Abstract
Background
Staphylococcus aureus is the predominant bacterium responsible for various diseases in animals and humans. Preventive strategies could be better implemented by understanding the prevalence, genetic patterns, and the presence of enterotoxin and biofilm-producing genes along with the antibiotic susceptibility of this organism. This study was conducted in Rajasthan, the northwestern state of India, holding the largest population of cattle that makes it the second largest milk producer in India and no such prior information is available on these aspects.
Methods
A total of 368 individual quarter bovine raw milk samples were collected from 13 districts of Rajasthan, and screened for the presence of S. aureus. Microbiological and molecular approaches were followed for bacterial identification. Genetic diversity was determined by polymerase chain reaction-restriction fragment length polymorphism (PCR–RFLP) of coagulase gene (coa), whereas enterotoxin and biofilm-producing genes were studied by PCR analysis. Antibiotic strips were employed to study the antibiotic resistance among strains.
Results
In all, 73 S. aureus strains were obtained from 368 bovine raw milk samples out of that only 30 showed the presence of coa. Nine types of coa patterns ranging from 730 to 1130 bp were observed among these isolates. PCR–RFLP of coa distinguished the isolates into 15 genotypic patterns, of which patterns I, IV, V, and VI were predominant. Of the isolates, 30% were positive for sec, 10% for sea, and 3.3% for seb; these genes are responsible for enterotoxin production, whereas all isolates were found positive for icaAD and eno. The prevalence rates of other biofilm-producing genes fnbA, clfB, ebpS, sasG, fnbB, sasC, cna, bap, fib and, bbp were 97, 93, 90, 80, 80, 77, 53, 27, 10, and 6.6%, respectively. Twenty-seven (90%) strains were multidrug resistant, of which 15 were methicillin resistant. Maximum sensitivity was reported for kanamycin and it could be considered as a drug of choice for controlling S. aureus mediated cattle infections in the studied regions.
Conclusions
Overall, these strains could cause several diseases to humans, insisting the need for developing a stricter hygiene program for improving milking practices and animal health.
Keywords: Staphylococcus aureus, Coagulase gene polymorphism, RFLP, Biofilm, Enterotoxin, Antibiotic susceptibility
Background
The dairy sector in India has grown substantially over the years, making India the world’s largest producer of milk [1], and Rajasthan, the northwestern state of India, has the largest number of cattle that makes it the second largest milk producer in India [2]. However, the yield of animals within the state is not satisfactory due to various production diseases and is understood as the major factor responsible for economic losses to the dairy industry in India [3, 4].
Subclinical mastitis (SCM) with no sign of inflammation is more severe than clinical forms and is responsible for a huge loss to the dairy industry [5, 6]. Staphylococcus aureus is a part of the normal gut microbiota [7] but is also a predominant microorganism implicated in clinical mastitis and SCM [8] and is difficult to eliminate. The pathogenic strains of S. aureus are generally coagulase positive and reported to cause illness in their host around the world [9]. From the infected udders, S. aureus directly passes into the raw milk and affects the quality of milk and makes the milk less suitable for further product formulations (e.g., cheese, paneer [a soft, white cheese]). In India, 85% of total milk produced is handled by the noncommercial sector [1]. Therefore, the direct consumption and transformation of such milk into traditional dairy foods increase the chances of associated infections because of improper storage and transportation facilities. The hygiene present in dairies and the literacy of handlers about safe milk production in Rajasthan are below satisfactory level [10]. For the effective control measures of S. aureus, its genetic characterization is necessary [11, 12]. Coagulase gene (coa) typing has been proven to be the most successful method to discriminate isolates at the strain level, recovered from different regions, mainly because of simple, accurate, and reproducible results obtained with this technique over other methods [13–15].
Multiple virulence factors have been suspected for the pathogenicity of S. aureus, among which the production of enterotoxins and toxic shock syndrome toxin–1 and the ability to form biofilms are important and worrisome for the food industry [16, 17]. Staphylococcal enterotoxins (SEs) are heat-stable; therefore, they may retain their biological activity even after pasteurization and various processing steps. An accidental ingestion of SEs causes several gastrointestinal disorders in the host [18].
Biofilm-producing S. aureus strains, especially the enterotoxigenic and antibiotic-resistant ones, are the major cause of several persistent bacterial diseases in the livestock sector, including mastitis [19]. During biofilm formation, initially, the bacteria adhere to each other by polysaccharide intercellular adhesion (PIA) and then propagate. The PIA is under the control of the icaADBC operon, and the strains possessing this gene cluster have been reported as strong biofilm producers [20]. In addition, several other surface markers have been reported to play a crucial role in biofilm formation, specifically called as microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) such as collagen-binding protein (Cna) and elastin-binding protein (Ebps). The formation of biofilm also provides a shielding effect to the bacterium from antibiotic attacks.
To tackle S. aureus-mediated infections, antibiotics are used by veterinary professionals but their therapeutic outcomes are nearly insignificant because of the stubborn nature of the pathogen [21]. The frequent use of antibiotics leads to the evolution of antibiotic-resistance genes in S. aureus that can be easily transferred among healthier commensals and to other animals and humans by close interactions [9]. Studying the response of S. aureus to different relevant antibiotics gives the present drug resistance scenario from a region and the measure of associated risk factors. In addition, it may also help in selecting a more effective drug from a veterinary clinical point of view.
Therefore, the aims of this study were to check the prevalence of S. aureus from bovine raw milk samples and determine the genetic heterogeneity, virulence potential, and antimicrobial susceptibility of S. aureus from different regions of Rajasthan. These tasks were performed to generate useful information for field veterinarians and policy makers to form strategies for controlling S. aureus-mediated infections and ultimately improving the quality of milk.
Methods
Milk sampling and microbiological analysis
From May 2014 to November 2015, 368 individual quarter bovine raw milk samples were collected from 13 districts of Rajasthan, India (Fig. 1). The samples were randomly taken from clinically healthy animals. The samples were collected from different dairy herds, and before sampling, the teats of animals were cleaned with cotton balls soaked with 70% alcohol and the first few milliliters of milk were discarded. The samples were maintained at 4 °C before processing in a laboratory. The samples were serially diluted in 2% peptone water, and 100 µL of each sample was poured onto S. aureus chromogenic agar plates (HiMedia Laboratories, Mumbai, India) containing polymyxin B (50 unit/mL). After 24-h of incubation at 37 °C, Staphylococcus colonies showing blue-green color from each sample were transferred to brain heart infusion (BHI) broth. The chromogenic mixture present in the medium is specifically cleaved by S. aureus to produce blue-green colonies, which are clearly visible against the opaque background. For reconfirmation, the cultures were streaked on Mannitol salt agar and Baird Parker agar plates (Himedia Laboratories). Next, the colonies were examined for morphology after Gram staining and were confirmed using the API Staph kit (bioMeriux, Marcy-l’Etoile, France). Confirmed S. aureus colonies were examined for coagulase activity by tube plasma agglutination test. The presumptively identified S. aureus colonies were maintained at −80 °C in glycerol stocks for further analysis. A single colony was picked per positive sample and used to determine the prevalence rate among different samples. The isolates were coded conforming to their origin.
Fig. 1.
Map of Rajasthan India showing the districts from where sampling was carried out
DNA extraction and purification
Genomic DNA was extracted from a freshly grown culture in BHI broth by using a modified method [22]. Briefly, 2 mL of overnight grown culture was centrifuged at 12,000×g for 10 min to form pellets. Afterwards, the pellets were resuspended in 200 µL of sucrose EDTA Tris (SET) buffer containing lysostaphin (100 µg/mL; Sigma, St. Louis, Missouri, USA) and lysozyme (100 µg/mL; Amersco, Solon, USA) for 1 h at 37 °C. The remaining procedure was similar as described elsewhere [22]. The extracted DNA was examined for purity and concentration using a microplate reader at 260 nm and thereafter stored at −20 °C.
All conventionally identified S. aureus were ascertained by species-specific primers (F 5′-CCATGTGTAGCGGTGAAATG–3′ and R 5′-TAAGGTTCTTCGCGTTGCTT–3′) obtained from the 16S rRNA region of S. aureus (GenBank accession number: KF993676.1) by using Primer 3 plus software. A 25-µL PCR reaction mixture consisting of 12.5 µL of 2X DreamTaq Green PCR Master Mix (Thermo Scientific, Mumbai, India), 0.5 µM of each primer, and 1 µL of template DNA was prepared and performed in a SureCycler 8800 Thermal Cycler (Agilent Technologies, New Delhi, India). Following program was used for PCR cycles: 2 min at 95 °C, followed by 35 cycles of 30 s at 95 °C, 45 s at 57 °C, and 60 s at 72 °C with a final extension of 10 min at 72 °C. A PCR product of approximately 306 bp signifies the presence of S. aureus isolates.
Using an earlier protocol, coa was identified in the S. aureus isolates [23]. The amplified products were electrophoresed on 2.0% agarose gel at 100 V for 1 h and photographed in a gel documentation system (BioRad Laboratories, Gurgaon, India). A suitable ladder was used as the size marker during electrophoresis.
Restriction enzyme digestion of PCR-amplified coa product
Of the PCR-amplified coa product, 10 µL was mixed with 17 µL of nuclease-free water, 2 µL of buffer Tango, and 1 µL of AluI (Thermo Scientific) and incubated at 37 °C for 1 h for digestion. The digested products were resolved on 2.0% agarose gel and examined in a gel documentation system. A 100–1500-bp ladder (Promega, Wisconsin, USA) was used as the size marker for analyzing the digested coa fragments.
The generated Coa-RFLP patterns were analyzed by the BioNumerics (version 7.6) software package (Applied Maths, Sint-Martens-Latem, Belgium). A dendrogram was prepared by using the Dice-coefficient and the unweighted pair group method with arithmetic averages as the cluster analysis method.
Reproducibility testing
PCR reproducibility was analyzed by testing five isolates for five consecutive days. The PCR–RFLP reproducibility of coa was determined by digesting four products twice with the AluI enzyme.
Calculating numerical index of discrimination
A numerical index equation was used to determine the discriminatory power of the typing method as described previously [24]. The following formula was used:
where D = discriminatory index, s = total number of different types, nj = number of isolates representing each type, and N = total number of isolates in the sample population.
Detection of enterotoxin and biofilm genes
The amplification of the genes coding for enterotoxins and biofilm production (Table 1) was performed using the multiplex PCR method [25, 26].
Table 1.
Primers used for detection of S. aureus enterotoxin and biofilm genes
| Gene | Primer sequence (5′–3′) | Product size (bp) | Multiplex PCR set |
|---|---|---|---|
| sea a | GGTTATCAATGTGCGGGTGG CGGCACTTTTTTCTCTTCGG |
102 | A |
| seb a | GTATGGTGGTGTAACTGAGC CCAAATAGTGACGAGTTAGG |
164 | A |
| sec a | AGATGAAGTAGTTGATGTGTATGG CACACTTTTAGAATCAACCG |
451 | A |
| sed a | CCAATAATAGGAGAAAATAAAAG ATTGGTATTTTTTTTCGTTC |
278 | A |
| see a | AGGTTTTTTCACAGGTCATCC CTTTTTTTTCTTCGGTCAATC |
209 | A |
| eta a | GCAGGTGTTGATTTAGCATT AGATGTCCCTATTTTTGCTG |
93 | B |
| etb a | ACAAGCAAAAGAATACAGCG GTTTTTGGCTGCTTCTCTTG |
226 | B |
| tst a | ACCCCTGTTCCCTTATCATC TTTTCAGTATTTGTAACGCC |
326 | B |
| bap b | GAGCCAAGACAAAGGTGAAG GTAGCCATAGCACGGAACAT |
873 | |
| bbp b | CTTAGCAGTTCAACAGGGTG TTGGCTTTATTGTGATGGTC |
1662 | |
| cna b | CGATAACATCTGGGAATAAA ATAGTCTCCACTAGGCAACG |
716 | |
| clfA b | AGTACCAAATGAGGCTGTTC AAATGCTACTTCGTTGTCCC |
796 | |
| clfB b | CACTTACTTTACCGCTACTTTC AACGAGCAATACCACTACAACAG |
968 | |
| ebpS b | GGTGAACCTGAACCGTAG CTGGCAAGGCGAATAACT |
661 | |
| eno b | GTCGTGCATTAGTACCATCAGG TTTCCAACCATCCCAGTCG |
812 | |
| fib b | AGATGCGAGCGAAGGGTA TAAACGAAACTAAGTTGACTGC |
347 | |
| fnbpA b | TCCGCCGAACAACATACC TCAAGCACAAGGACCAAT |
952 | |
| fnbpB b | TCTGCGTTATGAGGATTT ACAGTAGAGGAAAGTGGG |
452 | |
| icaAD b | TGGCTACTGGGATACTGATA TGGAAATGCGACAAGAACTA |
520 | |
| icaBC b | GCCTATCCTTATGGCTTGA TGGAATCCGTCCCATCTC |
182 | |
| sasC b | AGAATGAAGTCCGATAGAGT AATCATACAGATGGCAATAC |
936 | |
| sasG b | TATCAACACTTCCGTAACCTTC CGTCAGTCACTCATAACGCAGA |
159 |
aRepresents the enterotoxin genes while b depicts the biofilm genes
Determination of minimum inhibitory concentration
The minimum inhibitory concentration (MIC) of all S. aureus isolates was determined using E-strips as per the manufacturer’s instructions after adjusting their OD to McFarland 0.5 as disclosed previously [27]. The results obtained were analyzed according to CLSI guidelines [28]. The MIC E-strips (0.016–256 µg/mL) (HiMedia Laboratories) were used for susceptibility observation. The antimicrobials used were according to the information collected from the field veterinarians and practicing clinicians. S. aureus ATCC 29213 was used as the reference strain for quality control. Methicillin-resistant S. aureus (MRSA)-positive strains were further confirmed on HiCrome MeReSa Agar (HiMedia Laboratories) and validated by the amplification of mecA by PCR [29].
Results
Out of 368 bovine raw milk samples, S. aureus contamination was present in 73 (19.84%) samples (Table 2) as revealed by media plating analysis. All 73 isolates were found as S. aureus in API analysis. Out of the 73 isolates, 30 (41.1%) showed coagulase activity and were further confirmed as S. aureus by PCR. Furthermore, all these phenotypically coagulase-positive S. aureus scored positive for coa during PCR analysis (Fig. 2). Nine types of coa patterns ranging from 730 to 1130 bp were observed (Table 3), and the sizes, 820 and 920 bp, were the most predominantly noticed product sizes, accounting altogether for 67% of the total coa-positive isolates (Table 3). Twenty-seven isolates depicted one PCR band while three isolates (15, 17, and 19) depicted double bands (Fig. 2). The polymorphism in coa was studied by RFLP (Fig. 3), where fifteen (I–XV) RFLP patterns (Table 3 and Fig. 4) were observed. RFLP patterns I, IV, V, and VI were the dominant patterns being noticed in three (10%), three (10%), six (20%), and four (13.3%) isolates, respectively (Table 3). The isolates with the most predominant pattern (V) displayed two DNA fragments of 540 and 260 bp. The isolates with pattern VI had DNA fragments of 565 and 270 bp. The isolates with patterns I and IV had DNA fragments of 430 and 260 bp and 528 and 250 bp, respectively. Rest all other remaining patterns were shown by a single (II, VII, X, XI, XII, and XIV) or two (VIII, IX, and XIII) isolates. The reproducibility of the PCR products and coa PCR–RFLP was demonstrated with 100% of the repeatedly tested isolates; however, the results showed some variations in the intensity of bands.
Table 2.
Details of samples contaminated with S. aureus and their prevalence
| Sampling districts/regions | No. of raw milk samples collected | No. of positive samples (% prevalence) |
|---|---|---|
| Ajmer | 28 | 4 (1.09) |
| Bharatpur | 28 | 7 (1.90) |
| Bhilwara | 28 | 3 (0.82) |
| Bikaner | 28 | 2 (0.54) |
| Chittorgarh | 28 | 4 (1.09) |
| Hanumangarh | 28 | 7 (1.90) |
| Jaipur | 30 | 5 (1.36) |
| Mount Abu | 28 | 8 (2.17) |
| Pali | 28 | 6 (1.63) |
| Sirohi | 30 | 13 (3.53) |
| Sikar | 28 | 5 (1.36) |
| Shri-Ganganagar | 28 | 6 (1.63) |
| Udaipur | 28 | 3 (0.82) |
| Total | 368 | 73 (19.84) |
Fig. 2.
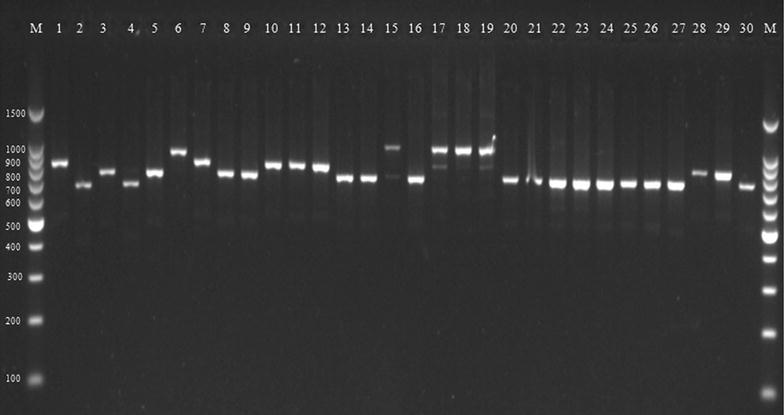
Agarose gel electrophoresis of the coagulase gene (coa). Lane M; 100–1500 bp molecular marker, 1-STRMCOR7 (920 bp), 2-STRMPL47(730 bp), 3-STRMSH56 (820 bp), 4-STRMBHL57 (750 bp), 5-STRMSH58 (820 bp), 6-STRMHMO59 (1060 bp), 7-STRMUDZ70 (920 bp), 8-STRMSH73 (820 bp), 9-STRMSH74 (820 bp), 10-STRMHMO79 (920 bp), 11-STRMSH37 (920 bp), 12-STRMSH42 (920p), 13-STRMMA11 (820 bp), 14-STRMMA20 (820p), 15-STRMUDZ2 (1130 bp) 16-STRMMA3 (820 bp), 17-STRMSGNR64 (1130 bp), 18-STRMSGNR19 (1130 bp), 19-STRMUDZ6 (1130 bp), 20-STRMBKN5 (820 bp), 21-STRMSK96 (820 bp), 22-STRMSK98 (820 bp), 23-STRMSK100 (820 bp), 24-STRMBP11 (820 bp), 25-STRMB12 (820 bp), 26-STRMJ102 (820 bp), 27-STRMJ3 (820 bp), 28-STRMP23 (920 bp), 29-STRMSGNR9 (880 bp), 30-STAJM33 (800 bp)
Table 3.
Genotype distributions of S. aureus cultures obtained from different regions of Rajasthan and their RFLP patterns
| Coa gene PCR product (bp) | RFLP pattern | n | Isolate name | Isolated region | Genotype code/distinct RFLP pattern |
|---|---|---|---|---|---|
| 730 | 430-260 | 1 | STRMPL47 | PL; Pali | I |
| 750 | 620 | 1 | STRMBHL57 | BHL; Bhilwara | II |
| 800 | 420-240 | 1 | STAJM33 | AJM; Ajmer | III |
| 820 | 528-250 | 3 | STRMBP12, STRMJ102, STRMJ3 | BP; Bharatpur, J; Jaipur | IV |
| 820 | 540-260 | 6 | STRMMA20, STRMMA3, STRMSK96, STRMSK 98, STRMSK100, STRMBP11 | SH; Sirohi MA; Mount abu, SK; Sikar, BP; Bharatpur | V |
| 820 | 565-270 | 4 | STRMSH56, STRMSH58, STRMSH73, STRMSH74 | SH; Sirohi | VI |
| 820 | 430-260 | 1 | STRMMA11 | MA; Mount abu | I |
| 820 | 440-360 | 1 | STRMBKN5 | BKN; Bikaner | VII |
| 880 | 420-330 | 1 | STRMSGNR9 | SGNR; Shri-Ganganagar | VIII |
| 920 | 440-260-170 | 2 | STRMSH37, STRMSH42 | SH; Sirohi | IX |
| 920 | 420-330 | 1 | STRMPL23 | PL: Pali | VIII |
| 920 | 462-373 | 1 | STRMHMO79 | HMO; Hanumangarh | X |
| 920 | 425-260 | 1 | STRMCOR7 | COR; Chittorgarh | I |
| 940 | 452-273-186 | 1 | STRMUDZ70 | UDZ; Udaipur | XI |
| 1060 | 540-350 | 1 | STRMHMO59 | HMO; Hanumangarh | XII |
| 1130 | 445-360-170 | 2 | STRMSGNR19, STRMUDZ6 | SGNR; Shri-Ganganagar, UDZ; Udaipur | XIII |
| 1130 | 452-370-175 | 1 | STRMSGNR64 | SGNR; Shri-Ganganagar | XIV |
| 1130 | 460-380-182 | 1 | STRMUDZ2 | UDZ; Udaipur | XV |
Fig. 3.
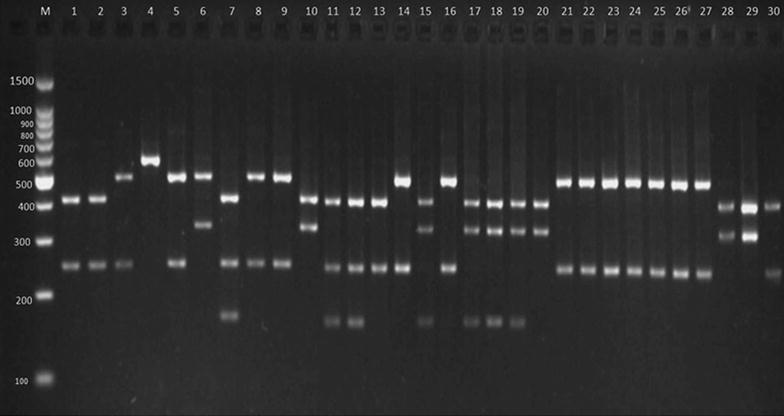
RFLP of the coagulase gene (coa) after digestion with AluI. Lane M; 100–1500 bp molecular marker, 1-STRMCOR7, 2-STRMPL47, 3-STRMSH56, 4-STRMBHL57, 5-STRMSH58, 6-STRMHMO59, 7-STRMUDZ70, 8-STRMSH73, 9-STRMSH74, 10-STRMHMO79, 11-STRMSH37, 12-STRMSH42, 13-STRMMA11, 14-STRMMA20, 15-STRMUDZ2, 16-STRMMA3, 17-STRMSGNR64, 18-STRMSGNR19, 19-STRMUDZ6, 20-STRMBKN5, 21-STRMSK96, 22-STRMSK98, 23-STRMSK100, 24-STRMBP11, 25-STRMB12, 26-STRMJ102, 27-STRMJ3, 28-STRMP23, 29-STRMSGNR9, 30-STAJM33
Fig. 4.
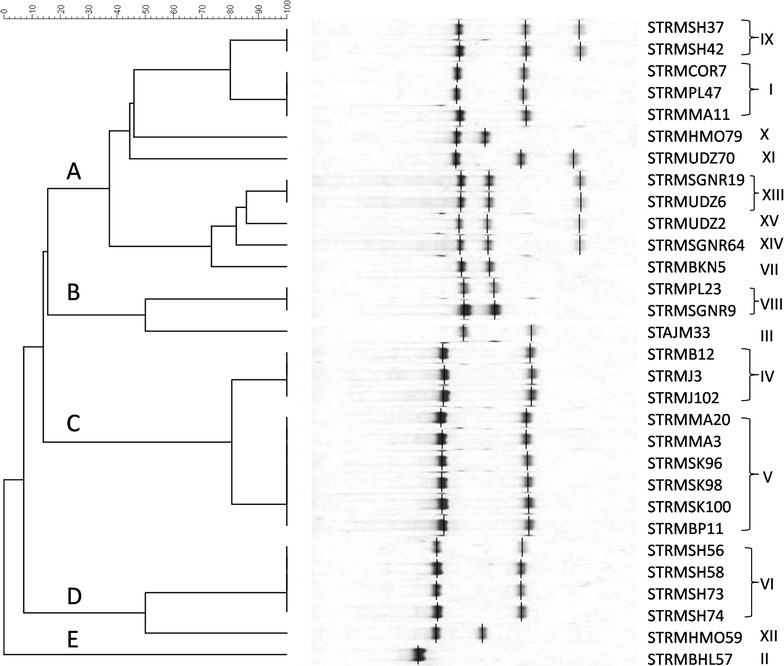
Dendrogram showing genetic heterogeneity among S. aureus isolates on the basis of the coa gene polymorphism
The discriminatory index for the AluI RFLP method was about 0.80, indicating a fair discriminatory power of this method for typing S. aureus. Further RFLP analysis classified the 30 S. aureus strains into five major clusters designated as A, B, C, D, and E (Fig. 4). The dendrogram depicted that cluster A was 40% similar to other clusters (B-D) and was composed of isolates obtained from eight regions. Cluster B was 52% identical with clusters C to E and was composed of isolates obtained from three regions. Similarly, cluster C with approximately 82% identity with clusters D and E included isolates from four regions. Cluster D was composed of five isolates obtained from two regions. The remaining cluster E was composed of only one isolate that was derived from a totally different region.
Of the analyzed enterotoxin genes, 30% isolates were positive for sec, 10% for sea, and 3.3% for seb (Table 4). No isolate was detected harboring the remaining enterotoxin genes.
Table 4.
Results of enterotoxins genes in S. aureus isolates
| Isolates (n = 30) | sea | seb | sec | sed | see | eta | etb | tst |
|---|---|---|---|---|---|---|---|---|
| STRMPL47 | – | + | – | – | – | – | – | – |
| STRMPL23 | – | – | – | – | – | – | – | – |
| STRMBHL57 | + | – | + | – | – | – | – | – |
| STAJM33 | – | – | – | – | – | – | – | – |
| STRMSH56 | – | – | + | – | – | – | – | – |
| STRMSH58 | – | – | – | – | – | – | – | – |
| STRMSH73 | – | – | – | – | – | – | – | – |
| STRMSH74 | – | – | + | – | – | – | – | – |
| STRMSH37 | – | – | – | – | – | – | – | – |
| STRMSH42 | – | – | – | – | – | – | – | – |
| STRMMA20 | – | – | – | – | – | – | – | – |
| STRMMA3 | – | – | – | – | – | – | – | – |
| STRMMA11 | – | – | + | – | – | – | – | – |
| STRMSK96 | – | – | – | – | – | – | – | – |
| STRMSK 98 | – | – | – | – | – | – | – | – |
| STRMSK100 | – | – | – | – | – | – | – | – |
| STRMBP11 | + | – | + | – | – | – | – | – |
| STRMBP12 | – | – | – | – | – | – | – | – |
| STRMJ102 | – | – | – | – | – | – | – | – |
| STRMJ3 | – | – | – | – | – | – | – | – |
| STRMBKN5 | – | – | + | – | – | – | – | – |
| STRMSGNR9 | – | – | – | – | – | – | – | – |
| STRMSGNR19 | – | – | – | – | – | – | – | – |
| STRMSGNR64 | + | – | + | – | – | – | – | – |
| STRMCOR7 | – | – | – | – | – | – | – | – |
| STRMHMO79 | – | – | – | – | – | – | – | – |
| STRMHMO59 | – | – | + | – | – | – | – | – |
| STRMUDZ70 | – | – | + | – | – | – | – | – |
| STRMUDZ2 | – | – | – | – | – | – | – | – |
| STRMUDZ6 | – | – | – | – | – | – | – | – |
| Occurrence (%) | 10 | 3.3 | 30 | 0 | 0 | 0 | 0 | 0 |
Regarding biofilm-encoding genes, all isolates typed 100% positive for icaAD, icaBC, and eno (Table 5). The prevalence rates of fnbA, clfB, ebpS, sasG, fnbB, sasC, cna, bap, fib, and bbp were 97, 93, 90, 80, 80, 53, 77, 27, 10, and 6.6%, respectively. Low prevalence was noticed for clfA (3.3%) as only one isolate was found positive for this gene.
Table 5.
Results of adhesion genes typed in S. aureus isolates
| Isolates (n = 30) | icaAD | icaBC | clfB | clfA | fnbA | fnbB | ebpS | eno | fib | sasG | sasC | bbp | bap | cna |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| STRMPL47 | + | + | + | – | + | + | + | + | – | – | – | – | – | – |
| STRMPL23 | + | + | + | – | + | + | + | + | – | + | – | – | – | + |
| STRMBHL57 | + | + | + | – | + | + | + | + | + | + | + | + | + | + |
| STAJM33 | + | + | + | – | + | + | + | + | – | + | – | – | – | – |
| STRMSH56 | + | + | + | – | + | + | + | + | – | + | + | – | – | + |
| STRMSH58 | + | + | + | – | + | + | + | + | – | + | + | – | – | + |
| STRMSH73 | + | + | + | – | + | + | + | + | – | + | – | – | – | + |
| STRMSH74 | + | + | + | – | + | + | + | + | – | + | + | – | + | + |
| STRMSH37 | + | + | + | – | + | + | + | + | – | + | + | – | – | + |
| STRMSH42 | + | + | + | – | + | – | – | + | – | + | – | – | – | – |
| STRMMA20 | + | + | + | – | + | + | + | + | – | + | – | – | – | + |
| STRMMA3 | + | + | + | – | + | – | + | + | – | – | – | – | – | – |
| STRMMA11 | + | + | + | – | + | + | + | + | – | + | + | – | – | + |
| STRMSK96 | + | + | + | – | + | + | + | + | – | + | + | – | – | + |
| STRMSK 98 | + | + | + | – | + | + | + | + | – | + | – | – | – | + |
| STRMSK100 | + | + | + | – | + | + | + | + | – | + | + | – | – | + |
| STRMBP11 | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| STRMBP12 | + | + | + | – | + | + | + | + | – | + | + | – | + | + |
| STRMJ102 | + | + | – | – | – | – | – | + | – | – | – | – | – | – |
| STRMJ3 | + | + | + | – | + | + | + | + | – | + | + | – | – | + |
| STRMBKN5 | + | + | + | – | + | – | + | + | – | + | – | – | – | + |
| STRMSGNR9 | + | + | + | – | + | + | + | + | + | + | + | – | + | + |
| STRMSGNR19 | + | + | + | – | + | + | + | + | – | + | + | – | + | + |
| STRMSGNR64 | + | + | + | – | + | – | + | + | – | – | – | – | – | + |
| STRMCOR7 | + | + | + | – | + | + | + | + | – | + | + | – | – | + |
| STRMHMO79 | + | + | + | – | + | + | + | + | – | + | + | – | + | + |
| STRMHMO59 | + | + | – | – | + | – | – | + | – | – | – | – | – | – |
| STRMUDZ70 | + | + | + | – | + | + | + | + | – | + | – | – | – | + |
| STRMUDZ2 | + | + | + | – | + | + | + | + | – | + | + | – | + | + |
| STRMUDZ6 | + | + | + | – | + | + | + | + | – | – | – | – | – | – |
| Occurrence (%) | 100 | 100 | 93 | 3.3 | 97 | 80 | 90 | 100 | 10 | 80 | 53 | 6.6 | 27 | 77 |
Antibiotic susceptibility test revealed that 27 (90%) isolates were resistant to penicillin, 23 (77%) to ampicillin, 17 (57%) to amikacin, 15 (50%) to oxacillin, 12 (40%) to ciprofloxacin, 9 (30%) to azithromycin, and 7 (23%) to piperacillin (Table 6). Five (17%) isolates showed resistance to linezolid and tetracycline; 3 (10%) to gatifloxacin and rifampicin; 2 (7%) to ceftazidime, chloramphenicol, and norfloxacin; and 1 (3%) to gentamicin, teicoplanin, and vancomycin (Table 6). Intermediate resistance to amikacin, azithromycin, ceftazidime, ceftriaxone, chloramphenicol, ciprofloxacin, gatifloxacin, gentamicin, teicoplanin, and vancomycin was shown by 1 (3%), 1 (3%), 2 (7%), 3 (10%), 2 (7%), 4 (13%), 3 (10%), 1 (3%), 1 (3%), and 3 (10%) isolates, respectively. Twenty-seven (90%) of the strains tested showed multiple drug resistance (complete or intermediate) to three or more antibiotics. All isolates were completely (100%) sensitive to kanamycin. Additionally, 15 strains were resistant to oxacillin, penicillin, and ampicillin along with other antibiotics and were considered as MRSA strains [9]. All 15 strains further showed their presence on MeReSa Agar and found positive for mecA (Fig. 5).
Table 6.
Antibiotic susceptibility patterns of S. aureus isolates
| Isolates (n = 30) | Antibiotics | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amikacin | Amoxyclav | Ampicillin | Azithromycin | Ceftazidime | Ceftriaxone | Chloramphenicol | Ciprofloxacin | Gatifloxacin | Gentamicin | Kanamycin | Linezolid | Norfloxacin | Oxacillin | Penicillin | Piperacillin | Rifampcin | Teicoplanin | Tetracyclin | Vancomycin | |
| STRMPL47 | R | R | S | S | S | S | S | R | S | S | S | R | S | S | R | R | S | S | S | I |
| STRMPL23 | S | S | R | S | S | S | S | S | S | S | S | S | S | S | R | S | R | S | S | S |
| STRMBHL57 | I | R | R | S | S | S | S | S | S | S | S | R | S | S | R | R | S | S | S | S |
| STAJM33 | R | R | R | S | S | S | S | I | S | R | S | S | S | S | R | R | S | S | S | S |
| STRMSH56 | R | S | R | S | S | S | S | I | I | S | S | S | S | S | R | R | S | S | S | S |
| STRMSH58 | S | S | S | S | S | S | S | S | S | S | S | S | S | S | R | R | S | S | S | S |
| STRMSH73 | R | S | R | R | S | S | S | R | S | S | S | R | S | S | R | S | S | S | S | S |
| STRMSH74 | R | S | R | S | R | S | S | R | S | S | S | S | S | R | R | S | S | S | R | R |
| STRMSH37 | R | S | R | S | S | S | R | S | S | S | S | S | S | R | R | S | S | S | R | S |
| STRMSH42 | S | S | S | S | S | S | S | S | R | S | S | S | S | S | R | S | S | S | S | S |
| STRMMA20 | R | S | R | S | S | S | S | R | R | S | S | S | S | R | R | S | S | S | S | S |
| STRMMA3 | R | S | R | R | I | S | S | R | S | S | S | S | S | R | R | S | S | R | S | S |
| STRMMA11 | R | S | R | R | S | S | S | I | S | S | S | S | S | R | R | S | S | S | S | S |
| STRMSK96 | S | S | S | S | S | S | S | R | S | S | S | S | S | S | R | S | S | I | S | S |
| STRMSK 98 | R | S | R | R | S | S | S | R | S | S | S | S | S | R | R | S | S | S | S | S |
| STRMSK100 | R | S | R | I | S | I | S | R | S | S | S | S | R | S | R | S | S | S | S | S |
| STRMBP11 | R | S | R | S | S | S | S | S | S | S | S | R | R | R | R | S | S | S | S | S |
| STRMBP12 | R | R | R | R | R | S | I | R | I | S | S | S | S | R | R | S | S | S | S | S |
| STRMJ102 | R | S | S | S | S | S | R | R | S | S | S | S | S | S | S | R | S | S | S | I |
| STRMJ3 | R | S | R | R | I | I | S | I | S | S | S | S | S | R | R | S | S | S | R | S |
| STRMBKN5 | R | S | R | R | S | S | S | S | S | S | S | S | S | R | R | S | S | S | S | S |
| STRMSGNR9 | S | S | R | R | S | S | S | S | S | S | S | S | S | S | S | S | S | S | R | S |
| STRMSGNR19 | S | S | R | S | S | I | S | S | S | S | S | S | S | R | R | S | S | S | S | S |
| STRMSGNR64 | S | S | R | S | S | S | S | S | S | S | S | R | S | R | R | S | R | S | S | S |
| STRMCOR7 | S | R | R | S | S | S | S | S | I | S | S | S | S | R | R | S | S | S | S | S |
| STRMHMO59 | R | S | S | S | S | S | S | R | S | I | S | S | S | S | R | S | S | S | S | S |
| STRMHMO79 | S | R | R | S | S | S | S | S | S | S | S | S | S | S | R | S | R | S | S | I |
| STRMUDZ70 | S | R | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| STRMUDZ2 | S | S | R | S | S | S | I | R | R | S | S | S | S | R | R | S | S | S | R | S |
| STRMUDZ6 | S | R | R | R | S | S | S | S | S | S | S | S | S | R | R | R | S | S | S | S |
| Sensitive | 12 | 22 | 07 | 20 | 26 | 27 | 26 | 14 | 24 | 28 | 30 | 25 | 28 | 15 | 03 | 23 | 27 | 28 | 25 | 26 |
| Intermediate | 01 | 0 | 0 | 01 | 02 | 03 | 02 | 04 | 03 | 01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 01 | 0 | 03 |
| Resistant | 17 | 08 | 23 | 09 | 02 | 0 | 02 | 12 | 03 | 01 | 0 | 05 | 02 | 15 | 27 | 07 | 03 | 01 | 05 | 01 |
Fig. 5.
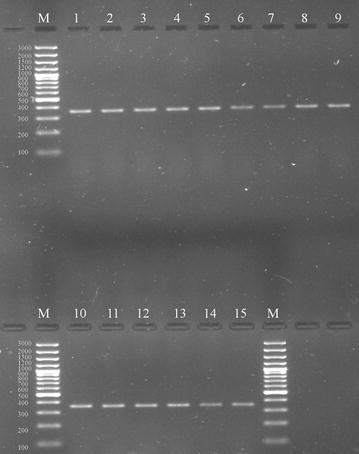
Agarose gel photograph showing the MRSA positive samples. Lane M; 100–3000 bp molecular marker, 1-STRMSH74, 2-STRMSH37, 3-STRMMA20, 4-STRMMA3, 5-STRMMA11, 6-STRMSK98, 7-STRMBP11, 8-STRMBP12, 9-STRMJ3, 10-STRMBKN5, 11-STRMSGNR19, 12-STRMSGNR64, 13-STRMUDZ2, 14-STRMUDZ2, 15-STRMUDZ6
Discussion
In this study, the prevalence, genetic patterns, pathogenic potential, and antibiotic susceptibility of S. aureus recovered from 13 districts of Rajasthan, India, were discussed. Our results indicated that 19.84% of milk samples were positive for S. aureus, and to the best of our knowledge, this is the first comprehensive study from Rajasthan. Similar findings concerning the prevalence of S. aureus in bovine raw milk samples were reported from different sub-parts of India; however, the prevalence rate varied greatly among the studies [30–36]. Of note, moderate prevalence was reported in the present study. We posit that inadequate hygiene and inferior management practices facilitate the entry of microorganisms to udders and concomitantly the shedding of microorganisms into milk.
The phenotypic method is not sufficient for categorizing between coagulase-positive and -negative S. aureus isolates [37]. Therefore, we examined the isolates for the coa presence by PCR. Nine coa PCR types were obtained, signifying considerable variation among the S. aureus types and this finding is in agreement with others [14, 23, 38]. The precise reason for this high rate of polymorphism observed among the strains is debatable. However, because of the presence of a series of 81-bp tandem repeats in the 3′-end region of coa, the high rate of polymorphism possibly alters the antigenic properties of the gene. Therefore, the gene’s conformity is possibly recognized as the topmost reason behind the failure in the action of neutralizing antibodies [39]. A double band for coa was also visualized among three isolates, indicating the existence of distinct allelic forms of coa. The appropriate reason behind this distinct coa pattern is linked to the production of more than one immunologic form of the coagulase protein by some S. aureus isolates [23], although this pattern is rarely observed and only a few authors have reported it [38, 40].
In the present study, the digestion of the different coa amplicons with AluI reveals considerable genetic variability as it classifies them into 15 patterns. The findings are similar to those of others [14, 23, 41] who found a considerable degree of variability in coa RFLP patterns after digestion with the same enzyme, although the sizes of PCR products obtained here were almost different from those in earlier studies. Besides, the typing method opted for the discrimination was found full-proof as it displayed a fairly good discriminatory power, i.e., of 0.80 and is supported by the findings of Sarvari et al. [15] who obtained a discriminatory index of 0.82 among S. aureus isolates.
As mentioned earlier, the study segregated all 30 S. aureus isolates into five clusters (A–E), among which patterns I, IV, V, and VI were predominant. Most patterns belonged to clusters A and C. This indicates weak to no heterogeneity in the AluI recognition sites among the isolates of these predominant patterns. Momtaz et al. [14] found that out of 42 coa-positive S. aureus isolates, 31 (73.8%) belonged to genotype pattern I and 11 (26%) to II. Likewise, in another study [39], pattern I was found predominant. These findings support our results and justify that in a particular geographical region a predominant genotype is present because of prevailing favorable environmental conditions. Besides, the internal resistance of strains toward the host immune system also substantially plays an important role in the strain’s dominance. The predominant isolates from a region have possibly evolved mechanisms to bypass the host phagocytosis than the isolates with rare genotypes [42, 43]. Furthermore, the existence of a similar RFLP pattern for intra-regional strains in genotypes, I, IV, V, VI, VIII, IX, and XIII suggests the possibility of transmission of some strains from one region to another during the transportation of milk.
Next, we tested all isolates for enterotoxin-producing genes and none of them from Ajmer, Jaipur, and Chittorgarh regions scored positive for these genes. In our study, the genes for toxins (sed, see, eta, etb, and tst) were not amplified in any of the 30 isolates. Enterotoxins contribute to the pathogenesis of S. aureus by modulating immunity and susceptibility to antibiotics resulting in the onset of many diseases [44]. The reports concerning the presence of enterotoxin-producing genes (sea, seb, sec, sed, and see) among S. aureus strains isolated from milk of animals with bovine mastitis concur with our findings [44]. Here, we did not find the presence of eta and etb exfoliative genes, and these results are in agreement with previous studies [11, 44, 45], which inferred that S. aureus strains isolated from animals with mastitis were rarely found positive for exfoliative toxins. Similarly, the presence of tsst–1 was not observed in any isolate, although its prevalence in isolates from cattle with mastitis was reported in conjunction with sec and sed [44].
Most information about the ability of S. aureus to form biofilms is restricted to the isolates recovered from clinical studies or medical devices, and insufficient information is available for bovine milk-derived isolates. Therefore, we studied MSCRAMM genes to understand their prevalence. The products of these genes aid the bacterium to adhere to the host components and produce biofilm. Previous studies on MSCRAMM genes in Enterococcus faecium, S. saprophyticus, and S. aureus mention that instead of their active role in biofilm development, some gene products are also involved in pathogenesis. Here, a great diversity in the presence of biofilm-producing genes was observed among the isolates recovered from different regions. The complete dominance was observed for icaAD and eno responsible for polysaccharide intracellular adhesion and laminin-binding proteins that have a crucial role in bovine mastitis pathogenesis [46]. These findings are in agreement with previous studies [26, 45, 47] that reported higher prevalence rates for these genes. fnbA and fnbB that are usually associated with invasive diseases [48] were also found abundantly (97 and 80%, respectively) among the isolates in this study; these rates are comparable to those in other studies [20, 26]. Similarly, clfB encoding an Fn-binding protein that facilitates S. aureus to establish nasal colonization [49] was also present in most (93%) strains. Proteins EbpS, SasG, and SasC, which play a considerable role in maintaining cell density, biofilm accumulation, and intercellular adhesion, were present in 90, 80, and 53% of isolates, respectively. These findings are comparable and in complete agreement with the findings of Tang et al. [26] who noticed a high prevalence of these genes. Another collagen-binding protein, Cna, which has been associated with virulence potential in rabbit models [48], was identified in 77% of isolates; this value is comparably higher than that reported by Pereyra et al. [50] but lower than others [20, 26]. Gene bbp, which is often found in the clinical strains of S. aureus and linked with bone infections, was only typed in a few (6.6%) isolates. The result is in accordance with the findings of Kot et al. [51] and Puacz et al. [52] who detected the presence of bbp in a few isolates of mastitis origin. Gene bap, whose protein was probably the first protein elucidated to have a role in biofilm formation in S. aureus and associated with chronic bovine mastitis, was present in 7% of isolates. This finding is consistent with the observations of Darwish and Asfour [45], who observed the presence of bap in 2.5 and 4.4% of S. aureus and coagulase-negative S. aureus isolates, respectively. Another gene fib, elucidated to have a crucial role in binding to extracellular matrix fibrinogen, was also detected in 10% of isolates. This finding is different from the findings of Pereyra et al. [50] and Zuniga et al. [53] who reported a higher prevalence of 90 and 71.7%, respectively. Altogether, our results suggest that most strains had the biofilm-forming ability, which may facilitate in establishing intramammary gland infections in cattle. Moreover, this property varies from strain to strain and depends on the genetic composition and geographical evolution.
In the present study, the observed antibiotic resistance level was relatively high (90%) and two strains (STRMSH74 and STRMBP12) from Sirohi and Bharatpur, respectively, were found to be most resistant. MRSA-positive strains were observed from each area except for Pali, Bhilwara, and Hanumangarh; this finding is in agreement with studies reporting varied MRSA prevalence in bovine milk [9, 43, 54]. No similarities in resistance patterns were observed among the strains from different regions except for one strain each from Jaipur (STRMJ3) and Bikaner (STRMBKN5). The inconsistencies in the resistance profile observed might be due to the excessive, indiscriminate use of a particular antibiotic in each geographical region for combating infections. This fact was established by similar findings in studies undertaken in the United States and European countries, where a lower resistance profile among S. aureus isolates was noticed because of stricter use of antibiotics in veterinary practice [54]. In Rajasthan, β-lactams, macrolides, aminopenicillins, cephalosporins, aminoglycosides, fluoroquinolones, tetracyclines, and amphenicols are frequently used in veterinary clinical practices. However, resistance toward the less commonly used antibiotics such as vancomycin, teicoplanin, and rifampicin in veterinary practice is not fully understood. This is possibly due to the contamination from human handlers [18], as strains with rare and high antibiogram patterns were generally isolated from hospital settings because of a high antimicrobial pressure [54]. Earlier, vancomycin-resistant enterococci strains were isolated from tertiary care hospitals in Rajasthan [55, 56], and it was proved that the gene cluster VanA was transferable from E. faecalis to S. aureus [57]. Similarly, teicoplanin-resistant S. aureus strains were reported from a hospital in Jaipur, Rajasthan [57]. Thus, considerable attention is needed for treating the vancomycin-resistant MRSA-induced infection. Based on our results, all strains were sensitive to kanamycin, and it could be the drug of choice to treat S. aureus mediated infections.
Overall, our results clearly indicate that bovine raw milk samples collected from different regions of Rajasthan were contaminated with coagulase-positive strains of S. aureus. Considerable variability in coa was observed, and genotypes I, IV, V, and VI were predominant. The close similarity in the RFLP pattern among the isolates of different regions suggested the transmission of isolates from one place to another by various means. The presence of enterotoxin and biofilm-producing genes among these strains poses a potential public health risk, as these strains may be implicated in milk-borne intoxications. Furthermore, the presence of multidrug-resistant and MRSA strains indicates the improper use of antibiotics for mastitis control and is a growing health concern globally. However, all strains were found susceptible to kanamycin, and it can be referred to as a drug of choice in case of mastitis infections in the regions included in the study. The information generated here might be useful for concerned veterinarians in improving cattle health and designing strategies for better and safe milk production. This will not only lower the risk of associated food poisoning but also prevent the spread of antibiotic resistance in the regions. Future research is warranted to establish the relationship between the presence of these strains in milk and their ability to cause infections in humans.
Authors’ contributions
SS and VS provide the financial support, proof-read the manuscript. DKD conceived the idea and wrote the manuscript and proof-read it. AK, MM and AS perform the experiments and help in proof-reading. All authors read and approved the final manuscript.
Acknowledgements
This study was supported by Rashtriya Krishi Vikas Yojana (RKVY), Government of Rajasthan, India.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data supporting the conclusions.
Consent for publication
All authors agreed for publication.
Ethics approval and consent to participate
Before collecting milk samples from cattle, oral consent was obtained from the farmers or dairy owners. As per the Indian laws, there is no requirement to seek any permission for milk sample collection from animals. Besides, milk samples were collected by a trained veterinary professional. In addition, we confirmed that the study does not involve any endangered species and no animals were involved in the experiments.
Funding
We did not receive any specific funding for conducting this research.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Vishnu Sharma, Email: drvishnus@yahoo.com.
Sanjita Sharma, Phone: +91-0141-6598997, Email: drsanjitas@gmail.com.
Dinesh Kumar Dahiya, Email: dineshlucky31@gmail.com.
Aarif Khan, Email: khan.aarif09@gmail.com.
Manisha Mathur, Email: matmanisha@gmail.com.
Aayushi Sharma, Email: aayushisharma103@gmail.com.
References
- 1.FAO. A report on milk and milk products. Food and agricultural organization, United States. 2013. www.milkproduction.com/Library/Editorial-articles/FAO-Food-Outlook-Nov-2013—Milk-and-Milk-products/.
- 2.Ministry of Agricultural. State/UT-wise estimates of Milk Production, Government of India. 2015. https://data.gov.in/catalog/stateut-wise-estimates-milk-production.
- 3.Reshi AA, Husain I, Bhat S, Rehman MU, Razak R, Bilal S, Mir MR. Bovine mastitis as an evolving disease and its impact on the dairy industry. Int J Curr Res Rev. 2015;7(5):48–55. [Google Scholar]
- 4.Motwani KT. Mastitis in dairy cattle in India. In Social Science Research Network. 2011 [Google Scholar]
- 5.Kader M, Samad M, Saha S, Taleb M. Prevalence and etiology of subclinical mastitis with antibiotic sensitivity to isolated organisms among milch cows in Bangladesh. Indian J Dairy Sci. 2002;55(4):218–223. [Google Scholar]
- 6.Seegers H, Fourichon C, Beaudeau F. Production effects related to mastitis and mastitis economics in dairy cattle herds. Vet Res. 2003;34(5):475–491. doi: 10.1051/vetres:2003027. [DOI] [PubMed] [Google Scholar]
- 7.Dahiya DK, Renuka MP, Puniya M, Shandilya UK, Dhewa T, Kumar N, Kumar S, Puniya AK, Shukla P. Gut microbiota modulation and its relationship with obesity using prebiotic fibers and probiotics: a review. Front Microbiol. 2017;8:563. doi: 10.3389/fmicb.2017.00563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sahu B, Mukherjee R, Kumar A, Kumar A, Sahu J. Prevalence of coagulase gene positive Staphylococcus aureus bovine mastitis in three distinct geoclimatic regions of India. Buffalo Bull. 2014;33(2):208–214. [Google Scholar]
- 9.Daka D, Yihdego D. Antibiotic-resistance Staphylococcus aureus isolated from cow’s milk in the Hawassa area, South Ethiopia. Ann Clin Microbiol Antimicrob. 2012;11(1):26. doi: 10.1186/1476-0711-11-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma S, Sharma V, Dahiya DK, Khan A, Mathur M, Sharma A. Prevalence, virulence potential, and antibiotic susceptibility profile of Listeria monocytogenes isolated from bovine raw milk samples obtained from Rajasthan, India. Foodborne Pathog Dis. 2017;14(3):132–140. doi: 10.1089/fpd.2016.2118. [DOI] [PubMed] [Google Scholar]
- 11.Salasia SIO, Lammler C. Comparative studies on pheno-and genotypic properties of Staphylococcus aureus isolated from bovine subclinical mastitis in central Java in Indonesia and Hesse in Germany. J Vet Sci. 2004;5(2):103–109. [PubMed] [Google Scholar]
- 12.Omuse G, Zyl KN, Hoek K, Abdulgader S, Kariuki S, Whitelaw A, Revathi G. Molecular characterization of Staphylococcus aureus isolates from various healthcare institutions in Nairobi, Kenya: a cross sectional study. Ann Clin Microbiol Antimicrob. 2016;15(1):51. doi: 10.1186/s12941-016-0171-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Acosta A, Guelfer E, Ruiz A, Mota RA, Uffo O, Gomes-Filho MA, Barbosa SBP. Molecular subtyping of Staphylococcus aureus isolated from bovine mastitis in Pernambuco State, Brazil. Comp Clin Path. 2014;23(4):1037–1041. doi: 10.1007/s00580-013-1739-z. [DOI] [Google Scholar]
- 14.Momtaz H, Tajbakhsh E, Rahimi E, Momeni M. Coagulase gene polymorphism of Staphylococcus aureus isolated from clinical and sub-clinical bovine mastitis in Isfahan and Chaharmahal va Bakhtiari provinces of Iran. Comp Clin Path. 2011;20(5):519–522. doi: 10.1007/s00580-010-1029-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarvari J, Bazargani A, Kandekar-Ghahraman MR, Nazari-Alam A, Motamedifar M. Molecular typing of methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates from Shiraz teaching hospitals by PCR-RFLP of coagulase gene. Iran J Microbiol. 2014;6(4):246–252. [PMC free article] [PubMed] [Google Scholar]
- 16.Oliver SP, Jayarao BM, Almeida RA. Foodborne pathogens in milk and the dairy farm environment: food safety and public health implications. Foodborne Pathog Dis. 2005;2(2):115–129. doi: 10.1089/fpd.2005.2.115. [DOI] [PubMed] [Google Scholar]
- 17.Srey S, Jahid IK, Ha S-D. Biofilm formation in food industries: a food safety concern. Food Control. 2013;31(2):572–585. doi: 10.1016/j.foodcont.2012.12.001. [DOI] [Google Scholar]
- 18.Al-Ashmawy MA, Sallam KI, Abd-Elghany SM, Elhadidy M, Tamura T. Prevalence, molecular characterization, and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus isolated from milk and dairy products. Foodborne Pathog Dis. 2016;13(3):156–162. doi: 10.1089/fpd.2015.2038. [DOI] [PubMed] [Google Scholar]
- 19.Fox L, Zadoks R, Gaskins C. Biofilm production by Staphylococcus aureus associated with intramammary infection. Vet Microbiol. 2005;107(3):295–299. doi: 10.1016/j.vetmic.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 20.Tang J, Chen J, Li H, Zeng P, Li J. Characterization of adhesin genes, staphylococcal nuclease, hemolysis, and biofilm formation among Staphylococcus aureus strains isolated from different sources. Foodborne Pathog Dis. 2013;10(9):757–763. doi: 10.1089/fpd.2012.1474. [DOI] [PubMed] [Google Scholar]
- 21.J-p Li, H-j Zhou. Yuan L, He T, Hu S-h. Prevalence, genetic diversity, and antimicrobial susceptibility profiles of Staphylococcus aureus isolated from bovine mastitis in Zhejiang Province, China. J Zhejiang Univ Sci B. 2009;10(10):753–760. doi: 10.1631/jzus.B0920072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dahiya DK, Puniya AK. Isolation, molecular characterization and screening of indigenous lactobacilli for their abilities to produce bioactive conjugated linoleic acid (CLA) J Food Sci. 2017;54(3):792–801. doi: 10.1007/s13197-017-2523-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goh SH, Byrne S, Zhang J, Chow A. Molecular typing of Staphylococcus aureus on the basis of coagulase gene polymorphisms. J Clin Microbiol. 1992;30(7):1642–1645. doi: 10.1128/jcm.30.7.1642-1645.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26(11):2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mehrotra M, Wang G, Johnson WM. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J Clin Microbiol. 2000;38(3):1032–1035. doi: 10.1128/jcm.38.3.1032-1035.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang J, Chen J, Liu J, Zhang R, Yang R, Chen L. Effects of different cultivation conditions on Staphylococcus aureus biofilm formation and diversity of adhesin genes. J Food Saf. 2012;32(2):210–218. doi: 10.1111/j.1745-4565.2012.00370.x. [DOI] [Google Scholar]
- 27.Dahiya DK, Puniya AK. Evaluation of survival, free radical scavenging and human enterocyte adherence potential of lactobacilli with anti-obesity and anti-inflammatory cla isomer-producing attributes. J Food Process Preserv. 2015;39(6):2866–2877. doi: 10.1111/jfpp.12538. [DOI] [Google Scholar]
- 28.CLSI. M100-S25 performance standards for antimicrobial susceptibility testing; Twenty-fifth informational supplement; 2015.
- 29.Stegger A, Andersen P, Kearns A, Pichon B, Holmes M, Edwards G, Laurent F, Teale C, Skov R, Larsen A. Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecALGA251. Clin Microbiol Infect. 2012;18(4):395. doi: 10.1111/j.1469-0691.2011.03715.x. [DOI] [PubMed] [Google Scholar]
- 30.Lingathurai S, Vellathurai P. Bacteriological quality and safety of raw cow milk in Madurai. South India. Webmed Cent Microbiol. 2013;1:1–10. [Google Scholar]
- 31.Sadashiv S, Kaliwal B. Antibiotic resistance of Staphyloccocus aureus and Coagulase negative Staphylococci (CNS) isolated from bovine mastitis in the region of North Karnataka, India. World J Pharm Res. 2013;3:571–586. [Google Scholar]
- 32.Sharma S, Khan A, Dahiya DK, Jain J, Sharma V. Prevalence, identification and drug resistance pattern of Staphylococcus aureus and Escherichia coli isolated from raw milk samples of jaipur city of Rajasthan. J Pure Appl Microbiol. 2015;9(1):341–348. [Google Scholar]
- 33.Sudhanthiramani S, Swetha CS, Bharathy S. Prevalence of antibiotic resistant Staphylococcus aureus from raw milk samples collected from the local vendors in the region of Tirupathi, India. Vet world. 2015;8(4):478–481. doi: 10.14202/vetworld.2015.478-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar R, Prasad A. Detection of E. coli and Staphylococcus in milk and milk products in and around Pantnagar. Pak J Nutr. 2010;1(3):151–152. [Google Scholar]
- 35.Thaker H, Brahmbhatt M, Nayak J, Thaker HC. Isolation and identification of Staphylococcus aureus from milk and milk products and their drug resistance patterns in Anand, Gujarat. Vet World. 2013;6(1):10–13. doi: 10.5455/vetworld.2013.10-13. [DOI] [Google Scholar]
- 36.Vieira-da-Motta O, Folly MM, Sakyiama CCH. Detection of different Staphylococcus aureus strains in bovine milk from subclinical mastitis using PCR and routine techniques. Braz J Microbiol. 2001;32(1):27–31. doi: 10.1590/S1517-83822001000100007. [DOI] [Google Scholar]
- 37.da Silva ER, da Silva N. Coagulase gene typing of Staphylococcus aureus isolated from cows with mastitis in southeastern Brazil. Can J Vet Res. 2005;69(4):260–264. [PMC free article] [PubMed] [Google Scholar]
- 38.Saei HD, Ahmadi M, Mardani K, Batavani R. Molecular typing of Staphylococcus aureus isolated from bovine mastitis based on polymorphism of the coagulase gene in the north west of Iran. Vet Microbiol. 2009;137(1):202–206. doi: 10.1016/j.vetmic.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 39.Schwarzkopf A, Karch H. Genetic variation in Staphylococcus aureus coagulase genes: potential and limits for use as epidemiological marker. J Clin Microbiol. 1994;32(10):2407–2412. doi: 10.1128/jcm.32.10.2407-2412.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishino K, Tsuchizaki N, Ishikawa J, Hotta K. Usefulness of PCR-restriction fragment length polymorphism typing of the coagulase gene to discriminate arbekacin-resistant methicillin-resistant Staphylococcus aureus strains. J Clin Microbiol. 2007;45(2):607–609. doi: 10.1128/JCM.02099-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abdulghany HM, Khairy RM. The Frequency of methicillin-resistant Staphylococcus aureus and coagulase gene polymorphism in Egypt. Int J Bacteriol. 2014;2014:1–6. doi: 10.1155/2014/680983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moon J, Lee A, Kang H, Lee E, Joo Y, Park YH, Kim M, Koo H. Antibiogram and coagulase diversity in staphylococcal enterotoxin-producing Staphylococcus aureus from bovine mastitis. J Dairy Sci. 2007;90(4):1716–1724. doi: 10.3168/jds.2006-512. [DOI] [PubMed] [Google Scholar]
- 43.Kumar R, Yadav B, Singh R. Antibiotic resistance and pathogenicity factors in Staphylococcus aureus isolated from mastitic Sahiwal cattle. J Biosci. 2011;36(1):175–188. doi: 10.1007/s12038-011-9004-6. [DOI] [PubMed] [Google Scholar]
- 44.El-Sayed A, Alber J, Lammler C, Jager S, Woter W, Vázquez H. Comparative study on genotypic properties of Staphylococcus aureus isolated from clinical and subclinical mastitis in Mexico. Vet Mex. 2006;37(2):165–179. [Google Scholar]
- 45.Darwish SF, Asfour HA. Investigation of biofilm forming ability in Staphylococci causing bovine mastitis using phenotypic and genotypic assays. Sci World J. 2013;2013:1–9. doi: 10.1155/2013/378492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raza A, Muhammad G, Sharif S, Atta A. Biofilm producing Staphylococcus aureus and bovine mastitis: a review. Mol Microbiol Res. 2013;3(1):1–8. [Google Scholar]
- 47.Salgado-Ruiz T, Rodríguez A, Gutiérrez D, Martínez B, García P, Espinoza-Ortega A, Martínez-Campos A, Lagunas-Bernabé S, Vicente F, Arriaga-Jordán C. Molecular characterization and antimicrobial susceptibility of Staphylococcus aureus from small-scale dairy systems in the highlands of Central México. Dairy Sci Technol. 2015;95(2):181–196. doi: 10.1007/s13594-014-0195-0. [DOI] [Google Scholar]
- 48.Mulcahy ME, Geoghegan JA, Monk IR, O’Keeffe KM, Walsh EJ, Foster TJ, McLoughlin RM. Nasal colonisation by Staphylococcus aureus depends upon clumping factor B binding to the squamous epithelial cell envelope protein loricrin. PLoS Pathog. 2012;8(12):e1003092. doi: 10.1371/journal.ppat.1003092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakakido M, Aikawa C, Nakagawa I, Tsumoto K. The staphylococcal elastin-binding protein regulates zinc-dependent growth/biofilm formation. J Biochem. 2014;156(3):155–162. doi: 10.1093/jb/mvu027. [DOI] [PubMed] [Google Scholar]
- 50.Pereyra EA, Picech F, Renna MS, Baravalle C, Andreotti CS, Russi R, Calvinho LF, Diez C, Dallard BE. Detection of Staphylococcus aureus adhesion and biofilm-producing genes and their expression during internalization in bovine mammary epithelial cells. Vet Microbiol. 2016;183:69–77. doi: 10.1016/j.vetmic.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 51.Kot B, Szweda P, Frankowska-Maciejewska A, Piechota M, Wolska K. Virulence gene profiles in Staphylococcus aureus isolated from cows with subclinical mastitis in eastern Poland. J Dairy Res. 2016;83(2):228–235. doi: 10.1017/S002202991600008X. [DOI] [PubMed] [Google Scholar]
- 52.Puacz E, Ilczyszyn WM, Kosecka M, Buda A, Dudziak W, Polakowska K, Panz T, Białecka A, Kasprowicz A, Lisowski A. Clustering of Staphylococcus aureus bovine mastitis strains from regions of Central-Eastern Poland based on their biochemical and genetic characteristics. Pol J Vet Sci. 2015;18(2):333–342. doi: 10.1515/pjvs-2015-0043. [DOI] [PubMed] [Google Scholar]
- 53.Zuniga E, Melville PA, Saidenberg AB, Laes MA, Gonsales FF, Salaberry SR, Gregori F, Brandao PE, Santos FG, Lincopan NE. Occurrence of genes coding for MSCRAMM and biofilm-associated protein Bap in Staphylococcus spp. isolated from bovine subclinical mastitis and relationship with somatic cell counts. Microb Pathog. 2015;89:1–6. doi: 10.1016/j.micpath.2015.08.014. [DOI] [PubMed] [Google Scholar]
- 54.Li L, Zhou L, Wang L, Xue H, Zhao X. Characterization of methicillin-resistant and-susceptible staphylococcal isolates from bovine milk in Northwestern China. PLoS ONE. 2015;10(3):e0116699. doi: 10.1371/journal.pone.0116699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Atray D, Sharma A, Atray M. Prevalence of enterococci and its antibiotic resistance in various clinical samples at tertiary care hospital in Southern Rajasthan, India. Int J Res Med Sci. 2017;4(8):3413–3416. [Google Scholar]
- 56.Sood S, Gupta R. Antibiotic resistance pattern of community acquired uropathogens at a tertiary care hospital in Jaipur, Rajasthan. Indian J Comm Med. 2012;37(1):39–44. doi: 10.4103/0970-0218.94023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharma KD, Saini RP, Karthik L. Current trends of antibiotic resistance in clinical isolates of Staphylococcus aureus. Front Biol. 2014;9(4):287–290. doi: 10.1007/s11515-014-1317-z. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data supporting the conclusions.


