Abstract
Background
The human genome is hierarchically organized into local and long-range structures that help shape cell-type-specific transcription patterns. Transfer RNA (tRNA) genes (tDNAs), which are transcribed by RNA polymerase III (RNAPIII) and encode RNA molecules responsible for translation, are dispersed throughout the genome and, in many cases, linearly organized into genomic clusters with other tDNAs. Whether the location and three-dimensional organization of tDNAs contribute to the activity of these genes has remained difficult to address, due in part to unique challenges related to tRNA sequencing. We therefore devised integrated tDNA expression profiling, a method that combines RNAPIII mapping with biotin-capture of nascent tRNAs. We apply this method to the study of dynamic tRNA gene regulation during macrophage development and further integrate these data with high-resolution maps of 3D chromatin structure.
Results
Integrated tDNA expression profiling reveals domain-level and loop-based organization of tRNA gene transcription during cellular differentiation. tRNA genes connected by DNA loops, which are proximal to CTCF binding sites and expressed at elevated levels compared to non-loop tDNAs, change coordinately with tDNAs and protein-coding genes at distal ends of interactions mapped by in situ Hi-C. We find that downregulated tRNA genes are specifically marked by enhanced promoter-proximal binding of MAF1, a transcriptional repressor of RNAPIII activity, altogether revealing multiple levels of tDNA regulation during cellular differentiation.
Conclusions
We present evidence of both local and coordinated long-range regulation of human tDNA expression, suggesting the location and organization of tRNA genes contribute to dynamic tDNA activity during macrophage development.
Electronic supplementary material
The online version of this article (doi:10.1186/s13059-017-1310-3) contains supplementary material, which is available to authorized users.
Keywords: tDNA, tRNAome, Hi-C, Biotin-capture, CTCF, Topologically associating domains, TFIIIC
Background
The role of transfer RNAs (tRNAs) in deciphering the genetic code is universal to cell biology. The trinucleotide anticodon sequence of each tRNA-type decodes specific codons employed by messenger RNAs (mRNAs). Overall, the number of genes encoding each tRNA-type and the relative cellular abundance of each tRNA-type have been shown to correlate with the frequency of codon usage in species-specific and tissue-specific contexts, respectively [1–3]. In eukaryotes, changes in tRNA abundance have been reported across proliferative and senescent cell types and in response to specific perturbations, such as exposure to oxidation and alkylation-related stress [3–5]. Several important extra-translational functions for tRNA and tRNA-derived fragments have also become apparent, such as interfering with transposon reactivation and antagonizing the stability of oncogenic transcripts in breast cancer cells [6–8]. Thus, adjusting the level of cellular tRNA molecules, through both transcriptional and post-transcriptional mechanisms, may be important for modulating translation and potential ancillary activities.
Deciphering the mechanisms by which nascent tRNA levels are dynamically regulated, however, remains difficult to address, due in part to the unique challenges related to tRNA sequencing and alignment, as well as the unique complexity of tRNA biology [9]. In regard to sequencing, cellular tRNAs are heavily modified and consequently difficult to reverse transcribe during library preparation. In recent years, studies have tackled specific challenges associated with tRNA sequencing, or utilized alternate means, such as mapping RNA polymerase III as readout for tDNA expression [10–12]. The use of a dealkylating enzyme, ALKB, improves the fraction of full-length tRNA reads by demethylating sites that block reverse transcription [13–16]. However, sequencing of cellular tRNA levels alone provides little information about the transcriptional activity of tRNA genes, as nascent tRNAs undergo a complex maturation process [17]. Mapping of RNA polymerase III, meanwhile, represents an imperfect measure of tRNA gene activity that does not directly assay the level of nascently transcribed RNA. To this end, biotin-capture based genomic run-on experiments, such as BioGRO and precision nuclear run-on sequencing (PRO-seq), allow quantitative transcriptional profiling and mapping of RNA polymerases [18–21]. Thus, leveraging both RNA polymerase III occupancy with biotin-capture of nascent, demethylated tRNAs may provide a more accurate measure of tRNA gene expression in growing cells.
Interaction-based studies profiling the structure of eukaryotic chromosomes have identified highly self-interacting topological domains, a unit of three-dimensional (3D) organization that divides the genome into local neighborhoods of similar gene activity and restricts the ability of enhancers to influence non-target genes [22–29]. Recent studies mapping global interaction frequencies by in situ high-throughput chromosome conformation capture (in situ Hi-C) have further improved the resolution of physical domain identification and suggest that these contact domains are largely stable across cell types [30–32]. These short-range structures are often established within loops connected by inward oriented CTCF binding sites, an architectural protein originally described by its ability to function as an insulator, and by the cohesin complex and factors that control its association with DNA [30, 33–40]. tRNA genes, which are also enriched at the boundaries of topological domains and, in certain contexts, have been shown to function as insulator elements in the classical sense, have also been reported to play a role in the organization of eukaryotic chromosomes [28, 41–44]. However, to what degree tRNA genes are involved in long-range interactions in humans, and whether the 3D organization of tRNA genes contributes to the activity of these genes themselves remains unknown.
We have recently profiled the 3D organization and long-range interactome of human THP-1 monocytes and THP-1-derived macrophages through deeply sequenced in situ Hi-C experiments. High-resolution mapping of DNA loops identified both static and dynamic loop-based regulation of key macrophage genes during cellular differentiation [45]. THP-1 monocytes were differentiated into macrophages by treating with phorbol myristate acetate (PMA), which induces significant changes in cellular morphology and expression of cell surface markers characteristic of macrophages [46–48]. THP-1 cells, which typically grow in suspension, become adherent within 72 h post PMA treatment, providing a straightforward method for isolating relatively pure populations of non-differentiated monocytes and THP-1-derived macrophages [49]. Isolation of homogeneous cell populations is particularly appealing for the study of tRNA gene dynamics during cellular differentiation, as tDNAs, which are essential for biosynthesis, are likely to exhibit comparatively subtle changes in transcription.
Here we present integrated tDNA expression profiling, a method that combines RNAPIII occupancy mapping with biotin-capture of nascent, demethylated tRNAs. We apply this method to the study of dynamic tRNA gene regulation during macrophage development and further integrate these data with our recently described maps of 3D chromatin structures in the same cell types. Integrated tDNA expression profiling reveals domain-level and loop-based organization of transcription during cellular differentiation, as well as dynamic transcription factor (TF) binding coincident with changes in tDNA transcription, altogether revealing novel features of tRNA gene regulation.
Results
Integrated tDNA expression profiling in THP-1 monocytes
To directly measure tRNA gene transcription in human THP-1 cells, we combined a previously described tRNA demethylation strategy with a biotin-capture based method for isolating nascently transcribed RNAs. With this approach, the complex cytoplasmic mixture of intermediate, mature aminoacyl-tRNA and tRNA fragments that are captured by conventional RNA-sequencing (RNA-seq) methods do not preclude our ability to specifically query nascent tRNA levels. Mapping of tRNA fragments, nevertheless, remains problematic due to the multi-copy nature of tRNA genes; multiple alignment and analysis strategies have been proposed [50, 51]. For tDNA transcription profiling, we chose to map nascent RNA reads to the entire human genome space to avoid false positives arising from sequence reads that are unrelated to tRNAs [50]. Additionally, the presence of non-templated “CCA” at the 3’ terminus of mature tRNAs were not considered within the context of tRNA gene transcription. As an independent assay, we mapped the occupancy of RNA polymerase III genome-wide by chromatin immunoprecipitation sequencing (ChIP-seq), targeting the POLR3D (RPC4) subunit as previously described [21]. Overall, biotin-capture and RNA polymerase III mapping experiments show strong correlation in estimated tDNA expression values for individual genes (Fig. 1a). Because tRNA alignment is an imperfect process and RNA polymerase III mapping indirectly defines tDNA transcription, we chose to integrate both measures of tDNA expression for all downstream analyses (see “Methods”). Importantly, inspection of integrated tDNA expression estimates demonstrates high correlation and reproducibility across biological replicates (Additional file 1: Figure S1a, Pearson correlation coefficient = 0.978; p < 10^-16).
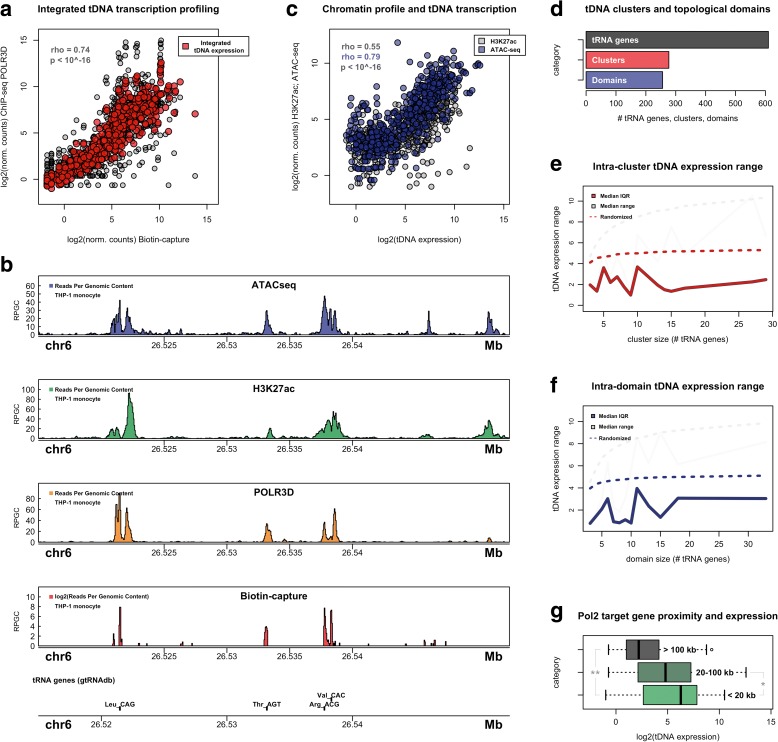
Fig. 1.
Integrated tDNA expression and chromatin profiling in THP-1 monocytes. a Correlation between tRNA gene expression as measured by biotin-capture of nascent, demethylated tRNAs and by RNA polymerase III occupancy mapping by ChIP-seq (black; Spearman’s rank correlation coefficient = 0.74; p < 10^-16). Integrated tDNA expression profile (red) utilizes the mean normalized count for each tRNA gene. b Example signal track representation of the chromatin accessibility (ATAC-seq, blue), active histone signature H3K27 acetylation (ChIP-seq, green), RNA polymerase III occupancy (ChIP-seq, orange), and nascently transcribed RNA (Biotin-capture, red) at a tDNA cluster located on chromosome 6. RPGC mean normalized reads per genomic content. c Correlation between integrated tDNA expression profile with H3K27ac ChIP-seq levels surrounding tRNA genes (black; Spearman’s rank correlation coefficient = 0.55; p < 10^-16) and with chromatin accessibility at tRNA genes as measured by ATAC-seq (blue; Spearman’s rank correlation coefficient = 0.79; p < 10^-16). d Number of tRNA genes, tDNA clusters, and physical contact domains containing tRNA genes in human THP-1 cells. e Median intra-cluster tDNA expression range (gray) and interquartile range (IQR; red) as a function of tDNA cluster size (number of tRNA genes present within each cluster). Dotted lines represent median range and IQR for randomly shuffled tRNA genes within tDNA clusters (100,000 permutations). f Median intra-domain tDNA expression range (gray) and IQR (blue) as a function of tDNA contact domain size (number of tRNA genes located within each contact domain). Dotted lines represent median range and IQR for randomly shuffled tRNA genes within contact domains (100,000 permutations). In the case of overlapping contact domains, tRNA genes were assigned to the single smallest resident domain. g Distribution of integrated tDNA expression values segregated by proximity to nearest RNA polymerase II-transcribed gene (>100 Kb; n = 81; 20–100 Kb; n = 232; < 20 Kb; n = 295; **p = 1.18^-10, *p = 0.02, Wilcoxon rank-sum test)
In total, we estimate the expression for 610 tRNA genes currently present in the human genomic tRNA database (gtRNAdb, hg19) [52, 53]. Integrated tDNA expression levels show a bimodal distribution consistent with previous reports suggesting that nearly half of all tRNA genes are not occupied by RNA polymerase III, resulting in little or no transcription (Additional file 1: Figure S1b) [54–56]. To better characterize the environmental context of tRNA gene transcription, we further profiled chromatin accessibility at tRNA genes using an assay for transposase accessible chromatin (ATAC-seq), as well as the level of histone H3K27 acetylation (H3K27ac), a histone modification positively associated with transcription levels at both RNA polymerase II and RNA polymerase III genes (Fig. 1b) [3, 57–60]. Correlation analysis reveals statistically significant relationships for both H3K27ac and chromatin accessibility with integrated tDNA expression levels at individual tRNA genes (Fig. 1c). The strong relationship observed between tDNA transcription and ATAC-seq (Spearman’s rank correlation coefficient = 0.79, p < 10^-16) suggests that DNA accessibility may uniquely capture the activity of short, non-coding tRNA genes, which are depleted of nucleosomes within the gene body and promoter regions [61]. Together, measures of tDNA accessibility and H3K27ac appropriately reflect the transcriptional activity of human tRNA genes and indicate that integrated tDNA expression profiling accurately captures nascent tRNA gene transcription levels.
Domain-level organization of human tRNA genes
We next considered whether the linear arrangement of tRNA genes into clusters and physical contact domains plays a role in the organization of tRNA gene transcription. Contact domains were annotated in THP-1 cells using the arrowhead algorithm as previously reported [30], which in total identifies more than 12,000 domains with enhanced contact frequency [45]. Altogether, we identify 256 physical contact domains containing one or more tRNA genes, with an average size of 2.3 resident tRNA genes per domain and a maximum size of 33 tRNA genes per domain (Fig. 1d). However, domain mapping alone does not encompass the entire human genome and thus we performed parallel analysis of tDNA clustering to comprehensively compare the expression of all human tRNA genes (Additional file 1: Figure S1c). Clusters were analogously defined as regions of DNA with one or more tDNAs, using a maximum tDNA-tDNA distance threshold of 20 Kb. In total, we identify 277 individual tDNA clusters with an average size of 2.2 tRNA genes per cluster and a maximum size of 29 tRNA genes per cluster (Fig. 1d).
We find that the median expression level for tRNA genes increases with the number of neighboring tDNAs, leveling off at a cluster size of approximately four tRNA genes (Additional file 1: Figure S1d). Inspection of individual multi-tDNA clusters and physical contact domains further suggests that proximal tRNA genes may be expressed at similar levels (Additional file 1: Figure S1e, f). To systematically determine whether tRNA genes present in the same cluster or domain do share similar gene activity, we compared the range and interquartile range (IQR) of tDNA transcription with a model in which tRNA genes are randomly assigned with respect to cluster and domain occupancy. Indeed, we find significantly lower spread in tRNA gene expression values across all cluster and domain sizes (Fig. 1e, f, Additional file 1: Figure S1g), suggesting cluster- and domain-level organization of human tDNAs group genes with similar transcriptional activity. We further find that tRNA gene expression positively associates with the proximity of tDNAs to genes transcribed by RNA polymerase II (Fig. 1g). These results agree with previous observations indicating correlative proximity and activity between neighboring RNA polymerase II and RNA polymerase III genes [60, 62, 63], and altogether argue that the surrounding context is an important aspect of tRNA gene transcription.
tDNA organization, transcription, and codon usage in THP-1 cells
Recent studies suggest that significant copy number variation may exist for tRNA genes across the human population [64]. We therefore performed a read-depth approach for estimating tRNA gene copy number in several deeply and moderately sequenced whole-genome datasets available through the 1000 Genomes Project [65]. Of note, we observe a high degree of variation across individuals at specific tDNAs, particularly at a set of genes encoding five distinct tRNA anticodon families that are located within a recently described variable number tandem repeat (VNTR) on chromosome 1 (Fig. 2a, b; asterisks, Additional file 1: Figure S2a, b) [66]. As expected, tDNA copy number estimates scale appropriately with VNTR number and, overall, our median tRNA gene copy number estimates show improved correlation between gene count and aggregate nascent tRNA levels for multi-tDNAs in THP-1 monocytes (Fig. 2c, Spearman’s rank correlation coefficient = 0.55; p = 0.0001, Additional file 1: Figure S2c, d).
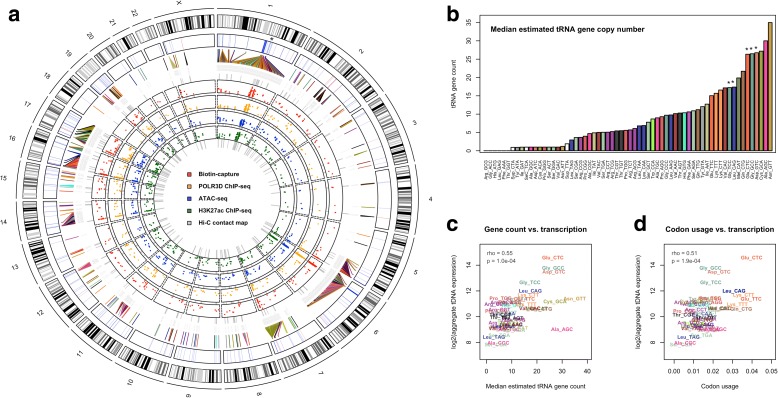
Fig. 2.
Organization and transcription of multicopy tRNA genes in humans. a Circular visualization of human tRNA gene coordinates across human chromosomes. Track descriptions from outermost moving inward: (1) Individual chromosome ideograms and cytogenetic band positions. (2) Location of tRNA genes (blue) and nuclear-encoded mitochondrial nmt-tRNA genes (red). *Asterisk represents approximate location of variable number tandem repeat (VNTR) on chromosome 1. (3) tRNA genes labeled by tRNA anticodon family and colored by amino acid isoacceptor family (note: not all tDNAs are directly labeled due to size and legibility constraints). (4) Biotin-capture based assay measuring nascent demethylated tRNAs (red). (5) ChIP-seq experiments mapping RNA polymerase III occupancy on tRNA genes (orange). (6) ATAC-seq experiments measuring chromatin accessibility at tRNA genes (blue). (7) ChIP-seq experiments capturing H3K27 acetylation levels at tRNA genes (green). (8) Link-plot of DNA loops identified by in situ Hi-C experiments proximal to tRNA genes (gray). b Median estimated human tRNA gene count for each anticodon tRNA family, colored by amino acid, as determined using a read-depth approach over several deeply sequenced whole-genome datasets from the 1000 Genomes Project. *Asterisks represent tRNA-types located on VNTR that show significant variation across sampled individuals. c Correlation between integrated tDNA expression, collapsed by anticodon tRNA family, with multi-copy tRNA gene count (Spearman’s rank correlation coefficient = 0.55; p = 1.0e-04). d Correlation between integrated tDNA expression, collapsed by anticodon tRNA family, and the frequency of cognate codon usage in the THP-1 transcriptome for multicopy tRNA genes (Spearman’s rank correlation coefficient = 0.51; p = 1.9e-04)
We observe a similar correlation between aggregate tRNA levels and the codon usage of the THP-1 transcriptome (Fig. 2d), further serving to validate the accuracy and quality of our integrated tDNA expression profiles. Transcription of tRNA genes is particularly better adjusted towards codon usage than gene count with respect to high copy number tDNAs (Additional file 1: Figure S2e). For example, tRNAAla-AGC and tRNAAsn-GTT represent two of the highest anticodon families in terms of estimated tRNA gene count, yet they decode moderately employed codons in THP-1 cells (Fig. 2c). The aggregate levels of nascent tRNAAla-AGC and tRNAAsn-GTT are instead better adjusted to codon usage (Fig. 2d), suggesting transcription of human tRNA genes is regulated beyond gene count.
Visualization of tRNA gene coordinates with respect to tRNA-type illustrates the level of overlap between distinct anticodon families across all human chromosomes (Fig. 2a). Hierarchical clustering of tRNA families by overlap frequencies, that is the number of times genes encoding two distinct tRNA-types are located within the same tDNA cluster, reveals preferential proximity between specific pairs of anticodon tRNA families (Additional file 1: Figure S2f). The tDNAs located within the VNTR on chromosome 1, for example, exhibit strong overlap frequencies and together represent several of the highest expressed tRNA species in THP-1 monocytes (Fig. 2b–d). Genes encoding tRNA-types with strong overlap frequencies tend to segregate by tDNA expression levels, reaffirming the important relationship between tRNA gene organization and transcription (Additional file 1: Figure S2f).
Dynamic transcription of tRNA genes during macrophage differentiation
The temporal dynamics of PMA-induced THP-1 differentiation include a distinct early response after 6 h, followed by a transition towards differentiation completion at 48–96 h, at which point THP-1 cells become adherent and express macrophage-related cell surface markers [48, 49]. We therefore profiled tDNA transcription, chromatin accessibility, and long-range interactions in adherent, THP-1-derived macrophages 72 h post PMA treatment (Fig. 3a). Comparison of tDNA expression before and after PMA treatment suggests that transcription of tRNA genes generally decreases during macrophage differentiation. This trend is consistently observed with respect to chromatin accessibility, RNA polymerase III occupancy, and nascent tRNA levels, both in terms of read density and normalized differential count-based analysis over all tRNA genes (Fig. 3b, Additional file 1: Figure S3a). Differential analysis of integrated tDNA expression profiles similarly identifies a bias of downregulation at most individual tRNA genes 72 h post PMA treatment (Fig. 3c). Nevertheless, transcription of several nuclear-encoded mitochondrial tRNA genes (nmt-tRNAs) increase in THP-1-derived macrophages (Fig. 3c, Additional file 1: Figure S3b–d). This finding is congruous with an observed increase in mitochondrial numbers during macrophage development and in PMA-stimulated THP-1 cells [48], suggesting a potential increase in the demand for mitochondrial protein synthesis.
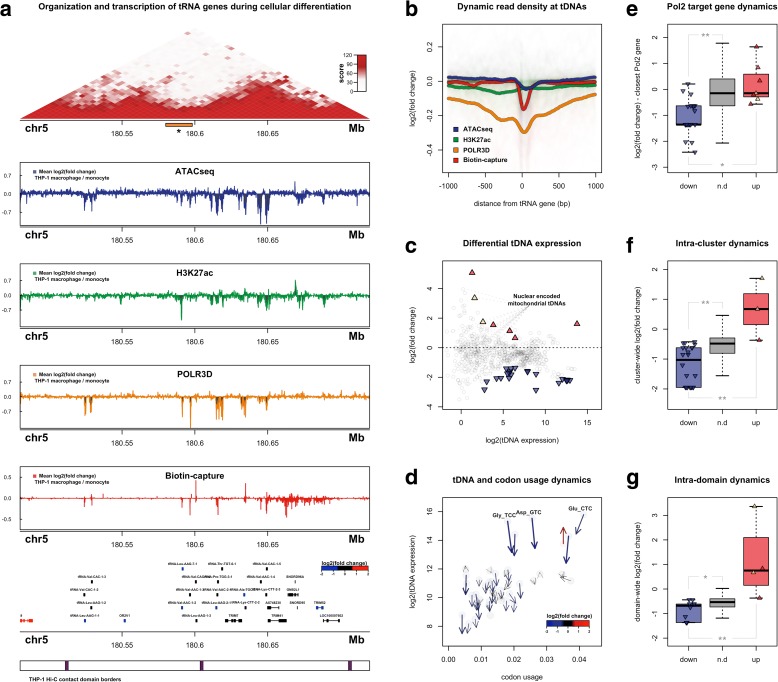
Fig. 3.
Dynamic domain-level regulation of tRNA gene transcription during macrophage differentiation. a Visualization of chromatin and transcriptional dynamics at an example tDNA locus on chromosome 5. Top: in situ Hi-C contact frequency matrix in THP-1 monocytes. *Asterisk represents long-range loop anchor region presented in Fig. 4c. Middle: mean log2(fold change) signal tracks for chromatin accessibility (ATAC-seq; blue), H3K27 acetylation (ChIP-seq; green), RNA polymerase III occupancy (ChIP-seq; orange), and nascent RNA (Biotin-capture RNA-seq; red) across two adjacent contact domains and neighboring tDNA clusters. Bottom: gene structure and physical contact domain border locations. log2(fold change) represents ± 72 h PMA treatment. Gene structure includes both tRNA genes and proximal RNAPII-transcribed genes. Lower panel depicts corresponding contact domain borders in THP-1 monocytes. b Mean log2(fold change) in normalized signal track read density for ATAC-seq, H3K27ac, POLR3D, and Biotin-capture RNA-seq across all human tRNA genes ± 1 Kb (−1000 upstream, +1000 downstream). c Mean log2(fold change) of integrated tDNA expression values across individual tRNA genes and mean log2(integrated tDNA expression) comparing THP-1 cells ± 72 h PMA treatment. Triangles represent individual tRNA genes that are upregulated (red) and downregulated (blue) at an FDR threshold of 0.15, including significant nmt-tDNAs (gold). d Arrow plot representation of aggregate nascent tRNA dynamics collapsed by tRNA-type vs. codon usage frequency (labeled by anticodon). Arrow-head represents log2(tDNA expression) and cognate codon usage after 72 h PMA treatment; arrow-bottom represents log2(tDNA expression) and cognate codon usage in untreated THP-1 monocytes. e log2(fold change) in closest RNAPII-transcribed genes for downregulated (blue), upregulated (red), and non-differential (n.d., gray) tRNA genes (top **p = 1.58^-5; bottom *p = 0.047, Wilcoxon rank-sum test). f Median cluster-wide log2(fold change) for tDNA clusters harboring downregulated (blue), upregulated (red), and non-differential (gray) tRNA genes (top **p = 0.0056, bottom **p = 6.41^-6, Wilcoxon rank-sum test). g Median contact domain-wide log2(fold change) for tDNA clusters harboring downregulated (blue), upregulated (red), and non-differential (gray) tRNA genes (top *p = 0.016; bottom **p = 0.0022, Wilcoxon rank-sum test)
The collective decrease in non-mitochondrial tDNA expression is consistent with previous comparisons of RNA polymerase III occupancy in human embryonic stem cells and induced pluripotent stem cells, which suggest that differentiation leads to a constricted RNAPIII repertoire [56]. We find that downregulation of nascent tRNA levels is most pronounced for the highest expressed tRNA-types, suggesting macrophage differentiation decreases the dynamic range of tRNA availability in THP-1 cells (Fig. 3c, d). This decrease in the most abundant tRNA anticodon families correlates with a decline in codon usage for several of the most frequently employed codons (Fig. 3d, Additional file 1: Figure S3h), suggesting a potentially coordinated decrease in the dynamic range for both tRNA supply and mRNA demand. Thus, we speculate that cell-type-specific tRNA levels may be adjusted in a manner that complements the dynamic range of codon usage rather than specific codon frequency optimization.
tDNA dynamics mirror the surrounding transcriptional environment of tRNA genes
Given the relationship between tDNA activity and proximity to RNA polymerase II genes, we next asked whether changes in tRNA gene expression coincide with the transcriptional environment surrounding differential tRNA genes. Indeed, tDNAs that decrease or increase significantly in THP-1-derived macrophages show enrichment for similar changes in nearby protein-coding genes (Fig. 3e, Additional file 1: Figure S3e). Beyond RNAPII transcribed genes, tDNAs also share similar transcriptional dynamics with tRNA genes that reside within the same cluster or topological domain. When tDNA transcription is compared with the median fold change across tDNA clusters and domains, increasing and decreasing tRNA genes again are biased towards genes the behave similarly, both for tDNA clusters (Fig. 3f) and for physical contact domains containing tRNA genes (Fig. 3g, Additional file 1: Figure S3f, g). Visual inspection of transcriptional and chromatin dynamics within specific tRNA domains and clusters illustrates that dynamic tDNA expression corresponds to changes in the surrounding environment. tRNA genes co-residing in clusters and contact domains within 200 Kb on chromosome 5, for example, exhibit similar transcriptional dynamics, both with nearby tRNA genes encoding distinct tRNA-types and with RNAPII-transcribed genes (Fig. 3a). A general decline in chromatin accessibility, H3K27 acetylation, and occupancy by RNA polymerase III is also observed across this locus, together suggesting that the topological organization of tDNAs within these physical contact domains may contribute to their expression and dynamic regulation during cellular differentiation.
Coordinated long-range regulation of tRNA genes during cellular differentiation
In addition to interactions enriched within physical contact domains, Hi-C captures DNA loops connecting distant loci, ranging between several hundred kilobases (Kb) to megabase (Mb) in size [30]. More than 16,000 DNA loops were identified in THP-1 cells at 10-Kb resolution [45]. Most tDNAs are within 100 Kb of an identified loop anchor and nearly 20% of all tRNA genes are directly located at the end of a DNA loop (Additional file 1: Figure S4a–c). We find that transcription of tRNA genes is significantly higher for tDNAs directly intersecting loop anchors compared to tRNA genes within 100 Kb or tRNA genes that are located beyond 100 Kb from a loop end (Fig. 4a). These results suggest that the 3D organization of tRNA genes, which might include interactions that bring tRNA genes together in RNAPIII transcription factories [67], may also be important for regulating tDNA transcription during cellular differentiation.
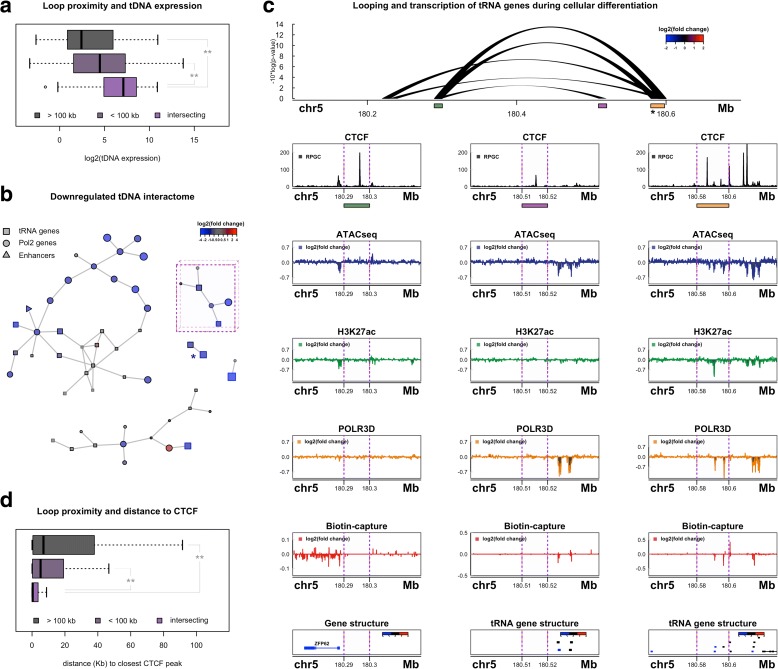
Fig. 4.
Coordinated long-range regulation of tRNA genes during cellular differentiation. a Distribution of integrated tDNA expression levels for tRNA genes > 100 Kb from a DNA loop end, within 100 Kb of a DNA loop end, and for tRNA genes that directly intersect DNA loop ends (left **p = 1.12^-6, right **p = 2.49^-11, Wilcoxon rank-sum test). b Network analysis of long-range interactions connecting tRNA genes downregulated in THP-1-derived macrophages. Each edge represents a DNA loop connecting two vertices (DNA loop anchors) that contain tRNA genes (square), RNAPII-transcribed genes (circle), or intergenic enhancers marked by H3K27 acetylation (triangle). Vertices with black frames represent loop anchors in which the identified feature (i.e. tRNA gene(s)) directly intersects the DNA loop end. Vertices without black frames represent loop anchors in which the identified feature is proximal to the DNA loop end (within 20 Kb). Both the size and color of each vertex is scaled by the mean log2(fold change) for resident feature(s). Purple outline marks the sub-community example further depicted in Fig. 4c. *Asterisk represents sub-community example further depicted in Additional file 1: Figure S5. c Visualization of chromatin and transcriptional dynamics at an example tDNA loop community located on chromosome 5. Colored rectangles define loop anchor regions further depicted below. *Asterisk represents loop anchor region depicted in Fig. 3a. Bottom left: signal track representation of CTCF binding sites (black, RPGC mean normalized reads per genomic content) and mean log2(fold change) for ATAC-seq (blue), H3K27ac (green), RNAPIII (orange), and nascent RNA (red) at the far-left loop anchor (green rectangle). Gene structure below includes RNAPII-transcribed gene ZFP62. Vertical dotted lines demarcate the actual loop anchor region. Bottom middle: analogous signal tracks depicting chromatin and transcriptional landscape at the middle loop anchor (purple rectangle) and proximal tDNA cluster. Bottom right: analogous signal tracks for the far-right loop anchor (orange rectangle) and intersecting tRNA genes. d Nearest distance to a CTCF binding site for tRNA genes that intersect DNA loop anchors, are within 100 Kb of a DNA loop anchor, or farther than 100 Kb from a DNA loop anchor (left **p = 2.31^-8, right **p = 6.61^-9, Wilcoxon rank-sum test)
We next asked what features are connected to tRNA genes by DNA loops and whether dynamic tDNA expression levels are coordinated by long-range interactions. Loop anchors identified by Hi-C often interact with more than one distant locus, forming multi-interaction networks or “hubs” that, in certain cases, connect multiple enhancers to target genes [45]. Analysis of all loop-associated downregulated tDNAs reveals a multi-interaction network that connects tRNA genes to other tDNAs as well as RNAPII-transcribed genes and intergenic enhancers marked by H3K27 acetylation (Fig. 4b). Features connected to tRNA genes by DNA loops show strikingly similar changes in transcription, suggesting tDNA dynamics are coordinated with other genes through long-range interactions during macrophage development (Fig. 4b). Visualization of individual tDNA interaction communities illustrates this phenomenon more clearly. For example, tRNA genes located at the boundary of adjacent tDNA contact domains on chromosome 5 (Fig. 3a) are directly located at a loop anchor that connects these tRNA genes to ZFP62, an RNA polymerase II-transcribed gene approximately 300 Kb upstream that concomitantly decreases in transcription in differentiating THP-1 cells (Fig. 4c, left and right signal track). DNA loops also bring ZFP62 in close spatial proximity to additional tDNAs that decrease in transcription and RNA polymerase III occupancy (Fig. 4c, middle signal track), altogether illustrating coordinated long-range transcriptional downregulation after treatment with PMA.
In contrast to downregulated tRNA genes, we find that tDNAs that increase in transcription during cellular differentiation form interaction networks with comparatively few genes and with genes and regulatory elements that do not show significant changes after treatment with PMA. However, we find that nuclear-encoded mitochondrial tRNA genes, which generally increase in expression in developing macrophages, are proximal to DNA loops that connect genes and enhancers that behave similarly after differentiation, further reaffirming an important relationship between long-range interactions and coordinated dynamic transcription of human tRNA genes (Additional file 1: Figure S4f, g).
Loop-based tRNA genes are connected by CTCF binding sites
Previous reports have identified enrichment for tRNA genes at the boundaries of topological domains, as well as a proximity relationship between specific tRNA genes and CTCF binding sites in eukaryotes [28, 62, 68–70]. Analysis of CTCF proximity with respect to tDNAs demonstrates that loop-associated tRNA genes are located near CTCF binding sites in THP-1 cells, consistent with recent models suggesting most long-range interactions are established by a loop extrusion complex that requires convergent CTCF binding sites (Fig. 4d) [36, 37, 40]. Inspection of DNA loops that bridge together tRNA genes clearly identifies CTCF binding at individual loop anchors, further suggesting that distal tRNA genes are brought together by CTCF (Fig. 4c). Supporting evidence for a functional role of CTCF in connecting tRNA genes is illustrated by an example in which two distinct clusters of tRNA genes, separated by more than 1.2 Mb on chromosome 6, interact via a DNA loop that is lost during macrophage differentiation, coincident with a loss of CTCF binding (Additional file 1: Figure S5). The tDNA clusters that are released by this long-range interaction are marked by decreasing tRNA gene transcription in THP-1 macrophages (Fig. 4b, asterisk), suggesting loss of DNA looping may be related to dynamic transcription regulation of tRNA genes.
Despite the observed relationship between tDNA transcription and CTCF-mediated long-range interactions, we do not identify any enrichment for differential tRNA genes at loop ends and, overall, changes in tDNA-associated long-range interactions are no more dynamic than non-tDNA loops (Additional file 1: Figure S4b, e). This suggests that while looping may be an important feature underlying tDNA expression levels, differentiation of THP-1 monocytes does not induce any widespread perturbation of the tDNA interactome in these cells. Indeed, regulation of tRNA genes is likely predominantly controlled by dynamic binding of TFs proximal to tDNAs within the framework of chromatin architecture. We therefore sought to further identify factors that might directly regulate tRNA gene expression during macrophage development.
Downregulation of tRNA genes coincides with enhanced MAF1 occupancy
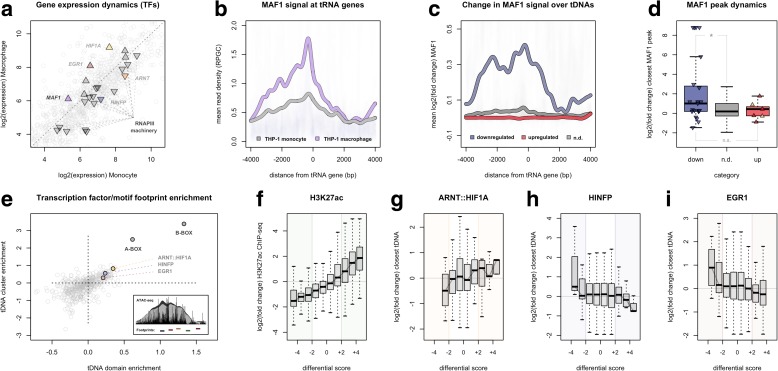
We find that the expression of TFs involved in RNA polymerase III initiation and transcription elongation moderately decreases after differentiation of THP-1 monocytes (Fig. 5a). This result is consistent with the downward bias for RNAPIII levels at tRNA genes after cellular differentiation and agrees with the expectation that tDNA regulation may be largely dictated through binding of TFIIIC and RNA polymerase III-related TFs. In contrast, nascent transcription of MAF1, a negative effector of RNA polymerase III activity, moderately increases after treatment with PMA (Fig. 5a). ChIP-seq experiments for MAF1 in monocytes and THP-1-derived macrophages confirm enhanced binding in the upstream promoter region for tRNA genes, particularly for tDNAs that decrease significantly after cellular differentiation (Fig. 5b, c). Downregulated tRNA genes also show significantly higher upregulation of MAF1 at the closest MAF1 peak (Fig. 5d, Additional file 1: Figure S6a), altogether consistent with the recently reported role of MAF1 in preventing RNA polymerase III recruitment and transcription initiation in humans [71]. The enhanced binding of MAF1 at a subset of tDNAs suggests that dynamic expression and binding of MAF1 is directed towards repressing specific tRNA genes during macrophage development and may therefore play an important role in controlling the dynamic range of nascent tRNA levels in humans. Collectively, these results agree with studies in yeast, in which deletion of the Maf1 transcriptional repressor results in differential sensitivity with respect to tRNA gene expression [72].
Fig. 5.
Differential transcription of tRNA genes coincides with enhanced MAF1 occupancy and dynamic TF binding. a Comparison of log2(expression) levels for TFs in THP-1 monocytes (x-axis) and THP-1 macrophages (y-axis). Triangles represent relevant genes encoding RNAPIII transcription machinery (black), MAF1 (purple), ARNT (orange), HIF1A (yellow), HINFP (blue), and EGR1 (red). Triangles do not denote statistical significance. b Mean read density of MAF1 ChIP-seq (RPGC mean normalized read per genomic content) at all tRNA genes in THP-1 cells before (gray) and after treatment with PMA (purple). c Mean log2(fold change) of MAF1 ChIP-seq read density at all downregulated (blue), upregulated (red), and non-differential (gray, n.d.) tRNA genes. d log2(fold change) of the closest MAF1 ChIP-seq peak for all downregulated (blue), upregulated (red), and non-differential (gray) tRNA genes (*p = 0.024, n.s. not statistically significant; Wilcoxon rank-sum test). e Enrichment of TF footprints identified by PIQ [75] within tDNA domains (x-axis) and clusters (y-axis). Enrichment measured as log10(observed/expected). Inset: illustration of ATAC-seq based footprinting analysis. Read ends represent hyperaccessible DNA adjacent to protected TF binding sites. f Comparison of dynamic H3K27ac levels at all footprints identified by PIQ. Differential accessibility scores were binned by the number of standard deviations from the average differential score across more than 2 million footprints (Additional file 1: Figure S6b). (+) denotes increased accessibility score, (−) denotes decreased accessibility score. Colored overlay represents strong differential accessibility score (≥2 standard deviations from the mean differential score) in THP-1-derived macrophages. g Comparison of dynamic ARNT::HIF1A footprint accessibility with the change in integrated tDNA expression at the nearest tDNA. h Comparison of dynamic HINFP footprint accessibility with the change in integrated tDNA expression at the nearest tDNA. i Comparison of dynamic EGR1 footprint accessibility with the change in integrated tDNA expression at the nearest tDNA
Transcription factor footprinting uncovers candidate regulators of tDNA expression
The ends of read fragments generated by ATAC-seq can be used to identify regions of DNA that are directly bound by TFs and protected against fragmentation [73, 74]. To this end, we applied the Protein Interaction Quantification (PIQ) footprinting algorithm on ATAC-seq data generated in THP-1 cells, identifying in total more than 2 million footprints for 516 distinct TF motifs (Additional file 1: Figure S6b) [75, 76]. TF binding analysis identifies enrichment for specific regulatory elements within tDNA domains and tDNA clusters. As expected, both the A-BOX and B-BOX motifs, which represent internal tDNA regulatory elements bound by the TF complex TFIIIC [77], are substantially more enriched in tDNA clusters and tDNA contact domains than any other regulatory motif (Fig. 5e). Beyond TFIIIC binding sites, we also identify enrichment for several non-tDNA related regulatory elements. We therefore sought to further compare changes in TF binding during cellular differentiation with dynamic transcription of tRNA genes.
We observe a strong correlation between dynamic H3K27ac, a mark of active enhancers and promoters, with changes in chromatin accessibility over all TF footprints identified by PIQ (Fig. 5f) [78, 79]. ChIP-seq measurements of CTCF binding at intersecting motifs further validates the ability of this approach to capture dynamic binding of specific TFs (Additional file 1: Figure S6c). Analysis of dynamic chromatin accessibility for footprints enriched within tDNA domains and clusters uncovers strong correlations between TF occupancy and changes in nearby tRNA genes for specific motifs. For example, differential accessibility at ARNT::HIF1A footprints correlates with changes in tDNA transcription at proximal tRNA genes (Fig. 5g). Bound ARNT::HIF1A elements are strongly enriched in both tDNA clusters and contact domains, supporting a potential role for these factors in tDNA transcription regulation (Fig. 5e). We additionally observe correlations between tDNA transcription and footprint dynamics at HINFP and EGR1 regulatory elements (Fig. 5h, i), which are also enriched within tDNA clusters and domains. The expression of HIF1A and EGR1, both of which have been shown to significantly increase in PMA-treated THP-1 cells [80, 81], are also upregulated in our system after cellular differentiation (Fig. 5a), further validating the dynamic binding captured at these specific regulatory elements and supporting a possible role for these factors in dynamic tRNA gene regulation.
Discussion
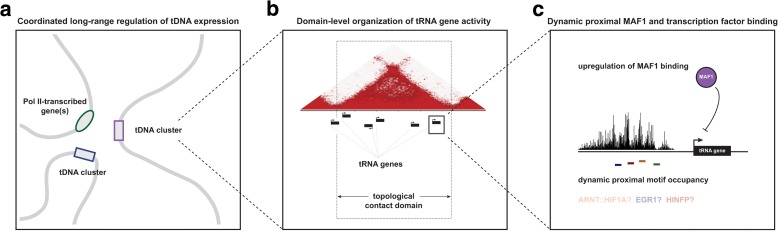
Integrated tDNA expression profiling uncovers dynamic transcription of individual tRNA genes during macrophage development in human THP-1 cells. By leveraging high-resolution maps of DNA interactions, chromatin accessibility, and histone and TF occupancy, we provide evidence of multi-level regulation of human tRNA genes during cellular differentiation, including long-range coordination of dynamic tDNA expression (Fig. 6a), cluster- and domain-level organization of tRNA gene activity (Fig. 6b), and canonical promoter-proximal regulation of individual tRNA genes (Fig. 6c). The systematic skew towards downregulation of most tRNA genes during monocyte-to-macrophage differentiation is consistent with previous RNA polymerase III mapping studies in other, distinct models of cellular differentiation. The repertoire of RNAPIII-bound and expressed tDNAs is more robust in human embryonic stem cells and induced pluripotent stem cells, for example, when compared to several differentiated cell types [56]. In the present study, we demonstrate for the first time that tDNA downregulation is organized across physical contact domains and through DNA loops connecting tRNA genes to distal tDNAs and RNAPII-transcribed genes. On the other hand, nuclear-encoded mitochondrial tDNAs are more likely to be upregulated and interact with genomic features that also increase after differentiation, a result that is congruous with an increasing number of mitochondria observed in THP-1-derived macrophages [48].
Fig. 6.
Model of multi-level tRNA gene regulation during macrophage development. a DNA loops bring together distal tRNA genes, RNAPII-transcribed genes, and enhancers that are coordinately downregulated during macrophage differentiation. b tDNAs are locally organized into clusters and physical contact domains that share similar activity and differential expression patterns in THP-1 cells. c Dynamic tDNA transcription correlates with proximal motif occupancy for specific, putative regulatory factors, such as HIF1A and EGR1. Downregulation of tRNA genes coincides with enhanced promoter-proximal binding of MAF1, a negative effector of RNAPIII activity
At the local level, downregulation of specific tRNA genes coincides with enhanced binding by MAF1, a transcriptional repressor that actively targets the promoter of RNAPIII genes [71, 82–88]. Though ectopic knock-down of MAF1 in human IMR90 fibroblasts leads to enhanced RNAPIII occupancy at all expressed tRNA genes, we show that MAF1 shows preferential enrichment after THP-1 differentiation at specific, downregulated tDNAs, consistent with differential sensitivity of tRNA genes to Maf1 in yeast [71, 72]. TF footprinting analyses in THP-1 monocytes and macrophages also reveal enrichment for specific DNA motifs located near tRNA genes (Fig. 5e). Furthermore, dynamic binding at certain motifs correlates with differential tDNA expression patterns during differentiation, suggesting a potential role in tRNA gene activity. Regulatory elements targeted by ARNT::HIF1A, for example, are enriched within tDNA clusters and domains and binding dynamics at these elements correlate with tRNA gene dynamics in THP-1 cells. Hypoxic stress, which induces the accumulation and nuclear translocation of ARNT and HIF1A, has been shown to suppress RNAPIII recruitment and tDNA expression in cardiomyocytes and increase levels of tRNA-derived fragments in breast cancer and mammary epithelial cells [7, 89–91]. Thus, whether and to what degree these factors directly and actively influence the level of nascent cellular tRNAs remains an intriguing subject for future research [80].
Our study suggests that the 3D organization of the human genome is an important underlying feature of tRNA gene expression. A subset of tDNAs participates in long-range interactions and is expressed at significantly elevated levels compared to non-loop tDNAs. Importantly, tRNA genes that are proximal or intersect DNA loop anchors are strongly enriched for CTCF and inspection of several tDNA-associated interaction sites reveals that CTCF, rather than the tRNA genes themselves, are centrally enriched at DNA loop anchors. This result suggests that while tRNA genes and sites bound by the TFIIIC TF complex may contribute to long-range chromosome organization and subnuclear localization in eukaryotes, in humans, loop-based cis-interactions appear to be predominantly determined by CTCF and factors controlling cohesin association with DNA [35, 41, 42, 44, 92–97].
Overall, DNA loops bridging tRNA genes to distal tDNAs and RNAPII transcribed genes are relatively stable after PMA-induced differentiation. We find that downregulated tDNAs interact with genomic features that are similarly downregulated, suggesting tDNAs and non-tRNA genes are coordinately regulated within the framework of a stable chromatin architecture established by CTCF. This result is consistent with a minimal change in CTCF binding observed during THP-1 differentiation and the predominant role of static rather than dynamic loops in bridging dynamic enhancers to key regulatory genes during macrophage development [45]. Nevertheless, we do observe specific examples of dynamic tDNA-associated loops, such as a tDNA–tDNA interaction on chromosome 6 that is concomitantly lost and marked by decreasing tRNA gene expression during differentiation (Addition file 5: Figure S5). Loss of this long-range interaction coincides with diminished CTCF binding, reaffirming the important role of CTCF in establishing DNA loops connecting tRNA genes. This result is consistent with a recent manuscript suggesting that knockdown of either CTCF or cohesin subunit Rad21 leads to dynamic expression of human tRNA genes in mouse tail fibroblasts [98].
The dynamic expression of individual tRNA genes during cellular differentiation supports a more complicated structure governing nascent tRNA levels than simply tDNA copy number. Instead, differential transcription of human tRNA genes may provide a system for refining cellular tRNA pools towards cell-type-specific demands, consistent with recent examples of tRNA dynamics in response to specific perturbations [4, 5, 99]. In THP-1 cells, the decrease in nascent tRNA levels is likely harmonized with a slow-down in biosynthesis, as differentiation is accompanied by a loss in cellular proliferation and decreasing levels of regulatory kinases that control entry and progression through the cell cycle [100, 101]. It is worth noting that we do not observe any significant correlation between changes in the tRNA-type abundance and codon usage, as might be expected if tDNA dynamics were specially tuned toward cell-type-specific codon usage (Additional file 1: Figure S7a). This result is perhaps consistent with a recent report suggesting translational efficiency is stable across diverse cell types, regardless of cellular tRNA pools [102]. Instead, we find that decreasing tDNA expression most significantly diminishes the level of highly expressed tRNAs, a phenomenon that appears to be coordinated with a decrease in codon usage for the most frequently employed codons (Additional file 1: Figure S7b, c). We therefore speculate that the range of tDNA expression may be coupled to the dynamic range in mRNA levels and resulting codon usage frequencies in different cell types, rather than specific anticodon-codon optimization. While the present study addresses dynamics of tRNA synthesis at the transcriptional level, to what degree these changes impact the cellular levels of mature, aminoacyl-tRNAs available for translation remains an important question for future research.
Conclusions
The transcription of tRNA genes, which are linearly organized into clusters and domains that share similar gene activity, generally decreases in developing macrophages, coincident with a decline in the dynamic range of transcriptomic codon usage. We find that downregulation of tRNA genes often occurs across topological domains and coordinately with other tRNA genes and RNA polymerase II-transcribed genes connected by DNA loops. We also show that MAF1, a negative effector of RNA polymerase III activity, increases at significantly downregulated tRNA genes during cellular differentiation, altogether revealing multiple levels of tDNA regulation during macrophage development.
Methods
THP-1 cell culture and differentiation
THP-1 cells were obtained from ATCC (Lot # 62454382) and grown for multiple passages in T-75 flasks of 2–8 × 105 cells/mL in growth medium containing RPMI-1640 (Corning), 10% fetal bovine serum, and 1% penicillin streptomycin. For differentiation of THP-1 cells, non-adherent cells were diluted to 2 × 105 cells/mL and grown overnight; a final concentration of 100 nM PMA was added the following morning. THP-1-derived macrophages were collected after 72-h exposure to PMA by aspirating media and any non-adherent cells and incubating adherent cells with TrypLE (ThermoFisher) for 15 min followed by cell wash in phosphate buffer saline (PBS) buffer.
Chromatin immunoprecipitation (ChIP)
Equal numbers of THP-1 monocytes and THP-1-derived macrophages were collected (~10 million cells per ChIP experiment) and resuspended in growth media at 1 × 106 cells/mL and cross-linked with rotation at room temperature in 1% formaldehyde for 10 min. Cross-linking was quenched with the addition of 200 mM glycine and an additional 5 min of rotation at room temperature. Cross-linked cells were then spun down and resuspended in 1× RIPA lysis buffer, followed by chromatin shearing via sonication (three cycles using a Branson sonicator: 30 s on, 60 s off; 15 additional cycles on a Bioruptor sonicator: 30 s on, 30 s off). Individual ChIP experiments were performed on pre-cleared chromatin using antibody-coupled Dynabead protein G (ThermoFisher) magnetic beads. Anti-histone H3 (acetyl K27) antibody was obtained from Abcam (ab4729), POLR3D antibody was obtained from abcam (ab86786; Lot# GR267691-1), MAF1 antibody was obtained from Santa Cruz Biotechnology (sc-365312 lot # G1411), and CTCF antibody was obtained from Millipore (07-729). A total of 3–5 ug of antibody per ChIP was coupled to 18-uL beads and rotated overnight with sheared chromatin at 4 °C. Beads were then washed 5× in ChIP wash buffer (Santa Cruz), 1× in TE, and chromatin eluted in TE + 1% SDS. Cross-linking was then reversed by incubation at 65 °C overnight, followed by digestion of RNA (30 min RNase incubation at 37 °C) and digestion of protein (30 min proteinase K incubation at 45 °C). ChIP DNA was then purified on a minElute column (Qiagen), followed by DNA library preparation and size selection of 350–550 bp fragments via gel extraction (Qiagen).
Assay for transposase accessible chromatin (ATAC-seq)
Equal numbers of THP-1 monocytes and THP-1-derived macrophages were collected (50,000 cells per ATAC-seq experiment) and washed with 1× ice-cold PBS. Cells were pelleted via centrifugation (500 × g, 5 min, 4 °C) and resuspended in cell lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% IGEPAL CA-630), and immediately spun down (500 × g, 10 min, 4 °C). The supernatant was then discarded, and transposition reaction carried out for 30 min at 37 °C with Tn5 transposase in transposition buffer (Illumina, cat#FC-121-1030). DNA was immediately purified on a minElute column (Qiagen), followed by PCR amplification using the NEBNext high-fidelity master mix (NEB cat#M0541) with nextera PCR primers and barcodes. PCR amplification was monitored as described [58], and gel purified to remove contaminating primer-dimer species.
Biotin-capture of nascent RNA
Equal numbers of THP-1 monocytes and THP-1-derived macrophages were collected (~5 million cells per experiment) and washed 3× in ice-cold PBS, followed by resuspension and 5 min incubation in 10 mL ice-cold swelling buffer (10 mM Tris-Cl pH 7.5, 2 mM MgCl2, 3 mM CaCl2). Cells were pelleted at 4 °C and resuspended in 1 mL lysis buffer (swelling buffer + 0.5% Igepal, 10% glycerol, 2 u/mL SUPERase In (Ambion), and gently mixed 20× by pipetting with p1000 (pipette tip cutoff to reduce shearing). Nuclei were then pelleted (1000 × g) and washed once with lysis buffer, pelleted (1000 × g), and resuspended in 1 mL nuclear storage buffer (50 mM Tris-Cl pH 8.3, 40% glycerol, 5 mM MgCl2, 100 nM EDTA). Nuclei were again pelleted and resuspended in 100 uL nuclear storage buffer. Nuclei were mixed with an equal volume of run-on reaction buffer (10 mM Tris-Cl pH 8.0, 5 mM MgCl2, 1 mM DTT, 300 mM KCl, 20 u SUPERase In (Ambion), 1% Sarkosyl, + 0.375 mM biotin-11-C/UTP (Perkin-Elmer)) + 0.0375 mM biotin-11-A/GTP (Perkin-Elmer)), and incubated for 3 min at 30 °C. RNA was then extracted and isolated using the mirVana small RNA isolation kit (AM1560; Lot# 1412093). A total of 1 ug of purified small RNA was then incubated for 2 h at room temperature in 100 uL demethylation reaction buffer (600 mM KCl, 4 mM MgCl2, 100 uM NH4FeSO4, 600 uM α-ketoglutarate, 4 mM L-ascorbic acid, 100 ug/mL bovine serum albumin [BSA], and 100 mM MES buffer [pH 5]) with 80 pmol ALKB, 160 pmol ALKB D135S. Expression and purification of tag-free ALKB (Lot# 716634S01) and ALKB D135S (Lot# 711466S04) was carried out by GenScript (Piscataway, NJ, USA) and stored at ~ 0.2–0.5 mg/mL in 50 mM Tris-HCl, 150 mM NaCl, 10% Glycerol, pH 8.0. Demethylation experiments were quenched in 10 mM EDTA, followed by pull-down of nascent biotinylated RNA via streptavidin magnetic beads (NEB #S1420S). Beads were washed in high salt buffer (2 M NaCl, 50 mM Tris-Cl pH 7.4, 0.5% Triton X-100), medium salt buffer (300 mM NaCl, 10 mM Tris-Cl pH 7.4, 0.1% Triton X-100), and low salt buffer (5 mM Tris-Cl pH 7.4, 0.1% Triton X-100), and RNA library preparation was subsequently carried out on beads, using the NEBnext small RNA library preparation kit (NEB #E7330S/L) with the minor modifications that RNA was denatured at 80 °C prior to adaptor ligation, and reverse transcription was carried out at increasing temperatures (50 °C for 1 h, 60 °C for 30 min, 70 °C for 15 min). Following PCR amplification (12 cycles), DNA library was purified on a minElute column and subsequently size selected to remove primer dimer contamination.
DNA sequencing and pre-processing
Biological replicates and experimental conditions (– PMA; + PMA) were sequenced together on an Illumina HiSeq2500 (paired-end, 100) for each individual experiment type (RNA-seq, ChIP-seq, biotin-capture) and sequencing reads trimmed using trim galore v. 0.4.0 (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/), before downstream sequence alignment and analyses.
Integrated tDNA expression profiling
tRNA gene annotation and coordinates were collected from the genomic tRNA database (gtRNAdb) for Homo sapiens (hg19 – NCBI Build 37.1 Feb 2009) by downloading the tRNAscan-SE results bed file http://gtrnadb.ucsc.edu/genomes/eukaryota/Hsapi19/ [52, 53]. Mapping of tRNA fragments to tDNA coordinates nevertheless remains an imperfect process due to the multi-copy nature of many tRNA genes. Multiple alignment and analysis strategies have been proposed with distinct decision-making trees [50, 51]. For our specific interest in tDNA transcription profiling, we chose to map nascent RNA reads to the entire genome space to avoid false positives arising from sequence reads that are unrelated to tRNAs, which may occur when aligned to a limited reference set of tRNA genes [50]. In addition, the biotin-capture protocol ensures that tRNA fragments represent nascently transcribed RNA and thus special consideration of pre-tRNA and processed tRNA fragments was unnecessary. The presence of non-templated “CCA” at the 3′ terminus of mature tRNAs was not considered within the context of tRNA gene transcription. We further chose to limit multi-mapping reads to a single “best” alignment location to avoid erroneously increasing tDNA transcription estimation (as would occur with multi-mapping) or decreasing tDNA transcription estimation (as would occur if all multi-mapping reads were discarded).
Specifically, sequencing reads were filtered to a minimum size of 18 bp and the first sequencing read aligned to the hg19 reference genome with bowtie version 1.1.1 using options –k 1 “best” [103]. Aligned nascent RNA sequence reads were extracted over the coordinates of all tRNA genes, ± 100 bp, in addition to all annotated genes. Normalized read counts for each condition replicate were determined with the DESeq package for differential expression, using the estimateSizeFactors function on count datasets and counts function with option normalized = TRUE [104]. For integrated tDNA expression measurement, the mean normalized counts over tRNA genes, determined independently by biotin-capture RNA-seq and POLR3D ChIP-seq experiments, were taken for each condition and replicate. Integration of these two independent measures, which shows strong agreement (Fig. 1a), was chosen to benefit from the unique advantages of each assay. Importantly, inspection of integrated tDNA expression estimates demonstrates high correlation and reproducibility across biological replicate both before and after treatment with PMA (Additional file 1: Figure S1a). Nevertheless, in some cases, discrepancies in RNAPIII occupancy and nascent tRNA levels are observed for a subset of tDNAs (Fig. 1a). These differences potentially arise from differences in mappability, technical challenges specific to tRNA-seq, or to the indirect nature between POLR3D occupancy mapping, which alone may not equate to productive or efficient transcription of a given (tRNA) gene. We believe these differences give merit to the need for integrated tDNA expression profiling. Changes in tRNA gene expression levels before and after PMA treatment were determined as the mean fold change of integrated tDNA transcription across two biological replicates. tRNA genes that were considered downregulated and upregulated after 72 h PMA treatment show congruent changes in independent biotin-capture and RNAPIII mapping experiments with a false discovery rate (FDR) threshold of 0.15 (FDR determined using the Benjamini and Hochberg corrected exact test p value against integrated tDNA expression estimates).
ChIP-seq analysis
Trimmed paired-end ChIP sequencing reads were mapped to the hg19 genome using bowtie version 1.1.1. with settings “bowtie –q –phred33-quals –X 2000 –fr –p 9 –S –chunkmbs 400” [103]. Mitochondrial reads were filtered and duplicate reads removed using Picard tools v. 1.92 (http://broadinstitute.github.io/picard). ChIP-seq peaks were identified for each condition using MACS2 v. 2.1.0 with options “macs2 callpeak –bdg –t –g hs” [105]. POLR3D ChIP-seq reads were extracted over tRNA genes, ± 100 bp, as well as all non-tRNA genes before normalization and integration with biotin-capture experiments for tDNA expression profiling. MAF1 binding dynamics were determined by differential count analysis over a merged MAF1 peak list using the exactTest function in the edgeR package for differential expression analysis [106, 107].
ATAC-seq and TF footprinting analysis
Trimmed ATAC sequencing reads were mapped to the hg19 genome using Bowtie v 2.2.4 with settings “bowtie2 –t –sensitive” [108]. Mapped reads were merged across several sequencing replicates, before filtering mitochondrial reads and removing duplicate reads with Picard tools v. 1.92 (http://broadinstitute.github.io/picard). ATAC-seq peaks were identified for each condition using MACS2 v. 2.1.0 with options “macs2 callpeak –bdg –nomdel –t –g hs” [105]. Changes in chromatin accessibility were determined by differential read count analysis using the glmTreat function in edgeR over a merged list of peaks identified in each condition and biological replicate. TF footprinting on ATAC-seq data was broken into two steps: identifying bound TF motifs and determining the differential binding score at motifs bound in either or both conditions. Bound TF motifs were identified using the PIQ pipeline against motifs annotated in the Jaspar Core Vertebrate Database (http://jaspar.genereg.net) [75, 76]. Motif matches against the hg19 reference genome were identified using the PIQ package pwmmatch.exact.r script. TF footprint scores were then determined for each motif using the PIQ package pertf.bg.r and common.r scripts with default settings. Motifs that were considered bound were filtered at a minimum positive prediction value (PPV) of 0.7, as previously described for bound motif calling [75]. Differential TF binding scores for each motif were then determined using the Wellington-bootstrap algorithm for differential footprinting [78], using the bootstrap.py script for differential footprinting with command-line option “-A” for ATAC-seq input, followed by the pyDNase dnase_ddhs_scorer.py script for differential score calling over footprints identified by PIQ. Differential accessibility scores were median normalized and subsequently binned by standard deviation.
Signal track and data visualization
Signal track data were generated from post-filtering read alignment bam files using the deeptools bamCompare tool [109, 110]. For individual sample conditions, normalized read per genomic count (RPGC) signal tracks were created by taking the mean ratio between biological replicates with bamCompare options “--ratio mean --normalizeTo1x 2451960000 --binSize 20 --smoothLength 60”. For ± PMA treatment comparison tracks, signal files were generated with bamCompare options “--pseudocount 30 --normalizeTo1x 2451960000 --binSize 20 --smoothLength 60”. Read density plots were generated using the deeptools computeMatrix tool with specified distances from strand-oriented tRNA genes [109, 110]. Signal track visualization and in situ Hi-C integration plots were generated using the Sushi package for genomic visualization [111]. Genome-wide tRNA gene circle visualization plots were generated using the RCircos package for Circos 2D track plots [112]. tDNA interactome network maps were generated using the R package iGraph for network analysis and visualization [113].
tDNA cluster, domain, and loop calling
Clusters of tRNA genes were defined as a stretch of tRNA genes separated by a maximum tDNA-tDNA distance of 20 Kb. In other words, tDNAs within 20 Kb of another tRNA gene were grouped, with cluster size increasing until no remaining tRNA genes were within the specified distance. In situ Hi-C contact domains were defined in THP-1 monocytes using the previously described arrowhead algorithm at 5-Kb resolution with default Juicer parameters [30]. In total, 12,272 contact domains were identified [45]. tDNA domains were defined as any Hi-C contact domain, profiled in THP-1 cells by high-throughput chromosome conformation capture [45], that contains any tRNA gene(s). tRNA clusters and domains were scored by the number of resident tRNA genes. In some cases, contact domains are located within a larger overlapping contact domain. Thus, certain analyses avoid redundant tDNA-domain calling by assigning individual tRNA genes to the smallest resident tDNA contact domain. On the other hand, several tRNA genes are not within an identifiable contact domain and thus parallel analysis of tDNA clustering presents an analogous means of determining the role of proximal tDNA gene regulation. Loop-associated tRNA genes were defined as tRNA genes that intersect either end of a loop (10-Kb resolution), and comparisons of distance between tRNA genes and long-range interactions calculated as the shortest distance between individual tDNAs and loop ends (bedtools).
Intra-cluster, intra-domain, and tDNA-interactome expression analyses
Intra-cluster range and IQR of integrated tDNA expression levels were determined for each unique tDNA cluster. tRNA genes were then randomly shuffled with respect to tDNA cluster assignment and the range and IQR permuted 100,000 times. Observed and expected ranges were compared across all clusters and domains (Additional file 1: Figure S1g) and with respect to cluster and domain size (Fig. 1e, f). In situ Hi-C contact domains containing tRNA genes were analyzed for intra-domain tDNA expression range and IQR in the same manner, with the exception that in cases where tRNA genes are present in multiple overlapping contact domains, these tDNAs were assigned to the single, smallest intersecting domain to avoid redundancy. tDNA interactome network analysis was generated using the iGraph R package for network analysis and visualization [113]. A graph object was created for all DNA loops mapped in THP-1 cells [45] with vertices representing loop anchors connected by edges (loops). All sub-network looping communities were detected using the fast greedy algorithm; communities that contain tRNA genes of interest (i.e. downregulated, upregulated, nmt-tDNA, etc.) were extracted for further analysis. All extracted community vertices were then characterized by intersecting or proximal features assigned by a ranking system: (1) intersecting tRNA genes; (2) proximal tRNA genes within four 10-Kb bins of a vertex; (3) non-tRNA genes that intersect a vertex; (4) non-tRNA genes that are proximal within two 10-Kb bins of a vertex; and (5) intergenic H3K27 acetylation peaks (putative enhancers) that directly intersect a vertex. Vertex shapes were determined by the highest ranked feature (tRNA genes = squares; non-tRNA genes = circles; putative enhancers = triangles), and the vertex shape and color scaled by the median log2(fold change) of the highest ranked feature(s).
Calculation of median haploid tRNA gene copy number
We utilized a read depth approach to estimate the median tRNA gene copy number in humans. Specifically, whole-genome sequencing data for 15 individuals in the 1000 Genomes Project (six high-coverage genomes: NA12878, NA12891, NA12892, NA19240, NA19239, NA19238; nine moderate-coverage genomes: NA10847, NA12890, NA18489, NA18499, NA18504, NA18505, NA18519, NA19098, and NA19099) were mapped to the hg19 reference genome. Read coverage over individual tRNA genes were compared to 1000 randomly shuffled blocks of the same length to determine background coverage. Read coverage was then collapsed by anticodon tRNA families and tRNA gene count estimated by comparison to collapsed randomized background coverage (Additional file 1: Figure S2a).
Acknowledgements
Illumina sequencing services were performed by the Stanford Center for Genomics and Personalized Medicine. We thank members of the Snyder lab for insightful discussion and feedback.
Funding
KVB is supported by National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant F32DK107112. DHP is supported by National Institutes of Health, National Human Genome Research Institute (NHGRI) grant R00HG008662. This research was supported by NIH Centers of Excellence in Genomic Science (CEGS) grant 5P50HG00773502, NIH NHGRI grant 5U01HG007919-03, and NIH grant 3U54HG00699604S1.
Availability of data and materials
The sequencing data from this study are publicly available under GEO series number GSE96800 through the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/). Previously reported in situ Hi-C data [45] is publicly available through Sequencing Read Archive (SRA) bioproject number PRJNA385337. Processed Hi-C data are available through the visualization software tool Juicebox (http://www.aidenlab.org/juicebox/). TF footprinting scripts were obtained from https://bitbucket.org/thashim/piq-single (PIQ R scripts pwmmatch.exact.r, pairedbam2rdata.r, pertf.bg.r and common.r) and http://pythonhosted.org/pyDNase/ (Wellington-bootstrap python scripts wellington-bootstrap.py and dnase_ddhs_scorer.py).
Additional file
Domain and cluster-level organization of tDNA transcription (related to Fig. 1). Figure S2. tDNA copy number estimation and transcription in THP-1 cells (related to Fig. 2). Figure S3. Dynamic chromatin and tRNA transcription profiles during differentiation (related to Fig. 3). Figure S4. Dynamic chromatin and tDNA transcription profiles during differentiation (related to Fig. 4). Figure S5. Concurrent downregulation of tDNA looping and transcription (related to Fig. 4). Figure S6. MAF1 binding and differential TF footprinting during THP-1 differentiation (related to Fig. 5). Figure S7. Dynamic range of codon usage and aggregate tDNA expression levels during THP-1 differentiation. (PDF 8237 kb)
Authors’ contributions
KVB, DHP, and MPS conceived the research project. KVB and DHP carried out molecular experiments and bioinformatic analyses. KVB and MPS wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Competing interests
MPS is a founder and member of the science advisory board of Personalis and Qbio and a science advisory board member of Genapsys and Epinomics.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s13059-017-1310-3) contains supplementary material, which is available to authorized users.
References
- 1.Qian W, Yang JR, Pearson NM, Maclean C, Zhang J. Balanced codon usage optimizes eukaryotic translational efficiency. PLoS Genet. 2012;8:e1002603. doi: 10.1371/journal.pgen.1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Novoa EM, Ribas de Pouplana L. Speeding with control: codon usage, tRNAs, and ribosomes. Trends Genet. 2012;28:574–81. doi: 10.1016/j.tig.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 3.Gingold H, Tehler D, Christoffersen NR, Nielsen MM, Asmar F, Kooistra SM, et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell. 2014;158:1281–92. doi: 10.1016/j.cell.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 4.Pelechano V, Wei W, Steinmetz LM. Widespread co-translational RNA decay reveals ribosome dynamics. Cell. 2015;161:1400–12. doi: 10.1016/j.cell.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pang YL, Abo R, Levine SS, Dedon PC. Diverse cell stresses induce unique patterns of tRNA up- and down-regulation: tRNA-seq for quantifying changes in tRNA copy number. Nucleic Acids Res. 2014;42:e170. doi: 10.1093/nar/gku945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schorn AJ, Gutbrod MJ, LeBlanc C, Martienssen R. LTR-Retrotransposon control by tRNA-derived small RNAs. Cell. 2017;170:61–71. doi: 10.1016/j.cell.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, Tavazoie SF. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015;161:790–802. doi: 10.1016/j.cell.2015.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodarzi H, Nguyen HCB, Zhang S, Dill BD, Molina H, Tavazoie SF. Modulated expression of specific tRNAs drives gene expression and cancer progression. Cell. 2016;165:1416–27. doi: 10.1016/j.cell.2016.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24:1832–60. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kutter C, Brown GD, Goncalves A, Wilson MD, Watt S, Brazma A, et al. Pol III binding in six mammals shows conservation among amino acid isotypes despite divergence among tRNA genes. Nat Genet. 2011;43:948–55. doi: 10.1038/ng.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmitt BM, Rudolph KL, Karagianni P, Fonseca NA, White RJ, Talianidis I, et al. High-resolution mapping of transcriptional dynamics across tissue development reveals a stable mRNA-tRNA interface. Genome Res. 2014;24:1797–807. doi: 10.1101/gr.176784.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts DN, Stewart AJ, Huff JT, Cairns BR. The RNA polymerase III transcriptome revealed by genome-wide localization and activity-occupancy relationships. Proc Natl Acad Sci U S A. 2003;100:14695–700. doi: 10.1073/pnas.2435566100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cozen AE, Quartley E, Holmes AD, Hrabeta-Robinson E, Phizicky EM, Lowe TM. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat Methods. 2015;12:879–84. doi: 10.1038/nmeth.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng G, Qin Y, Clark WC, Dai Q, Yi C, He C, et al. Efficient and quantitative high-throughput tRNA sequencing. Nat Methods. 2015;12:835–7. doi: 10.1038/nmeth.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu F, Clark W, Luo G, Wang X, Fu Y, Wei J, et al. ALKBH1-mediated tRNA demethylation regulates translation. Cell. 2016;167:1897. doi: 10.1016/j.cell.2016.11.045. [DOI] [PubMed] [Google Scholar]
- 16.Hrabeta-Robinson E, Marcus E, Cozen AE, Phizicky EM, Lowe TM. High-throughput small RNA sequencing enhanced by AlkB-facilitated RNA de-methylation (ARM-Seq) Methods Mol Biol. 2017;1562:231–43. doi: 10.1007/978-1-4939-6807-7_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phizicky EM, Hopper AK. tRNA processing, modification, and subcellular dynamics: past, present, and future. RNA. 2015;21:483–5. doi: 10.1261/rna.049932.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahat DB, Kwak H, Booth GT, Jonkers IH, Danko CG, Patel RK, et al. Base-pair-resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO-seq) Nat Protoc. 2016;11:1455–76. doi: 10.1038/nprot.2016.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jordan-Pla A, Gupta I, de Miguel-Jimenez L, Steinmetz LM, Chavez S, Pelechano V, et al. Chromatin-dependent regulation of RNA polymerases II and III activity throughout the transcription cycle. Nucleic Acids Res. 2015;43:787–802. doi: 10.1093/nar/gku1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jordan-Pla A, Miguel A, Serna E, Pelechano V, Perez-Ortin JE. Biotin-Genomic Run-On (Bio-GRO): A high-resolution method for the analysis of nascent transcription in yeast. Methods Mol Biol. 2016;1361:125–39. doi: 10.1007/978-1-4939-3079-1_8. [DOI] [PubMed] [Google Scholar]
- 21.James Faresse N, Canella D, Praz V, Michaud J, Romascano D, Hernandez N. Genomic study of RNA polymerase II and III SNAPc-bound promoters reveals a gene transcribed by both enzymes and a broad use of common activators. PLoS Genet. 2012;8:e1003028. doi: 10.1371/journal.pgen.1003028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Symmons O, Uslu VV, Tsujimura T, Ruf S, Nassari S, Schwarzer W, et al. Functional and topological characteristics of mammalian regulatory domains. Genome Res. 2014;24:390–400. doi: 10.1101/gr.163519.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lupianez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–25. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hnisz D, Day DS, Young RA. Insulated neighborhoods: structural and functional units of mammalian gene control. Cell. 2016;167:1188–200. doi: 10.1016/j.cell.2016.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159:374–87. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Symmons O, Pan L, Remeseiro S, Aktas T, Klein F, Huber W, et al. The Shh topological domain facilitates the action of remote enhancers by reducing the effects of genomic distances. Dev Cell. 2016;39:529–43. doi: 10.1016/j.devcel.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andrey G, Montavon T, Mascrez B, Gonzalez F, Noordermeer D, Leleu M, et al. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science. 2013;340:1234167. doi: 10.1126/science.1234167. [DOI] [PubMed] [Google Scholar]
- 28.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–5. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–80. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darrow EM, Huntley MH, Dudchenko O, Stamenova EK, Durand NC, Sun Z, et al. Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture. Proc Natl Acad Sci U S A. 2016;113:E4504–4512. doi: 10.1073/pnas.1609643113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–6. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuin J, Dixon JR, van der Reijden MI, Ye Z, Kolovos P, Brouwer RW, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci U S A. 2014;111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heidari N, Phanstiel DH, He C, Grubert F, Jahanbani F, Kasowski M, et al. Genome-wide map of regulatory interactions in the human genome. Genome Res. 2014;24:1905–17. doi: 10.1101/gr.176586.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Bortle K, Nichols MH, Li L, Ong CT, Takenaka N, Qin ZS, et al. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol. 2014;15:R82. doi: 10.1186/gb-2014-15-5-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanborn AL, Rao SS, Huang SC, Durand NC, Huntley MH, Jewett AI, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci U S A. 2015;112:E6456–6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nichols MH, Corces VG. A CTCF code for 3D genome architecture. Cell. 2015;162:703–5. doi: 10.1016/j.cell.2015.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nora EP, Goloborodko A, Valton AL, Gibcus JH, Uebersohn A, Abdennur N, et al. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell. 2017;169:930–44. doi: 10.1016/j.cell.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haarhuis JHI, van der Weide RH, Blomen VA, Yanez-Cuna JO, Amendola M, van Ruiten MS, et al. The cohesin release factor WAPL restricts chromatin loop extension. Cell. 2017;169:693–707. doi: 10.1016/j.cell.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. Formation of chromosomal domains by loop extrusion. Cell Rep. 2016;15:2038–49. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Snider CE, Stephens AD, Kirkland JG, Hamdani O, Kamakaka RT, Bloom K. Dyskerin, tRNA genes, and condensin tether pericentric chromatin to the spindle axis in mitosis. J Cell Biol. 2014;207:189–99. doi: 10.1083/jcb.201405028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raab JR, Chiu J, Zhu J, Katzman S, Kurukuti S, Wade PA, et al. Human tRNA genes function as chromatin insulators. EMBO J. 2012;31:330–50. doi: 10.1038/emboj.2011.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ebersole T, Kim JH, Samoshkin A, Kouprina N, Pavlicek A, White RJ, et al. tRNA genes protect a reporter gene from epigenetic silencing in mouse cells. Cell Cycle. 2011;10:2779–91. doi: 10.4161/cc.10.16.17092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noma K, Cam HP, Maraia RJ, Grewal SI. A role for TFIIIC transcription factor complex in genome organization. Cell. 2006;125:859–72. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 45.Phanstiel DH, Van Bortle K, Spacek DV, Hess G, Shamim MS, Machol I, et al. Static and Dynamic DNA Loops form AP-1-Bound Activation Hubs during Macrophage Development. Mol Cell. 2017. http://dx.doi.org/10.1016/j.molcel.2017.08.006. [DOI] [PMC free article] [PubMed]
- 46.Tsuchiya S, Kobayashi Y, Goto Y, Okumura H, Nakae S, Konno T, et al. Induction of maturation in cultured human monocytic leukemia cells by a phorbol diester. Cancer Res. 1982;42:1530–6. [PubMed] [Google Scholar]
- 47.Auwerx J. The human leukemia cell line, THP-1: a multifacetted model for the study of monocyte-macrophage differentiation. Experientia. 1991;47:22–31. doi: 10.1007/BF02041244. [DOI] [PubMed] [Google Scholar]
- 48.Daigneault M, Preston JA, Marriott HM, Whyte MK, Dockrell DH. The identification of markers of macrophage differentiation in PMA-stimulated THP-1 cells and monocyte-derived macrophages. PLoS One. 2010;5:e8668. doi: 10.1371/journal.pone.0008668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kouno T, de Hoon M, Mar JC, Tomaru Y, Kawano M, Carninci P, et al. Temporal dynamics and transcriptional control using single-cell gene expression analysis. Genome Biol. 2013;14:R118. doi: 10.1186/gb-2013-14-10-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Telonis AG, Loher P, Kirino Y, Rigoutsos I. Consequential considerations when mapping tRNA fragments. BMC Bioinf. 2016;17:123. doi: 10.1186/s12859-016-0921-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Selitsky SR, Sethupathy P. tDRmapper: challenges and solutions to mapping, naming, and quantifying tRNA-derived RNAs from human small RNA-sequencing data. BMC Bioinf. 2015;16:354. doi: 10.1186/s12859-015-0800-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chan PP, Lowe TM. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016;44:D184–189. doi: 10.1093/nar/gkv1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Canella D, Bernasconi D, Gilardi F, LeMartelot G, Migliavacca E, Praz V, et al. A multiplicity of factors contributes to selective RNA polymerase III occupancy of a subset of RNA polymerase III genes in mouse liver. Genome Res. 2012;22:666–80. doi: 10.1101/gr.130286.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Canella D, Praz V, Reina JH, Cousin P, Hernandez N. Defining the RNA polymerase III transcriptome: Genome-wide localization of the RNA polymerase III transcription machinery in human cells. Genome Res. 2010;20:710–21. doi: 10.1101/gr.101337.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alla RK, Cairns BR. RNA polymerase III transcriptomes in human embryonic stem cells and induced pluripotent stem cells, and relationships with pluripotency transcription factors. PLoS One. 2014;9:e85648. doi: 10.1371/journal.pone.0085648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–8. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. 2015;109:21–9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sagi D, Rak R, Gingold H, Adir I, Maayan G, Dahan O, et al. Tissue- and time-specific expression of otherwise identical tRNA genes. PLoS Genet. 2016;12:e1006264. doi: 10.1371/journal.pgen.1006264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raha D, Wang Z, Moqtaderi Z, Wu L, Zhong G, Gerstein M, et al. Close association of RNA polymerase II and many transcription factors with Pol III genes. Proc Natl Acad Sci U S A. 2010;107:3639–44. doi: 10.1073/pnas.0911315106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Helbo AS, Lay FD, Jones PA, Liang G, Gronbaek K. Nucleosome positioning and NDR structure at RNA polymerase III promoters. Sci Rep. 2017;7:41947. doi: 10.1038/srep41947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moqtaderi Z, Wang J, Raha D, White RJ, Snyder M, Weng Z, et al. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol. 2010;17:635–40. doi: 10.1038/nsmb.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.White RJ. Transcription by RNA polymerase III: more complex than we thought. Nat Rev Genet. 2011;12:459–63. doi: 10.1038/nrg3001. [DOI] [PubMed] [Google Scholar]
- 64.Iben JR, Maraia RJ. tRNA gene copy number variation in humans. Gene. 2014;536:376–84. doi: 10.1016/j.gene.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Genomes Project Consortium. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Darrow EM, Chadwick BP. A novel tRNA variable number tandem repeat at human chromosome 1q23.3 is implicated as a boundary element based on conservation of a CTCF motif in mouse. Nucleic Acids Res. 2014;42:6421–35. doi: 10.1093/nar/gku280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Melnik S, Deng B, Papantonis A, Baboo S, Carr IM, Cook PR. The proteomes of transcription factories containing RNA polymerases I, II or III. Nat Methods. 2011;8:963–8. doi: 10.1038/nmeth.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oler AJ, Alla RK, Roberts DN, Wong A, Hollenhorst PC, Chandler KJ, et al. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol. 2010;17:620–8. doi: 10.1038/nsmb.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smith ST, Wickramasinghe P, Olson A, Loukinov D, Lin L, Deng J, et al. Genome wide ChIP-chip analyses reveal important roles for CTCF in Drosophila genome organization. Dev Biol. 2009;328:518–28. doi: 10.1016/j.ydbio.2008.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yuen KC, Slaughter BD, Gerton JL. Condensin II is anchored by TFIIIC and H3K4me3 in the mammalian genome and supports the expression of active dense gene clusters. Sci Adv. 2017;3:e1700191. doi: 10.1126/sciadv.1700191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Orioli A, Praz V, Lhote P, Hernandez N. Human MAF1 targets and represses active RNA polymerase III genes by preventing recruitment rather than inducing long-term transcriptional arrest. Genome Res. 2016;26:624–35. doi: 10.1101/gr.201400.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Turowski TW, Lesniewska E, Delan-Forino C, Sayou C, Boguta M, Tollervey D. Global analysis of transcriptionally engaged yeast RNA polymerase III reveals extended tRNA transcripts. Genome Res. 2016;26:933–44. doi: 10.1101/gr.205492.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Song L, Zhang Z, Grasfeder LL, Boyle AP, Giresi PG, Lee BK, et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 2011;21:1757–67. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gusmao EG, Allhoff M, Zenke M, Costa IG. Analysis of computational footprinting methods for DNase sequencing experiments. Nat Methods. 2016;13:303–9. doi: 10.1038/nmeth.3772. [DOI] [PubMed] [Google Scholar]
- 75.Sherwood RI, Hashimoto T, O’Donnell CW, Lewis S, Barkal AA, van Hoff JP, et al. Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nat Biotechnol. 2014;32:171–8. doi: 10.1038/nbt.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mathelier A, Fornes O, Arenillas DJ, Chen CY, Denay G, Lee J, et al. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2016;44:D110–115. doi: 10.1093/nar/gkv1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Male G, von Appen A, Glatt S, Taylor NM, Cristovao M, Groetsch H, et al. Architecture of TFIIIC and its role in RNA polymerase III pre-initiation complex assembly. Nat Commun. 2015;6:7387. doi: 10.1038/ncomms8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Piper J, Assi SA, Cauchy P, Ladroue C, Cockerill PN, Bonifer C, et al. Wellington-bootstrap: differential DNase-seq footprinting identifies cell-type determining transcription factors. BMC Genomics. 2015;16:1000. doi: 10.1186/s12864-015-2081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.He HH, Meyer CA, Chen MW, Jordan VC, Brown M, Liu XS. Differential DNase I hypersensitivity reveals factor-dependent chromatin dynamics. Genome Res. 2012;22:1015–25. doi: 10.1101/gr.133280.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Oda T, Hirota K, Nishi K, Takabuchi S, Oda S, Yamada H, et al. Activation of hypoxia-inducible factor 1 during macrophage differentiation. Am J Physiol Cell Physiol. 2006;291:C104–113. doi: 10.1152/ajpcell.00614.2005. [DOI] [PubMed] [Google Scholar]
- 81.Akuzawa N, Kurabayashi M, Ohyama Y, Arai M, Nagai R. Zinc finger transcription factor Egr-1 activates Flt-1 gene expression in THP-1 cells on induction for macrophage differentiation. Arterioscler Thromb Vasc Biol. 2000;20:377–84. doi: 10.1161/01.ATV.20.2.377. [DOI] [PubMed] [Google Scholar]
- 82.Boguta M. Maf1, a general negative regulator of RNA polymerase III in yeast. Biochim Biophys Acta. 2013;1829:376–84. doi: 10.1016/j.bbagrm.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 83.Rideout EJ, Marshall L, Grewal SS. Drosophila RNA polymerase III repressor Maf1 controls body size and developmental timing by modulating tRNAiMet synthesis and systemic insulin signaling. Proc Natl Acad Sci U S A. 2012;109:1139–44. doi: 10.1073/pnas.1113311109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vannini A, Ringel R, Kusser AG, Berninghausen O, Kassavetis GA, Cramer P. Molecular basis of RNA polymerase III transcription repression by Maf1. Cell. 2010;143:59–70. doi: 10.1016/j.cell.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 85.Gajda A, Towpik J, Steuerwald U, Muller CW, Lefebvre O, Boguta M. Full repression of RNA polymerase III transcription requires interaction between two domains of its negative regulator Maf1. J Biol Chem. 2010;285:35719–27. doi: 10.1074/jbc.M110.125286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ciesla M, Boguta M. Regulation of RNA polymerase III transcription by Maf1 protein. Acta Biochim Pol. 2008;55:215–25. [PubMed] [Google Scholar]
- 87.Goodfellow SJ, Graham EL, Kantidakis T, Marshall L, Coppins BA, Oficjalska-Pham D, et al. Regulation of RNA polymerase III transcription by Maf1 in mammalian cells. J Mol Biol. 2008;378:481–91. doi: 10.1016/j.jmb.2008.02.060. [DOI] [PubMed] [Google Scholar]
- 88.Rollins J, Veras I, Cabarcas S, Willis I, Schramm L. Human Maf1 negatively regulates RNA polymerase III transcription via the TFIIB family members Brf1 and Brf2. Int J Biol Sci. 2007;3:292–302. doi: 10.7150/ijbs.3.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dengler VL, Galbraith MD, Espinosa JM. Transcriptional regulation by hypoxia inducible factors. Crit Rev Biochem Mol Biol. 2014;49:1–15. doi: 10.3109/10409238.2013.838205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ernens I, Goodfellow SJ, Innes F, Kenneth NS, Derblay LE, White RJ, et al. Hypoxic stress suppresses RNA polymerase III recruitment and tRNA gene transcription in cardiomyocytes. Nucleic Acids Res. 2006;34:286–94. doi: 10.1093/nar/gkj402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guimaraes-Camboa N, Stowe J, Aneas I, Sakabe N, Cattaneo P, Henderson L, et al. HIF1alpha represses cell stress pathways to allow proliferation of hypoxic fetal cardiomyocytes. Dev Cell. 2015;33:507–21. doi: 10.1016/j.devcel.2015.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mourad R, Li L, Cuvier O. Uncovering direct and indirect molecular determinants of chromatin loops using a computational integrative approach. PLoS Comput Biol. 2017;13:e1005538. doi: 10.1371/journal.pcbi.1005538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Van Bortle K, Corces VG. tDNA insulators and the emerging role of TFIIIC in genome organization. Transcription. 2012;3:277–84. doi: 10.4161/trns.21579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kirkland JG, Raab JR, Kamakaka RT. TFIIIC bound DNA elements in nuclear organization and insulation. Biochim Biophys Acta. 2013;1829:418–24. doi: 10.1016/j.bbagrm.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev. 2008;22:2204–14. doi: 10.1101/gad.1675908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Thompson M, Haeusler RA, Good PD, Engelke DR. Nucleolar clustering of dispersed tRNA genes. Science. 2003;302:1399–401. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, et al. A three-dimensional model of the yeast genome. Nature. 2010;465:363–7. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Woolnough JL, Schneider DA, Giles KE. The regulation of tDNA transcription during the directed differentiation of stem cells. bioRxiv. 2017. https://doi.org/10.1101/108472.
- 99.Van Bortle K, Nichols MH, Ramos E, Corces VG. Integrated tRNA, transcript, and protein profiles in response to steroid hormone signaling. RNA. 2015;21:1807–17. doi: 10.1261/rna.052126.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schwende H, Fitzke E, Ambs P, Dieter P. Differences in the state of differentiation of THP-1 cells induced by phorbol ester and 1,25-dihydroxyvitamin D3. J Leukoc Biol. 1996;59:555–61. [PubMed] [Google Scholar]
- 101.Richter E, Ventz K, Harms M, Mostertz J, Hochgrafe F. Induction of macrophage function in human THP-1 cells is associated with rewiring of MAPK signaling and activation of MAP3K7 (TAK1) protein kinase. Front Cell Dev Biol. 2016;4:21. doi: 10.3389/fcell.2016.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rudolph KL, Schmitt BM, Villar D, White RJ, Marioni JC, Kutter C, et al. Codon-driven translational efficiency is stable across diverse mammalian cell states. PLoS Genet. 2016;12:e1006024. doi: 10.1371/journal.pgen.1006024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nikolayeva O, Robinson MD. edgeR for differential RNA-seq and ChIP-seq analysis: an application to stem cell biology. Methods Mol Biol. 2014;1150:45–79. doi: 10.1007/978-1-4939-0512-6_3. [DOI] [PubMed] [Google Scholar]
- 107.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ramirez F, Dundar F, Diehl S, Gruning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Phanstiel DH, Boyle AP, Araya CL, Snyder MP. Sushi.R: flexible, quantitative and integrative genomic visualizations for publication-quality multi-panel figures. Bioinformatics. 2014;30:2808–10. doi: 10.1093/bioinformatics/btu379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang H, Meltzer P, Davis S. RCircos: an R package for Circos 2D track plots. BMC Bioinf. 2013;14:244. doi: 10.1186/1471-2105-14-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Csardi G, Nepusz T. The igraph software package for complex network research. Inter J Complex Syst. 2006;1695:1–9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequencing data from this study are publicly available under GEO series number GSE96800 through the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/). Previously reported in situ Hi-C data [45] is publicly available through Sequencing Read Archive (SRA) bioproject number PRJNA385337. Processed Hi-C data are available through the visualization software tool Juicebox (http://www.aidenlab.org/juicebox/). TF footprinting scripts were obtained from https://bitbucket.org/thashim/piq-single (PIQ R scripts pwmmatch.exact.r, pairedbam2rdata.r, pertf.bg.r and common.r) and http://pythonhosted.org/pyDNase/ (Wellington-bootstrap python scripts wellington-bootstrap.py and dnase_ddhs_scorer.py).