Abstract
Ixodes pacificus is a host of many bacteria including Rickettsia species phylotypes G021 and G022. As part of the overall goal of understanding interactions of phylotypes with their tick host, this study focused on molecular detection of rickettsiae in ovarian and midgut tissue of I. pacificus by fluorescent in situ hybridization (FISH), PCR, and ultrastructural analysis. Of three embedding media (Technovit 8100, Unicryl, and paraffin) tested for generating thin sections, tissues embedded in paraffin resulted in the visualization of bacteria with low autofluorescence in FISH. Digoxigenin-labeled probes were used in FISH to intensify bacterial hybridization signals using Tyramide Signal Amplification reaction. Using this technique, rickettsiae were detected in the cytoplasm of oocytes of I. pacificus. The presence of rickettsiae in the ovary and midgut was further confirmed by PCR and transmission electron microscopic analysis. Overall, the methods in this study can be used to identify locations of tick-borne bacteria in tick tissues and understand transmission routes of bacterial species in ticks.
Keywords: Rickettsia, Ixodes pacificus, ovary, fluorescent in situ hybridization, Tyramide Signal Amplification
INTRODUCTION
Ticks harbor a variety of bacterial species. Distributions of tick-borne bacteria in naturally infected tick tissues have been studied since the 1920s. Using Giemsa staining and light microscopy, Gram-negative intracellular bacteria were observed in Malpighian tubules of six species of ticks (Roshdy, 1968). Subsequently, a growing number of culture-independent techniques were developed that enabled the detection of more bacterial species in tick tissues. For example, common culture-independent techniques include indirect fluorescent antibody staining (Burgdorfer et al., 1985; Smith et al., 1976), restriction fragment length polymorphism analysis (Niebylski et al., 1997), DNA hybridization (Ambrosio et al., 1988), PCR (Niebylski et al., 1997; Persing et al., 1990), DNA sequencing (Whitworth et al., 2003), fluorescent in situ hybridization (FISH) (Matsuura et al., 2012; Sassera et al., 2006), and more recently quantitative real-time PCR (Piesman et al., 2001).
A great variety of tick-borne bacteria, due to the establishment of these techniques, were identified in different types of tick tissues (Brinton and Burgdorfer, 1971). However, many tick-borne bacteria were identified to be primarily distributed in the ovaries, midgut, salivary glands, Malpighian tubules, and hemocytes (Burgdorfer et al., 1985; el Shoura, 1990; Hart et al., 1991; Smith et al., 1976). Identification of bacteria in tick tissues is central for better understanding of the interactions between bacteria and ticks. Burgdorfer et al. (1985) reported a generalized infection of Borrelia burgdorferi in all tissues of Ixodes pacificus, implying a negative effect of the bacterium on the physiology of the tick host. Niebylski et al. (1997) reported that a Francisella species in D. andersoni is located primarily in the ovarian tissue but is absent in the salivary gland, suggesting that the bacterium is transovarially transmitted through host tick generations. The identification of tick-borne bacteria in the midgut and salivary gland suggests that the bacteria are horizontally transmitted to vertebrate hosts by ticks (Klyachko et al., 2007; Lynn et al., 2015). Furthermore, the identification of bacteria in hemocytes suggests possible dissemination of the bacteria from one tick organ to another (Hart et al., 1991).
I. pacificus is the vector of Borrelia burgdorferi and Anaplasma phagocytophilum, the causative agent of Lyme borreliosis and anaplasmosis, respectively (Burgdorfer et al., 1985; Foley et al., 2008; Piesman et al., 1999). The tick, which is mainly distributed in the West Pacific Coast, is inhabited with two Rickettsia species: phylotypes G021 and G022 (Phan et al., 2011). Phylotype G021 was detected with nearly 100% prevalence in I. pacificus collected from Northern California and was found to have a 100% efficiency for transovarial transmission and transstadial survival, respectively (Cheng et al., 2013a; Cheng et al., 2013b). The abundance and transmission of phylotype G021 in I. pacificus is indicative of a beneficial symbiotic relationship. As part of a study on the interactions of phylotypes G021/G022 with I. pacificus, we identified the five genes responsible for de novo folate biosynthetic pathway in the genome of phylotype G021, indicating that the nature of the symbiotic relationship between phylotype G021 and I. pacificus is nutrition-based (Hunter et al., 2015).
Identification of the localization of phylotype G021 in tissues of I. pacificus is an important step in understanding the symbiotic relationship and route of transmission of the bacterium in ticks. Here, we use fluorescence in situ hybridization (FISH), PCR, and transmission electron microscopic analysis to determine the distribution of this phylotype and other rickettsiae in the eggs, ovary and midgut of adult female I. pacificus.
MATERIALS AND METHODS
Tick collection
Eighteen engorged female Ixodes pacificus were collected from domestic dogs at the Mendocino County Animal Shelter, Ukiah, California, over a period of ten months (September 2010 and June 2011; UTM Coordinate: E483340 N4329309 Z10) to be used in this study. The genus and species of the ticks were identified using morphological features observed under a Leica MZ Apo dissecting microscope (Leica, Buffalo Grove, IL) (Furman and Loomis, 1984). Of the 18 engorged ticks collected, five ticks were dissected and their midguts and ovaries were collected for this study. The remaining ten engorged females were maintained in a glass desiccator at 25°C and 90% relative humidity for oviposition and collection of eggs for further analysis. Eight of the ten died and were excluded from the study. The remaining two that laid eggs were used for further analysis.
Genomic DNA isolation
Genomic DNA was successfully isolated from the midgut and ovarian tissues of five engorged female I. pacificus ticks. In addition, genomic DNA was isolated from two clutches of eggs. From each clutch, 10–15 eggs were used for the isolation of the DNA. The eggs, ovaries, and midgut tissue from engorged I. pacificus were transferred separately to Eppendorf tubes and placed in a container with liquid nitrogen. Frozen tissues were ground to a fine powder using sterile disposable pestles (Fisher Scientific, Houston, TX). Genomic DNA from these finely ground tissues were isolated using a Qiagen DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA), as described previously (Zhong et al., 2007). The concentration of the eluted DNA was determined using a Nanodrop spectrophotometer (ND-1000, Thermo Scientific, Wilmington, DE) and stored at −20°C for further analysis. In the mock extraction control, the tissues were replaced with sterile nuclease free water to check for possible contaminations in the DNA extraction.
Polymerase chain reaction (PCR)
To detect rickettsiae in tick tissues of I. pacificus, DNA isolated from the ovaries, midgut, and eggs were subsequently used in PCR using primers of the rickettsial outer membrane protein A (ompA) gene and 16S rRNA gene. The primers to amplify the ompA gene are 5′-ATGGCGAATATTTCTCCAAAA-3′ and 5′-GTTCCGTTAATGGCAGCATCT-3′ (Roux et al., 1996). The primers to amplify the 16S rRNA genes are 5′-TAAGGAGGTAATCCAGCC-3′ and 5′-CCTGGCTCAGAACGAA-3′ (Phan et al., 2011). These primers were targeted to conserved regions of the 16S rRNA gene (Phan et al., 2011) and the ompA gene of spotted fever group of Rickettsia species (Roux et al., 1996). The PCR procedure was done in 25 μL volume solutions containing 12.5 μl of 2× PCR Master Mix (Promega Corporation, Madison, WI), 0.4 μM of forward and reverse primers, appropriate volume of genomic DNA of I. pacificus (10–15 ng), and nuclease free water. The positive PCR reaction contained genomic DNA of Rickettsia montanensis as the template. The mock DNA extract was used as the negative control for PCR.
The ompA and 16S rRNA genes of Rickettsia species of I. pacificus were PCR amplified for thirty-five cycles on an ABI 2700 thermal cycler (Life Technologies, Foster City, CA) using the following parameters: denaturation at 92°C for 30 seconds, annealing at 59°C for 30 seconds for the ompA gene and 52°C for 1 minute for the 16S rRNA gene, respectively, and extension at 72°C for 30 seconds for the ompA gene and 2 minutes for the 16S rRNA gene, respectively. The PCR products were analyzed on a 0.8% agarose gel, stained with 0.5 μg/mL ethidium bromide, and imaged using Alpha Innotech Gel Documentation system (Alpha Innotech, Santa Clara, CA).
Tick tissue sample preparation for in situ hybridization
Ovaries of I. pacificus were cut into 3–4 mm pieces and fixed with 4% buffered paraformaldehyde at 4°C for 24–48 hours. The fixed samples were washed in 1× PBS, dehydrated using an ethanol series (25%, 50%, 70%, 90%, 100%), and embedded in Technovit 8100 resin (Heraeus Kulzer, Wehrheim, Germany), Unicryl resin (Electron Microscopy Services, Hatfield, PA), and low melting paraffin wax Histoplast LP (Thermo Scientific, Kalamazoo, MI). Blocks with tissues were sectioned at 5 μm by a microtome 820 (American Optical Company, Buffalo, NY). To determine the ideal embedding medium and conditions for in situ hybridization studies, E. coli in 2% agarose were embedded in the same three embedding media as the ovarian tissue. Both the tick and E. coli sample sections were then rehydrated by a graded series of ethanol in the reverse order (95%, 90%, 70%, and 50%) followed by DEPC treated sterile water.
Fluorescence in-situ hybridization (FISH)
Four oligonucleotide probes conjugated with fluorescent dyes were used in FISH for the direct detection of bacteria. EUB338 antisense probe is a domain-specific probe that detects all eubacteria (Amann et al., 1990). Rick_B1 antisense probe is a probe that is specific for the genus of Rickettsia (Kuchler et al., 2009; Perotti et al., 2006). Based on our BLAST search, the Rick_B1 probe binds to 16S rRNA of spotted fever group rickettsiae but can’t bind to other known bacteria in ticks. EUB II antisense probe binds to bacterial phylum of Planctomycetales (Daims et al., 1999). NONRickettsia probe is a sense probe with the nucleotide sequence that is complementary to Rick_B1 probe. The EUB II and NONRickettsia probes are used for checking nonspecific incorporation of probes into tick tissues (Table 1). The EUB338 and EUB II probes were conjugated with 6FAM, Cy3, Fluorescein (FITC), or Alexa 488 fluorophores, whereas the Rick_B1 and NONRickettsia probes were conjugated with Alexa 568. The 6FAM, Cy3, and FITC probes were custom synthesized by Integrated DNA Technologies (Coralville, IA). Alexa 488 and Alexa 568 probes were custom synthesized by Thermo Fisher Scientific (Eugene, OR). The EUB II and NONRickettsia probes, as well as hybridization buffer without probe, served as negative controls for hybridization.
Table 1.
Primers and probes used in fluorescent in-situ Hybridization (FISH) and Fluorescence in-situ Hybridization with Tyramide Signal Amplification (TSA-FISH).
| Oligonucleotide | Target gene | Specificity | Sequence | Reference |
|---|---|---|---|---|
| Probes for FISH: | ||||
| EUB338 (antisense) | 16S rRNA |
Eubacteria | 5′GCTGCCTCCCGTAGGAGT3′ | Amann et al., 1990 |
| EUB II (antisense) | 16S rRNA |
Planctomycetales | 5′GCAGCCACCCGTAGGTGT3′ | Amann et al., 1990 |
| Rick-B1 (antisense) | 16S rRNA |
Rickettsia | 5′CCATCATCCCCTACTACA3′ | Perotti et al., 2006 |
| NONRickettsia (sense) | 16S rRNA |
Rickettsia | 5′TGTAGTAGGGGATGATGG3′ | Kuchler et al., 2009 |
| Primers used for generating DIG-labeled ssDNA probes in TSA-FISH: | ||||
| Antisense primer | ompA | Rickettsia | 5′GTTCCGTTAATGGCAGCATCT3 | Roux et al., 1996 |
| Sense primer | ompA | Rickettsia | 5′ATGGCGAATATTTCTCCAAAA3′ | Roux et al., 1996 |
FISH was performed as described by Svendsen et al. (2009) with the following modifications. Prehybridization was carried out in the hybridization buffer for FISH (0.1 M Tris buffer pH 7.2 with 0.9 M NaCl, 0.1% SDS, 0.1% Poly A DNA [Roche Diagnostics, Indianapolis, IN] in 50% deionized fresh formamide [AMRESCO, Solon, OH]) for 2 hours at 50°C. Then the sections were hybridized with 50 ng/μL fluorescent-labeled probes in the hybridization buffer for FISH and incubated for 24–48 hours at 50°C. The sections were washed three times with wash buffer containing 0.1 M Tris buffer (pH 7.2) with 0.1 M NaCl and 0.01% SDS at 50°C, mounted with Prolong Gold mounting medium with DAPI (Thermo Fisher Scientific, Eugene, OR), and viewed using a Zeiss Axiophot epifluorescent microscope (Carl Zeiss, Jena, Germany). The images were captured using a SPOT RT camera using SPOT advanced image capture software (SPOT Imaging Solutions, Sterling Heights, MI).
Fluorescence in-situ hybridization with Tyramide Signal Amplification (TSA-FISH)
To increase sensitivity of FISH in detecting rickettsiae in I. pacificus, TSA-FISH was used to intensify bacterial fluorescent signals (Schonhuber et al., 1997). Sense and antisense single-stranded digoxigenin (DIG) labeled Rickettsia-specific ompA probes were generated by a PCR DIG probe synthesis kit (Roche, Indianapolis, IN) according to the manufacturer’s recommendations. The DNA templates for the asymmetric PCR reaction were generated by PCR amplification of the ompA gene from I. pacificus genomic DNA using the primers and conditions that were described earlier in this study. The resulting ompA PCR fragments were purified using a Wizard® SV Gel and PCR Clean-Up System (Promega, Madison, WI). The concentration of eluted DNA was determined using a Nanodrop Spectrophotometer. If the desired concentration of 200 ng/μL was not obtained, the purified DNA products were concentrated in a vacuum centrifuge (Savant, Hicksville, NY) until the desired concentration was reached. The DIG-labeled DNA probes were synthesized by an asymmetric PCR assay in 50 μL volume containing PCR buffer with 15 mM MgCl2, one of the two primers at 0.4 μM (Table 1), 200 μM dATP, dCTP, dGTP, 130 μM dTTP, 70 μM DIG-dUTP, 2.625 units of Taq DNA polymerase, and 100 pg of the purified ompA PCR fragment that was produced by conventional PCR. The asymmetric PCR amplifications were carried out for 40 cycles with the identical amplification conditions as the conventional PCR in an ABI 2700 thermal cycler (Life Technologies, Foster City, CA). The resulting DIG-labeled probes were precipitated by 1 M NaCl and 100% cold ethanol, washed with 70% ethanol, and resuspended in the hybridization buffer for TSA-FISH (5× SSC [15 mM sodium citrate, 150 mM NaCl, pH 7.5], 50% deionized formamide [AMRESCO, Solon, OH], 100 μg/mL autoclaved herring sperm DNA [Sigma-Aldrich, Carlsbad, CA], 50 μg/mL Heparin [Sigma-Aldrich, Carlsbad, CA] and 0.1% Tween-20 [Arcos, NJ]). The probe was aliquoted and stored at −80°C. 16S rRNA antisense DIG probe was also generated by the asymmetric PCR reaction using DIG nucleotides. However, since high nonspecific fluorescence was observed, the 16S rRNA antisense DIG probe was not used further in this study.
TSA-FISH was performed in accordance with previous reports with modification (Clay and Ramakrishnan, 2005). Tick ovary tissues were formaldehyde-fixed and paraffin-embedded as described previously in FISH. The tick sections on slides were prehybridized with hybridization buffer for TSA-FISH for 2 hours at 37°C. After the prehybridization was completed, the tick sections were hybridized with 1:10 diluted (in hybridization buffer), heat denatured DIG-labeled ompA probes for 24–48 hours at 37°C in the moist chamber in an incubator. At the end of the incubation period, the excess and non-specifically bound probes were removed by washing three times at 50°C in PBS with 1× PBS containing 0.1% Tween-20 (PBTw) for 45 minutes.
Posthybridization immunohistochemistry was performed to visualize hybridization signals. The sections hybridized with the DIG-labeled probe were blocked with PBTw containing 0.5 mg/mL Bovine Serum Albumin (BSA) for 15 minutes at 48°C to avoid non-specific hybridizations, incubated overnight in 1:1000 mouse anti-DIG primary antibody (Roche Applied Science, Indianapolis, IN) in PBTw with 0.5 mg/mL BSA at 4°C, and then incubated for two hours with 1:100 HRP-conjugated anti-mouse secondary antibody (Roche Applied Science, Indianapolis, IN) in PBTw at room temperature. The unbound antibody was removed by washing in PBTw.
Tyramide Signal Amplification was carried out as specified by the manufacturer using a Tyramide SuperBoost™ kit (Thermo Fisher, Eugene, OR). Following incubations with the primary and secondary antibody, the tick sections were treated with 1:100 Alexa Fluor™ 568-labeled Tyramide conjugate diluted in the amplification buffer (Thermo Fisher, Eugene, OR) containing 0.0015% hydrogen peroxide for 30 minutes at room temperature. The slides were then washed in PBTw, mounted with Prolong Gold mounting medium with DAPI (Thermo Fisher, Eugene, OR), and viewed using Zeiss Axiophot epifluorescent microscope. The images were captured with a SPOT RT camera using a PCI image capture program. There were two negative controls in the TSA-FISH assay in this study to detect any non-specific TSA amplification: Sense probes served as the first control and hybridization buffer for TSA-FISH without any probes was the second control.
Transmission electron microscopy
Transmission electron microscopy was performed as previously described to visualize bacteria in ticks and to confirm morphology of the bacterium (Phan et al., 2011). Briefly, dissected ovarian tissue samples were fixed in 2.5% buffered glutaraldehyde (Ted Pella, Redding, CA) in 100 mM cacodylate buffer (pH 7.4) and post-fixed in 1% osmium tetroxide. A Leica Thincut ultramicrotome (Leica, Buffalo grove, II) fitted with a diamond knife was used to cut 0.07 μm thin sections. The sections were mounted on clean glass slides and examined using a Phillips EM 208S transmission electron microscope (Phillips USA, Hillsboro, OR) operating at 60 kV. Images captured on photographic film were developed and printed.
RESULTS AND DISCUSSION
The goal of this study was to detect and directly visualize rickettsiae in tick tissues. Ovaries, midguts, and eggs were chosen for analysis because the efficiency of transovarial transmission and transstadial survival of phylotype G021 in I. pacificus was previously determined to be 100% (Cheng et al., 2013a) and because Rickettsia-like bacteria have been visualized inhabiting the midgut of I. pacificus (Phan et al., 2011). To determine the location of rickettsiae in tissues of I. pacificus, genomic DNA was isolated from midgut and ovarian tissue that had been dissected from five engorged female ticks as well as eggs from two engorged females of I. pacificus. PCR showed that both ompA and 16S rRNA genes were amplified from all DNA samples of I. pacificus (Table 2). The size of the ompA and 16S rRNA amplicons is approximately 630 base pairs (bp) and 1,480 bp, respectively (Figure 1). These results suggest that Rickettsia species are present in the ovaries, midgut, and eggs of I. pacificus and confirm that phylotype G021 is transovarially transmitted through generations of the tick host, I. pacificus (Cheng et al., 2013a).
Table 2.
Summary of PCR amplification of the ompA gene and 16S rRNA gene of rickettsiae using DNA extracts from eggs and engorged female Ixodes pacificus. The “+” symbol indicates amplification of the gene by PCR.
| Tick | Tissue | ompA gene | 16S rRNA gene |
|---|---|---|---|
| Engorged female | |||
| #1 | ovary | + | + |
| midgut | + | + | |
| Engorged female | |||
| #2 | ovary | + | + |
| midgut | + | + | |
| Engorged female | |||
| #3 | ovary | + | + |
| midgut | + | + | |
| Engorged female | |||
| #4 | ovary | + | + |
| midgut | + | + | |
| Engorged female | |||
| #5 | ovary | + | + |
| midgut | + | + |
Figure 1.
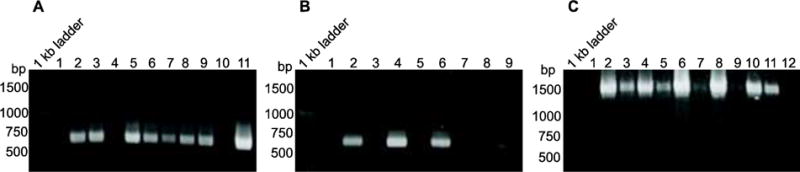
Representative images of PCR amplification of ompA and 16S rRNA genes of rickettsiae from Ixodes pacificus. Genomic DNA, which was isolated from ovary and midgut of engorged female ticks as well as from eggs of I. pacificus, was used in PCR. PCR fragments were separated on 0.8% agarose gel and stained with ethidium bromide. A) Representative images of Rickettsia specific ompA gene amplicon from four engorged female ticks. In the lane marked 1 kb ladder, Promega 1 kb benchtop ladder was loaded. Lanes marked 2, 3, 5 are the ompA amplicon from genomic DNA isolated from the ovary tissues of three engorged ticks (#1, #2, and #4 of the engorged ticks) and lanes 6–9 are ompA amplicon from genomic DNA isolated from the midgut tissues of four engorged ticks (#1–#4 ticks). Lane 10 is the negative control with DNA from mock extraction. Lane 11 is the positive control of the ompA amplicon amplified from Rickettsia montanensis. No samples were loaded in lane 1 and 4. B) Representative images of Rickettsia specific ompA gene amplicon from eggs of two engorged female ticks. Lanes marked 2, 4 and 6 are amplified ompA amplicon using genomic DNA isolated from two clutches of eggs (#9 and #10 of the engorged ticks, and the DNA mixture of #9 and #10 ticks). The lane marked 8 is a negative control with DNA from mock extraction. The lane marked 9 is the positive control of the ompA amplicon amplified from Rickettsia montanensis. No samples were loaded in lanes 1, 3, 5, and 7. C) Representative images of Rickettsia specific amplicon of 16S rRNA gene amplified from genomic DNA isolated from five different female engorged ticks. Lanes marked 2, 4, 6, 8, and 10 are amplified products from genomic DNA isolated from ovary tissues of five engorged ticks (#1–#5 of the engorged ticks) and lanes 3, 5, 7, and 9 are amplified products from genomic DNA isolated from the midgut tissues (#1–#4 of the engorged ticks). Lane 11 is the positive control of the 16S rRNA amplicon amplified from Rickettsia montanensis. The lane marked 12 is the negative control with DNA from mock extraction. No sample was loaded in lane 1.
PCR-based assays are routinely used for the identification of microorganisms in tick tissues (Parola et al., 2013; Richards, 2012). However, any contamination during the tick dissection or the DNA isolation procedure could result in false positives in the detection of bacteria in the tissues. In addition, the presence of bacteria in the hemolymph of the ticks could be another source of contamination (Lange et al., 1992). Therefore, direct evidence using microscopic methods is necessary to confirm the presence of rickettsiae, detected by PCR-based methods, in the tick tissues. The FISH assay is one of the methods for direct visualization of bacterial infection in situ (Svendsen et al., 2009).
One of the objectives of this study was to choose an embedding medium for the FISH assay that would generate sufficient fluorescent signal for clear identification of the bacteria within the tissues. Three different embedding media, Technovit 8100 resin, Unicryl resin, and paraffin wax, were used for hybridization analysis of E. coli as the test specimen to establish the optimum conditions for FISH using a 16S rRNA-targeted, FITC-labeled eubacterial EUB338 probe (Table 1). Of the two resins tested, the Technovit 8100 resin exhibited high autofluorescence that made it difficult to clearly distinguish the bacterial signal from the background. Even though this resin has been successfully used in immunocytochemical studies (De Jonge et al., 2005; Takechi et al., 1999), in this study no fluorescent bacteria could be obviously detected using eubacterial EUB338 probes conjugated to FITC or Cy3 or Alexa 568 (data not shown). Unicryl is a single-component resin that has been shown to be an effective embedding medium for in situ hybridization studies (Pantoja and Lightner, 2001). This resin has the added advantage that the same embedded tissue sample can be processed for both FISH and ultrastructural analysis (Pantoja and Lightner, 2001). E. coli were visualized clearly when bacteria were embedded in Unicryl resin and hybridized with the EUB338 probe that was conjugated to FITC (Supplemental S1). E. coli were also embedded in paraffin and hybridized specifically to the eubacterial EUB338 probe conjugated to FITC. The FISH results using paraffin-embedded E. coli showed that the bacteria were able to hybridize with probe EUB338, but not with probe EUB II, which indicated that our method of sample embedding in paraffin and hybridization was successful (Supplemental S1).
Thin sections of tick tissue embedded in Unicryl or paraffin resin were hybridized with Rick_B1, a Rickettsia genus specific 16S rRNA-targeted oligonucleotide probe conjugated to Cy3 fluorescent dye in FISH (Table 1). 16S rRNA was chosen for probing due to its abundant availability in bacteria; however, our FISH results showed that no fluorescent bacteria were unambiguously detected in any tick tissues. Specifically, the background fluorescence was too high to distinguish bacterial cells from the noise due to the presence of a strong autofluorescence signal from the tick tissues (data not shown). To decrease the amount of autofluorescence at the excitation wavelength of 550 nm for Cy3, the Rick_B1 probe conjugated to Alexa 488 was used for Unicryl or paraffin embedded tick tissue in FISH. Even though the autofluorescence using the Rick-B1 probe conjugated with Alexa 488 was not as high as that conjugated with Cy3, bacterial cells were still not clearly visible in Unicryl resin-embedded or paraffin-embedded samples (data not shown).
Our FISH results confirmed previous reports that the low signal to noise ratio due to autofluorescence of tick tissues made it difficult to visualize bacteria using fluorescently labeled probes (Epis et al., 2013; Hammer et al., 2001; Ment et al., 2012). The low signal to noise ratio in FISH could be due to weak emission signals of the probes and the low ribosome content of bacteria in tissues of field-collected samples (Lenaerts et al., 2007; Zwirglmaier, 2005). Both flat and engorged I. pacificus carry rickettsiae. However, the amount of ribosome content is still below the sensitivity level of FISH, although it has been reported the rickettsial burden in engorged I. pacificus has a 313-fold increase compared with that in flat I. pacificus (Cheng et al., 2013a). Several strategies have been developed to intensify signals of bacteria of field-collected samples, including insects, in the signal detection step of FISH (Kliot and Ghanim, 2016). One of the methods to improve the sensitivity in FISH is TSA-FISH, which is a technique that enables phylogenetic identification of low abundant bacteria using single probes without pre-amplification of target DNA sequences (Kawakami et al., 2012; Zordan, 2011; Zwirglmaier, 2005). Since FISH is coupled with Tyramide Signal Amplification and multiple DIG-labeled nucleotides are incorporated into the PCR products, better quality signals, specifically improved signal to noise ratio, are obtained than those generated by fluorescent oligonucleotide probes in conventional FISH (Kawakami et al., 2012; Kessler et al., 1990; Zordan, 2011; Zwirglmaier, 2005). FISH coupled with Tyramide Signal Amplification takes advantage of specific and strong interactions between DIG and anti-DIG antibody. DIG is a steroid that is found exclusively in digitalis plants. Nonspecific binding of the anti-DIG antibody to animal tissues is low in the FISH assay (Kessler, 1991).
To detect the presence of rickettsiae in paraffin-embedded tick tissues, DIG-labeled probes of Rickettsia specific ompA gene were generated for TSA-FISH. Using ompA specific antisense DIG-labeled probe, significant hybridization signals above the background fluorescence in the ovary tissue of I. pacificus were detected by TSA-FISH. Specifically, red coccobacilli were readily visualized in the periphery of the developing oocytes of I. pacificus ovary tissue (Figure 2A). The bacteria that were stained red by the ompA specific antisense DIG-labeled probe were also stained by DAPI (Figure 2B). The DAPI-stained ovary cells appeared blue, which showed that rickettsiae are within the cytoplasm of the oocytes and that most bacteria are in the periphery of the developing oocytes. A majority of the DAPI positive signal co-localized with the TSA amplified signal, indicating that Rickettsia species is the dominant bacteria inhabiting the oocytes of I. pacificus. However, the signal intensity of fluorescent bacteria probed by the ompA specific antisense DIG-labeled probe seems to vary among the ovary cells of I. pacificus, suggesting that the number of rickettsiae varied from cell to cell. This is consistent with a previous finding that Rickettsia species were present at the periphery of oocytes of ticks (da Silva Costa et al., 2011). The peripheral location may represent a stage of Rickettsia species within their life cycle. Direct visualization of Rickettsia species in ovary and other tick tissues has been reported by researchers in the past years. Using a histological analysis, da Silva Costa et al. (2011) reported that Rickettsia rickettsii is located in the oocytes of Rhipicephalus sanguineus. Whitworth et al. (2003) reported that R. honei are visualized in the cytosol of oocytes and eggs of Bothriocroton hydrosauri (Formerly; Aponomma hydrosauri) using an ultrastructural analysis. Last, Yano et al. (1993) reported that Rickettsia species are detected in midguts, Malpighian tubules, salivary glands, and ovaries of Dermacentor taiwanensis by an ultrastructural analysis.
Figure 2.
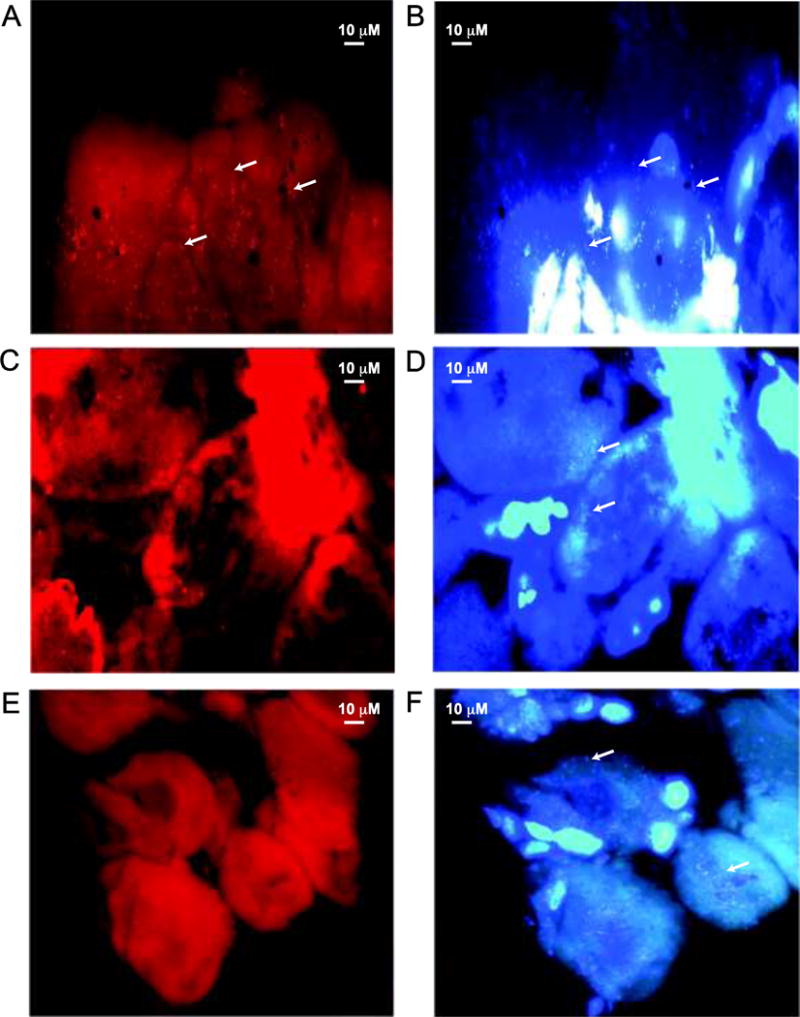
Fluorescence microscopic images of thin sections of Ixodes pacificus ovary tissue embedded in paraffin. Ovary tissue from I. pacificus was hybridized with the DIG-labeled Rickettsia-specific ompA probes in FISH. Hybridized DIG-labeled probes were detected using anti-DIG antibody and the signal was amplified using Tyramide Signal Amplification. 2A) Brightly red coccobacilli hybridized to the DIG-labeled Rick_B1, Rickettsia-specific ompA antisense probe, were visible on the fringes of the ovarian cells (arrows). 2B) Fluorescent image of the same ovary section in 2A counter-stained with DAPI showing DAPI positive bacteria (arrows) and host nuclei. 2C) Ovary tissue hybridized to the DIG-labeled NONRickettsia, a Rickettsia ompA sense probe, was served as a control. 2D) Fluorescent image of the same section in 2C counter-stained with DAPI showing DAPI positive bacteria (arrows) and host nuclei. 2E) Ovary tissue hybridized to the hybridization buffer for TSA-FISH without probes was served as another control. 2F) Fluorescent image of the same section in 2E counter-stained with DAPI showing DAPI positive bacteria (arrows) and host nuclei.
To verify that the ompA specific antisense DIG-labeled probe binds specifically to rickettsiae in I. pacificus, the ovary tissue sections were hybridized with either ompA sense DIG-labeled probe or hybridization buffer for TSA-FISH without probe as a control. TSA-FISH showed that neither ompA sense probes (Figures 2C and 2D) nor hybridization buffer without probe (Figures 2E and 2F) produced any signal above the background fluorescence. Since the bacterial fluorescence signals were visualized only with the antisense probe and co-localized to DAPI signals, it can be argued that the rickettsial specific signals represent bacteria and therefore, Rickettsia species in ticks. Furthermore, although the TSA-FISH method failed to detect rickettsiae in tick eggs in this study, the presence of rickettsiae within the developing oocytes of I. pacificus provides a good visual indication and is consistent with our previous report that Rickettsia species phylotype G021 is transmitted transovarially in I. pacificus (Cheng et al., 2013a).
Transmission electron microscopic analysis of tick ovarian tissue samples confirmed that bacteria are located within the developing oocytes and that bacteria are clustered at the periphery of the oocytes (Figure 3). The microorganisms appeared rod shaped with a distinct inner and outer membrane with an average length and width of 1.5 by 0.5 μm. The morphology of bacteria in I. pacificus resembles that of other Rickettsia species described previously (Nilsson et al., 1997; Phan et al., 2011). Similar to the observations that were made using FISH analysis (Figure 2), many of these bacteria appear to be in clusters. The functional significance of observed clusters of bacteria is not clear at this time. However, it can be argued that they may be indicative of cells undergoing active cell division in engorged ticks (Cheng et al., 2013a).
Figure 3.

Transmission electron microscopic image of Ixodes pacificus ovary tissue showing rod shaped bacterial cells with double membranes. Clusters of bacterial cells were observed in an oocyte (arrows). A double membrane, characteristic of gram-negative bacteria is clearly visible in many longitudinal sections of the bacteria. The darkly stained objects are believed to be lipid droplets. The thick outer layer of the oocyte is labeled as (O).
CONCLUSIONS
This study has shown that fluorescent in situ hybridization, PCR, and transmission electron microscopic data have confirmed that the developing oocytes of engorged I. pacificus are inhabited with Rickettsia species. Our data support our previous findings that Rickettsia species phylotype G021 is transovarially transmitted in I. pacificus. We have presented data that rickettsia in ticks can be directly visualized by TSA-FISH. This method can be used to study the location and transmission routes of rickettsiae and other bacteria in ticks and other arthropods.
Supplementary Material
Acknowledgments
We want to thank Dr. Jacob P. Varkey from Humboldt State University for developing the protocol of TSA-FISH and for providing technical support over the course of this study. We also want to thank Dr. David H. Walker from University of Texas Medical Branch, Galveston for donating genomic DNA of Rickettsia montanensis and Ms. Bliss Fisher from Mendocino County Animal Shelters for collecting all tick samples. We would like to thank Dr. Casey Lu from Humboldt State University for his support in performing TEM studies. This work was supported by the National Institutes of Health [1R15AI082515-01].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest in this study.
References
- Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosio RE, Visser ES, Koekhoven Y, Kocan KM. Hybridization of DNA probes to A. marginale isolates from different sources and detection in Dermacentor andersoni ticks. Onderstepoort J Vet Res. 1988;55:227–229. [PubMed] [Google Scholar]
- Brinton LP, Burgdorfer W. Fine structure of Rickettsia canada in tissues of Dermacentor andersoni Stiles. J Bacteriol. 1971;105:1149–1159. doi: 10.1128/jb.105.3.1149-1159.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertaux J, Schmid M, Hutzler P, Hartmann A, Garbaye J, Frey-Klett P. Occurrence and distribution of endobacteria in the plant-associated mycelium of the ectomycorrhizal fungus Laccaria bicolor. Environ Microbiol. 2005;7:1786–1795. doi: 10.1111/j.1462-2920.2005.00867.x. [DOI] [PubMed] [Google Scholar]
- Burgdorfer W, Lane RS, Barbour AG, Gresbrink RA, Anderson JR. The western black-legged tick, Ixodes pacificus: a vector of Borrelia burgdorferi. Am J Trop Med Hyg. 1985;34:925–930. doi: 10.4269/ajtmh.1985.34.925. [DOI] [PubMed] [Google Scholar]
- Cheng D, Lane RS, Moore BD, Zhong J. Host blood meal-dependent growth ensures transovarial transmission and transstadial passage of Rickettsia sp phylotype G021 in the western black-legged tick (Ixodes pacificus) Ticks Tick Borne Dis. 2013a;4:421–426. doi: 10.1016/j.ttbdis.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D, Vigil K, Schanes P, Brown RN, Zhong J. Prevalence and burden of two rickettsial phylotypes (G021 and G022) in Ixodes pacificus from California by real-time quantitative PCR. Ticks Tick Borne Dis. 2013b;4:280–287. doi: 10.1016/j.ttbdis.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay H, Ramakrishnan L. Multiplex fluorescent in situ hybridization in zebrafish embryos using tyramide signal amplification. Zebrafish. 2005;2:105–111. doi: 10.1089/zeb.2005.2.105. [DOI] [PubMed] [Google Scholar]
- da Silva Costa LF, Nunes PH, Soares JF, Labruna MB, Camargo-Mathias MI. Distribution of Rickettsia rickettsii in ovary cells of Rhipicephalus sanguineus (Latreille1806) (Acari: Ixodidae) Parasit Vectors. 2011;4:222. doi: 10.1186/1756-3305-4-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daims H, Brühl A, Amann R, Schleifer KH, Wagner M. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol. 1999;22:434–444. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- De Jonge HW, De Bakker MA, Verbeek FJ, Weijs WA. Embedding of large specimens in glycol methacrylate: prerequisites for multi-signal detection and high-resolution imaging. Microsc Res Tech. 2005;66:25–30. doi: 10.1002/jemt.20139. [DOI] [PubMed] [Google Scholar]
- el Shoura SM. Ultrastructure and distribution of intracellular rickettsia-like microorganisms in various organs of the laboratory-reared adult tick Argas (Persicargas) arboreus (Ixodoidea: Argasidae) Exp Appl Acarol. 1990;9:137–143. doi: 10.1007/BF01198992. [DOI] [PubMed] [Google Scholar]
- Epis S, Mandrioli M, Genchi M, Montagna M, Sacchi L, Pistone D, Sassera D. Localization of the bacterial symbiont Candidatus Midichloria mitochondrii within the hard tick Ixodes ricinus by whole-mount FISH staining. Ticks Tick Borne Dis. 2013;4:39–45. doi: 10.1016/j.ttbdis.2012.06.005. [DOI] [PubMed] [Google Scholar]
- Foley JE, Clueit SB, Brown RN. Differential exposure to Anaplasma phagocytophilum in rodent species in northern California. Vector Borne Zoonotic Dis. 2008;8:49–55. doi: 10.1089/vbz.2007.0175. [DOI] [PubMed] [Google Scholar]
- Furman DP, Loomis EC. The ticks of California (Acari:Ixodida) University of California Press; Berkeley: 1984. [Google Scholar]
- Hammer B, Moter A, Kahl O, Alberti G, Göbel UB. Visualization of Borrelia burgdorferi sensu lato by fluorescence in situ hybridization (FISH) on whole-body sections of Ixodes ricinus ticks and gerbil skin biopsies. Microbiology. 2001;147:1425–1436. doi: 10.1099/00221287-147-6-1425. [DOI] [PubMed] [Google Scholar]
- Hart A, Kocan KM, Bezuidenhout JD, Prozesky L. Ultrastructural morphology of Cowdria ruminantium in midgut epithelial cells of adult Amblyomma hebraeum female ticks. Onderstepoort J Vet Res. 1991;58:187–193. [PubMed] [Google Scholar]
- Hunter DJ, Torkelson JL, Bodnar J, Mortazavi B, Laurent T, Deason J, Thephavongsa K, Zhong J. The Rickettsia Endosymbiont of Ixodes pacificus Contains All the Genes of De Novo Folate Biosynthesis. PloS one. 2015;10:e0144552. doi: 10.1371/journal.pone.0144552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami S, Hasegawa T, Imachi H, Yamaguchi T, Harada H, Ohashi A, Kubota K. Detection of single-copy functional genes in prokaryotic cells by two-pass TSA-FISH with polynucleotide probes. J Microbiol Methods. 2012;88:218–223. doi: 10.1016/j.mimet.2011.11.014. [DOI] [PubMed] [Google Scholar]
- Kessler C, Holtke HJ, Seibl R, Burg J, Muhlegger K. Non-radioactive labeling and detection of nucleic acids. I. A novel DNA labeling and detection system based on digoxigenin: anti-digoxigenin ELISA principle (digoxigenin system) Biol Chem Hoppe Seyler. 1990;371:917–927. doi: 10.1515/bchm3.1990.371.2.917. [DOI] [PubMed] [Google Scholar]
- Kessler C. The digoxigenin:anti-digoxigenin (DIG) technology–a survey on the concept and realization of a novel bioanalytical indicator system. Mol Cell Probes. 1991;5:161–205. doi: 10.1016/0890-8508(91)90041-h. [DOI] [PubMed] [Google Scholar]
- Kliot A, Ghanim M. Fluorescent in situ hybridization for the localization of viruses, bacteria and other microorganisms in insect and plant tissues. Methods. 2016;98:74–81. doi: 10.1016/j.ymeth.2015.12.003. [DOI] [PubMed] [Google Scholar]
- Klyachko O, Stein BD, Grindle N, Clay K, Fuqua C. Localization and visualization of a coxiella-type symbiont within the lone star tick, Amblyomma americanum. Appl Environ Microbiol. 2007;73:6584–6594. doi: 10.1128/AEM.00537-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchler SM, Kehl S, Dettner K. Characterization and localization of Rickettsia sp in water beetles of genus Deronectes (Coleoptera: Dytiscidae) FEMS Microbiol Ecol. 2009;68:201–211. doi: 10.1111/j.1574-6941.2009.00665.x. [DOI] [PubMed] [Google Scholar]
- Lange JV, el Dessouky AG, Manor E, Merdan AI, Azad AF. Spotted fever rickettsiae in ticks from the northern Sinai Governate, Egypt. Am J Trop Med Hyg. 1992;46:546–551. doi: 10.4269/ajtmh.1992.46.546. [DOI] [PubMed] [Google Scholar]
- Lenaerts J, Lappin-Scott HM, Porter J. Improved fluorescent in situ hybridization method for detection of bacteria from activated sludge and river water by using DNA molecular beacons and flow cytometry. Appl Environ Microbiol. 2007;73:2020–2023. doi: 10.1128/AEM.01718-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn GE, Oliver JD, Nelson CM, Felsheim RF, Kurtti TJ, Munderloh UG. Tissue distribution of the Ehrlichia muris-like agent in a tick vector. PLoS one. 2015;10:e0122007. doi: 10.1371/journal.pone.0122007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura Y, Kikuchi Y, Meng XY, Koga R, Fukatsu T. Novel clade of alphaproteobacterial endosymbionts associated with stinkbugs and other arthropods. Appl Environ Microbiol. 2012;78:4149–4156. doi: 10.1128/AEM.00673-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ment D, Churchill AC, Gindin G, Belausov E, Glazer I, Rehner SA, Rot A, Donzelli BG, Samish M. Resistant ticks inhibit Metarhizium infection prior to haemocoel invasion by reducing fungal viability on the cuticle surface. Environ Microbiol. 2012;14:1570–1583. doi: 10.1111/j.1462-2920.2012.02747.x. [DOI] [PubMed] [Google Scholar]
- Niebylski ML, Peacock MG, Fischer ER, Porcella SF, Schwan TG. Characterization of an endosymbiont infecting wood ticks, Dermacentor andersoni, as a member of the genus Francisella. Appl Environ Microbiol. 1997;63:3933–3940. doi: 10.1128/aem.63.10.3933-3940.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson K, Jaenson TG, Uhnoo I, Lindquist O, Pettersson B, Uhlen M, Friman G, Pahlson C. Characterization of a spotted fever group Rickettsia from Ixodes ricinus ticks in Sweden. J Clin Microbiol. 1997;35:243–247. doi: 10.1128/jcm.35.1.243-247.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantoja CR, Lightner DV. Detection of hepatopancreatic parvovirus (HPV) of penaeid shrimp by in situ hybridization at the electron microscope level. Dis Aquat Organ. 2001;44:87–96. doi: 10.3354/dao044087. [DOI] [PubMed] [Google Scholar]
- Parola P, Paddock CD, Socolovschi C, Labruna MB, Mediannikov O, Kernif T, Abdad MY, Stenos J, Bitam I, Fournier PE, Raoult D. Update on tick-borne rickettsioses around the world: a geographic approach. Clin Microbiol Rev. 2013;26:657–702. doi: 10.1128/CMR.00032-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perotti MA, Clarke HK, Turner BD, Braig HR. Rickettsia as obligate and mycetomic bacteria. FASEB J. 2006;20:2372–2374. doi: 10.1096/fj.06-5870fje. [DOI] [PubMed] [Google Scholar]
- Persing DH, Telford SR, 3rd, Spielman A, Barthold SW. Detection of Borrelia burgdorferi infection in Ixodes dammini ticks with the polymerase chain reaction. J Clin Microbiol. 1990;28:566–572. doi: 10.1128/jcm.28.3.566-572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan JN, Lu CR, Bender WG, Smoak RM, 3rd, Zhong J. Molecular detection and identification of Rickettsia species in Ixodes pacificus in California. Vector Borne Zoonotic Dis. 2011;11:957–961. doi: 10.1089/vbz.2010.0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piesman J, Clark KL, Dolan MC, Happ CM, Burkot TR. Geographic survey of vector ticks (Ixodes scapularis and Ixodes pacificus) for infection with the Lyme disease spirochete, Borrelia burgdorferi. J Vector Ecol. 1999;24:91–98. [PubMed] [Google Scholar]
- Piesman J, Schneider BS, Zeidner NS. Use of quantitative PCR to measure density of Borrelia burgdorferi in the midgut and salivary glands of feeding tick vectors. J Clin Microbiol. 2001;39:4145–4148. doi: 10.1128/JCM.39.11.4145-4148.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards AL. Worldwide detection and identification of new and old rickettsiae and rickettsial diseases. FEMS Immunol Med Microbiol. 2012;64:107–110. doi: 10.1111/j.1574-695X.2011.00875.x. [DOI] [PubMed] [Google Scholar]
- Roshdy MA. A rickettsialike microorganism in the tick Ornithodoros savignyi: Observations on its structure and distribution in the tissues of the tick. J Invertebr Pathol. 1968;11:155–169. doi: 10.1016/0022-2011(68)90146-8. [DOI] [PubMed] [Google Scholar]
- Roux V, Fournier PE, Raoult D. Differentiation of spotted fever group rickettsiae by sequencing and analysis of restriction fragment length polymorphism of PCR-amplified DNA of the gene encoding the protein rOmpA. J Clin Microbiol. 1996;34:2058–2065. doi: 10.1128/jcm.34.9.2058-2065.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassera D, Beninati T, Bandi C, Bouman EA, Sacchi L, Fabbi M, Lo N. ‘Candidatus Midichloria mitochondrii’, an endosymbiont of the tick Ixodes ricinus with a unique intramitochondrial lifestyle. Int J Syst Evol Microbiol. 2006;56:2535–2540. doi: 10.1099/ijs.0.64386-0. [DOI] [PubMed] [Google Scholar]
- Schonhuber W, Fuchs B, Juretschko S, Amann R. Improved sensitivity of whole-cell hybridization by the combination of horseradish peroxidase-labeled oligonucleotides and tyramide signal amplification. Appl Environ Microbiol. 1997;63:3268–3273. doi: 10.1128/aem.63.8.3268-3273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schriml LM, Padilla-Nash HM, Coleman A, Moen P, Nash WG, Menninger J, Jones G, Ried T, Dean M. Tyramide signal amplification (TSA)-FISH applied to mapping PCR-labeled probes less than 1 kb in size. Biotechniques. 1999;27:608–613. doi: 10.2144/99273pf01. [DOI] [PubMed] [Google Scholar]
- Smith RD, Sells DM, Stephenson EH, Ristic MR, Huxsoll DL. Development of Ehrlichia canis, causative agent of canine ehrlichiosis, in the tick Rhipicephalus sanguineus and its differentiation from a symbiotic Rickettsia. Am J Vet Res. 1976;37:119–126. [PubMed] [Google Scholar]
- Svendsen CB, Boye M, Struve C, Krogfelt KA. A novel fluorescent in situ hybridization technique for detection of Rickettsia spp. in archival samples. J Microbiol Methods. 2009;76:301–304. doi: 10.1016/j.mimet.2008.10.012. [DOI] [PubMed] [Google Scholar]
- Takechi K, SAKAMOTO W, KATSUHARA M, MURATA M, MOTOYOSHI F. In situ RNA hybridization using Technovit resin in Arabidopsis thaliana. Plant Mol Biol Rep. 1999;17:43–51. [Google Scholar]
- Whitworth T, Popov V, Han V, Bouyer D, Stenos J, Graves S, Ndip L, Walker D. Ultrastructural and genetic evidence of a reptilian tick, Aponomma hydrosauri, as a host of Rickettsia honei in Australia: possible transovarial transmission. Ann NY Acad Sci. 2003;990:67–74. doi: 10.1111/j.1749-6632.2003.tb07339.x. [DOI] [PubMed] [Google Scholar]
- Yano Y, Takada N, Fujita H. Ultrastructure of spotted fever rickettsialike microorganisms observed in tissues of Dermacentor taiwanensis (Acari: Ixodidae) J Med Entomol. 1993;30:579–585. doi: 10.1093/jmedent/30.3.579. [DOI] [PubMed] [Google Scholar]
- Zhong J, Jasinskas A, Barbour AG. Antibiotic treatment of the tick vector Amblyomma americanum reduced reproductive fitness. PloS one. 2007;2:e405. doi: 10.1371/journal.pone.0000405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zordan A. Fluorescence in situ hybridization on formalin-fixed, paraffin-embedded tissue sections. Methods Mol Biol. 2011;730:189–202. doi: 10.1007/978-1-61779-074-4_14. [DOI] [PubMed] [Google Scholar]
- Zwirglmaier K. Fluorescence in situ hybridisation (FISH)–the next generation. FEMS Microbiol Lett. 2005;246:151–158. doi: 10.1016/j.femsle.2005.04.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


