Bacteria (9) emphasizes that efforts are needed to advance the development of new antibiotics and alternative therapies to fight resistance and the diseases associated with antimicrobial use. As a result, a number of emerging therapies are being investigated, including probiotics, immunotherapies, toxin binding agents, defined microbial therapy, and nontoxigenic C. difficile strains (7, 10).
Antibiotic disruption of a healthy microbiota leaves the host susceptible to CDI. Probiotics are a promising alternative therapy and are proposed to combat antimicrobial-associated diseases by preventing pathogen invasion and protecting the healthy microbiota. Some promising adjunct therapies include the supplementation of staggered antibiotic withdrawal with Lifeway kefir in recurrent CDI cases (11) or the coadministration of Bio-K+ with antibiotics to decrease the incidence of primary CDI (12, 13). In the United States, adjunct therapy is not yet recommended by the Society for Healthcare Epidemiology of America or the Infectious Diseases Society of America, despite a growing body of evidence in the literature supporting probiotic use for CDI prevention (14–16). Nevertheless, next-generation probiotics are actively being investigated for use in CDI prevention (17). Lactic acid bacteria have long been considered important protectors of gut health, with many probiotic organisms belonging to the genus Lactobacillus. Several Lactobacillus spp. are intrinsically resistant to antibiotics, and this is an important feature when probiotics are considered for adjunct treatment to antimicrobial therapy (18).
Naturally occurring antimicrobial production by host-specific probiotic bacteria is a rich area for further innovation for the development of next-generation therapies targeting drug-resistant pathogens. Certain human-derived probiotic Lactobacillus reuteri strains produce an isomeric mixture of 3-hydroxypropionaldehyde (3-HPA) (19), a three-carbon secondary metabolite commonly known as reuterin (Fig. 1) with broad-spectrum in vitro antimicrobial activity against enteric pathogens and other intestinal bacteria (20, 21). The vitamin B12-dependent production of reuterin occurs when L. reuteri ferments the substrate glycerol. This process is driven by the horizontally acquired 57-gene pdu-cbi-hem-cob cluster containing genes important for (i) de novo vitamin B12 synthesis, (ii) the generation of microcompartments where reuterin production occurs, and (iii) glycerol fermentation to reuterin (22–24). Indicative of its probiotic character, reuterin does not typically interfere with the growth of commensal lactic acid bacteria (20), and preliminary reports indicate that this compound may promote microbiota diversity, a potential consideration in CDI (25).
FIG 1.
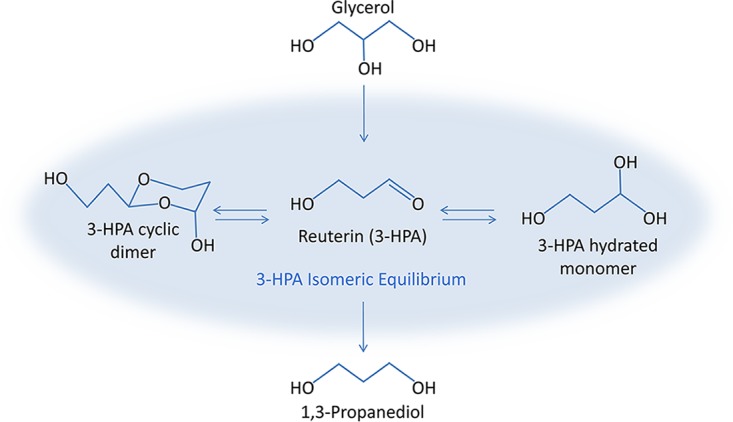
Reuterin synthesis pathway. Microbial fermentation of glycerol by L. reuteri results in the synthesis of reuterin (3-HPA) and the corresponding by-product, 1,3-propanediol.
In this study, we investigated the survival of select probiotic candidates under antibiotic pressure. Using drugs marketed for treating CDI, our screening highlighted L. reuteri as the organism that was the least susceptible to antibiotics. Further analysis showed that the production of reuterin through the active fermentation of glycerol prohibited C. difficile invasion of antibiotic-disrupted microbial community models, making this precursor-directed antimicrobial biosynthesis strategy a front-runner for the development of next-generation probiotics as adjunct therapy in CDI prevention.
RESULTS
L. reuteri is intrinsically resistant to antibiotics used to treat CDI.
Intrinsic resistance to multiple antibiotics is common in Lactobacillus spp., although the taxonomic complexity of the genus has made it difficult to define the antimicrobial susceptibilities of its members (26). To identify probiotics potentially useful in combating recurrent CDI, we determined the MICs of vancomycin, metronidazole, and fidaxomicin, drugs currently used to treat CDI, for specific human-derived strains of candidate probiotics, L. casei, L. gasseri, L. rhamnosus, and L. reuteri (Table 1). A recent U.S.-based surveillance study of the drug susceptibilities of diarrhea-associated C. difficile isolates showed the MIC90 of vancomycin, metronidazole, and fidaxomicin to be 4, 2, and 0.5 μg/ml, respectively (27). L. casei LC-39 and L. rhamnosus LR-34, known to produce factors that modulate the inflammation stimulated by C. difficile in vitro (28), demonstrated substantial resistance to vancomycin and metronidazole, with an MIC value of each drug >256 μg/ml, a concentration 64- or 128-fold greater than the vancomycin or metronidazole MIC90 for C. difficile, respectively. However, these strains were susceptible to fidaxomicin at 2 μg/ml, which is a concentration only 4-fold greater than the fidaxomicin MIC90 for C. difficile and much lower than the estimated colonic concentrations of fidaxomicin in patients (29). L. gasseri LG-3, an isolate from the feces of a human infant, was resistant to metronidazole (MIC > 256 μg/ml) and susceptible to vancomycin and fidaxomicin at 1 and 2 μg/ml, respectively. Human-derived L. reuteri strains 17938 and 6475 demonstrated the most consistent and robust resistance to all three drugs, with the MICs being from 64- to 128-fold greater than the corresponding MIC90 for C. difficile. In all, the L. reuteri strains exhibited the lowest susceptibility to the antibiotics used to treat CDI, potentiating their survival capabilities if given simultaneously with these drugs, and were further screened as potential probiotics for use in CDI prevention.
TABLE 1.
MICs of Lactobacillus spp. indicate resistance to antibiotics associated with an increased risk of development of CDI at clinically relevant concentrations
| Strain | Source | MICa (μg/ml) |
||
|---|---|---|---|---|
| Vancomycin | Metronidazole | Fidaxomicin | ||
| C. difficile R20291b | CDI patient stool | 4 | 2 | 0.5 |
| L. casei LC-39 | Infant feces | 256 | >256 | 2 |
| L. gasseri LG-3 | Infant feces | 1 | >256 | 2 |
| L. rhamnosus LR-34 | Infant feces | 256 | >256 | 2 |
| L. reuteri 17938 | Breast milk | 256 | >256 | >32 |
| L. reuteri 6475 | Breast milk | 256 | 128 | >32 |
Strain-specific, reuterin-dependent antimicrobial effects of L. reuteri on C. difficile growth in vitro.
Many human-derived L. reuteri strains metabolize glycerol into the antimicrobial three-carbon aldehyde reuterin with broad-spectrum activity toward enteric pathogens (30). Reuterin production is modulated by PocR, an AraC-like transcriptional regulator present in the 57-gene pdu-cbi-hem-cob cluster. Inactivation of the pocR gene inhibits both reuterin and vitamin B12 synthesis (31). We previously demonstrated strain-dependent reuterin production by L. reuteri strains 17938 and 6475, showing that the former strain produces 3-fold more reuterin (30, 32), leading us to hypothesize that an L. reuteri strain with a greater capacity for reuterin production would be more efficient at inhibiting C. difficile growth in vitro.
The strain-dependent effects of L. reuteri on C. difficile growth were evaluated using an agar spot overlay assay optimized for reuterin production (32). We tested wild-type L. reuteri strains 17938 and 6475 alongside isogenic pocR mutants 17938::pocR and 6475::pocR, which are incapable of producing reuterin (31), for activity against multiple C. difficile strains and ribotypes, including clinically relevant isolates. Specifically, we tested ribotype 027 C. difficile strains CD2015 and R20291, ribotype 087 strain VPI 10463, and ribotype 012 strain 630 (Fig. 2). A disc of vancomycin (5 μg) was included as a positive control for inhibition of C. difficile growth. All C. difficile strains tested were susceptible to wild-type L. reuteri strains 17938 and 6475. No inhibition of the growth of the reuterin-deficient pocR mutants was seen, and inhibition of C. difficile was restored and exceeded wild-type inhibition when strain 6475::pocR was complemented with the wild-type pocR gene (6475::pocR/pJKS102) but not the empty vector (6475::pocR/pJKS100) (Fig. 2A). To ensure that inhibition of C. difficile growth was not due to other factors affected by the pocR mutation, we tested isogenic glycerol dehydratase (gdh) mutants of L. reuteri 17938 and 6475. Glycerol dehydratase is the vitamin B12-dependent enzyme located in the pdu-cbi-hem-cob cluster that converts glycerol to reuterin (33, 34), and mutations in this gene do not affect expression of other genes in the cluster. As expected, no inhibition of C. difficile growth by the reuterin-deficient gdh mutants was seen (see Fig. S1 in the supplemental material). Differential inhibitory activity between L. reuteri strains was observed, with strain 17938 exhibiting the greatest inhibition of growth of all C. difficile strains tested compared to that by strain 6475 and the vancomycin control (P < 0.05) (Fig. 2B). Assays conducted in the absence of glycerol did not result in C. difficile growth inhibition (data not shown). Taken together, these data show that in the presence of glycerol, human-derived L. reuteri strains have antimicrobial activity against C. difficile in vitro and reuterin production is required for this phenotype.
FIG 2.
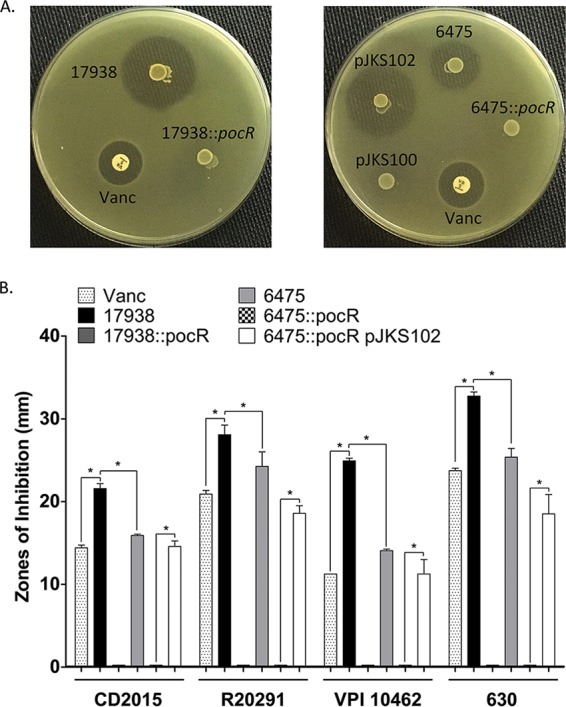
Zones of inhibition show that L. reuteri inhibits C. difficile in a strain-specific and reuterin-dependent manner in vitro. A vancomycin (Vanc) disc (5 μg) was placed and L. reuteri strains were spotted and developed on BHI medium with 20 mM glucose. C. difficile strains were overlaid in BHI soft agar containing 2% glycerol and incubated for growth. Clear zones of inhibition (in millimeters) were measured. (A) Representative images of pathogen overlays showing clear zones where the growth of C. difficile R20291 was inhibited. Wild-type L. reuteri strains 17938 and 6475, isogenic pocR insertion mutants (17938::pocR and 6475::pocR), and complemented strains (pJKS100 is the vector control, pJKS102 is the vector expressing the wild-type strain 6475 pocR gene) are shown. (B) Bar graph representing zone of inhibition measurements for L. reuteri strains tested against C. difficile. Results represent the means ± SEMs (n = 3). Inhibitory zones significantly larger (unpaired, 2-tailed t test with equal variances) than the zones for the corresponding vancomycin control, the 6475 wild type, or the 6475::pocR strain are indicated (*, P < 0.05).
Glycerol fermentation by L. reuteri 17938 prevents C. difficile invasion in antibiotic-treated human fecal MBRAs.
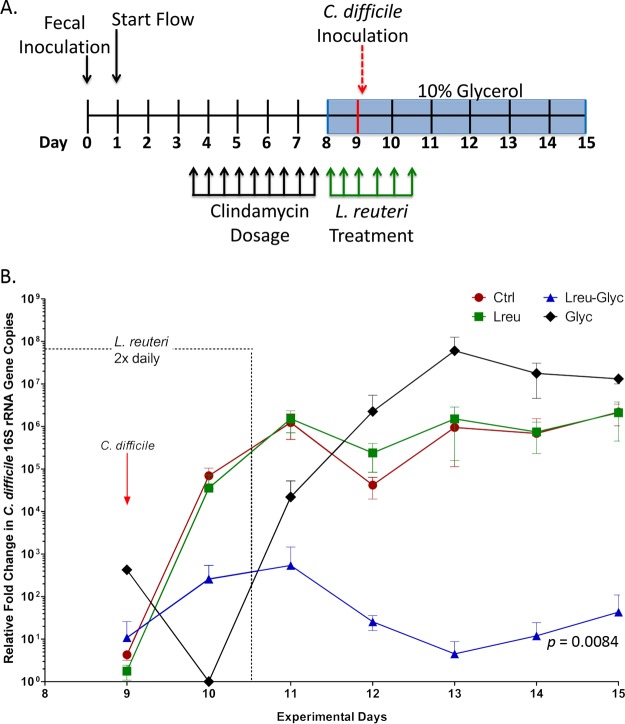
Fecal minibioreactor arrays (MBRAs) were established by Robinson et al. (35) to model C. difficile invasion of antibiotic-disrupted microbial communities. In this model, fecal samples from toxigenic C. difficile-negative adult donors were inoculated into multiple continuous-flow MBRAs. After microbial communities were allowed to establish in anaerobic culture, communities were disrupted by antibiotic treatment and infected with C. difficile CD2015 (Fig. 3A). The MBRA model of C. difficile infection was initially optimized using the clinically relevant isolate CD2015 (ribotype 027). The antibiotic clindamycin predisposes intestinal microbial communities to C. difficile invasion (36) and is used to disrupt the fecal MBRAs; those not treated with clindamycin are resistant to C. difficile colonization (35). We utilized this model to determine if the production of reuterin by L. reuteri could prevent C. difficile invasion of an antibiotic-treated microbial community in MBRAs. Additionally, next-generation sequencing of 16S rRNA genes and a global metabolomics approach were employed to determine how the microbial communities in the MBRA model of C. difficile infection compared to the microbial communities in patients with CDI.
FIG 3.
Precursor-directed reuterin production by L. reuteri suppresses the growth of C. difficile in a human fecal microbial community. (A) Time line depicting the experimental design of the MBRA experiments. (B) Quantities of 16S rRNA gene copies of C. difficile relative to the total number of 16S rRNA gene copies in bioreactor samples over time. Data are represented as means ± SDs. Significant differences in relative C. difficile 16S rRNA gene copy numbers over seven time points between the Ctrl and Lreu-Glyc groups are indicated (repeated-measures ANOVA).
We modified the MBRA model to study the effects of L. reuteri and reuterin production on C. difficile invasion of antibiotic-treated microbial communities (Fig. 3A). Specifically, we tested whether the addition of glycerol alone (Glyc), L. reuteri alone (Lreu), or L. reuteri and glycerol together (Lreu-Glyc) altered the C. difficile invasion dynamics compared to those achieved in untreated reactors (Ctrl). We monitored the abundance of C. difficile relative to the total bacterial load in MBRAs using quantitative PCR (qPCR) with primers specific to the C. difficile 16S rRNA gene and broad-spectrum 16S rRNA gene primers on days 9 to 15 (Fig. 3B) (data for day 9 were collected prior to C. difficile CD2015 addition). We found that the overall levels of C. difficile in glycerol-treated reactors with L. reuteri decreased significantly (they were ∼105-fold lower than the levels in untreated reactors; P = 0.0084). L. reuteri treatment alone did not significantly alter the C. difficile levels compared to those attained after the control treatment, whereas glycerol alone resulted in ∼10-fold greater concentrations of C. difficile, although this increase was not statistically significant. Furthermore, the direct addition of reuterin to MBRAs had no effect on C. difficile growth (data not shown), indicating the requirement of viable L. reuteri, substrate availability, and active reuterin production for C. difficile growth inhibition.
Consistent with our results on the MBRA, ex vivo inhibition of C. difficile germination and growth was observed in the cecal contents from germfree mice simultaneously treated with L. reuteri and glycerol. The codelivery of L. reuteri and glycerol resulted in an ∼104-fold suppression of the numbers of C. difficile CFU per milliliter (P < 0.01). Treatment with phosphate-buffered saline (PBS), glycerol, or L. reuteri alone supported the germination and growth of C. difficile ex vivo (Fig. S2).
Changes in microbial community dynamics in the MBRA in response to treatments.
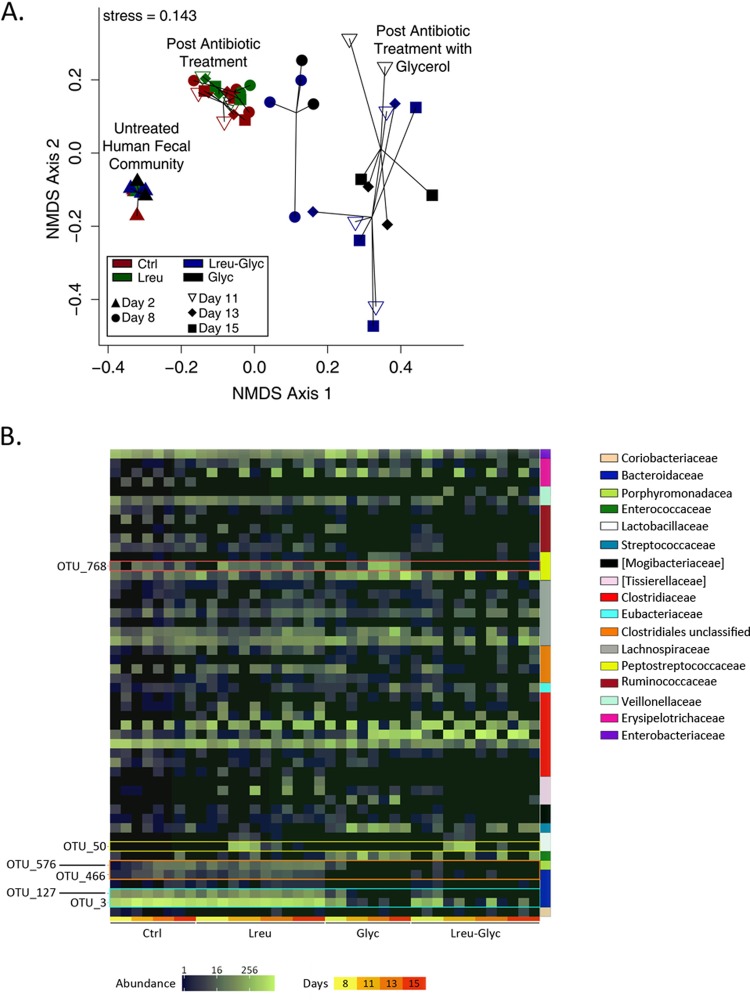
To examine the effects of substrate-based L. reuteri supplementation on the microbial community structure and composition, we sequenced the V3V5 variable region of the 16S rRNA genes from DNA isolated from MBRA samples at days 2, 8, 11, 13, and 15. The sequences of the variable regions of microbial communities collected before (day 2) and after (day 8) antibiotic treatment were compared to those of the same variable regions from communities of stool samples from patients with CDI and healthy control subjects published previously (37). In agreement with the findings of an earlier MBRA characterization study (35), clindamycin treatment resulted in a 52% loss (P = 0.0027, Mann-Whitney U test) in the number of observed species-level operational taxonomic units (OTUs; clustered at ≥97% sequence identity), paralleling the findings observed in specimens from patients treated with antibiotics (Fig. S3A). The OTUs lost to clindamycin treatment in the MBRAs were distributed across multiple bacterial families, similar to what we observed when we compared samples from healthy control subjects and patients with CDI (Fig. S3B).
Longitudinal comparisons of 16S rRNA gene sequence data demonstrated coordinated shifts in the structure of the microbial community from that of the initially established communities (day 2) in response to antibiotic treatment and glycerol supplementation. Overall changes in composition were assessed using Bray-Curtis dissimilarities and visualized through nonmetric multidimensional scaling (NMDS) (Fig. 4A). Testing of the statistical significance of the differences in community structure (using analysis of similarities [ANOSIM]) demonstrated that (i) the communities present prior to antibiotic treatment were significantly different from those present following antibiotic treatment (R = 1, P < 0.001) and (ii) treatment of reactors with glycerol for a single day initiated a shift in the community structure compared to that in non-glycerol-treated MBRAs after antibiotic treatment (R = 0.41, P = 0.009) (Fig. 4A; compare the blue [Lreu-Glyc] and black [Glyc] circles to the green [Lreu] and maroon [Ctrl] circles) that became more pronounced after longer growth in glycerol-containing medium (R = 0.83, P < 0.001) (Fig. 4A; compare the maroon circles to the white inverted open triangles [day 11], black diamonds [day 13], and black squares [day 15]). Comparison of OTU abundances between communities grown in the presence of glycerol (Glyc and Lreu-Glyc) and those grown in the absence of glycerol (Ctrl and Lreu) on each day in culture (days 8, 11, 13, and 15) revealed significant declines in two abundant Bacteroides OTUs (OTU_3 and OTU_127) by day 8 in culture that decreased further during prolonged cultivation in glycerol (P < 0.01, Mann-Whitney U test) (Fig. 4B; OTUs outlined in the teal box), as well as Bacteroides OTU_466 and Parabacteroides OTU_576, which failed to expand in glycerol-treated cultures (P < 0.01, Mann-Whitney U test) (Fig. 4B; OTUs outlined in the orange box). No statistically significant increases in OTU abundance were observed in communities cultivated in the presence of glycerol; rather, there was a trend toward an increased abundance of variable OTUs in the Firmicutes phylum (Fig. 4B).
FIG 4.
Reuterin actively targets C. difficile with minimal consequences to the overall microbial community structure. (A) Temporal analysis of microbial community composition and structure using the 16S rRNA gene sequence data generated from bioreactor samples. Pairwise relationships between samples were determined using the Bray-Curtis dissimilarity measure and plotted with nonmetric multidimensional scaling (NMDS). (B) Distribution of the abundance of MBRA OTUs represented as a heatmap, with family-level classifications being represented by the colored bars on the right. OTUs significantly differing in abundance between groups at each time point are labeled on the left.
Somewhat surprisingly, the changes in community structure following treatment with L. reuteri in the presence of glycerol were modest and not statistically significant (R = 0.12, P = 0.131) (Fig. 4A; compare the blue [Lreu-Glyc] and black [Glyc]) symbols), even though in the presence of glycerol L. reuteri exhibited potent antimicrobial activity against C. difficile. Changes in community structure following treatment with L. reuteri in the absence of glycerol were similarly modest and not statistically significantly different from those in the presence of glycerol (R = 0.12, P = 0.17) (Fig. 4A; compare the green [Lreu] and maroon [Ctrl] symbols). Comparison of OTU abundances between communities treated with L. reuteri (Lreu and Lreu-Glyc) and those not treated with L. reuteri (Ctrl and Glyc) on each day in culture following treatment (days 11, 13, and 15; the day 8 sample was collected prior to treatment) revealed potentially significant differences only on day 11 (P < 0.05, Mann-Whitney U test) just following the cessation of L. reuteri administration. OTU_50 (Fig. 4B; outlined in the yellow box) classified to the genus Lactobacillus, and its abundance increased significantly in treated samples on day 11. OTU_50 was absent in the starting fecal material, it was most abundant in L. reuteri-treated reactors at day 11 (Lreu-Glyc, 473 ± 294 counts; Lreu, 380 ± 170.2 counts), and its abundance quickly declined to less than 5 counts at days 13 and 15 in the L. reuteri-treated groups following the cessation of treatment. The representative sequence for OTU_50 had 98% identity to the L. reuteri sequence by BLAST analysis against the sequences in the NCBI 16S rRNA database. Because continuous-flow cultivation of L. reuteri alone (in the absence of fecal communities) in MBRA bioreactor medium (BRM2) resulted in poor growth (2.5 × 106 CFU/ml) compared to the growth after cultivation in a medium preferential for L. reuteri growth (deMan, Rogosa, Sharpe [MRS] medium; ∼1 × 109 CFU/ml), it was not surprising that L. reuteri levels declined quickly following the cessation of treatment.
To examine whether L. reuteri treatment in the presence of glycerol significantly impacted the levels of any OTUs in addition to C. difficile, we compared the OTU abundances in these treated reactors (Lreu-Glyc) to those in all other reactors (Ctrl, Lreu, and Glyc) on days 11, 13, and 15. These comparisons identified a single OTU, OTU_768 (Fig. 4B; outlined in the red box), whose abundance was statistically significantly different between these two populations (P = 0.017, Mann-Whitney U test) on days 11, 13, and 15. OTU_768, classified in the Peptostreptococcaceae family, was present in both the untreated control group and the group treated with L. reuteri without glycerol (22.3 ± 18.6 counts) and was present at increased levels in the glycerol-grown reactors (150 ± 173.6 counts), and its levels were reduced to 0.1 ± 0.3 count in the glycerol-grown, L. reuteri-treated (Lreu-Glyc) MBRAs. The representative sequence of OTU_768 had 99.2% identity to the C. difficile sequence by BLAST analysis against the sequences in the NCBI 16S rRNA gene database, and its relative abundance patterns between MBRA groups were consistent with the C. difficile qPCR results reported in Fig. 3B.
Metabolic profiling provides indirect evidence of reuterin production in MBRAs.
Global unbiased metabolomics analysis was used to determine whether metabolic profiles and/or specific metabolites were associated with L. reuteri-mediated suppression of C. difficile. After data processing and normalization, the Bray-Curtis dissimilarity of normalized metabolite concentrations visualized through NMDS showed that the overall changes in metabolic profiles (Fig. 5) were consistent with the shifts in microbial communities (Fig. 4A), and the primary factors driving the greatest change in both the microbial community composition and metabolomic profiles along NMDS axis 1 were antibiotic treatment and the addition of glycerol. The addition of glycerol to the MBRAs significantly affected biochemical pathways important for glycerol metabolism; the concentrations of glycerophospholipids and the corresponding monacyl- and diacylglycerol derivatives positively correlated (Spearman rho [rs] = 0.61 to 0.95, P < 0.05) (Table 2) with metabolic shifts along NMDS axis 1 (Fig. 5, bottom).
FIG 5.
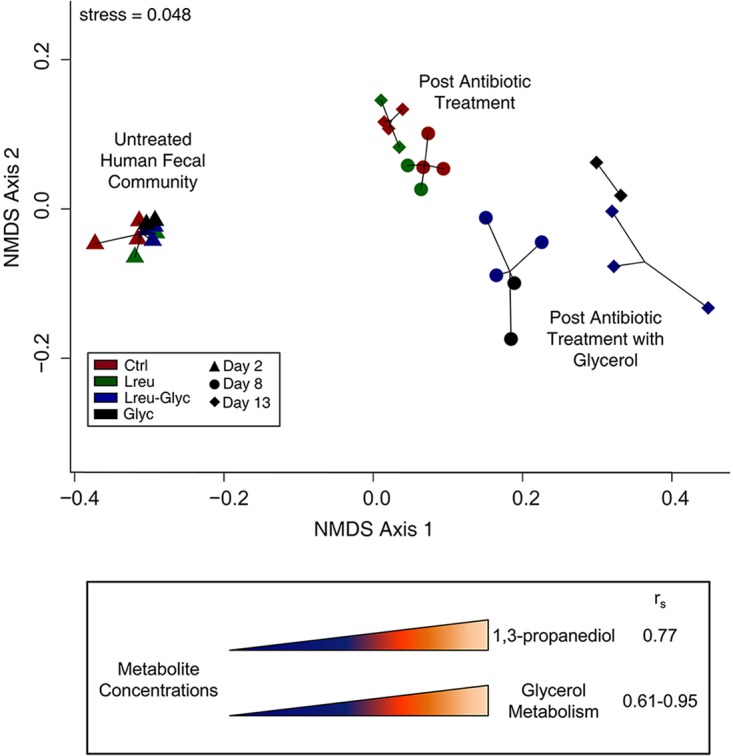
Global metabolomic changes correlate with glycerol fermentation and associated by-products. (Top) Analysis of metabolic profiles from MBRAs using the normalized metabolite concentrations generated by Metabolon. Pairwise relationships between samples were determined using the Bray-Curtis dissimilarity measure and plotted with NMDS. (Bottom) Glycerol-associated metabolite concentrations that positively correlated (Spearman rho [rs] > 0.6, P < 0.05) with NMDS axis 1 loadings.
TABLE 2.
Spearman correlations of reuterin- and glycerol-associated metabolites to global metabolite NMDS axis 1 loadings
| Biochemicala | Subpathway | rs | P valueb |
|---|---|---|---|
| 1,3-Propanediol in reuterin pathway | Chemical | 0.765 | 1.271E−02 |
| Biochemicals in glycerol metabolism | |||
| 1-Palmitoyl-2-oleoyl-GPC | Phospholipid metabolism | 0.952 | 7.892E−13 |
| 1-Palmitoleoyl-2-linoleoyl-GPE | Phospholipid metabolism | 0.897 | 2.167E−09 |
| 1-Palmitoylglycerol | Monoacylglycerol | 0.783 | 9.006E−06 |
| Glycerophosphorylcholine | Phospholipid metabolism | 0.770 | 1.167E−04 |
| 1-Stearoyl-2-linoleoyl-GPC | Phospholipid metabolism | 0.801 | 1.327E−04 |
| Palmitoyl-oleoyl-glycerol | Diacylglycerol | 0.675 | 4.877E−04 |
| Trimethylamine N-oxide | Phospholipid metabolism | 0.668 | 5.937E−04 |
| Choline phosphate | Phospholipid metabolism | 0.674 | 6.665E−04 |
| Glycerol-3-phosphate | Glycerolipid metabolism | 0.643 | 1.079E−03 |
| Choline | Phospholipid metabolism | 0.642 | 1.104E−03 |
| Glycerophosphoethanolamine | Phospholipid metabolism | 0.831 | 3.872E−03 |
| Glycerol | Glycerolipid metabolism | 0.736 | 5.007E−03 |
| 1-Palmitoyl-GPC | Lysolipid | 0.812 | 5.560E−03 |
| 1-Palmitoyl-2-linoleoyl-GPC | Phospholipid metabolism | 0.605 | 2.793E−02 |
| 1-Stearoyl-2-oleoyl-GPC | Phospholipid metabolism | 0.618 | 4.629E−02 |
GPC, glycerophosphocholine; GPE, glycerophosphoethanolamine.
Benjamin-Hochberg method-corrected P values.
One arm of glycerol fermentation produces 1,3-propanediol via the transient reuterin (3-HPA) intermediate (38), as illustrated in Fig. 1. In L. reuteri, the 3-HPA intermediate accumulates for reasons that have yet to be elucidated. Although reuterin (3-HPA) was not present in the global metabolite library and is unable to be detected directly due to its high reactivity and transient nature, the primary by-product derived from reuterin synthesis, 1,3-propanediol, was detected in reactors receiving glycerol (Lreu-Glyc and Glyc). The Spearman correlations of the 1,3-propanediol concentrations to the NMDS axis loadings showed that the presence of 1,3-propanediol positively correlated with the global metabolite shift across NMDS axis 1 (rs = 0.77, P = 0.01) (Fig. 5, bottom), providing indirect evidence that the reuterin pathway is active in MBRAs receiving glycerol.
DISCUSSION
Here we show the integration of Lactobacillus and precursor-directed antimicrobial therapy as a prototypical next-generation probiotic strategy targeting CDI. We demonstrate that L. reuteri 17938, which is a commercially available probiotic strain currently undergoing a clinical trial in children receiving antibiotic therapy (39) and which is also clinically indicated to reduce viral and bacterial diarrhea in children (40–42) and respiratory and gastrointestinal (GI) disease in adults (43, 44), shows promise as preventative therapy in patients with CDI when coadministered with glycerol. The codelivery of L. reuteri and glycerol prevented C. difficile from establishing high concentrations in a number of antibiotic-treated microbial communities, and the suppression of C. difficile growth remained after the termination of L. reuteri supplementation. Although reuterin is characterized as having broad-spectrum activity against a wide variety of pathogens, the cumulative effects of reuterin production on microbial community dynamics were virtually indistinguishable from the effects of glycerol alone. The narrow-spectrum effects of reuterin on the microbial community structure are supported in part by previous studies indicating less sensitivity by commensal families of intestinal bacteria (21, 45). While C. difficile viability was notably diminished by reuterin in our in vitro and ex vivo studies, in vivo efficacy studies are under way to assess whether coadministration of L. reuteri and glycerol can be used to treat C. difficile infections in vivo.
Success with adjunct probiotic therapy relies on maintenance of the viability of the probiotic when it is paired with antimicrobial administration. Antimicrobial resistance either can be acquired or is natural (intrinsic). The evolution of acquired bacterial resistance is propagated through antibiotic exposure, which can force spontaneous genetic mutations or the transfer of resistance genes via mobile genetic elements that ensure bacterial survival. In an effort to comply with recommendations from the European Union PROSAFE project that probiotics not harbor known antibiotic resistance traits (46), L. reuteri 55730 was cured of two plasmids (pLR581 and pLR585) that possessed the transferrable antimicrobial resistance genes tet(W) and lnu(A), resulting in daughter strain L. reuteri 17938 (47). Upon plasmid removal, L. reuteri 17938 no longer carried the transferrable resistance genes tet(W) and lnu(A) and retained all known probiotic characteristics (47). Nontransferable intrinsic resistance is generally common to all members of a species, is independent of antibiotic selective pressure (48), and, therefore, is a desired phenotype in adjunct probiotic therapy (18). Parent strain L. reuteri ATCC 55730 is intrinsically resistant to β-lactams (47), vancomycin (49, 50), metronidazole (51), and several other antibiotics to which resistance is collectively shared within this species (52, 53). Intrinsic resistance to fidaxomicin has yet to be documented, although little to no activity of fidaxomicin against Gram-negative aerobic and anaerobic bacteria is common (26). Susceptibility to fidaxomicin has been demonstrated in some lactic acid bacteria (Enterococcus spp. and Lactobacillus casei). However, the MIC for Lactobacillus acidophilus is similar to that for L. reuteri at >32 μg/ml (26), indicating that these species do not fall within the drug's narrow spectrum of activity. With the knowledge that fidaxomicin targets bacterial RNA polymerase (54) and Lactobacillus spp. are usually naturally resistant to nucleic acid synthesis inhibitors (18, 51), it is plausible that the high MIC values demonstrated by L. reuteri in this study are due to intrinsic mechanisms.
Glycerol, a common dietary component, is used as a sweetener, solvent, thickener, and preservative in pharmaceutical and food products and is a natural component of triglycerides. Glycerol absorption primarily occurs in the small intestine, although unabsorbed glycerol, free fatty acids, and undigested glycerides do transit to the colon. Gut microbes produce bacterial lipases that hydrolyze glycerides into free fatty acids and glycerol (55). Other sources of colonic glycerol are luminal fat digestion, intestinal clearing of endogenous plasma glycerol, desquamated epithelial cells, and luminal microbial fermentation (20). Microbial fermentation of glycerol can occur by organisms in the class Clostridia and the families Enterobacteriaceae and Lactobacillaceae and can result in many products, including butyrate or 1,3-propandiol, that can either stimulate or selectively inhibit glycerol absorption in the colon (38, 56, 57). Batch fermentation studies have shown glycerol metabolism to be highly variable between human fecal microbiota communities (58), and our own study emphasized glycerol-induced changes to the microbial communities in the MBRAs. Here we clearly demonstrated reuterin-dependent L. reuteri-mediated inhibition of C. difficile growth both in vitro and ex vivo. Pathogen killing by reuterin relied on the immediate bioavailability of glycerol to L. reuteri in close proximity with C. difficile.
In summary, our results show that probiotic strain L. reuteri 17938 is intrinsically resistant to the antimicrobial drugs used to treat antibiotic-associated CDI and has reuterin-mediated antimicrobial activity against C. difficile. The codelivery of L. reuteri 17938 and glycerol interferes with C. difficile growth within an antibiotic-treated human fecal microbial community in vitro without significantly affecting the overall microbial community composition. These findings indicate that substrate bioavailability may represent an important patient variable and a determinant of efficacy in clinical probiotic trials. Future efforts will be aimed at identifying the mechanisms evolved by the gut microbiota to evade reuterin cytotoxicity and optimizing the delivery of L. reuteri, targeting the site of infection with the intention of preventing recurrent episodes, as adjunct therapy to antibiotics used to treat CDI.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The strains used in this study are listed in Table S1 in the supplemental material. Routine culturing of Lactobacillus spp. in deMan, Rogosa, Sharpe (MRS) medium (Difco, Franklin Lakes, NJ) and C. difficile strains in brain heart infusion (BHI) medium (BD Biosciences, Franklin Lakes, NJ) was carried out at 37°C in an anaerobic chamber (catalog number AS-580; Anaerobe Systems, Morgan Hill, CA) supplied with a mixture of 10% CO2, 5% H2, and 85% N2 for 16 to 18 h. Erythromycin (Erm; 10 μg/ml) or chloramphenicol (Cm; 10 μg/ml) was added when necessary for plasmid or chromosomal insertion maintenance (Table S1). The specific culture conditions used for individual experiments are detailed throughout.
Antibiotic susceptibility testing.
The susceptibility of the Lactobacillus spp. to antibiotics was determined using standard broth microdilution procedures (59). Briefly, bacteria (1 × 106 CFU/ml) were inoculated into a 96-well plate containing serial dilutions of vancomycin (0.125 to 256 μg/ml), metronidazole (0.125 to 256 μg/ml), or fidaxomicin (0.016 to 32 μg/ml) in MRS medium. The plates were incubated anaerobically at 37°C for 24 h. Optical density measurements (600 nm) were recorded using a Synergy H1 hybrid multimode microplate reader (BioTek Instruments, Inc., Winooski, VT). The results were compared to those for a growth control (with lactobacillus and vehicle only), and the MIC endpoint was the concentration of antibiotic at which a ≥90% reduction in growth (MIC90) was observed. Significance was determined using Student's t test with equal variance.
Pathogen inhibition assay.
C. difficile susceptibility to reuterin was measured using an agar spot test optimized to promote reuterin production by L. reuteri. Assays were performed as previously described (32) with minimal modifications. Briefly, overnight cultures of L. reuteri were spotted (2 μl) onto BHI medium supplemented with 20 mM glucose and developed by anaerobic incubation at 37°C for 24 h. Overnight cultures of C. difficile strains were inoculated (107 to 108 cells/ml) in 7 ml soft agar (BHI broth, 2% glycerol, 0.7% technical agar), the C. difficile-inoculated soft agar was layered over the L. reuteri spots, and the culture was incubated anaerobically at 37°C for 24 h. Clear zones of inhibition (≥1 mm) around each spot were scored. Significance was determined using Student's t test with equal variance.
MBRA preparation and operation and sample collection.
The Institutional Review Board from Michigan State University reviewed and approved fecal sample collection. The effects of reuterin on C. difficile growth in a human fecal microbial community were tested in minibioreactor arrays (MBRAs). Replicate MBRAs (12 independent 15-ml reactors) and bioreactor medium (BRM2) were prepared as previously described (60). The reactors were inoculated with an anaerobic preparation of a 25% fecal slurry from an anonymous healthy donor (final concentration, 5% [wt/vol]) and were operated in an anaerobic chamber with a 5% H2, 5% CO2, 90% N2 atmosphere. After 16 h of outgrowth in batch culture mode, continuous-flow cultivation was initiated at a flow rate of 0.94 ml/h (retention time, 16 h). After 36 h of flow, the reactor communities were treated twice daily with clindamycin (final concentration, 500 mg/liter) for 4.5 days. The reactors were transitioned to fresh medium on day 4 of clindamycin treatment, with six reactors receiving standard BRM2 and six reactors receiving BRM2 supplemented with glycerol (final concentration, 10% [vol/vol]). The reactors were continued on the respective medium throughout the duration of the experiment. Thirteen hours after the cessation of antibiotic treatment, six reactors were treated with L. reuteri 17938 (three reactors in BMR2 and three reactors in BRM2 plus glycerol), prepared as described in “L. reuteri cultivation and dosing for MBRAs” below. L. reuteri treatment continued twice daily for 3 days. After the first day of L. reuteri treatment, all 12 reactors were challenged with ∼1 × 106 vegetative cells of C. difficile CD2015 (clinical ribotype 027) that had been propagated in BRM2 prior to inoculation (35).
Samples (1 ml) were collected from each MBRA as described previously (35) prior to the initiation of continuous flow and then daily through the duration of the experiment. Samples were centrifuged at 20,000 × g for 1 min, and then the supernatants and pellets were separated and stored at −80°C. Microbial community and metabolite analysis and quantification of C. difficile levels by quantitative PCR (qPCR) were performed as described below.
L. reuteri cultivation and dosing for MBRAs.
L. reuteri colonies growing anaerobically at 37°C on MRS agar (≤2 days) were inoculated into 10 ml of MRS broth and incubated anaerobically at 37°C for 8 to 12 h. Two 3-ml aliquots of culture were removed, concentrated by centrifugation for 5 min at 1,753 × g, and washed in BRM2 or BRM2 plus 10% glycerol. Bacterial cell pellets were resuspended in 1 ml BRM2 or BRM2 plus 10% glycerol and incubated at 37°C anaerobically for 15 min prior to dosing of 300 μl into reactors. Aliquots of the L. reuteri inocula were serially diluted, plated on MRS agar, and incubated anaerobically overnight at 37°C to determine the number of CFU per milliliter of inoculum. The counts were 5.2 ± 4.2 × 109 CFU/ml and 3.4 ± 3.9 × 109 CFU/ml for cultures incubated in BRM2 and BRM2 plus glycerol, respectively.
Ex vivo germination and outgrowth studies in GI luminal contents.
Animal protocols were approved by the Baylor College of Medicine Institutional Animal Care and Use Committee. The susceptibility of C. difficile to L. reuteri in mouse cecal contents was determined using methods similar to those previously described (61, 62). Cecal content was harvested from germfree 7- to 10-week-old C57BL/6 male mice, resuspended 1:2 in PBS, and aliquoted into 200-μl samples. Each aliquot was inoculated with C. difficile VPI 10463 spores (104/ml) and then treated with PBS, L. reuteri 17938 cells (107/ml), 10% glycerol, or L. reuteri 17938 cells (107/ml) plus 10% glycerol. Suspensions were incubated anaerobically at 37°C for 24 h. Samples were taken at 0 and 24 h, and C. difficile bacteria were quantified by overnight culture on prereduced selective cycloserine-cefoxitin-fructose agar with sodium taurocholate (TCCFA) medium. The percentage of spores was determined by dilution plating before and after heating to 65°C for 20 min. Plates were incubated anaerobically for 48 h at 37°C. Significance was determined using two-way repeated-measures analysis of variance (ANOVA) with the Bonferroni multiple-comparison correction.
Microbial DNA extraction.
DNA was extracted from samples from intestinal contents and MBRAs using methods described previously, with modifications (63, 64). Briefly, samples were suspended in preheated lysis buffer (65°C) and subjected to two cycles of homogenization using a BeadBlaster tissue homogenizer (Benchmark Scientific, Melrose, MA) for 20 s at 6.00 meters/s. Remaining intact cells and debris were pelleted by centrifugation for 5 min at 5,000 × g, and the supernatant was collected. An additional 300 μl of preheated lysis buffer was added to the pellet, the homogenization and centrifugation steps were repeated, and the supernatants were pooled. A mixture of ammonium acetate (final concentration, 2 M) and pooled supernatant was incubated on ice for 10 min and then centrifuged at 4°C for 10 min at 14,000 × g. The supernatant was collected and mixed with an equal volume of isopropanol, and the mixture was incubated overnight at −20°C. Precipitation, washing, and removal of RNA and protein were performed as described by Yu and Morrison (63). DNA was purified using a Zymo Research DNA Clean and Concentrator 25 kit (Irvine, CA) according to the manufacturer's instructions and stored at −20°C until further analysis by 16S rRNA gene sequencing or qPCR.
Quantitation of C. difficile by qPCR.
Real-time qPCR was used to determine the quantity of C. difficile 16S rRNA gene copies relative to the total quantity of bacterial 16S rRNA gene copies in samples from MBRAs or intestinal contents as previously reported (65–67). Template DNA was extracted as described above, and 10 ng was used in reactions with SYBR green PCR master mix (Applied Biosystems, Waltham, MA) according to the manufacturer's instructions. Bacterial 16S rRNA gene primers were used to assess the total bacterial 16S rRNA gene (forward primer, 5′-GCA GGC CTA ACA CAT GCA AGT C; reverse primer, 5′-CTG CTG CCT CCC GTA GGA GT) (68) or C. difficile 16S rRNA gene (forward primer, 5′-TTG AGC GAT TTA CTT CGG TAA AGA; reverse primer, 5′-CCA TCC TGT ACT GGC TCA CCT) content (69). Real-time quantification was carried out on an MJ Research PTC-200 thermocycler (Bio-Rad, Hercules, CA) with cycling conditions of an initial cycle of 95°C for 5 min and then 41 cycles of 95°C for 20 s, 60°C for 1 min, and 84°C for 1 s, followed by a melting curve. The relative amount of the C. difficile 16S rRNA gene was calculated by the 2−ΔΔCT threshold cycle (CT) method (65–67). The significance of the difference in the relative abundance of C. difficile between groups was determined using repeated-measures ANOVA over 7 time points and pairwise comparisons of the results for three treatments versus those for the control.
16S rRNA gene sequencing.
Microbial DNA extracted as outlined above was sequenced as previously described by the Human Microbiome Project (70). Sequencing of the V3V5 region of the 16S rRNA gene was accomplished on the 454 GSFLX platform (Roche) with the forward primer 357F (CCTACGGGAGGCAGCAG) and adapter-tagged reverse primer 534R (CCGTCAATTCMTTTRAGT).
Microbiome analysis.
The UPARSE (v8.1) (71) platform for 16S rRNA gene sequence analysis was used to quality filter reads and de novo cluster sequences into operational taxonomic units (OTUs) at 97% sequence identity. The reads were filtered according to quality by the maximum number of expected errors (E_max) of 0.5 and trimmed to a length of 250 bp, and singletons were discarded. The diversity and composition of the bacterial community were evaluated using the QIIME (v1.8) (72) sequence analysis platform. Assignment of the taxonomy of a representative sequence for each OTU was completed using the UCLUST consensus taxonomy classifier algorithm and the Greengenes reference database (v13.8). Libraries were randomly subsampled to a depth of 2,300 sequences prior to the calculation of diversity metrics or assessment of the differences in the relative abundances of taxons. Significant differences in alpha diversity and OTU abundances were assessed using the Mann-Whitney U test. Differences in OTU abundances across treatment conditions were calculated for each culture day independently. Bray-Curtis dissimilarities were calculated from untransformed abundance data using the vegan package in R. Stable nonmetric multidimensional scaling (NMDS) coordinates were determined with the metaMDS function (in R) and plotted in R. The significance of changes in community structure between treatment groups (before clindamycin treatment, following clindamycin treatment, following clindamycin treatment and treatment with L. reuteri, following clindamycin treatment and treatment with glycerol, and following clindamycin treatment and treatment with glycerol plus L. reuteri) was evaluated by analysis of similarities (ANOSIM), calculated using the mothur (v1.36.1) program (79). In addition to the Greengenes classification outlined above, specific representative OTU sequences of interest were analyzed by BLASTn analysis against the sequences in the NCBI 16S Microbial database to determine the top hit.
Measurement of metabolite composition of MBRA supernatants.
Supernatants generated from the MBRA experiments were shipped on dry ice to Metabolon, Inc., for metabolite quantification as described previously (73, 74). The supernatants (100 μl) used for extraction were prepared using a MicroLab Star automated system (Hamilton Company). A recovery standard was added before the extraction process for quality control (QC) purposes. Samples were extracted using an aqueous methanol extraction process to remove the protein fraction while allowing the maximum recovery of small molecules. The resulting extract was divided into fractions for analysis by ultraperformance liquid chromatography (UPLC)-tandem mass spectroscopy (MS/MS) (75) in the positive mode and by UPLC-MS/MS (75) in the negative mode. Samples were placed briefly on a TurboVap concentration evaporator workstation (Zymark) to remove the organic solvent. Each sample was then frozen and dried under vacuum and prepared to run on the UPLC-MS/MS (75).
Extracted samples were processed as described previously (76, 77). For quality assurance (QA)/QC, additional samples were included with the experimental samples in each day's analysis. Samples used for QC were spaced evenly among the injections, and all experimental samples were randomly distributed throughout the run. A selection of compounds for QC analysis was added to every sample for chromatographic alignment, including those being tested.
Raw data were extracted, and the peak was identified and processed for QC using Metabolon's hardware and software. Metabolon maintains a library based on authenticated standards that contain the retention time/retention index (RI), mass-to-charge ratio (m/z), and chromatographic data (including MS/MS spectral data) on all molecules present in the library. Biochemical identifications are based on three criteria: the retention index within a narrow RI window of the proposed identification, a nominal mass match to the library ±0.2 atomic mass unit, and the MS/MS forward and reverse scores between the experimental data and authentic standards. MS/MS scores are based on a comparison of the ions present in the experimental sample spectrum to the ions present in the library spectrum. While there may be similarities between these molecules on the basis of one of these factors, the use of all three data points can be used to distinguish and differentiate biochemicals. More than 2,400 commercially available purified standard compounds have been acquired and registered in the Laboratory Information Management System for determination of their analytical characteristics.
Metabolomic data analysis.
Metabolite abundance data were processed to (i) remove metabolites with >10% missing values across all samples, (ii) impute missing values with 10% of the minimum observed value for each metabolite, and (iii) determine the minimum and maximum values and the difference between maximum and minimum values for each metabolite for minimum-maximum value normalization. Bray-Curtis dissimilarities were calculated from normalized metabolite abundance data using the vegan package in R (80). Stable NMDS coordinates were determined with metaMDS (in R) and plotted in R. Spearman correlations with P values were calculated for the metabolites against NMDS axis loadings using the rcorr package in RStudio. To control for the false discovery rate, the P values were corrected for multiple testing by the Benjamini-Hochberg method. Statistically significant biochemicals (corrected P ≤ 0.05) with a Spearman rho (rs) value of >0.6 were analyzed further.
Statistics.
Results are presented as mean values ± standard deviations (SDs), mean values ± standard errors of the means (SEMs), or a median rank, as indicated in the figure legends. Statistical significance was determined using the t test, repeated-measures ANOVA, the Mann-Whitney U test, the Kruskal-Wallis test, ANOSIM, and the Spearman correlation with correction for multiple testing where indicated; the precise test is specified where appropriate. Three biological replicates were assayed unless otherwise stated; a P value of <0.05 was considered statistically significant.
Accession number(s).
The sequences (Table S2) have been deposited with the NCBI Short Read Archive under BioProject accession number PRJNA395577.
Supplementary Material
ACKNOWLEDGMENTS
We thank Eamonn Connolly (Biogaia AB, Stockholm, Sweden) for providing L. reuteri 17938 and 6475 for use in this study and Toni-Ann Mistretta and Emily B. Hollister for bioinformatics and statistical analysis expertise.
This work was supported by the National Institutes of Health, including a pilot funding award through the Texas Medical Center Digestive Disease Center (P30 DK56338) (J.K.S.), the National Institute of Allergy and Infectious Diseases (NIAID) R01 AI10094001 (T.C.S.), U01 AI124290-01 (T.C.S.), and 5U19AI090872-02 (R.A.B.), and the Institute for Translational Sciences at the University of Texas Medical Branch through Clinical and Translational Science Award UL1TR000071 (S.M.D.).
The authors disclose the following. J.K.S., R.A.B., and J.V. receive unrestricted research support from Biogaia AB. The remaining authors disclose no conflicts.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00303-17.
REFERENCES
- 1.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 2.Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, Wilson LE, Winston LG, Cohen JA, Limbago BM, Fridkin SK, Gerding DN, McDonald LC. 2015. Burden of Clostridium difficile infection in the United States. N Engl J Med 372:825–834. doi: 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Desai K, Gupta SB, Dubberke ER, Prabhu VS, Browne C, Mast TC. 2016. Epidemiological and economic burden of Clostridium difficile in the United States: estimates from a modeling approach. BMC Infect Dis 16:303. doi: 10.1186/s12879-016-1610-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garey KW, Sethi S, Yadav Y, DuPont HL. 2008. Meta-analysis to assess risk factors for recurrent Clostridium difficile infection. J Hosp Infect 70:298–304. doi: 10.1016/j.jhin.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Kelly CP. 2012. Can we identify patients at high risk of recurrent Clostridium difficile infection? Clin Microbiol Infect 18(Suppl 6):S21–S27. doi: 10.1111/1469-0691.12046. [DOI] [PubMed] [Google Scholar]
- 6.McFarland LV, Elmer GW, Surawicz CM. 2002. Breaking the cycle: treatment strategies for 163 cases of recurrent Clostridium difficile disease. Am J Gastroenterol 97:1769–1775. doi: 10.1111/j.1572-0241.2002.05839.x. [DOI] [PubMed] [Google Scholar]
- 7.Ofosu A. 2016. Clostridium difficile infection: a review of current and emerging therapies. Ann Gastroenterol 29:147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Food and Drug Administration (FDA). Guidance for industry. Enforcement policy regarding investigational new drug requirements for use of fecal microbiota for transplantation to treat Clostridium difficile infection not responsive to standard therapies. FDA, Rockville, MD. [Google Scholar]
- 9.Task Force for Combatting Antibitotic-Resistant Bacteria. 2015. National action plan for combating antibiotic-resistant bacteria. The White House, Washington, DC. [Google Scholar]
- 10.McFarland LV. 2016. Therapies on the horizon for Clostridium difficile infections. Expert Opin Investig Drugs 25:541–555. doi: 10.1517/13543784.2016.1161025. [DOI] [PubMed] [Google Scholar]
- 11.Bakken JS. 2014. Staggered and tapered antibiotic withdrawal with administration of kefir for recurrent Clostridium difficile infection. Clin Infect Dis 59:858–861. doi: 10.1093/cid/ciu429. [DOI] [PubMed] [Google Scholar]
- 12.Maziade PJ, Andriessen JA, Pereira P, Currie B, Goldstein EJ. 2013. Impact of adding prophylactic probiotics to a bundle of standard preventative measures for Clostridium difficile infections: enhanced and sustained decrease in the incidence and severity of infection at a community hospital. Curr Med Res Opin 29:1341–1347. doi: 10.1185/03007995.2013.833501. [DOI] [PubMed] [Google Scholar]
- 13.Maziade PJ, Pereira P, Goldstein EJ. 2015. A decade of experience in primary prevention of Clostridium difficile infection at a community hospital using the probiotic combination Lactobacillus acidophilus CL1285, Lactobacillus casei LBC80R, and Lactobacillus rhamnosus CLR2 (Bio-K+). Clin Infect Dis 60(Suppl 2):S144–S147. doi: 10.1093/cid/civ178. [DOI] [PubMed] [Google Scholar]
- 14.Spinler JK, Ross CL, Savidge TC. 2016. Probiotics as adjunctive therapy for preventing Clostridium difficile infection—what are we waiting for? Anaerobe 41:51–57. doi: 10.1016/j.anaerobe.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen SH, Gerding DN, Johnson S, Kelly CP, Loo VG, McDonald LC, Pepin J, Wilcox MH. 2010. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA). Infect Control Hosp Epidemiol 31:431–455. doi: 10.1086/651706. [DOI] [PubMed] [Google Scholar]
- 16.Sinclair A, Xie X, Saab L, Dendukuri N. 2016. Lactobacillus probiotics in the prevention of diarrhea associated with Clostridium difficile: a systematic review and Bayesian hierarchical meta-analysis. CMAJ Open 4:E706–E718. doi: 10.9778/cmajo.20160087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pamer EG. 2016. Resurrecting the intestinal microbiota to combat antibiotic-resistant pathogens. Science 352:535–538. doi: 10.1126/science.aad9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jose NM, Bunt CR, Hussain MA. 2015. Implications of antibiotic resistance in probiotics. Food Rev Int 31:52–62. doi: 10.1080/87559129.2014.961075. [DOI] [Google Scholar]
- 19.Vollenweider S, Grassi G, Konig I, Puhan Z. 2003. Purification and structural characterization of 3-hydroxypropionaldehyde and its derivatives. J Agric Food Chem 51:3287–3293. doi: 10.1021/jf021086d. [DOI] [PubMed] [Google Scholar]
- 20.Casas IA, Dobrogosz WJ. 2000. Validation of the probiotic concept: Lactobacillus reuteri confers broad-spectrum protection against disease in humans and animals. Microb Ecol Health Dis 12:247–285. [Google Scholar]
- 21.Cleusix V, Lacroix C, Vollenweider S, Duboux M, Le Blay G. 2007. Inhibitory activity spectrum of reuterin produced by Lactobacillus reuteri against intestinal bacteria. BMC Microbiol 7:101. doi: 10.1186/1471-2180-7-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morita H, Toh H, Fukuda S, Horikawa H, Oshima K, Suzuki T, Murakami M, Hisamatsu S, Kato Y, Takizawa T, Fukuoka H, Yoshimura T, Itoh K, O'Sullivan DJ, McKay LL, Ohno H, Kikuchi J, Masaoka T, Hattori M. 2008. Comparative genome analysis of Lactobacillus reuteri and Lactobacillus fermentum reveal a genomic island for reuterin and cobalamin production. DNA Res 15:151–161. doi: 10.1093/dnares/dsn009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santos F, Vera JL, van der Heijden R, Valdez G, de Vos WM, Sesma F, Hugenholtz J. 2008. The complete coenzyme B12 biosynthesis gene cluster of Lactobacillus reuteri CRL1098. Microbiology 154:81–93. doi: 10.1099/mic.0.2007/011569-0. [DOI] [PubMed] [Google Scholar]
- 24.Sriramulu DD, Liang M, Hernandez-Romero D, Raux-Deery E, Lunsdorf H, Parsons JB, Warren MJ, Prentice MB. 2008. Lactobacillus reuteri DSM 20016 produces cobalamin-dependent diol dehydratase in metabolosomes and metabolizes 1,2-propanediol by disproportionation. J Bacteriol 190:4559–4567. doi: 10.1128/JB.01535-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobsen CN, Rosenfeldt Nielsen V, Hayford AE, Moller PL, Michaelsen KF, Paerregaard A, Sandstrom B, Tvede M, Jakobsen M. 1999. Screening of probiotic activities of forty-seven strains of Lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl Environ Microbiol 65:4949–4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldstein EJ, Tyrrell KL, Citron DM. 2015. Lactobacillus species: taxonomic complexity and controversial susceptibilities. Clin Infect Dis 60(Suppl 2):S98–S107. doi: 10.1093/cid/civ072. [DOI] [PubMed] [Google Scholar]
- 27.Snydman DR, McDermott LA, Jacobus NV, Thorpe C, Stone S, Jenkins SG, Goldstein EJ, Patel R, Forbes BA, Mirrett S, Johnson S, Gerding DN. 2015. U.S.-based national sentinel surveillance study for the epidemiology of Clostridium difficile-associated diarrheal isolates and their susceptibility to fidaxomicin. Antimicrob Agents Chemother 59:6437–6443. doi: 10.1128/AAC.00845-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boonma P, Spinler JK, Venable SF, Versalovic J, Tumwasorn S. 2014. Lactobacillus rhamnosus L34 and Lactobacillus casei L39 suppress Clostridium difficile-induced IL-8 production by colonic epithelial cells. BMC Microbiol 14:177. doi: 10.1186/1471-2180-14-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sears P, Crook DW, Louie TJ, Miller MA, Weiss K. 2012. Fidaxomicin attains high fecal concentrations with minimal plasma concentrations following oral administration in patients with Clostridium difficile infection. Clin Infect Dis 55(Suppl 2):S116–S120. doi: 10.1093/cid/cis337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spinler JK, Sontakke A, Hollister EB, Venable SF, Oh PL, Balderas MA, Saulnier DM, Mistretta TA, Devaraj S, Walter J, Versalovic J, Highlander SK. 2014. From prediction to function using evolutionary genomics: human-specific ecotypes of Lactobacillus reuteri have diverse probiotic functions. Genome Biol Evol 6:1772–1789. doi: 10.1093/gbe/evu137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santos F, Spinler JK, Saulnier DM, Molenaar D, Teusink B, de Vos WM, Versalovic J, Hugenholtz J. 2011. Functional identification in Lactobacillus reuteri of a PocR-like transcription factor regulating glycerol utilization and vitamin B12 synthesis. Microb Cell Fact 10:55. doi: 10.1186/1475-2859-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spinler JK, Taweechotipatr M, Rognerud CL, Ou CN, Tumwasorn S, Versalovic J. 2008. Human-derived probiotic Lactobacillus reuteri demonstrate antimicrobial activities targeting diverse enteric bacterial pathogens. Anaerobe 14:166–171. doi: 10.1016/j.anaerobe.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaefer L, Auchtung TA, Hermans KE, Whitehead D, Borhan B, Britton RA. 2010. The antimicrobial compound reuterin (3-hydroxypropionaldehyde) induces oxidative stress via interaction with thiol groups. Microbiology 156:1589–1599. doi: 10.1099/mic.0.035642-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Talarico TL, Dobrogosz WJ. 1990. Purification and characterization of glycerol dehydratase from Lactobacillus reuteri. Appl Environ Microbiol 56:1195–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson CD, Auchtung JM, Collins J, Britton RA. 2014. Epidemic Clostridium difficile strains demonstrate increased competitive fitness compared to nonepidemic isolates. Infect Immun 82:2815–2825. doi: 10.1128/IAI.01524-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bartlett JG. 1981. Antimicrobial agents implicated in Clostridium difficile toxin-associated diarrhea of colitis. Johns Hopkins Med J Suppl 149:6–9. [PubMed] [Google Scholar]
- 37.Schubert AM, Rogers MA, Ring C, Mogle J, Petrosino JP, Young VB, Aronoff DM, Schloss PD. 2014. Microbiome data distinguish patients with Clostridium difficile infection and non-C. difficile-associated diarrhea from healthy controls. mBio 5:e01021-14. doi: 10.1128/mBio.01021-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biebl H, Menzel K, Zeng AP, Deckwer WD. 1999. Microbial production of 1,3-propanediol. Appl Microbiol Biotechnol 52:289–297. doi: 10.1007/s002530051523. [DOI] [PubMed] [Google Scholar]
- 39.Kolodziej M, Szajewska H. 2017. Lactobacillus reuteri DSM 17938 in the prevention of antibiotic-associated diarrhoea in children: protocol of a randomised controlled trial. BMJ Open 7:e013928. doi: 10.1136/bmjopen-2016-013928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dinleyici EC, Dalgic N, Guven S, Metin O, Yasa O, Kurugol Z, Turel O, Tanir G, Yazar AS, Arica V, Sancar M, Karbuz A, Eren M, Ozen M, Kara A, Vandenplas Y. 2015. Lactobacillus reuteri DSM 17938 shortens acute infectious diarrhea in a pediatric outpatient setting. J Pediatr (Rio J) 91:392–396. doi: 10.1016/j.jped.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 41.Dinleyici EC, Vandenplas Y. 2014. Lactobacillus reuteri DSM 17938 effectively reduces the duration of acute diarrhoea in hospitalised children. Acta Paediatr 103:e300–e305. doi: 10.1111/apa.12617. [DOI] [PubMed] [Google Scholar]
- 42.Shornikova AV, Casas IA, Isolauri E, Mykkanen H, Vesikari T. 1997. Lactobacillus reuteri as a therapeutic agent in acute diarrhea in young children. J Pediatr Gastroenterol Nutr 24:399–404. doi: 10.1097/00005176-199704000-00008. [DOI] [PubMed] [Google Scholar]
- 43.Cimperman L, Bayless G, Best K, Diligente A, Mordarski B, Oster M, Smith M, Vatakis F, Wiese D, Steiber A, Katz J. 2011. A randomized, double-blind, placebo-controlled pilot study of Lactobacillus reuteri ATCC 55730 for the prevention of antibiotic-associated diarrhea in hospitalized adults. J Clin Gastroenterol 45:785–789. doi: 10.1097/MCG.0b013e3182166a42. [DOI] [PubMed] [Google Scholar]
- 44.Tubelius P, Stan V, Zachrisson A. 2005. Increasing work-place healthiness with the probiotic Lactobacillus reuteri: a randomised, double-blind placebo-controlled study. Environ Health 4:25. doi: 10.1186/1476-069X-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cleusix V, Lacroix C, Vollenweider S, Le Blay G. 2008. Glycerol induces reuterin production and decreases Escherichia coli population in an in vitro model of colonic fermentation with immobilized human feces. FEMS Microbiol Ecol 63:56–64. doi: 10.1111/j.1574-6941.2007.00412.x. [DOI] [PubMed] [Google Scholar]
- 46.Vankerckhoven VV, Van Autgaerden T, Geert H, Vancanneyt M, Swings J, Goossens H. 2004. Establishment of the PROSAFE collection of probiotic and human lactic acid bacteria. Microb Ecol Health Dis 16:131–136. doi: 10.1080/08910600410032349. [DOI] [Google Scholar]
- 47.Rosander A, Connolly E, Roos S. 2008. Removal of antibiotic resistance gene-carrying plasmids from Lactobacillus reuteri ATCC 55730 and characterization of the resulting daughter strain, L. reuteri DSM 17938. Appl Environ Microbiol 74:6032–6040. doi: 10.1128/AEM.00991-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cox G, Wright GD. 2013. Intrinsic antibiotic resistance: mechanisms, origins, challenges and solutions. Int J Med Microbiol 303:287–292. doi: 10.1016/j.ijmm.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 49.Klein G, Hallmann C, Casas IA, Abad J, Louwers J, Reuter G. 2000. Exclusion of vanA, vanB and vanC type glycopeptide resistance in strains of Lactobacillus reuteri and Lactobacillus rhamnosus used as probiotics by polymerase chain reaction and hybridization methods. J Appl Microbiol 89:815–824. doi: 10.1046/j.1365-2672.2000.01187.x. [DOI] [PubMed] [Google Scholar]
- 50.van Pijkeren JP, Britton RA. 2012. High efficiency recombineering in lactic acid bacteria. Nucleic Acids Res 40:e76. doi: 10.1093/nar/gks147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ammor MS, Florez AB, Mayo B. 2007. Antibiotic resistance in non-enterococcal lactic acid bacteria and bifidobacteria. Food Microbiol 24:559–570. doi: 10.1016/j.fm.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 52.Klare I, Konstabel C, Werner G, Huys G, Vankerckhoven V, Kahlmeter G, Hildebrandt B, Muller-Bertling S, Witte W, Goossens H. 2007. Antimicrobial susceptibilities of Lactobacillus, Pediococcus and Lactococcus human isolates and cultures intended for probiotic or nutritional use. J Antmicrob Chemother 59:900–912. doi: 10.1093/jac/dkm035. [DOI] [PubMed] [Google Scholar]
- 53.Egervarn M, Danielsen M, Roos S, Lindmark H, Lindgren S. 2007. Antibiotic susceptibility profiles of Lactobacillus reuteri and Lactobacillus fermentum. J Food Prot 70:412–418. doi: 10.4315/0362-028X-70.2.412. [DOI] [PubMed] [Google Scholar]
- 54.Venugopal AA, Johnson S. 2012. Fidaxomicin: a novel macrocyclic antibiotic approved for treatment of Clostridium difficile infection. Clin Infect Dis 54:568–574. doi: 10.1093/cid/cir830. [DOI] [PubMed] [Google Scholar]
- 55.Jaeger KE, Ransac S, Dijkstra BW, Colson C, van Heuvel M, Misset O. 1994. Bacterial lipases. FEMS Microbiol Rev 15:29–63. doi: 10.1111/j.1574-6976.1994.tb00121.x. [DOI] [PubMed] [Google Scholar]
- 56.Kato T, Hayashi Y, Inoue K, Yuasa H. 2005. Glycerol absorption by Na+-dependent carrier-mediated transport in the closed loop of the rat small intestine. Biol Pharm Bull 28:553–555. [DOI] [PubMed] [Google Scholar]
- 57.Fujimoto N, Inoue K, Ohgusu Y, Hayashi Y, Yuasa H. 2007. Enhanced uptake of glycerol by butyrate treatment in HCT-15 human colon cancer cell line. Drug Metab Pharmacokinet 22:195–198. doi: 10.2133/dmpk.22.195. [DOI] [PubMed] [Google Scholar]
- 58.De Weirdt R, Possemiers S, Vermeulen G, Moerdijk-Poortvliet TC, Boschker HT, Verstraete W, Van de Wiele T. 2010. Human faecal microbiota display variable patterns of glycerol metabolism. FEMS Microbiol Ecol 74:601–611. doi: 10.1111/j.1574-6941.2010.00974.x. [DOI] [PubMed] [Google Scholar]
- 59.National Committee for Clinical Laboratory Standards. 2004. Methods for antimicrobial susceptibility testing of anaerobic bacteria; approved standard, 6th ed. National Committee for Clinical Laboratory Standards, Wayne, PA. [Google Scholar]
- 60.Auchtung JM, Robinson CD, Britton RA. 2015. Cultivation of stable, reproducible microbial communities from different fecal donors using minibioreactor arrays (MBRAs). Microbiome 3:42. doi: 10.1186/s40168-015-0106-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koenigsknecht MJ, Theriot CM, Bergin IL, Schumacher CA, Schloss PD, Young VB. 2015. Dynamics and establishment of Clostridium difficile infection in the murine gastrointestinal tract. Infect Immun 83:934–941. doi: 10.1128/IAI.02768-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MR, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG. 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu Z, Morrison M. 2004. Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques 36:808–812. [DOI] [PubMed] [Google Scholar]
- 64.Salonen A, Nikkila J, Jalanka-Tuovinen J, Immonen O, Rajilic-Stojanovic M, Kekkonen RA, Palva A, de Vos WM. 2010. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J Microbiol Methods 81:127–134. doi: 10.1016/j.mimet.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 65.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 66.Savidge TC, Pan WH, Newman P, O'Brien M, Anton PM, Pothoulakis C. 2003. Clostridium difficile toxin B is an inflammatory enterotoxin in human intestine. Gastroenterology 125:413–420. doi: 10.1016/S0016-5085(03)00902-8. [DOI] [PubMed] [Google Scholar]
- 67.Spinler JK, Brown A, Ross CL, Boonma P, Conner ME, Savidge TC. 2016. Administration of probiotic kefir to mice with Clostridium difficile infection exacerbates disease. Anaerobe 40:54–57. doi: 10.1016/j.anaerobe.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Castillo M, Martin-Orue SM, Manzanilla EG, Badiola I, Martin M, Gasa J. 2006. Quantification of total bacteria, enterobacteria and lactobacilli populations in pig digesta by real-time PCR. Vet Microbiol 114:165–170. doi: 10.1016/j.vetmic.2005.11.055. [DOI] [PubMed] [Google Scholar]
- 69.Rinttila T, Kassinen A, Malinen E, Krogius L, Palva A. 2004. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J Appl Microbiol 97:1166–1177. doi: 10.1111/j.1365-2672.2004.02409.x. [DOI] [PubMed] [Google Scholar]
- 70.Aagaard K, Petrosino J, Keitel W, Watson M, Katancik J, Garcia N, Patel S, Cutting M, Madden T, Hamilton H, Harris E, Gevers D, Simone G, McInnes P, Versalovic J. 2013. The Human Microbiome Project strategy for comprehensive sampling of the human microbiome and why it matters. FASEB J 27:1012–1022. doi: 10.1096/fj.12-220806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 72.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Montrose DC, Zhou XK, Kopelovich L, Yantiss RK, Karoly ED, Subbaramaiah K, Dannenberg AJ. 2012. Metabolic profiling, a noninvasive approach for the detection of experimental colorectal neoplasia. Cancer Prev Res 5:1358–1367. doi: 10.1158/1940-6207.CAPR-12-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chumpitazi BP, Hollister EB, Oezguen N, Tsai CM, McMeans AR, Luna RA, Savidge TC, Versalovic J, Shulman RJ. 2014. Gut microbiota influences low fermentable substrate diet efficacy in children with irritable bowel syndrome. Gut Microbes 5:165–175. doi: 10.4161/gmic.27923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Beerenwinkel N, Antal T, Dingli D, Traulsen A, Kinzler KW, Velculescu VE, Vogelstein B, Nowak MA. 2007. Genetic progression and the waiting time to cancer. PLoS Comput Biol 3:e225. doi: 10.1371/journal.pcbi.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reitman ZJ, Jin G, Karoly ED, Spasojevic I, Yang J, Kinzler KW, He Y, Bigner DD, Vogelstein B, Yan H. 2011. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc Natl Acad Sci U S A 108:3270–3275. doi: 10.1073/pnas.1019393108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. 2009. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem 81:6656–6667. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- 78.Goldstein EJ, Citron DM, Tyrrell KL, Merriam CV. 2013. Comparative in vitro activities of SMT19969, a new antimicrobial agent, against Clostridium difficile and 350 Gram-positive and Gram-negative aerobic and anaerobic intestinal flora isolates. Antimicrob Agents Chemother 57:4872–4876. doi: 10.1128/AAC.01136-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dixon P. 2003. VEGAN, a package of R functions for community ecology. J Veg Sci 14:927–930. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.