Abstract
Genetically encoded voltage indicators create an opportunity to monitor electrical activity in defined sets of neurons as they participate in the complex patterns of coordinated electrical activity that underlie nervous system function. Taking full advantage of genetically encoded voltage indicators requires a generalized strategy for targeting the probe to genetically defined populations of cells. To this end, we have generated a mouse line with an optimized hybrid voltage sensor (hVOS) probe within a locus designed for efficient Cre recombinase-dependent expression. Crossing this mouse with Cre drivers generated double transgenics expressing hVOS probe in GABAergic, parvalbumin, and calretinin interneurons, as well as hilar mossy cells, new adult-born neurons, and recently active neurons. In each case, imaging in brain slices from male or female animals revealed electrically evoked optical signals from multiple individual neurons in single trials. These imaging experiments revealed action potentials, dynamic aspects of dendritic integration, and trial-to-trial fluctuations in response latency. The rapid time response of hVOS imaging revealed action potentials with high temporal fidelity, and enabled accurate measurements of spike half-widths characteristic of each cell type. Simultaneous recording of rapid voltage changes in multiple neurons with a common genetic signature offers a powerful approach to the study of neural circuit function and the investigation of how neural networks encode, process, and store information.
SIGNIFICANCE STATEMENT Genetically encoded voltage indicators hold great promise in the study of neural circuitry, but realizing their full potential depends on targeting the sensor to distinct cell types. Here we present a new mouse line that expresses a hybrid optical voltage sensor under the control of Cre recombinase. Crossing this line with Cre drivers generated double-transgenic mice, which express this sensor in targeted cell types. In brain slices from these animals, single-trial hybrid optical voltage sensor recordings revealed voltage changes with submillisecond resolution in multiple neurons simultaneously. This imaging tool will allow for the study of the emergent properties of neural circuits and permit experimental tests of the roles of specific types of neurons in complex circuit activity.
Keywords: Cre-lox, genetically encoded voltage indicators, interneuron, mossy cells, neural circuits, voltage imaging
Introduction
Neural networks function through the collective action of large assemblies of neurons to generate complex forms of electrical activity as they encode and process sensory inputs, interact with other networks, and ultimately generate behaviors. Traditional electrical recording techniques generally fail to reveal the emergent properties of neural networks due to their lack of cell-type specificity and limitations in the number of cells that can be monitored simultaneously. Optical techniques permit the investigation of large numbers of neurons and can thus reveal the higher-order operations of neural networks. Voltage-sensitive dyes enable the imaging of voltage changes in many neurons simultaneously (Salzberg et al., 1977; Cohen and Salzberg, 1978), but synthetic dyes label neurons and glia indiscriminately, and in intact mammalian brain preparations have been limited to the study of population activity. The development of genetically encoded voltage indicators (GEVIs) (Siegel and Isacoff, 1997; Sakai et al., 2001; Ataka and Pieribone, 2002; Knöpfel et al., 2006; Gong et al., 2015; Storace et al., 2016) creates the opportunity to target probes to specific classes of neurons. Not only does this approach enable one to focus on distinct cell types, but the sparser labeling often allows signals from individual cells to be resolved. Despite recent improvements in performance (Gong et al., 2014; St-Pierre et al., 2014), GEVIs have only recently begun to meet the challenge of reporting voltage changes in genetically defined populations (Lou et al., 2016). Thus, researchers have only scratched the surface of the vast potential of GEVI targeting using powerful methods, such as Cre-loxP technology (Madisen et al., 2010, 2012).
Here we present imaging of voltage changes from targeted cell types with high temporal resolution using a hybrid voltage sensor (hVOS) (Chanda et al., 2005). This method uses a membrane-targeted fluorescent protein, which interacts optically with dipicrylamine (DPA), a small hydrophobic anion. Voltage-induced movement of DPA through the membrane alters a Förster resonance energy transfer interaction with the fluorescent protein to produce robust and rapid changes in fluorescence. An optimized hVOS probe with cerulean fluorescent protein tethered to the inner face of the plasma membrane by a truncated h-ras farnesylation motif produces a fractional fluorescence change (ΔF/F) of ∼25% per 100 mV (Wang et al., 2010). This sensitivity is comparable with that of other widely used GEVIs (Storace et al., 2016); but because DPA transits the membrane in only 100 μs (Bradley et al., 2009), the temporal resolution of hVOS is >1 order of magnitude better than GEVIs of comparable brightness and sensitivity. In brain slices, hVOS imaging can detect subthreshold synaptic potentials from many neurons simultaneously in a single trial, and action potentials produce fluorescence changes with a signal-to-noise ratio of ∼≥10 (Ghitani et al., 2015). To extend hVOS imaging to genetically defined cell types, we generated a mouse with an hVOS probe inserted in the ROSA26 locus following a floxed stop cassette, in a vector designed for high-performance Cre recombinase-dependent expression (Madisen et al., 2010, 2012). This Cre reporter mouse (designated Ai35-hVOS) provides a general vehicle for targeting hVOS probe to cells expressing Cre recombinase. Crossing Ai35-hVOS mice with 6 different established Cre driver mice enabled us to image voltage changes in a wide variety of genetically defined subpopulations of cells in slices of hippocampus and cortex. We imaged voltage in GABAergic inhibitory neurons, astrocytes, parvalbumin interneurons, calretinin interneurons, hilar mossy cells, newborn granule cells, and neurons that had been activated by exposure to a novel environment. These experiments included imaging in cells expressing probe under temporal control with tamoxifen-induced Cre recombinase expression. This work illustrates a powerful general approach for studying neural circuits, and for elucidating the mechanisms by which the nervous system generates complex patterns of electrical activity.
Materials and Methods
Cre reporter mouse generation.
Our targeting construct incorporated hVOS1.5 (Addgene: plasmid 45261), an optimized hVOS probe consisting of cerulean fluorescent protein with a truncated h-ras motif appended to the C terminus (Wang et al., 2010), into the Ai35 vector (Madisen et al., 2012) (Addgene: plasmid 34882) built from a Rosa26 locus modified to include a CAG promoter, a floxed stop-cassette, and a woodchuck hepatitis virus post-transcriptional regulatory element to optimize Cre-mediated recombination (Madisen et al., 2010). The hVOS coding sequence was PCR-amplified from hVOS1.5 encoding DNA using a forward primer (5′-TAATACGCGTACCGGTCGCCACCATGGTGAGCAGGGGCGAGGA-3′) and a reverse primer (5′-AGGCACGCGTTCAGGAGAGCACACACTTGC-3′), each containing a MluI restriction site. The Ai35 backbone vector was restriction digested with MluI and dephosphorylated. The purified PCR-amplified hVOS DNA was then digested using MluI and ligated to the MluI-cut Ai35 backbone vector. The hVOS targeting vector was confirmed by sequencing. The vector was then linearized with KpnI enzyme for electroporation into JM8A3 ES cells. ES clones were picked and screened for homologous recombination using PCR and then confirmed by southern blot analysis. ES cells were injected into C57BL/6J blastocysts to generate mice harboring hVOS probe in the ROSA26 locus. Chimeras were bred to uniformity on a C57BL/6J background, resulting in the hVOS Cre reporter mouse line (Ai35-hVOS) used in this study.
Cre drivers.
Cre driver mice for parvalbumin (B6.129P2-Pvalbtm1(cre)Arbr/J, 017320), GAD2 (B6N.Cg-Gad2tm2(cre)Zjh/J, 019022), calbindin 2 (B6(Cg)-Calb2tm2.1(cre/ERT2)Zjh/J, 013730), calcitonin receptor-like receptor (C57BL/6N-Tg(CalCrl,cre)4688Nkza/J, 023014), and Fos (B6.129(Cg)-Fostm1.1(cre/ERT2)Luo/J) were purchased from the The Jackson Laboratory. The Nestin-CreERT2 mouse was described previously (Lagace et al., 2007) and provided by Dr. Amelia Eisch. Female Ai35-hVOS Cre reporter mice were bred with male Cre driver mice, generating double-transgenic Ai35-hVOS::Pvalbtm1(cre)Arbr (simplified as hVOS::Parv), Ai35-hVOS::GAD2tm2(cre)Zjh (simplified as hVOS::GAD2), Ai35-hVOS::Calb2tm2.1(cre/ERT2)Zjh (simplified as hVOS::Calb2), hVOS::Tg(CalCrl,cre)4688Nkza (simplified as hVOS::CalCrl), Ai35-hVOS::Nestin-CreERT2 (simplified as hVOS::Nestin), and Ai35-hVOS::Fostm1.1(cre/ERT2)Luo (simplified as hVOS::Fos) mice.
Nestin-CreERT2 and Calb2-CreERT2 mice are tamoxifen-inducible Cre drivers. Offspring from these animals were injected 5 times on 5 consecutive days with tamoxifen (100–160 mg/kg i.p. injections 30 mg/ml in 10% EtOH mixed with sunflower oil). Experiments were performed 7–28 d following the last injection. All mice were genotyped by standard protocols.
Novel environment.
hVOS::Fos-ERT2 is also tamoxifen-inducible, and Fos is an immediate-early gene that becomes active when a neuron is electrically active (Sheng and Greenberg, 1990). We used an established novel environment protocol to activate the Fos gene in hippocampal neurons (Guenthner et al., 2013). hVOS::Fos-ERT2 mice were transferred to a cage in a dimly lit room (<2 lux) containing an exercise wheel, a plastic tunnel, wood chips, and buried food. After 1 h, mice were injected with 100–160 mg/kg tamoxifen and returned to this cage for an additional hour. Mice were then returned to their home cage and slices were harvested 1 week later. Mice not exposed to a novel environment showed no probe expression in hippocampal slices.
Slice preparation.
Mice were anesthetized by isoflurane inhalation and killed by cervical dislocation in accordance with the recommendations of the Panel of Euthanasia of the Veterinary Medical Association and the United States Public Health Service policy on the humane care and use of laboratory animals. All protocols were approved by the Animal Care and Use Committee at the University of Wisconsin, Madison. Brains were quickly removed and placed in ice-cold cutting solution (composition, in mm as follows: 124 NaCl, 3.2 KCl, 26 NaHCO3, 1.25 NaH2PO4, 1 CaCl2, 6 MgSO4, and 10 glucose), bubbled with 95% O2/5% CO2. Brains were blocked, and 300 μm slices cut in either the coronal or horizontal plane using a Leica VT1200S tissue slicer. Slices were transferred to a recovery chamber containing ACSF (same composition as cutting solution but with 2.5 mm CaCl2 and 1.3 mm MgSO4).
Voltage imaging.
DPA was added to the slice recovery chamber (4 μm), and slices were stained for ∼45 min at room temperature (21°C–24°C) before imaging. Slices were transferred to a submerged recording chamber perfused continuously with 95% O2/5% CO2-bubbled ACSF and 4 μm DPA at room temperature. Slices were viewed using a BX51 microscope (Olympus) with infrared differential interference contrast optics and a C2400 CCD camera (Hamamatsu). Fluorescence illumination was provided by a 435 nm 29 W LED light source (Prizmatix) filtered through a 435 ± 5 nm bandpass filter (Chroma). Images were acquired using a CCD-SMQ camera (RedShirt Imaging) with images collected at 2 kHz with a spatial resolution of 80 × 80. Some images were acquired at 5 kHz at 16 × 16 resolution. Images were read into a Windows-based PC running Neuroplex version 9.0 (software provided with the camera).
Electrophysiology.
Extracellular stimulation was applied with ACSF-filled electrodes fabricated from KG-33 glass capillaries (King Precision Glass) with 3–10 μm tips. Single pulses (50–300 μA, 180 μs) were applied using a stimulus isolator (model A365; WPI) controlled through Neuroplex. The microscope was on a translation stage that allowed movement of the preparation after electrode placement, so that we could routinely stimulate outside the field of view.
Patch-clamp electrodes were fabricated using a PC-10 vertical puller (Narishige) from KG-33 glass capillaries and had resistances of 3–5 mΩ when filled with intracellular solution consisting of 130 mm K-gluconate, 10 mm HEPES, 7 mm KCl, 1 mm EGTA, 2 mm Na2ATP, and 2 mm MgATP, pH 7.2. Neurons expressing hVOS probe were visualized under fluorescence, and patch pipettes were positioned with a Sutter MP225 motorized micromanipulator (Sutter Instruments) with visual guidance under infrared differential interference contrast optics. After establishing whole-cell recordings, current or voltage was recorded by an Axopatch 200B amplifier, digitized with a Digidata 1200 interface, and acquired onto a Windows-based PC running pCLAMP 9 (Molecular Devices).
Immunohistochemistry and morphological processing.
Brain sections were processed as described previously (Gao et al., 2017). Briefly, adult mice were transcardially perfused with saline and then 4% PFA. Brains were removed, postfixed overnight in 4% PFA, and then equilibrated in 30% sucrose. Coronal brain sections (40 μm) were prepared using a sliding microtome and stored at −20°C as floating sections in 96-well plates filled with cryoprotectant solution (glycerol, ethylene glycol, and 0.1 m phosphate buffer, pH 7.4, 1:1:2 by volume). For immunohistochemical analysis, floating brain sections were blocked in TBS containing 3% normal serum and 0.25% Triton X-100. As primary antibodies, we used rabbit anti-parvalbumin antibody (1:1000, Abcam, ab11427), mouse anti-GFAP (1:1000, Millipore, MAB360), chicken anti-GFP (1:1000, Invitrogen, A10262), mouse anti-GAD67 (GAD2) (1:800, Millipore, MAB5406), mouse anti-calretinin (Calb2) (1:500, Millipore, MAB1568), and chicken anti-Nestin (1:500, Aves, NES0407). For fluorescent secondary antibodies, we used goat anti-rabbit AlexaFluor-568 (1:1000, Invitrogen, A11036), goat anti-chicken AlexaFluor-488 (1:1000, Invitrogen, A11039), and goat anti-mouse AlexaFluor-568 (1:1000, Invitrogen, A11031). For GAD67 staining only, we used a detergent-free solution and incubated with primary antibody at room temperature for 2 d (Taniguchi et al., 2011). After staining, all sections were counterstained with DAPI, mounted, coverslipped, and maintained at 4°C in the dark until analysis.
To evaluate hVOS expression and determine colocalization with immunohistochemically labeled cells, images were acquired using an Ultima II 2-photon microscope (Bruker) illuminated by a Chameleon Ti-Sapphire laser (Coherent) tuned to 860 nm. Images were captured using a 20× Olympus XLUMPlanFLN water-immersion objective (NA 1.0). Emitted light was filtered at 525 ± 35 nm for hVOS probe and at 607 ± 45 nm (Chroma Technology) for Alexa-568.
Data processing and analysis.
Voltage imaging data were analyzed using Neuroplex. Regions of interest (ROIs) encompassing individual neurons were selected for spatial averaging. Signals were divided by the resting light intensity and low-pass filtered at 600 Hz. Slow decays in fluorescence were subtracted in Origin 8.6 (Origin Labs) by fitting to a fourth-order polynomial. Spike amplitude, width, and time to half-peak amplitude were determined in Igor Pro 6 using a macro that enabled manual selection of spike onset and repolarization, with baseline calculated as the average of the preceding 80 points.
Patch-clamp records were analyzed in Clampfit 9 (Molecular Devices), and traces were exported as Axon text files. Individual traces were smoothed using an unweighted sliding-average algorithm (m = 3) using Volumetry version G6a (provided by Grant W. Hennig).
Difference maps were constructed from 80 × 80 raw images acquired by the CCD-SMQ camera. Data were temporally low-pass filtered at 300 Hz and inverted and encoded as brightness, with inversion to make positive-going voltage changes appear as bright spots. Images were spatially filtered using a 3 × 3 center-weighed filter. Maps represent the difference between the maximal response in the 10 ms interval following the stimulus and the average fluorescence of the 40 ms interval preceding the stimulus. Two-photon images and image stacks were processed using ImageJ 1.43u (National Institutes of Health).
Results
hVOS probe expression
Crossing our Ai35-hVOS Cre reporter mouse with various Cre drivers produced offspring that were normal in weight, appearance, and behavior. In offspring harboring Cre and hVOS genes, the targeted cell type expressed Cre recombinase, which removed the stop codon in front of the hVOS locus to initiate expression in the intended cell type (Fig. 1, top). For example, crossing Ai35-hVOS with a parvalbumin (Parv) Cre driver, which expresses Cre recombinase in inhibitory interneurons defined by the Ca2+-binding protein Parv, produced hVOS::Parv mice expressing hVOS probe in Parv interneurons. Cortical slices from these animals showed sparse probe expression in individual cells in layers 2/3 and 5 (Fig. 1A). Labeled cell bodies are surrounded by extensive axonal arbors characteristic of Parv interneurons. Parv-expressing interneurons in the neocortex include basket cells and chandelier cells (DeFelipe et al., 1989; Wang et al., 2002; Markram et al., 2004), and the observed locations of the hVOS-positive cells correspond well with the established distribution of these interneurons. Immunohistochemical staining against Parv revealed hVOS expression with 99.2% specificity (defined as the number of hVOS-expressing cells that coexpress an immunohistochemical marker; 378 total cells; Fig. 2A).
Figure 1.
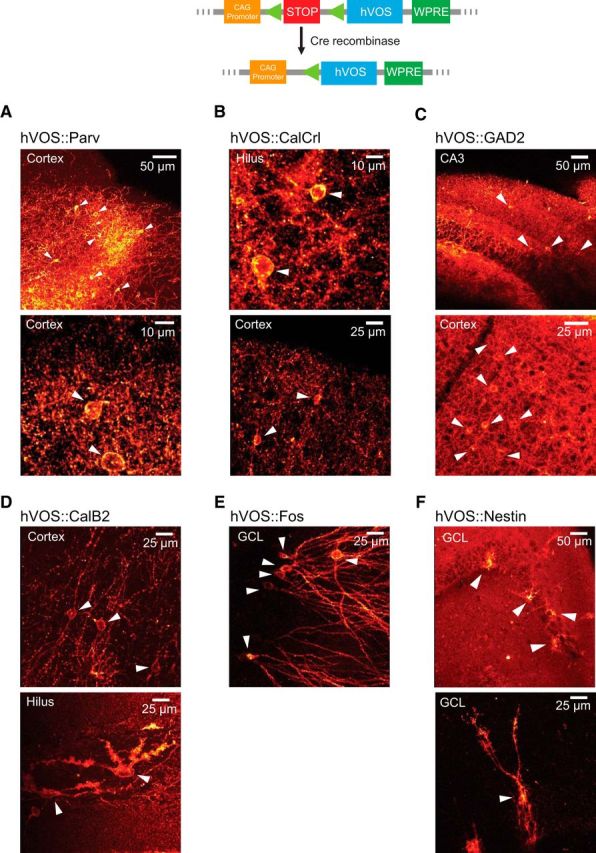
hVOS probe expression in Cre-targeted neurons. Top diagram, The general approach of Cre recombinase-dependent expression; Cre recombinase excises a stop cassette after the CAG promoter to enable hVOS probe transcription (probe expression is enhanced by the downstream woodchuck hepatitis virus post-transcriptional regulatory element) (Madisen et al., 2010, 2012). A–F, Probe fluorescence in brain sections from mice obtained by crossing Ai35-hVOS mice with various Cre drivers. A, hVOS::Parv double-transgenic mice express probe in layers 2/3 and 5 of the somatosensory cortex. White arrowheads indicate probe-expressing cells. B, hVOS::CalCrl mice express probe in hilar mossy cells of the DG (top) and in interneurons of layer 2/3 of the somatosensory cortex (bottom). C, hVOS::GAD2 mice express probe in the CA3 region of the hippocampus (top) and inner molecular layer of the DG (bottom). D, hVOS::Calb2 mice express probe in interneurons in layer 5 of the somatosensory cortex (top) and in a hilar mossy cell in the DG (bottom). The image was enhanced by GFP immunofluorescence to show the finer structure (thorny excrescences) of mossy cell dendrites. E, hVOS::Fos mice exposed to a novel environment express probe in neurons scattered through the granule cell layer of the DG. F, hVOS::Nestin mice express probe in newborn neurons in the granule cell layer of the DG.
Figure 2.
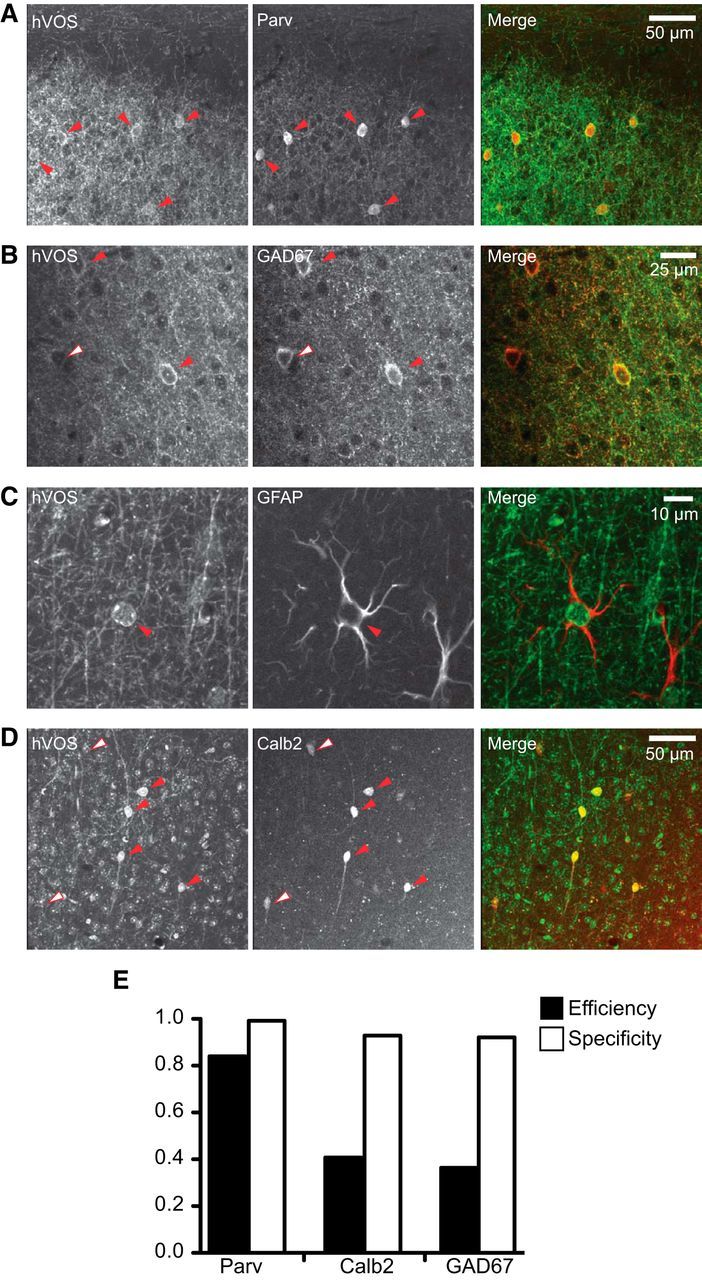
hVOS probe coexpression with the targeted gene product in the relevant Cre driver. A, Two-photon image from the somatosensory cortex (layer 2/3) of an hVOS::Parv mouse. hVOS probe expression, augmented by GFP immunofluorescence (left), immunohistochemical staining for Parv (middle), and merged image (right). B, hVOS probe expression in a section from an hVOS::GAD2 mouse (left), GAD67 immunofluorescence (middle), and merged (right) in layer 5 of the somatosensory cortex. C, Coexpression of hVOS probe (left) and GFAP (right) in the hippocampal CA1 region of an hVOS::GAD2 mouse. D, hVOS probe expression in a section from an hVOS::Calb2 mouse (left, GFP-augmented), immunofluorescence for Calb2 (calretinin) (middle), and merged (right) from layer 2/3 in the somatosensory cortex. E, Specificity (hVOS probe expressing cells that coexpress marker) and efficiency (marker expressing cells that express hVOS probe) for Parv, Calb2, and GAD67.
Glutamic acid decarboxylase (GAD) broadly defines GABAergic inhibitory neurons. hVOS::GAD2 mice were found to express hVOS probe throughout the somatosensory cortex and hippocampus (Fig. 1C). Immunostaining with an antibody against GAD67 (the GAD2 gene product) revealed 92.1% specificity (of 178 cells) in hVOS::GAD2 mice (Fig. 2B). Previous reports had shown that the GAD2 Cre driver also directs expression to a small percentage of astrocytes (Taniguchi et al., 2011), consistent with reports that astrocytes contain and release GABA (Yoon and Lee, 2014). Colabeling with a GFAP antibody revealed a small percentage (15.4%) of hVOS-expressing cells (of 188 total) that were positive for GFAP (Fig. 2C). Crosses with other Cre drivers produced analogous results. The CalCrl Cre driver was generated using the calcitonin receptor-like receptor promoter, which in the hippocampus directs expression to hilar mossy cells in the dentate gyrus (DG) (Jinde et al., 2012). hVOS::CalCrl mice were found to express hVOS probe in large neurons in the hilus with multiple mossy dendrites, as expected (Fig. 1B, top). We also observed sparser expression through various layers in the cortex (Fig. 1B, bottom), which was not described in the initial report of CalCrl targeting (Jinde et al., 2012).
Ai35-hVOS offspring also expressed probe conditionally with Cre recombinase on a modified estrogen receptor promoter (ERT2) that could be activated by tamoxifen. Calbindin 2 (Calb2, also known as calretinin) is a Ca2+-binding protein expressed in subpopulations of interneurons in the cortex as well as in rodent hilar mossy cells (Gulyás et al., 1992; Rogers, 1992). In slices from hVOS::Calb2-CreERT2 mice harvested following tamoxifen injection, we observed hVOS probe in cells in layers 2/3 and layer 5 of somatosensory cortex, as well as in large neurons in the hilus with mossy dendrites (Fig. 1D). Immunostaining for calretinin in sections from hVOS::Calb2-CreERT2 mice showed a high specificity of hVOS expression (92.9%, 196 cells; Fig. 2D). Fos, the protein encoded by the early proto-oncogene c-Fos, serves as a marker for recent electrical activity (Dragunow and Robertson, 1987; Hunt et al., 1987; Morgan et al., 1987), and studies have shown that it is expressed in granule cells of the hippocampus when an animal is placed in a novel environment (Guenthner et al., 2013). One week after exposure to a novel environment (see Materials and Methods), mice expressed hVOS probe in a number of granule cells of the DG (Fig. 1E), whereas granule cells in animals not exposed to a novel environment showed no labeling (data not shown). We also labeled DG granule cells with a Nestin Cre driver. Nestin, an intermediate filament protein expressed in neuroblasts and immature neurons (Bonaguidi et al., 2012), serves as a marker for neural stem and progenitor cells during adult neurogenesis. The DG generates newborn neurons throughout adult life (Cameron and McKay, 2001; Fukuda et al., 2003); and in hippocampal slices from hVOS::Nestin-CreERT2 mice 28 d after tamoxifen injections, hVOS probe-expressing cells were observed scattered through the granule cell layer (Fig. 1F).
In summary, we observed appropriate hVOS probe expression in each of the double transgenics generated by crossing various Cre drivers with our Ai35-hVOS Cre reporter mouse. Probe colocalized with high specificity with the product of the endogenous gene used to create the Cre driver. Expression patterns showed general agreement with the Allen Brain Atlas, with labeled cells of the expected morphology in the expected brain region and layer.
Simultaneous hVOS imaging and voltage recording
To test the ability of hVOS imaging to report voltage changes in Cre-targeted cells, we performed simultaneous whole-cell patch-clamp recording and imaging in slices of somatosensory cortex. Brief current pulses (200 μs, 5 nA) evoked action potentials, and these action potentials were accompanied by clear hVOS signals. Action potentials were clear in single trials in patch-clamped neocortical neurons from hVOS::Parv mice (Fig. 3A), hVOS::Calb2 mice (Fig. 3B), and hVOS::GAD2 mice (Fig. 2C). In each case, the optical response was synchronous with the action potential. In the recording from the hVOS::Parv neuron (Fig. 3A), we showed additional traces from two neighboring cells in which the fluorescence did not change. Because Parv neurons are inhibitory, neighboring cells will not be depolarized even if they are synaptically coupled. Thus, the voltage change in the patch-clamped neuron does not produce stray fluorescence changes in neighboring cells. This indicates that imaging with this expression system provides single-cell resolution.
Figure 3.
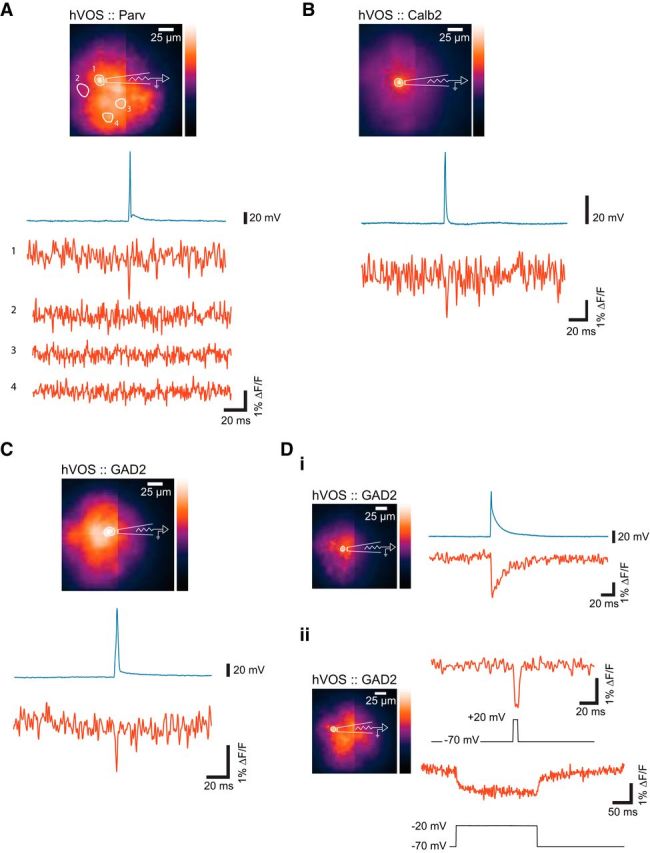
Simultaneous imaging and electrical recording, with resting light images, whole-cell patch-clamp recordings (above), and hVOS responses (below). Action potentials were evoked by current injection under current clamp. A, Action potential and hVOS signal from a neuron in cortical layer 2/3 of a slice from an hVOS::Parv mouse. No fluorescence changes were seen in 3 other nearby cells numbered 2–4. B, Action potential and hVOS signal from a neuron in cortical layer 5 from an hVOS::Calb2 mouse. C, Action potential and hVOS signal from a layer 2/3 cortical neuron from an hVOS::GAD2 mouse. Di, hVOS and voltage responses to current injection in a GAD2 glial cell in layer 5 of the somatosensory cortex of an hVOS::GAD2 mouse. The cell was held at −30 mV. Dii, Glial cell hVOS responses to voltage steps (under voltage clamp) from −70 to 20 mV (5 ms, top) and −70 to −20 mV (200 ms, bottom traces) in layer 5 of the somatosensory cortex of an hVOS::GAD2 mouse. A–C, All hVOS recordings of neuronal responses are single trials. Glial responses are 5 trial averages, due to the significantly smaller signal amplitudes.
As noted above, the GAD2 Cre driver directs expression in a small percentage of astrocytes in the cortex (Fig. 1C) (Taniguchi et al., 2011). We patch-clamped several cells in slices from hVOS::GAD2 animals that were not excitable. These cells never spiked but responded to current injection with small, rapid depolarizations, followed by prolonged repolarizations (Fig. 3Di). These voltage changes are consistent with the presence of delayed rectifier K+ current in astrocytes (Sontheimer and Waxman, 1993), and the hVOS signals tracked these voltage changes with high temporal fidelity. Under voltage clamp, stepping from −70 to 20 mV for 5 ms elicited an hVOS signal with a ΔF/F amplitude of ∼1.5% (Fig. 3Dii, top). A longer voltage step (200 ms) from −70 mV to −20 mV produced a longer, steady hVOS signal with a lower amplitude (Fig. 3Dii, bottom), consistent with the approximately linear relationship between voltage and hVOS signals in this range of voltage (Chanda et al., 2005; Wang et al., 2010).
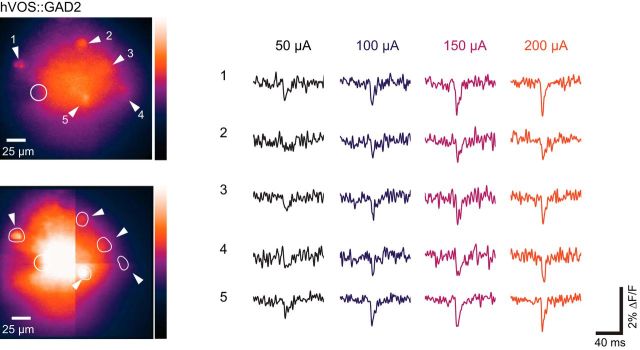
Responses to afferent activation
To evaluate the ability of Cre-targeted probe to report depolarizations from multiple neurons, we selected fields of view with several labeled cells and activated synaptic inputs using extracellular electrical stimulation. We observed electrically evoked responses in multiple neurons in all six double transgenics tested in this study. Figure 4A–G presents fields of view of various slices containing 3–12 cells from which evoked responses were recorded in a single trial. In these images, the plane of focus could be varied and labeled cell bodies could be recognized. The plane of focus selected for imaging in each of the experiments in Figure 4 had some cells in focus, but other cells were out of focus to varying degrees. The ROIs displayed contain neurons that we located by changing the focus. We also presented traces from locations that did not appear to contain a neuron (Fig. 4B,D,E, blue traces). One of the displayed traces in Figure 4B shows a small fluorescence change that probably reflects voltage changes in processes. This suggests that labeled processes make a significant contribution to the diffuse fluorescence seen in many of our images. The responses of most neurons were spike-like and resembled the action potentials evoked by current injection in patch-clamped cells (Fig. 3). Interestingly, CalCrl cells in the hilus (mossy cells) spiked (Fig. 4E), but CalCrl cells in the somatosensory cortex responded to extracellular stimulation with rapidly rising, slowly decaying EPSPs that were easily distinguished from spikes (Fig. 4F). Figure 4G presents responses in the granule cell layer of the DG of an hVOS::Fos mouse that had been exposed to a novel environment following tamoxifen injection. Twelve labeled cells responded to the stimulus, as illustrated with the fluorescence traces from the corresponding ROIs.
Figure 4.
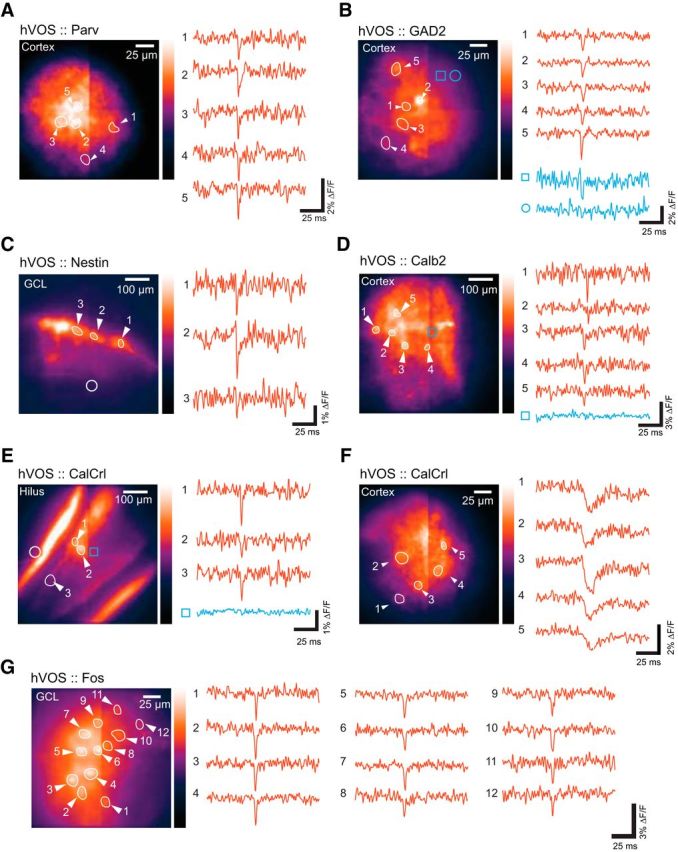
Simultaneous single-trial recordings from multiple neurons. Left, Resting light image. Right, hVOS traces. Selected cells were numbered to correspond with the traces and the ROIs for each cell were outlined. A, hVOS responses in five different parvalbumin interneurons in layer 2/3 of the somatosensory cortex. Stimulus site is just dorsal to (below) the field of view. B, hVOS responses in five interneurons of layer 5 in the somatosensory cortex of an hVOS::GAD2 mouse. Blue traces from two additional ROIs represent that the background fluorescence produces no fluorescence change (circle) or a small fluorescence change (square). The small fluorescence change likely arises from voltage changes in processes from that location. Stimulus site is dorsal to (above) the field of view. C, Newborn granule cells in the DG of an hVOS::Nestin mouse responding to outer molecular layer stimulation (site indicated by open circle). D, hVOS responses in five neurons in layer 2/3 of the entorhinal cortex of an hVOS::Calb2 mouse. A trace from a region without a labeled neuron (blue) shows no fluorescence change. Stimulus site is caudal to the field of view (left). E, hVOS responses in three neurons from the hilus in the DG of an hVOS::CalCrl mouse. A trace from a region without a labeled neuron (blue) shows no fluorescence change. Stimulus site is in the middle molecular layer, indicated by an open circle. The intense fluorescence in the inner molecular layer arises from mossy cell axons. F, hVOS responses in five neurons from layer 2/3 the somatosensory cortex of an hVOS::CalCrl mouse. Stimulation in layer 2/3 (left) evoked EPSPs. G, hVOS responses in 12 neurons in the DG of a hippocampal slice from an hVOS::Fos mouse expressing hVOS probe in granule cells. Stimulation was applied to the outer molecular layer. The mouse had been placed in a novel environment as described in Materials and Methods. These neurons were presumably active in that environment. All traces are single trials; stimulus: 200 μA, 180 μs.
In slices containing multiple labeled neurons, the responsive cells can be highlighted by creating a spatial map of the maximal signal (the peak response) minus the resting light intensity (RLI), as described in Data processing and analysis. An example of such a map from an hVOS::Parv cortical slice (Fig. 5A) shows a number of foci of activity corresponding spatially to the neurons that fired in response to the stimulus. In this experiment, we were able to locate 7 neurons that responded. Additional bright spots in the map may be other cells that were difficult to discern or responses in processes as in one of the traces in Figure 4B. Figure 5B presents resting light and difference maps for the hVOS::Fos experiment from which Figure 4G was prepared, showing the 12 responsive cells as bright spots in the difference map. Figure 5C presents the RLI and difference maps for an entorhinal cortex slice from an hVOS::Calb2 mouse, with 5 responsive cells highlighted. The three response maps in Figure 5A–C all contain holes. These dark spots are locations where fluorescence changes are conspicuously absent and indicate the somata of cells not responding to the particular stimulus applied. The diffuse responses in the difference maps are likely to be voltage changes both in cells that are difficult to discern and in processes of responsive cells.
Figure 5.
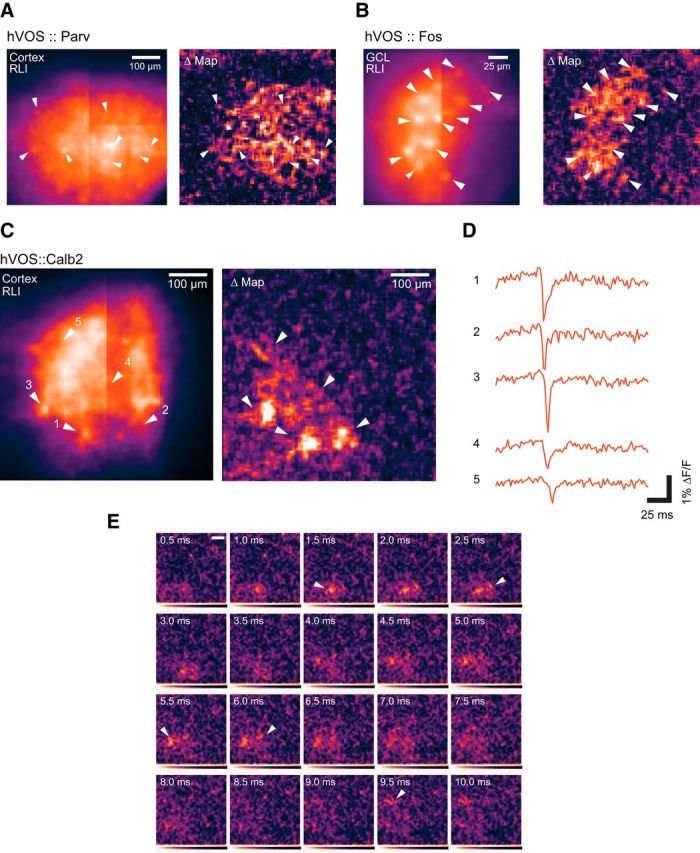
Network response to extracellular stimulation illustrated with spatial maps of peak response. A, A cortical slice from an hVOS::Parv mouse with RLI (left) illustrating probe fluorescence. The difference map of responses (Δ, right), with fluorescence inverted to render responses as brightness increases, was prepared as described in Data processing and analysis. Seven labeled cells where response peaks were evident are highlighted with arrowheads in the two maps. Stimulus was applied to layer 2/3 above the field of view. B, The DG slice from the hVOS::Fos experiment of Figure 4G was used to prepare a maximal response map with the same procedure used to prepare the map in A for the hVOS::Parv slice. The 12 responsive cells highlighted in Figure 4G produced foci in the maximal response map and are highlighted with arrowheads. C, In a slice of medial entorhinal cortex from an hVOS::Calb2 mouse, a stimulus was applied in layer 2/3 outside the field of view below and to the left. An RLI image (left) in layer 2/3 illustrates the distribution of hVOS probe. A difference map of the peak signal minus resting light (Δ, right) illustrates the spatial distribution of responses. Arrowheads indicate 5 responsive cells. D, Fluorescence versus time from ROIs containing the five neurons indicated in A show differences in response latency. E, Intensity maps at 500 μs intervals reveal the sequence of activation of neurons following stimulation. Arrowheads indicate the peak response of each cell in the frame where the peak occurs. The neurons are activated in the sequence of their labels. Displayed data were from five trial averages. The stimulus was 200 μA, 180 μs in all experiments.
When the intensity maps are presented as a succession over time rather than using the response maximum, these experiments revealed the spread of activity through the network as individual neurons respond in succession. The fluorescence traces from the ROIs containing responsive neurons (Fig. 5D) reveal differences in response latency as activity propagates through the network. This propagation can be visualized directly in a sequence of images with fluorescence intensity encoded as brightness (Fig. 5E). One neuron in the lower left corner of the field of view responded first, and the other neurons responded over the next several milliseconds, with the last neuron of the group responding 9.5 ms after stimulation and 8 ms after the first neuron.
Trial-to-trial variation
Although averaging multiple trials is common in GEVI imaging, this practice obscures trial-to-trial variations. Single-trial hVOS responses from a Calb2 neuron revealed latencies that varied by as much as 3 ms for the same stimulation current (Fig. 6A). As a result, the average of these trials appeared much smaller and broader than any of the individual trials. hVOS responses from another Calb2 neuron showed large variations in amplitude and frequent failures (Fig. 6B). Again, the average had a smaller amplitude and longer duration. In Figure 6B, only one of the single-trial stimulations resulted in a spike (red trace), one may have shown a small response (blue trace), and the other three failed to generate a response (black traces). The averaged response thus reported only a minimal (∼1% ΔF/F) spike, compared with the larger (∼4.5% ΔF/F) response seen in the one trial where the spike occurred.
Figure 6.
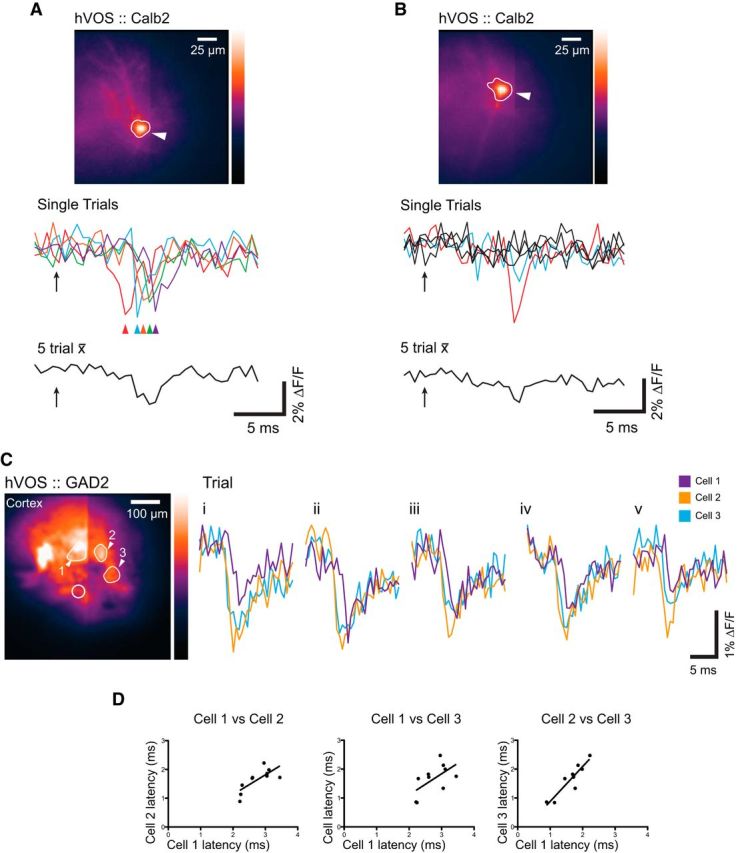
Single-trial recordings reveal trial-to-trial variations. A, Responses of a Calb2 cell in layer 2/3 of the somatosensory cortex. Five separate trials (multiple-colored traces) were taken at 10 s intervals. Colored arrows indicate the time of the observed spike peak in each trial. The 5 trial average is shown below. Arrow indicates stimulus time. The stimulus electrode was positioned above and left, just outside the field of view in layer 2/3. B, Another Calb2 cell in layer 2/3 of the somatosensory cortex clearly spiked in only 1 of 5 trials (red, blue, and black traces). The 5 trial average is shown below. A stimulus of 200 μA, 180 μs (at the arrow) was applied in layer 2/3, above and outside the field of view. C, Single-trial responses of 3 GAD neurons (indicated in the image by arrowheads and numbers) in the somatosensory cortex show variations in response latency. Colored traces represent five single-trial spikes from cells 1–3. Open circle represents site of stimulation. D, Response latencies (times from stimulus to half-peak amplitude) in 10 successive single-trial recordings from the three neurons in C, plotted against each other to evaluate correlations. Solid lines indicate the linear regression fits to the data.
The trial-to-trial variations in latency offer the possibility of exploring response timing across a circuit, and establishing correlations in the firing behavior of individual neurons. We analyzed the latency from stimulus to the time a response was half-maximal in three neurons from layer 2/3 of the somatosensory cortex of an hVOS::GAD2 mouse (Fig. 6C). All three neurons responded in each trial. Neuron 1 (Fig. 6C, purple traces) consistently responded later than neuron 2 (Fig. 6C, orange traces), whereas neuron 3 had a latency close to that of neuron 2, but with some jitter in the difference (Fig. 6C, blue traces). Plotting the individual latencies of pairs of the three neurons over 10 trials enabled us to quantify these relations (Fig. 6D). Linear regression of latency plots revealed that neurons 2 and 3 were highly correlated (p = 0.0002), neurons 1 and 2 were significantly correlated (p = 0.013), and neurons 1 and 3 may be weakly correlated, with p = 0.067 slightly above the common threshold for statistical significance. The very tight correlation between neurons 2 and 3 suggests that these cells are part of a functional unit of circuitry. By contrast, the weaker correlations between the other pairs of neurons suggest that these cells belong to different functional units of interneuron circuitry. Analyzing latencies at this level provides an approach that identifies correlations arising from circuit architecture, as distinguished from intrinsic fluctuations in the synaptic delay (Barrett and Stevens, 1972). The sequence of events that contributes to an observed response latency will vary in a complex manner that depends on stimulus current and location. When one particular event that contributes to the latency in two different neurons varies, we will expect that the latencies of their responses will covary. Correlations can thus provide clues about common and distinct elements of the circuitry that neurons within a field of view may share. These data suggest that multicell recording with high temporal resolution provides a window into unexplored aspects of connectivity within networks.
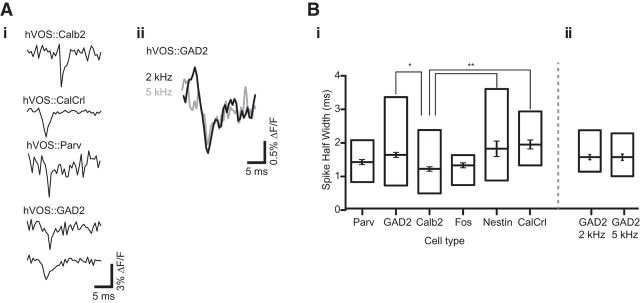
Cell-type-specific spike properties
Spike-like responses had amplitudes (ΔF/F) in the range of 1.5% to 2.5% (Table 1). Calb2 and Fos neurons had the largest changes in fluorescence. The amplitudes of spike-like responses to extracellular stimulation were generally similar to the amplitudes of spikes evoked by current injection in simultaneous whole-cell recordings (Fig. 3), where we could confirm that a voltage change was associated with an action potential. In previous experiments in which the same probe was expressed by in utero electroporation or from the pan-neuronal thy-1 promoter, action potentials were accompanied by slightly larger fluorescence changes (2.5%–3.5%) (Ghitani et al., 2015). Probe expression with viral vector has also been found to result in better overall GEVI performance compared with Cre-loxP-based expression (Lou et al., 2016); and as noted in the previous report, these differences probably reflect higher levels of probe expression to provide more signal relative to background fluorescence.
Table 1.
Fractional fluorescence changes during action potentials, normalized to resting light intensity (ΔF/F)a
| Mouse line | Amplitude (ΔF/F × 100) | Animals/slices/cells |
|---|---|---|
| hVOS::Parv | 2.4 ± 0.15 | 4/7/20 |
| hVOS::GAD2 | 1.99 ± 0.09 | 5/11/40 |
| hVOS::CalClr | 1.6 ± 0.19 | 3/5/11 |
| hVOS::Calb2 | 2.69 ± 0.15 | 8/13/39 |
| hVOS::Nestin | 2.09 ± 0.23 | 4/8/13 |
| hVOS::Fos | 2.61 ± 0.16 | 3/4/10 |
SEM computed using cell numbers as N. Spike half-widths presented in Figure 7.
The different targeted cell types generated optical spikes with distinct dynamic properties. The half-widths of spikes recorded in single trials were largely in agreement with previously reported values based on intracellular recording. The fastest spikes were observed in Calb2 neurons (Fig. 7Ai, top; mean half-width: 1.23 ± 0.08 ms). The spikes of Parv interneurons were almost as fast (Fig. 7Ai, third trace; mean half-width: 1.43 ± 0.09 ms). By contrast, hippocampal CalCrl neurons (Fig. 7Ai, second trace) and Nestin neurons had significantly slower spikes (1.95 ± 0.14 ms and 1.83 ± 0.24 ms, respectively). GAD2 neurons were also found to have relatively slow spikes (1.64 ± 0.09 ms); but as GAD2 is a pan-interneuron marker, we found both fast-spiking and slow-spiking neurons (Fig. 7Ai, bottom two traces). The mean spike half-widths of each cell type are displayed in Figure 7B, along with the ranges (box). The ranges from the hVOS::Nestin and hVOS::GAD2 lines were clearly broader than from the hVOS::Parv, hVOS::Calb2, and hVOS::Fos lines. As Nestin-positive newborn neurons have been shown to have slower action potentials (Cabezas et al., 2013; Pedroni et al., 2014), this range likely represents neurons in different states of maturation. Fos and Nestin both target granule cells, and the differences can be attributed to differences in neuronal age.
Figure 7.
Spike characteristics in different Cre-targeted cell types. Ai, Single-trial spikes from a cortical Calb2 neuron, a CalCrl mossy cell in the hilus, a Parv neuron, and two different GAD2 neurons from the somatosensory cortex. Aii, Successive recordings of a spike from a GAD2 neuron with acquisition at 2 kHz (black trace) and 5 kHz (gray trace). Bi, Half-widths of spikes from different cell types. Boxes represent the ranges. Error bars indicate SE. One-way ANOVA indicated significant variation within the group (F = 5.32, p = 0.0002), and a Tukey post hoc test revealed significant differences (p < 0.05) between the GAD2 and Calb2 neuron spikes, the Nestin and Calb2 neuron spikes, and the CalCrl and Calb2 neuron spikes. Numbers of animals, slices, and spikes are shown in Table 1 for each type of neuron. Bii, Half-widths of GAD2 cortical neuron spikes recorded at 2 kHz and at 5 kHz. Paired t-test indicated that the values were indistinguishable (p = 0.98), thus indicating that our acquisition rate was adequate for accurate spike width determination.
For most of our spike recordings, we acquired images at 2 kHz. To evaluate spike half-width values at higher temporal resolution, we imaged hVOS::GAD2 cortical neurons from the same field of view at frame acquisition rates of both 2 and 5 kHz. The faster acquisition mode required reducing spatial resolution from 80 × 80 to 26 × 26, and yielded higher noise levels, but spikes from the same neurons appeared to have similar dynamics (Fig. 7Aii). Averaging responses in 16 neurons from 3 animals showed the spike half-widths to be indistinguishable when recording at either 2 or 5 kHz (Fig. 7Bii, p = 0.97). Thus, acquiring hVOS images at a frame rate of 2 kHz provides adequate temporal resolution for accurate determination of spike half-width. These measurements of spike duration illustrate a significant advantage of hVOS probes. The GEVI QuasAr2 reported patterns of ongoing electrical activity characteristic of different types of neurons; but because of the ∼2 ms response time of this probe, half-widths of spikes in fluorescence were longer than in direct electrical recordings from fast-spiking interneurons (Lou et al., 2016).
Graded responses
With low stimulus currents, the response amplitude varied with stimulus intensity. To see the smaller responses clearly at low stimulus current, we averaged 5 trials. In layer 2/3 in the somatosensory cortex of an hVOS::GAD2 mouse, we observed 5 GAD2 interneurons (Fig. 8, left). Stimulation with 50 μA evoked small, but visible, responses with rapid rises and slow decays (similar to responses shown in Fig. 4F from cortical CalCrl cells). Increasing the stimulus current increased the response amplitude; and in some cells, the larger amplitude responses had spike-like shapes similar to that seen in patch-clamped neurons firing an action potential (Fig. 3). Two neurons appeared to spike in response to only 100 μA (neurons 3 and 5), whereas two others (neurons 1 and 2) appeared to spike in response to 150 μA. Thus, hVOS imaging revealed heterogeneous responses among individual Cre-targeted neurons within a genetically defined population.
Figure 8.
Graded responses to electrical stimulation. Five GAD2 interneurons in layer 2/3 of the somatosensory cortex of an hVOS::GAD2 mouse are indicated in the high-resolution (480 × 640) CCD image (top left) and in the CCD-SMQ (80 × 80) camera image below. Five trial averages show that increasing stimulus current from 50, 100, 150, to 200 μA increased response amplitudes to varying degrees in the different cells. Open circle represents site of stimulation.
Intracellular dynamics
Voltage sensors can probe dynamic processes within neurons, such as dendritic integration and action potential propagation (Antić and Zecević, 1995; Palmer and Stuart, 2009; Ghitani et al., 2015; Popovic et al., 2015b). hVOS probe expression revealed dendritic arbors, often extending several hundreds of micrometers (Fig. 1). To evaluate the spatial resolution of hVOS imaging at the level of such fine structures, we selected ROIs over the cell soma and over regions directly adjacent to it without probe (in a slice from an hVOS::Parv mouse). In ROIs encompassing an entire soma (Fig. 9A, left) or small (2 × 2 pixel) ROIs around the edge of a labeled neuron (Fig. 9A, right), we detected action potentials from the soma and saw no discernible signals from adjacent locations. This demonstrates that hVOS imaging can resolve responses at subcellular spatial resolution.
Figure 9.
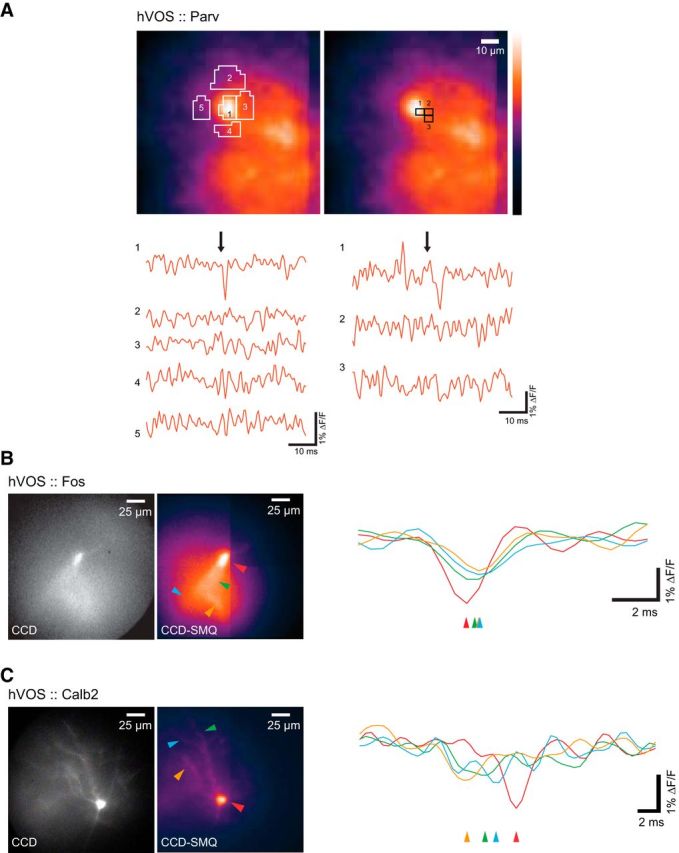
Voltage changes in different cellular compartments. A, Spatial resolution of hVOS responses in a somatosensory cortical slice from an hVOS::Parv mouse. Fluorescence traces from large ROIs (left) outlined, and small 2 × 2 pixel ROIs (right) boxed. B, A neuron in the granule cell layer of the DG from an hVOS::Fos mouse. A high-resolution CCD image (left) and CCD-SMQ image (middle) show a labeled neuron with a dendritic arbor. ROIs, including the cell soma (red) and proximal dendrites (green, orange, blue), are indicated with colors corresponding to the traces on the right. Arrowheads below the traces indicate the sequence of activation by outer molecular layer stimulation, demonstrating back-propagation from the soma to the dendrites. C, A cortical interneuron in layer 2/3 of the somatosensory cortex of an hVOS::Calb2 mouse. A high-resolution CCD image (left) and the CCD-SMQ image (middle) show a labeled cell with ROIs indicated. Stimulation (200 μA, 180 μs) generated hVOS responses in three dorsal dendrites (orange, green, and blue), which propagated to the soma (red). Arrowheads below traces indicate sequence of activation. Stimulus is in the layer 1/2 border, dorsal to the field of view. All traces are averages of five trials.
In a granule cell in the DG of an hVOS::Fos mouse, extracellular stimulation of the perforant path generated a back-propagating voltage response (Fig. 9B). Responses originated in the soma (red trace) and propagated through the apical dendrite (green trace) to more distal locations (blue and orange traces). In other neurons, voltage changes propagated in the opposite direction with dendritic responses preceding the somatic response. A cortical layer 2/3 interneuron from an hVOS::Calb2 mouse (Fig. 9C) responded to electrical stimulation with an initial, low-amplitude response in the dendrites (orange, green, and blue traces), followed by a spike in the soma (red trace). hVOS imaging thus revealed responses in dendrites as well as somata, with trends in latency that defined a direction of propagation. These data show that Cre-targeted hVOS probe reports intracellular voltage dynamics and thus provides information about the integration of synaptic inputs.
Discussion
In the present study, we generated an hVOS Cre-reporter mouse and used it to image electrical activity in a variety of genetically targeted populations of cells. The targeting of a probe to specific neuronal subtypes using Cre-loxP technology generalizes the expression of voltage sensors and has much to offer in addressing the complexities of neural circuit function. Using established Cre driver lines, we have targeted hVOS probe to various subpopulations of neurons with high specificity. In brain slices from double-transgenic mice, we imaged responses to electrical stimulation from multiple labeled cells in single trials. A large number of GEVIs are currently available with a very wide range of characteristics (Siegel and Isacoff, 1997; Sakai et al., 2001; Ataka and Pieribone, 2002; Knöpfel et al., 2006; Gong et al., 2015; Storace et al., 2016). A key advantage of hVOS imaging is the speed of response, which enables the detection of rapid depolarizations with high temporal fidelity (Chanda et al., 2005; Wang et al., 2010; Ghitani et al., 2015). The ability to monitor rapid changes in voltage in many individual neurons simultaneously in genetically defined populations represents an important milestone in the use of GEVIs to study neural network activity.
The expression patterns of hVOS probe in the double-transgenic mice generated here recapitulated the established distributions of the targeted cell types (Fig. 1) as well as the expression of the endogenous gene (Fig. 2). hVOS probe expression with this Cre reporter system confirmed the expression of other reporter probes used previously with these Cre drivers (Taniguchi et al., 2011; Jinde et al., 2012). This indicates that we can express hVOS probe based on a specific genetic signature and then explore the detailed microcircuitry of the targeted group of neurons. Imaging experiments in brains from the kinds of double-transgenic mice described here can provide insight into the spatiotemporal dynamics of distinct cell types. For example, hilar mossy cells of the DG have been recognized as important but poorly understood (Henze and Buzsáki, 2007; Scharfman, 2016). hVOS::CalCrl and hVOS::Calb2 mice express probe in these cells, enabling hVOS imaging to explore how they are activated by each element of hippocampal circuitry. Specific cell types are also likely to control spatial variations in the expression of LTP (Chang and Jackson, 2006), how LTP modulates information flow through the hippocampal trisynaptic circuit (Kleschevnikov and Routtenberg, 2003; Wright and Jackson, 2014), and how the hippocampus performs basic logical operations, such as pattern separation and completion (Leutgeb and Leutgeb, 2007; Jackson, 2013; Rolls and Kesner, 2016). The expression of hVOS probe in newborn granule cells using hVOS::Nestin double-transgenic mice offers the opportunity to explore the function of newborn neurons and how they integrate into circuits (Marín-Burgin et al., 2012). Using hVOS::Fos mice to express probe in neurons active during a prior experience offers an approach to the study of how neural circuitry encodes and stores information.
With each of the targeting strategies tested here, expression was sufficient to enable the recording of voltage changes in single trials without averaging. This opens up the possibility of detecting spontaneous activity and burst activity. Furthermore, the results presented here demonstrated that multiple trial averaging can distort rapid responses, reducing their amplitudes and increasing their durations. Single-trial recording more accurately represents electrophysiological dynamics and also reveals fluctuations in responses that may contain information about circuit mechanisms.
Genetically encoded Ca2+ indicators (Tian et al., 2009) have proven useful in the study of functional activity in neural circuits (Wachowiak et al., 2013; Sheffield and Dombeck, 2015; Smith et al., 2015). However, changes in Ca2+ concentration are substantially slower than changes in voltage (Koester and Sakmann, 2000; Canepari et al., 2008). Furthermore, very brief action potentials, or action potentials in cells with strong Ca2+ buffers, produce little, if any, change in cytosolic Ca2+. Ca2+ indicators also buffer Ca2+ to reduce the magnitude and slow the time course of cytosolic Ca2+ changes (Neher, 1995). hVOS imaging can record very brief spikes, thus providing a window into a range of important physiological processes that cannot be studied with Ca2+ probes. The hVOS probe used here has been shown to produce a ΔF/F of ∼25% per 100 mV in cultured cells (Wang et al., 2010), and in brain slices action potentials can be detected with a signal-to-noise ratio of 9–16 (Ghitani et al., 2015). Action potentials generally produced ΔF/F amplitudes of 1.6%–2.7% in the various cell types targeted here (Table 1). Furthermore, the rapid time response of hVOS imaging enabled us to determine action potential durations characteristic of different cell types, including fast-spiking interneurons (Fig. 7). This could be especially useful when the expression method targets multiple cell types, and interesting questions can be addressed by distinguishing them on the basis of their physiological properties. We also resolved the brief time intervals as voltage changes propagated rapidly between different cellular compartments (Fig. 9). This approach offers an alternative to the use of single-cell labeling with voltage-sensitive dye to study synaptic integration (Popovic et al., 2015a; Ross et al., 2015), with the added benefit of being able to observe dynamic changes in voltage in more distal processes as well as in multiple cells simultaneously.
Although Cre-loxP-based expression of hVOS probes opens up many opportunities for the study of neural circuitry, the quality of the signals reported here fell below that seen with probes expressed by in utero electroporation (Ghitani et al., 2015). A similar disparity was noted for Cre-loxP-based expression with another GEVI (Lou et al., 2016). The difference could be due to lower expression levels; but because Cre-loxP-based expression has a high targeting efficiency, labeled cells are often closer together and their processes intermingle. Labeled fine processes create ubiquitous diffuse fluorescence, which probably accounts for much of the background seen in our images. This background light reduces the signal-to-noise ratio. In addition, labeled processes can give rise to small signals in some locations (Fig. 4B). This problem would be greatly reduced if we had probes that targeted nerve cell bodies and did not localize to dendrites and axons. Indeed, a probe with reverse targeting, which preferentially localized to axons, was very useful in probing the dynamics of action potential conduction (Ma et al., 2017). The development of soma-targeting probes will likely significantly improve the quality of data produced by GEVIs. Light scattering also degrades the signal quality by increasing background. Scattered light would be greatly reduced with scanning illumination; but to exploit the rapid time response of hVOS probes, scanning rates of >kHz will be needed. There is great interest in using GEVIs to study neural circuitry in vivo (Gong et al., 2015); but because most GEVIs have thus far have been used with single-photon excitation, only labeled neurons near the brain surface can be studied. Imaging voltage in multiple neurons below the surface will require multiphoton illumination, which, in turn, will require GEVIs with larger fluorescence changes, as well as more rapid scanning technology.
The use of Cre-loxP-based expression to target an hVOS probe enables the exploration of neuronal circuit function in genetically defined cell types with single-cell resolution. The capability demonstrated here of detecting rapid voltage changes in single trials from multiple neurons will bring the study of reverberations, bursting activity, and information processing to a new level of detail. The ability to express hVOS probe with any Cre driver provides neuroscience with a general tool for studying the dynamic flow of information and the emergent network properties in virtually any neuronal circuit.
Footnotes
This work was supported by National Institutes of Health Grants NS093866 and DC014858 to M.B.J., MH080434, MH07897, and NS095632 to X.Z., and U54 HD090256 to the Waisman Center. We thank Dr. Kathleen Krentz and Dr. Manu John (University of Wisconsin Transgenic Animal Facility) for work on the development of the Ai35-hVOS Cre reporter mouse line; and Chung-Wei Chiang for help in generating the algorithm used for the analysis of spike half-widths.
The authors declare no competing financial interests.
References
- Antić S, Zecević D (1995) Optical signals from neurons with internally applied voltage-sensitive dyes. J Neurosci 15:1392–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ataka K, Pieribone VA (2002) A genetically targetable fluorescent probe of channel gating with rapid kinetics. Biophys J 82:509–516. 10.1016/S0006-3495(02)75415-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett EF, Stevens CF (1972) The kinetics of transmitter release at the frog neuromuscular junction. J Physiol 227:691–708. 10.1113/jphysiol.1972.sp010054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaguidi MA, Song J, Ming GL, Song H (2012) A unifying hypothesis on mammalian neural stem cell properties in the adult hippocampus. Curr Opin Neurobiol 22:754–761. 10.1016/j.conb.2012.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley J, Luo R, Otis TS, DiGregorio DA (2009) Submillisecond optical reporting of membrane potential in situ using a neuronal tracer dye. J Neurosci 29:9197–9209. 10.1523/JNEUROSCI.1240-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezas C, Irinopoulou T, Cauli B, Poncer JC (2013) Molecular and functional characterization of GAD67-expressing, newborn granule cells in mouse dentate gyrus. Front Neural Circuits 7:60. 10.3389/fncir.2013.00060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron HA, McKay RD (2001) Adult neurogenesis produces a large pool of new granule cells in the dentate gyrus. J Comp Neurol 435:406–417. 10.1002/cne.1040 [DOI] [PubMed] [Google Scholar]
- Canepari M, Vogt K, Zecević D (2008) Combining voltage and calcium imaging from neuronal dendrites. Cell Mol Neurobiol 28:1079–1093. 10.1007/s10571-008-9285-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda B, Blunck R, Faria LC, Schweizer FE, Mody I, Bezanilla F (2005) A hybrid approach to measuring electrical activity in genetically specified neurons. Nat Neurosci 8:1619–1626. 10.1038/nn1558 [DOI] [PubMed] [Google Scholar]
- Chang PY, Jackson MB (2006) Heterogeneous spatial patterns of long-term potentiation in rat hippocampal slices. J Physiol 576:427–443. 10.1113/jphysiol.2006.112128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen LB, Salzberg BM (1978) Optical measurement of membrane potential. Rev Physiol Biochem Pharmacol 83:35–88. [DOI] [PubMed] [Google Scholar]
- DeFelipe J, Hendry SH, Jones EG (1989) Visualization of chandelier cell axons by parvalbumin immunoreactivity in monkey cerebral cortex. Proc Natl Acad Sci U S A 86:2093–2097. 10.1073/pnas.86.6.2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragunow M, Robertson HA (1987) Kindling stimulation induces c-fos protein(s) in granule cells of the rat dentate gyrus. Nature 329:441–442. 10.1038/329441a0 [DOI] [PubMed] [Google Scholar]
- Fukuda S, Kato F, Tozuka Y, Yamaguchi M, Miyamoto Y, Hisatsune T (2003) Two distinct subpopulations of nestin-positive cells in adult mouse dentate gyrus. J Neurosci 23:9357–9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Wang F, Eisinger BE, Kelnhofer LE, Jobe EM, Zhao X (2017) Integrative single-cell transcriptomics reveals molecular networks defining neuronal maturation during postnatal neurogenesis. Cereb Cortex 27:2064–2077. 10.1093/cercor/bhw040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghitani N, Bayguinov PO, Ma Y, Jackson MB (2015) Single-trial imaging of spikes and synaptic potentials in single neurons in brain slices with genetically encoded hybrid voltage sensor. J Neurophysiol 113:1249–1259. 10.1152/jn.00691.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Wagner MJ, Zhong Li J, Schnitzer MJ (2014) Imaging neural spiking in brain tissue using FRET-opsin protein voltage sensors. Nat Commun 5:3674. 10.1038/ncomms4674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Huang C, Li JZ, Grewe BF, Zhang Y, Eismann S, Schnitzer MJ (2015) High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. Science 350:1361–1366. 10.1126/science.aab0810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenthner CJ, Miyamichi K, Yang HH, Heller HC, Luo L (2013) Permanent genetic access to transiently active neurons via TRAP: targeted recombination in active populations. Neuron 78:773–784. 10.1016/j.neuron.2013.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulyás AI, Miettinen R, Jacobowitz DM, Freund TF (1992) Calretinin is present in non-pyramidal cells of the rat hippocampus: I. A new type of neuron specifically associated with the mossy fibre system. Neuroscience 48:1–27. 10.1016/0306-4522(92)90334-X [DOI] [PubMed] [Google Scholar]
- Henze DA, Buzsáki G (2007) Hilar mossy cells: functional identification and activity in vivo. Prog Brain Res 163:199–216. 10.1016/S0079-6123(07)63012-X [DOI] [PubMed] [Google Scholar]
- Hunt SP, Pini A, Evan G (1987) Induction of c-fos-like protein in spinal cord neurons following sensory stimulation. Nature 328:632–634. 10.1038/328632a0 [DOI] [PubMed] [Google Scholar]
- Jackson MB. (2013) Recall of spatial patterns stored in a hippocampal slice by long-term potentiation. J Neurophysiol 110:2511–2519. 10.1152/jn.00533.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinde S, Zsiros V, Jiang Z, Nakao K, Pickel J, Kohno K, Belforte JE, Nakazawa K (2012) Hilar mossy cell degeneration causes transient dentate granule cell hyperexcitability and impaired pattern separation. Neuron 76:1189–1200. 10.1016/j.neuron.2012.10.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleschevnikov AM, Routtenberg A (2003) Long-term potentiation recruits a trisynaptic excitatory associative network within the mouse dentate gyrus. Eur J Neurosci 17:2690–2702. 10.1046/j.1460-9568.2003.02709.x [DOI] [PubMed] [Google Scholar]
- Knöpfel T, Díez-García J, Akemann W (2006) Optical probing of neuronal circuit dynamics: genetically encoded versus classical fluorescent sensors. Trends Neurosci 29:160–166. 10.1016/j.tins.2006.01.004 [DOI] [PubMed] [Google Scholar]
- Koester HJ, Sakmann B (2000) Calcium dynamics associated with action potentials in single nerve terminals of pyramidal cells in layer 2/3 of the young rat neocortex. J Physiol 529:625–646. 10.1111/j.1469-7793.2000.00625.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagace DC, Whitman MC, Noonan MA, Ables JL, DeCarolis NA, Arguello AA, Donovan MH, Fischer SJ, Farnbauch LA, Beech RD, DiLeone RJ, Greer CA, Mandyam CD, Eisch AJ (2007) Dynamic contribution of nestin-expressing stem cells to adult neurogenesis. J Neurosci 27:12623–12629. 10.1523/JNEUROSCI.3812-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leutgeb S, Leutgeb JK (2007) Pattern separation, pattern completion, and new neuronal codes within a continuous CA3 map. Learn Mem 14:745–757. 10.1101/lm.703907 [DOI] [PubMed] [Google Scholar]
- Lou S, Adam Y, Weinstein EN, Williams E, Williams K, Parot V, Kavokine N, Liberles S, Madisen L, Zeng H, Cohen AE (2016) Genetically targeted all-optical electrophysiology with a transgenic Cre-dependent Optopatch mouse. J Neurosci 36:11059–11073. 10.1523/JNEUROSCI.1582-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Bayguinov PO, Jackson MB (2017) Action potential dynamics in fine axons probed with an axonally targeted optical voltage sensor. eNeuro 4:ENEURO.0146–17.2017. 10.1523/ENEURO.0146-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H (2010) A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13:133–140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Mao T, Koch H, Zhuo JM, Berenyi A, Fujisawa S, Hsu YW, Garcia AJ 3rd, Gu X, Zanella S, Kidney J, Gu H, Mao Y, Hooks BM, Boyden ES, Buzsáki G, Ramirez JM, Jones AR, Svoboda K, Han X, et al. (2012) A toolbox of Cre-dependent optogenetic transgenic mice for light-induced activation and silencing. Nat Neurosci 15:793–802. 10.1038/nn.3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín-Burgin A, Mongiat LA, Pardi MB, Schinder AF (2012) Unique processing during a period of high excitation/inhibition balance in adult-born neurons. Science 335:1238–1242. 10.1126/science.1214956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markram H, Toledo-Rodriguez M, Wang Y, Gupta A, Silberberg G, Wu C (2004) Interneurons of the neocortical inhibitory system. Nat Rev Neurosci 5:793–807. 10.1038/nrn1519 [DOI] [PubMed] [Google Scholar]
- Morgan JI, Cohen DR, Hempstead JL, Curran T (1987) Mapping patterns of c-fos expression in the central nervous system after seizure. Science 237:192–197. 10.1126/science.3037702 [DOI] [PubMed] [Google Scholar]
- Neher E. (1995) The use of fura-2 for estimating Ca buffers and Ca fluxes. Neuropharmacology 34:1423–1442. 10.1016/0028-3908(95)00144-U [DOI] [PubMed] [Google Scholar]
- Palmer LM, Stuart GJ (2009) Membrane potential changes in dendritic spines during action potentials and synaptic input. J Neurosci 29:6897–6903. 10.1523/JNEUROSCI.5847-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedroni A, Minh do D, Mallamaci A, Cherubini E (2014) Electrophysiological characterization of granule cells in the dentate gyrus immediately after birth. Front Cell Neurosci 8:44. 10.3389/fncel.2014.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic MA, Carnevale N, Rozsa B, Zecević D (2015b) Electrical behaviour of dendritic spines as revealed by voltage imaging. Nat Commun 6:8436. 10.1038/ncomms9436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic M, Vogt K, Holthoff K, Konnerth A, Salzberg BM, Grinvald A, Antić SD, Canepari M, Zecević D (2015a) Imaging submillisecond membrane potential changes from individual regions of single axons, dendrites and spines. Adv Exp Med Biol 859:57–101. 10.1007/978-3-319-17641-3_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers JH. (1992) Immunohistochemical markers in rat cortex: co-localization of calretinin and calbindin-D28k with neuropeptides and GABA. Brain Res 587:147–157. 10.1016/0006-8993(92)91439-L [DOI] [PubMed] [Google Scholar]
- Rolls ET, Kesner RP (2016) Pattern separation and pattern completion in the hippocampal system. Neurobiol Learn Mem 129:1–3. 10.1016/j.nlm.2016.02.001 [DOI] [PubMed] [Google Scholar]
- Ross WN, Miyazaki K, Popovic MA, Zecević D (2015) Imaging with organic indicators and high-speed charge-coupled device cameras in neurons: some applications where these classic techniques have advantages. Neurophotonics 2:021005. 10.1117/1.NPh.2.2.021005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai R, Repunte-Canonigo V, Raj CD, Knöpfel T (2001) Design and characterization of a DNA-encoded, voltage-sensitive fluorescent protein. Eur J Neurosci 13:2314–2318. 10.1046/j.0953-816x.2001.01617.x [DOI] [PubMed] [Google Scholar]
- Salzberg BM, Grinvald A, Cohen LB, Davila HV, Ross WN (1977) Optical recording of neuronal activity in an invertebrate central nervous system: simultaneous monitoring of several neurons. J Neurophysiol 40:1281–1291. [DOI] [PubMed] [Google Scholar]
- Scharfman HE. (2016) The enigmatic mossy cell of the dentate gyrus. Nat Rev Neurosci 17:562–575. 10.1038/nrn.2016.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield ME, Dombeck DA (2015) Calcium transient prevalence across the dendritic arbour predicts place field properties. Nature 517:200–204. 10.1038/nature13871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, Greenberg ME (1990) The regulation and function of c-fos and other immediate early genes in the nervous system. Neuron 4:477–485. 10.1016/0896-6273(90)90106-P [DOI] [PubMed] [Google Scholar]
- Siegel MS, Isacoff EY (1997) A genetically encoded optical probe of membrane voltage. Neuron 19:735–741. 10.1016/S0896-6273(00)80955-1 [DOI] [PubMed] [Google Scholar]
- Smith GB, Sederberg A, Elyada YM, Van Hooser SD, Kaschube M, Fitzpatrick D (2015) The development of cortical circuits for motion discrimination. Nat Neurosci 18:252–261. 10.1038/nn.3921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer H, Waxman SG (1993) Expression of voltage-activated ion channels by astrocytes and oligodendrocytes in the hippocampal slice. J Neurophysiol 70:1863–1873. [DOI] [PubMed] [Google Scholar]
- Storace D, Sepehri Rad M, Kang B, Cohen LB, Hughes T, Baker BJ (2016) Toward better genetically encoded sensors of membrane potential. Trends Neurosci 39:277–289. 10.1016/j.tins.2016.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre F, Marshall JD, Yang Y, Gong Y, Schnitzer MJ, Lin MZ (2014) High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nat Neurosci 17:884–889. 10.1038/nn.3709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsani D, Fu Y, Lu J, Lin Y, Miyoshi G, Shima Y, Fishell G, Nelson SB, Huang ZJ (2011) A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron 71:995–1013. 10.1016/j.neuron.2011.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH, Petreanu L, Akerboom J, McKinney SA, Schreiter ER, Bargmann CI, Jayaraman V, Svoboda K, Looger LL (2009) Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods 6:875–881. 10.1038/nmeth.1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachowiak M, Economo MN, Díaz-Quesada M, Brunert D, Wesson DW, White JA, Rothermel M (2013) Optical dissection of odor information processing in vivo using GCaMPs expressed in specified cell types of the olfactory bulb. J Neurosci 33:5285–5300. 10.1523/JNEUROSCI.4824-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Zhang Z, Chanda B, Jackson MB (2010) Improved probes for hybrid voltage sensor imaging. Biophys J 99:2355–2365. 10.1016/j.bpj.2010.07.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Gupta A, Toledo-Rodriguez M, Wu CZ, Markram H (2002) Anatomical, physiological, molecular and circuit properties of nest basket cells in the developing somatosensory cortex. Cereb Cortex 12:395–410. 10.1093/cercor/12.4.395 [DOI] [PubMed] [Google Scholar]
- Wright BJ, Jackson MB (2014) Long-term potentiation in hilar circuitry modulates gating by the dentate gyrus. J Neurosci 34:9743–9753. 10.1523/JNEUROSCI.0814-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon BE, Lee CJ (2014) GABA as a rising gliotransmitter. Front Neural Circuits 8:141. 10.3389/fncir.2014.00141 [DOI] [PMC free article] [PubMed] [Google Scholar]