Abstract
Background
Kernel hardness, which has great influence on the end-use properties of common wheat, is mainly controlled by Puroindoline genes, Pina and Pinb. Using EcoTILLING platform, we herein investigated the allelic variations of Pina and Pinb genes and their association with the Single Kernel Characterization System (SKCS) hardness index in a diverse panel of wheat germplasm.
Results
The kernel hardness varied from 1.4 to 102.7, displaying a wide range of hardness index. In total, six Pina and nine Pinb alleles resulting in 15 genotypes were detected in 1787 accessions. The most common alleles are the wild type Pina-D1a (90.4%) and Pina-D1b (7.4%) for Pina, and Pinb-D1b (43.6%), Pinb-D1a (41.1%) and Pinb-D1p (12.8%) for Pinb. All the genotypes have hard type kernel hardness of SKCS index (>60.0), except the wild types of Pina and Pinb combination (Pina-D1a/Pinb-D1a). The most frequent genotypes in Chinese and foreign cultivars was Pina-D1a/Pinb-D1b (46.3 and 39.0%, respectively) and in Chinese landraces was Pina-D1a/Pinb-D1a (54.2%). The frequencies of hard type accessions are increasing from 35.5% in the region IV, to 40.6 and 61.4% in the regions III and II, and then to 77.0% in the region I, while those of soft type are accordingly decreasing along with the increase of latitude. Varieties released after 2000 in Beijing, Hebei, Shandong and Henan have higher average kernel hardness index than that released before 2000.
Conclusion
The kernel hardness in a diverse panel of Chinese wheat germplasm revealed an increasing of kernel hardness generally along with the latitude across China. The wild type Pina-D1a and Pinb-D1a, and one Pinb mutant (Pinb-D1b) are the most common alleles of six Pina and nine Pinb alleles, and a new double null genotype (Pina-D1x/Pinb-D1ah) possessed relatively high SKCS hardness index. More hard type varieties were released in recent years with different prevalence of Pin-D1 combinations in different regions. This work would benefit the understanding of the selection and molecular processes of kernel hardness across China and different breeding stages, and provide useful information for the improvement of wheat quality in China.
Electronic supplementary material
The online version of this article (10.1186/s12870-017-1101-8) contains supplementary material, which is available to authorized users.
Keywords: Common wheat, Kernel hardness, Puroindoline genes, EcoTILLING, Allelic variants
Background
Wheat (Triticum aestivum L.) is one of the most widely grown food crop all over the world with a wide array of food products for human consumption. The kernel hardness is a major determinant of end-use food properties of wheat grain. Kernel hardness refers to the texture of the grain (caryopsis), that is, whether the endosperm is physically hard or soft. This difference in grain texture is due to a 13–15 kDa marker protein, friabilin, which is highly present on the surface of water-washed starch of soft wheat, lower on hard wheat starch and absent on durum wheat starch [1]. The N-terminal sequence analysis of friabilin revealed a mixture of two or more distinct polypeptides [2, 3], which were found to be identical to the two lipid binding proteins, Puroindoline a (PINA) and b (PINB) [4]. The transcripts of PINA and PINB are controlled by two linked genes, Pina and Pinb, respectively, located on short arm of chromosome 5D [5]. The presence of wild type Pina-D1a and Pinb-D1a genes are both necessary to the grain softening of allohexaploid nature of wheat (AABBDD, 2n = 6× =42). However, homologous of the Pina and Pinb genes are absent on the wheat 5A and 5B chromosomes, and thus durum wheats (Triticum turgidum L.) (AABB, 2n = 4× = 28) lacking Pina and Pinb genes have hard textured grains [6].
Soft grain wheat varieties have wild type (WT) alleles of both Pina and Pinb genes and any mutation in WT alleles at one or both Pin genes gives rise to hard grain texture leading to changed food technological properties [7]. The variation in degree of grain texture hardness is due to a spectrum of alleles and their combinations at Pina and Pinb, and the number of alleles at Pina and Pinb detected so far has increased to 23 and 33, respectively [7–10]. Since the basic mechanism by which Puroindolines induce soften endosperm is not well known, genotype-phenotype associations are useful to estimate the kernel hardness effect of a Pin allele. The absence or altered primary structure of one of PINA and PINB will result in a hard grain texture, and among commercial wheat cultivars the most prevalent hard genotypes are the absence of PINA or the altered primary structure of PINB with null alleles of Pina and Pina-D1a/Pinb-D1b, respectively [11–13]. However, cultivars with null alleles of Pina have been proposed harder than those with Pina-D1a/Pinb-D1b [13–19]. Moreover, the genotype with Pina-D1b (the null allele of Pina) may have poor milling quality and relatively inferior processing quality for steamed bread, pan bread, and Chinese noodles [16]. This null allele has been molecular characterized with a large deletion of 15,380 bp through a primer walking strategy and a diagnostic sequence tagged site (STS) marker has also been developed spanning the deletion fragment [20]. Moreover, on chromosomes 7A, 7B and 7D in bread wheat, homologous Pinb genes have been found with more than 70.0% similarity, though their function is elusive [21–23]. Although most of the known hardness alleles confer large and somewhat similar changes in endosperm texture relative to soft wheat, the discovery of new alleles could broaden the genetic background for kernel hardness and provide industry with grains more suitable for a variety of end-uses. Furthermore, different combinations of Pina and Pinb alleles in common wheat determine the grain textural classes with diverse end-use characteristics [24]. Thus, knowledge on the Puroindoline allelic composition in a diverse panel of germplasm is prerequisite for the parental selection for developing varieties with desired kernel hardness.
The research herein presents the analysis of Puroindoline allelic variations, genotypes and their association with kernel hardness in a diverse panel of wheat accessions comprising 1539 Chinese cultivars, 107 Chinese landraces and 141 foreign accessions. A subset of 623 accessions was evaluated for two consecutive years to assess environmental effect on SKCS index of kernel hardness. Accessions collected in Beijing, Hebei, Shandong and Henan were analyzed in detail to reveal the hardness trends along with breeding stages and the prevalence of Pina and Pinb alleles. The knowledge generated in this study about the allelic effect on and dynamics of kernel hardness, and the allele distribution across China would enhance breeder’s choice for suitable parent selection to develop cultivars of desired kernel hardness, and improve the understanding on the patterns of kernel hardness across different wheat zones in China. The new double null mutants would provide an alternative option for kernel hardness improvement.
Results
High phenotypic variation in kernel hardness
To assess the kernel hardness in Chinese wheat germplasm, 1646 accessions were collected from nine wheat cultivation regions in China and grown in Beijing in 2009–2010 cropping season, along with 141 foreign accessions from USA, Australia, Europe and Japan (Table 1). Of the Chinese accessions, 1582 accessions were from cultivation region I (Northern winter wheat region), II (Yellow and Huai River Valley winter wheat region), III (Middle and Low Yangtze Valley winter wheat region) and IV (Southwestern winter wheat region), and the remaining 64 accessions were from other spring wheat cultivation regions since most spring wheat from these regions hardly survive through the cold winter season in Beijing. The kernel hardness of harvested samples in 2009–2010 growing season measured with SKCS showed that 1787 accessions contained a wide range of SKCS index varying from 1.4 to 102.7 (Table 2). The major class was of hard type including 1075 accessions (60.2%), which were from most wheat cultivation regions of China, except Qing-Tibetan Plateau spring-winter wheat region (cultivation region IX) (Table 3). The SKCS index of hard type ranged from 60.1 to 102.7 with mean value of 72.4, and about 93.0% accessions have a hardness index lower than 85.0. The soft type class consisted of 523 accessions (29.2%) with SKCS index ranging from 1.4 to 40.0 and mean value of 25.1, while the medium type class with medium kernel hardness included only 189 accessions (10.6%), ranging from 40.0 to 60.0 with mean value of 54.1 (Table 2).
Table 1.
Number and types of accessions for kernel hardness measurement and Puroindoline-D1 detection
| Regiona | Cultivars | Landraces | Total | |
|---|---|---|---|---|
| China | I | 244 | 4 | 248 |
| II | 1065 | 25 | 1090 | |
| III | 81 | 25 | 106 | |
| IV | 95 | 43 | 138 | |
| V | 2 | 1 | 3 | |
| VI | 9 | 3 | 12 | |
| VIII | 32 | 4 | 36 | |
| IX | 1 | 2 | 3 | |
| X | 10 | 0 | 10 | |
| Foreign | 141 | |||
| Total | 1539 | 107 | 1787 |
aDefinition of wheat cultivation regions: I (Northern winter wheat region), II (Yellow and Huai River Valley winter wheat region), III (Middle and Low Yangtze Valley winter wheat region), IV (Southwestern winter wheat region), V (Southern winter wheat region), VI (Northeastern spring wheat region), VIII (Northwestern spring wheat region), IX (Qing-Tibetan Plateau spring-winter wheat region) and X (Xinjiang winter-spring wheat region)
Table 2.
Distribution of kernel hardness in and percentage of soft, hard and medium wheats
| Type | Number | Freq. (%) | SKCS index | |
|---|---|---|---|---|
| Mean | Range | |||
| Soft | 523 | 29.2 | 25.1 | 1.4–40.0 |
| Hard | 1075 | 60.2 | 72.4 | 60.1–102.7 |
| Medium | 189 | 10.6 | 54.1 | 40.0–60.0 |
Table 3.
Region-wise percent distribution of soft, hard and medium wheats in two different origins and nine wheat cultivation regions in Chinaa
| China (1646) | Foreign (141) | Freq. (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | VI | VIII | IX | X | |||
| Soft | 10.5 | 27.6 | 51.9 | 55.8 | 66.7 | 66.7 | 30.6 | 100.0 | 30.0 | 25.5 | 29.2 (522) |
| Hard | 77.0 | 61.4 | 40.6 | 35.5 | 33.3 | 33.3 | 63.9 | 0.0 | 60.0 | 63.1 | 60.2 (1075) |
| Medium | 12.5 | 11.0 | 7.5 | 8.7 | 0.0 | 0.0 | 5.6 | 0.0 | 10.0 | 11.3 | 10.6 (190) |
aThe values in parentheses are the number of accessions and the wheat cultivation regions are defined in Table 1
To determine the environment effect on kernel hardness, 623 accessions were grown on the same station during 2009–2010 and 2010–2011 growing seasons using the same agronomic practices and phenotyped for SKCS. The ranges of SKCS index for the year 2009–2010 were of 1.4 to 97.8 with an average of 49.2 and these for 2010–2011 were 1.1 to 98.0 with an average of 48.0. For the environment effect, the analysis of variance (ANOVA) showed non-significant with P value of 0.39 (Additional file 1: Table S1) between 2 years’ data, and thus we considered the SKCS data of 2009–2010 valid for association analysis between SKCS index of kernel hardness and genotypes of Pina and Pinb.
Classified regional distribution of hard and soft kernel accessions
Based on the kernel hardness, accessions from nine wheat growing regions in China were analyzed, region I (Northern winter wheat region) had the highest number of hard textured type wheat accessions, 77.0%, and the regions II (Yellow and Huai River Valley winter wheat region), VIII (Northwestern spring wheat region), X (Xinjiang winter-spring wheat region) and foreign accessions were also dominantly represented by hard type wheat, around 60.0% (Table 3). Conversely, the regions III (Middle and Lower Yangtze Valley winter wheat region), IV (Southwestern winter wheat region), V (Southern winter wheat region) and VI (Northeastern spring wheat region) were represented by soft type wheat with about 50.0% prevalence (Table 3). All the regions and foreign accessions were represented by all three kernel hardness types, except regions V and VI that did not contain medium type accessions and IX did not have hard and medium types. This might be due to only a few available accessions collected from these regions (Table 3).
In China, the winter wheat growing regions IV (Southwestern winter wheat region), III (Middle and Low Yangtze Valley winter wheat region), II (Yellow and Huai River Valley winter wheat region) and I (Northern winter wheat region) extends from South to North along with the increase of latitude. Regarding to hardness type in different wheat regions, we found that the frequencies of hard type accessions are increasing from 35.5% in the region IV, to 40.6 and 61.4% in the regions III and II, and then to 77.0% in the region I, while those of soft type are accordingly decreasing along with the increase of latitude (Table 3). This phenomenon was also observed in landraces (Additional file 1: Table S2). More soft accessions present in the regions III and IV than that in the region II, and no soft accessions were detected in the region I (Additional file 1: Table S2).
Allelic variants of Pina and Pinb genes
The nested PCR and modified EcoTILLING analysis was performed with allele specific primers to detect allelic variants of Pina and Pinb genes (Fig. 1; Additional file 1: Table S3). Of the analyzed 1787 accessions, six Pina allelic variants including the wild type (Pina-D1a) and five mutants were characterized (Table 4). Except Pina-D1l has a cytosine deletion as comparing to Pina-D1a (Additional file 4: Figure S3A), the remaining mutants belong to different types of Pina null mutant (Table 4). The wild type Pina-D1a was the most common allele observed in 1616 accessions (90.4%) followed by Pina-D1b in 133 accessions (7.4%), while the remaining variants, Pina-D1l, Pina-D1r, Pina-D1s and Pina-D1x were rare alleles, present in a few samples (Table 4). Nine allelic variants were identified at Pinb, of which Pinb-D1b, Pinb-D1c, Pinb-D1d, Pinb-D1p, Pinb-D1q and Pinb-D1u have SNP mutations, and Pinb-D1ah and Pinb-D1ai belong to different types of Pinb null mutant (Table 4; Additional file 4: Figure S3B). Only three variants Pinb-D1b, Pinb-D1a and Pinb-D1p (43.6, 41.1 and 12.8%, respectively) were common in these allelic variants (Table 4). For the null alleles of Pina and Pinb, the Pina-D1x as a novel null variant was different from previously reported alleles with various fragment deletions (Pina-D1b, Pina-D1r and Pina-D1s) and detected in seven Chinese cultivars and three foreign accessions. There are two novel Pinb-null alleles, one is Pinb absent but Pina present, so this Pinb-null allele was designated as Pinb-D1ai, and detected only in two accessions; the other Pinb-null allele was named as Pinb-D1ah and observed in the ten accessions containing the Pina-D1x allele, in which the simultaneous deletion of the large segment containing Pina and Pinb genes might have occurred.
Fig. 1.
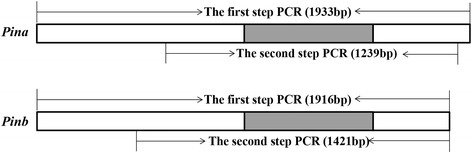
Diagrams of PCR amplification for Pina and Pinb. The gray boxes show coding regions for Pina and Pinb, respectively. The first step PCR reaction of Pina and Pinb (Pina-OutF/R, Pinb-OutF/R) used for amplifying Pina (1933 bp) and Pinb (1916 bp) genomic sequence. The second step PCR reaction of Pina and Pinb (Pina-InF/R, Pinb-InF/R) used for amplifying Pina (1239 bp) and Pinb (1421 bp) genomic sequence
Table 4.
Distribution of Pina and Pinb alleles in total 1787 accessions
| Pina a | Pinb a | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D1a | D1b | D1l | D1r | D1s | D1x | D1a | D1b | D1c | D1d | D1p | D1q | D1u | D1ah | D1ai | |
| Cultivars | 1414 | 113 | 1 | 2 | 2 | 7 | 589 | 721 | 0 | 13 | 201 | 7 | 0 | 7 | 1 |
| Landraces | 86 | 4 | 3 | 8 | 6 | 0 | 78 | 4 | 0 | 1 | 18 | 2 | 3 | 0 | 1 |
| Foreign | 116 | 16 | 0 | 1 | 5 | 3 | 68 | 55 | 2 | 4 | 9 | 0 | 0 | 3 | 0 |
| Total | 1616 | 133 | 4 | 11 | 13 | 10 | 735 | 780 | 2 | 18 | 228 | 9 | 3 | 10 | 2 |
aThe values are the number of accessions for each allele
Regarding the distribution of allelic variants across Chinese cultivars, landraces and foreign accessions, frequency of each allelic variant of Pina and Pinb genes was calculated. For Pina gene, the wild type allele Pina-D1a was predominant in Chinese cultivars, landraces and foreign accessions with frequencies of 91.9, 80.4 and 82.3% in the respective groups. Among the mutants, the most frequent allele was Pina-D1b in Chinese cultivars and foreign accessions with frequency of 7.3 and 11.3%, respectively, whereas Pina-D1r (7.5%) was the most in landraces, followed by Pina-D1s (5.6%) and Pina-D1b (3.7%). The variants Pina-D1l and Pina-D1x were missing in foreign and landrace accessions, respectively (Table 4). For Pinb gene, the highest frequency of the wild type allele Pinb-D1a was present in Chinese landraces (72.9%), followed by foreign accessions (48.2%) and Chinese cultivars (38.3%), which has an opposite pattern to that of Pina-D1a. Among the mutants, Pinb-D1b and Pinb-D1p had frequencies of 46.8 and 13.1% in Chinese cultivars, respectively, and other four variants were rarely present (totally less than 2.0%); In foreign accessions, Pinb-D1b and Pinb-D1p were also prevalent with frequencies of 39.0 and 6.4%, but other three variants (Pinb-D1c, Pinb-D1d and Pinb-D1ah) had relatively high frequencies as compared with those in Chinese accessions, whereas Pinb-D1b was present in only 3.7% Chinese landraces, and Pinb-D1p was the most present mutant (16.8%) in Chinese landraces (Table 4). Interestingly, the allele Pinb-D1c was missing in Chinese cultivars and landraces but was found in foreign accessions, while the allele Pinb-D1u was completely absent in Chinese cultivars and foreign accessions and was only detected in Chinese landraces, Zhushimai, Zhugoumai and Dajinbaihuazi (Table 4). Those three landraces are found in Yunnan and Sichuan provinces, belonging to Southwestern winter wheat region (IV), and they could be exploited to incorporate this novel allele into Chinese wheat cultivars. Moreover, the variants Pina-D1l, Pinb-D1q and Pinb-D1ai were missing in foreign accessions, and the double mutant Pina-D1x/Pinb-D1ah was not detected in Chinese landraces. The different allelic frequencies in three type accessions suggested that the Pina and Pinb were artificially selected by breeders.
To assess the diversity at Pina and Pinb genes, Nei’s diversity index (H) was calculated in the total collection of 1787 accessions. The H value at Pina and Pinb were 0.18 and 0.62, respectively. At Pina, the highest diversity (0.34) was observed in 107 Chinese landraces, followed by 0.31 in the group of 141 foreign accessions, and the least (0.15) was observed in 1539 Chinese cultivars. At Pinb, Chinese cultivars and foreign accessions had similar high diversity (0.62), and Chinese landraces possessed a lower diversity (0.43), which was different from that of Pina. These data demonstrated that more allelic variants of Pinb with higher frequencies were retained in breeding process than those of Pina.
Association analysis between genotype and kernel hardness
To investigate the effect of Pina and Pinb on kernel hardness, genotypes were assayed on six Pina and nine Pinb variants. Of the 15 Pina and Pinb combinations (Table 5), eight were the Pina wild type (Pina-D1a) with eight Pinb variants, occupying a 90.4% accessions, three were Pina-D1b and three Pinb variants with a total frequency of 7.4%, and the remaining four combinations were formed individually by four Pina variants with corresponding Pina variants. At Pinb genotypes, five were formed by the wild type (Pinb-D1a) with five Pina variants, and two were Pinb-D1b and two Pina variants, in which Pinb-D1a and Pinb-D1b genotypes occurred in 41.1 and 43.6% accessions, respectively (Table 5). The most abundant combination was Pina-D1a/Pinb-D1b (43.1%), followed by Pina-D1a/Pinb-D1a (32.8%), Pina-D1a/Pinb-D1p (12.5%) and Pina-D1b/Pinb-D1a (6.7%) with corresponding average kernel hardness of 67.7, 31.0, 68.0 and 79.3. The other 11 combinations were rare (≤1.0%) with a total frequency of 4.8%, however these combinations had higher average kernel hardness (≥60.0), of which that of Pina-D1x/Pinb-D1ah was 91.1, at least 10 hardness index higher than all the remaining genotypes (Table 5).
Table 5.
Association of Puroindoline-D1 alleles with SKCS index and their distributions in Chinese cultivars, Landraces and foreign accessions
| Genotype | SKCS index | Cultivars | Landraces | Foreign | |
|---|---|---|---|---|---|
| Mean | Range | ||||
| Pina-D1a/Pinb-D1a | 31.0 | 1.4–92.5 | 483 | 58 | 46 |
| Pina-D1a/Pinb-D1b | 67.7 | 11.0–102.7 | 712 | 4 | 55 |
| Pina-D1a/Pinb-D1c | 60.0 | 45.1–74.9 | 0 | 0 | 2 |
| Pina-D1a/Pinb-D1d | 63.6 | 48.5–73.6 | 13 | 1 | 4 |
| Pina-D1a/Pinb-D1p | 68.0 | 16.1–88.6 | 198 | 17 | 9 |
| Pina-D1a/Pinb-D1q | 66.2 | 57.0–78.3 | 7 | 2 | 0 |
| Pina-D1a/Pinb-D1u | 65.9 | 58.4–78.4 | 0 | 3 | 0 |
| Pina-D1a/Pinb-D1ai | 78.6 | 65.6–91.6 | 1 | 1 | 0 |
| Pina-D1b/Pinb-D1a | 79.3 | 19.7–95.6 | 101 | 3 | 16 |
| Pina-D1b/Pinb-D1b | 68.9 | 19.9–75.5 | 9 | 0 | 0 |
| Pina-D1b/Pinb-D1p | 71.1 | 66.3–82.4 | 3 | 1 | 0 |
| Pina-D1l/Pinb-D1a | 74.3 | 68.8–84.2 | 1 | 3 | 0 |
| Pina-D1r/Pinb-D1a | 76.7 | 59.9–94.1 | 2 | 8 | 1 |
| Pina-D1s/Pinb-D1a | 67.1 | 53.4–86.2 | 2 | 6 | 5 |
| Pina-D1x/Pinb-D1ah | 91.1 | 77.1–97.8 | 7 | 0 | 3 |
| Total | 1539 | 107 | 141 | ||
Regarding to the distribution of 15 combinations of Pina/Pinb alleles in three groups, 13, 12 and 9 were observed in Chinese cultivars, Chinese landraces and foreign accessions, respectively (Table 5). In Chinese cultivars, Pina-D1a/Pinb-D1b (46.3%), Pina-D1a/Pinb-D1a (31.4%), Pina-D1a/Pinb-D1p (12.9%) and Pina-D1b/Pinb-D1a (6.6%) were predominant, and those predominant combinations also had high frequencies in foreign accessions, while Pina-D1r/Pinb-D1a (7.5%) and Pina-D1s/Pinb-D1a (5.6%) were predominant in Chinese landraces, along with Pina-D1a/Pinb-D1a (54.2%) and Pina-D1a/Pinb-D1p (15.9%) (Table 5). In addition, some rare combinations were observed in some specific samples, e.g. Pina-D1a/Pinb-D1u (2.8%) in Chinese landraces, Pina-D1a/Pinb-D1c (1.4%) in foreign accessions, and Pina-D1b/Pinb-D1b (0.6%) in Chinese cultivars. All in together, the Nei’s diversity, allelic and genotype frequency results show the promising use of Chinese landraces and foreign accessions to broaden the genetic base of modern Chinese cultivars, and the frequency difference of combinations in three groups suggested that the Pina and Pinb genes might have been broadly selected during the breeding process (Table 5).
Selection of kernel hardness and allele preference during breeding process
To investigate the hardness trend and allelic variant preference during Chinese wheat breeding process, 1108 cultivars bred in Beijing, Hebei, Shandong and Henan with clear released time were selected (Table 6). In each province, based on the released year, cultivars were separated into two groups, before year 2000 and after 2000 (including 2000), for the kernel hardness was considered as one of important criterions for new hard wheat varieties released in north China in year 2000 and later. The average kernel hardness was calculated in each group, along with numbers and hardness of three kernel categories (hard, medium and soft) (Table 6). Varieties released after 2000 in Beijing, Hebei, Shandong and Henan had an average kernel hardness index of 69.2, 68.3, 63.7 and 61.1, respectively, significantly higher than that released before 2000 in all four provinces, which demonstrated that the kernel hardness has been greatly improved in the new century. The increase of hardness index after 2000 might be due to the higher frequency of hard type varieties, e.g. 90.4 Vs 65.1% in Beijing, 86.5 Vs 42.2% in Hebei,73.2 Vs 48.7% in Shandong, and 67.3 Vs 30.7% in Henan. Interestingly, the frequencies of hard type varieties released after 2000 were increasing along with the latitude, resulting in an increase of the average hardness index from Henan (generally lower latitude) to Beijing (higher latitude). Moreover, slight differences of the average hardness index were observed in hard type varieties in different groups and provinces, and this could be attributed mainly to the frequencies of combinations of Pina and Pinb alleles since they preserved different effects on kernel hardness (Table 5). In the hard type varieties, Pina-D1a/Pinb-D1b, Pina-D1a/Pinb-D1p and Pina-D1b/Pinb-D1a were three dominant genotypes, occupying 90.0% or more varieties in each groups. However, these genotypes were differentially selected by breeders in different stages and provinces. For example, Pina-D1a/Pinb-D1p was a dominant genotype with a percentage of 44.0% in the group of varieties released after 2000 in Beijing, while the preference of Pina-D1a/Pinb-D1b was observed before 2000; the frequency of Pina-D1b/Pinb-D1a has been greatly improved in the group after 2000 in Shandong; Pina-D1b/Pinb-D1a was seldom detected in varieties from Hebei and released before 2000 in Shandong. These data demonstrated that combinations of Pina and Pinb genes have been widely selected in different breeding stages in different regions.
Table 6.
Frequencies of hard, medium and soft wheats bred in different periods in Beijing, Hebei, Shandong and Henan, and their prevalent genotypes in hard wheat
| Location | Perioda (year 2000) | Number | SKCS index | Freq. (%) | Aver. Hardness Freq. (%) | Genotype freq. in hard wheat | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | Range | Hard | Medium | Soft | Hard | Medium | Soft | Pina-D1a/ Pinb-D1b | Pina-D1a/ Pinb-D1p | Pina-D1b/ Pinb-D1a | |||
| Beijing | after | 83 | 69.2 | 10.0–85.5 | 90.4 | 4.8 | 4.8 | 72.5 | 57.1 | 20.7 | 37.3 | 44.0 | 10.7 |
| before | 106 | 58.8 | 12.1–92.5 | 65.1 | 16.0 | 18.9 | 69.7 | 52.6 | 26.7 | 59.4 | 20.3 | 14.5 | |
| Hebei | after | 281 | 68.3 | 13.0–94.2 | 86.5 | 8.2 | 5.3 | 72.0 | 56.1 | 26.6 | 64.2 | 16.9 | 9.9 |
| before | 128 | 48.4 | 11.0–90.4 | 42.2 | 18.8 | 39.1 | 68.4 | 53.0 | 24.5 | 61.1 | 29.6 | 1.9 | |
| Shandong | after | 108 | 63.7 | 1.4–95.6 | 73.2 | 6.5 | 20.4 | 76.5 | 55.0 | 20.5 | 55.7 | 13.9 | 20.3 |
| before | 156 | 50.4 | 11.1–92.7 | 48.7 | 10.3 | 41.0 | 71.6 | 52.9 | 24.7 | 75.0 | 17.1 | 1.3 | |
| Henan | after | 107 | 61.1 | 6.7–95.0 | 67.3 | 13.1 | 19.6 | 72.8 | 53.2 | 26.3 | 75.0 | 8.3 | 12.5 |
| before | 140 | 42.2 | 1.5–83.4 | 30.7 | 13.6 | 55.7 | 71.1 | 52.5 | 23.7 | 67.4 | 11.6 | 16.3 | |
aafter year 2000 means accessions or varieties bred and released in 2000 or later, and before year 2000 means released in 1999 or before
The novel alleles at Pina and Pinb
Several new types of alleles of Pina and Pinb were also detected in this study. According to the catalogue of gene symbols [6, 9], a synonymous allele ‘C 321 T’ of Pina-D1a was designated as Pina-D1y (Additional file 2: Figure S3), and its kernel hardness index was 20.5 ± 12.7, categorized to the soft wheat class, which indicated that the synonymous allele of Pina-D1a had non-significant effect on kernel hardness. The null alleles at both Pina and Pinb loci (Pina-D1x/Pinb-D1ah) were observed in ten accessions (0.6%), of which three were detected in foreign accessions and seven were from Chinese cultivars, including five from Yellow and Huai River Valley winter wheat region (II), and one each from region III and VI (Table 5; Additional file 2: Figure S1; Additional file 3: Figure S2). The SKCS index of these double null mutants ranged from 77.1 to 97.8 with mean of 91.1, which had highest average kernel hardness among 15 combinations of Pina and Pinb genes and were comparable with that of durum wheat (88.1 ± 15.4) (Additional file 1: Table S4; Table 5).
In effort to further illustrate the deletion size and position in these double null mutants, nine primer pairs (Additional file 1: Table S3) specific to chromosome 5D were designed surrounding the Pina and Pinb genes with the availability of the BAC sequence (CT009735) in NCBI. Based on the primer walking strategy, amplicons with expected fragment size could be observed with primer sets Pina-1, Pina-7, Pina-8 and Pina-9, whereas no targeted fragment was amplified with primer sets through Pina-2 to Pina-6 in four Chinese cultivars Yunfengzao 21, 06–01216, Kelao 4 and Shan 150 and one foreign accession NIL-Novos 67 (Additional file 1: Table S4; Additional file 2: Figure S1). These absent amplicons revealed a 25-kb deletion at least from −435 to +24,592 bp (reference to ATG of Pina) on these five accessions compared with the BAC sequence of Chinese Spring (CT009735) containing the Pina and Pinb genes. However, no expected fragments were detected with all the primer sets through Pina-1 to Pina-9 in three Chinese accessions Hedong TX-008, Xinong 8925–13, and 91G149/Chang 128,865, and one foreign accession Vendvr, which provided a large fragment deletion in these accessions (at least 90-kb, from −21,803 to +68,481 bp referring to ATG of Pina in Chinese Spring BAC sequence CT009735 in NCBI). Moreover, expected fragments of amplicons were only gained with primer sets Pina-7, Pina-8 and Pina-9, but not through Pina-1 to Pina-6 in foreign accession Victory, demonstrating an at least 63-kb deletion from −21,803 to 41,844 bp of Chinese Spring BAC sequence (CT009735) on chromosome 5DS (Additional file 1: Table S4). All these data with primer walking strategy suggested that the Pina and Pinb gene regions were completely deleted in these double null mutants, which confirmed the EcoTILLING data and the durum-like kernel hardness of these accessions, though further analysis is needed to elucidate the exact deletion sizes. However, these double null mutants with extremely high kernel hardness provided an elite germplasm resource for kernel hardness improvement in wheat breeding though molecular marker assisted selection.
Double null mutants have durum-like starch granules and protein matrix
SEM has been considered as a straightforward method for determining the adhesion between protein matrix and starch granules [25, 26]. In this study, SEM was performed with one wild type accession (Chinese Spring), two double null accessions (Xinong-8925-13 and Yunfengzao 21) and one durum wheat accession (Neixiang 4184). The SEM images revealed large, medium and small round to spherical starch granules in all four accessions (Fig. 2a-d), and the basic differences were of the adhesion between starch granules and protein matrix among the wild type (Pina-D1a/Pinb-D1a) (Fig. 2a) and the double null mutants (Pina-D1x/Pinb-D1ah) (Fig. 2c, d). Starch granules and protein matrix are clearly separated without significant adhesions in-between in the wild type soft textured Chinese Spring, while starch granules were strongly adhered to protein matrix in durum wheat Neixiang 4184 (Fig. 2a, b). More medium and small starch granules were observed in durum wheat, which was prone to be adhered to protein matrix, resulting in higher kernel hardness. SEM images of the double null mutants Xinong 8925-13 (Pina-D1x/Pinb-D1ah) and Yunfengzao 21 (Pina-D1x/Pinb-D1ah) revealed that both double null mutants had strong adhesion between protein matrix and starch granules, narrow space between starch granules, and more medium and small starch granules, which were similar to that of durum wheat but completely different from that of Chinese Spring. This phenomena and extreme high kernel hardness confirmed the deletion of both Pina and Pinb genes in Xinong 8925-13 and Yunfengzao 21 for durum wheat lacking both Pina and Pinb genes used to have high kernel hardness. Moreover, the protein matrix appeared differently between hexaploid and durum wheat, which has more flake-shape fragments on the surface of starch granules in Chinese Spring and both mutants (Fig. 2a-d). The SKCS and SEM analysis conclusively demonstrated that high kernel hardness was attributed to durum-like grain structure of double null mutants, and both Pina and Pinb genes might play a vital role in kernel hardness development.
Fig. 2.
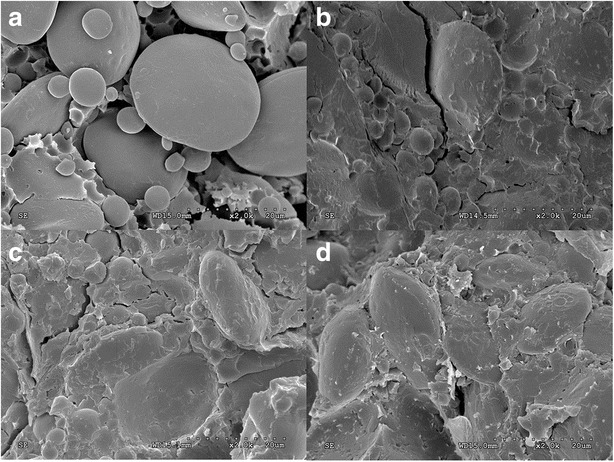
Scanning electron micrographs of the central endosperm region of double null mutants of Pina and Pinb genes. a Freeze-fractured grain of wild type accession (Chinese Spring, soft type control). b Durum wheat accession (Neixiang 4184; Pin null: both genes deleted, very hard control). c Xinong 8925–13 (Pina-D1x/Pinb-D1ah). d Yunfengzao 21 (Pina-D1x/Pinb-D1ah)
Discussion
Due to their important roles in determining kernel hardness, the allelic variation of Pina and Pinb genes have been widely investigated using wheat germplasms from different countries and regions [5, 9–11, 13, 24, 27]. The methods used for characterizing Pina and Pinb alleles were improved from SDS-PAGE analysis, cloning and sequencing of the alleles in a few lines to EcoTILLING [5]. The optimized EcoTILLING approach was exploited to investigate Pina and Pinb alleles in the 225 micro-core collection (MCC) accessions of Chinese wheat germplasm [10]. Compared to previous techniques, EcoTILLING approach is highly efficient on time and cost for largely reducing the repetitive work of sequencing the most frequent Pina-D1a, Pinb-D1a and Pinb-D1b alleles. On the other hand, the detection of novel alleles was directly based on the nucleotide sequences, which improved the discovery of novel variants with electrophoretic mobility similar to previously known alleles in SDS-PAGE analysis. The differentiation of Pinb-D1x allele (14.5 kDa) from Pinb-D1ab (14.4 kDa) was due to the distinct advantage of EcoTILLING approach [10] over SDS-PAGE analysis that might had regarded the two alleles as identical. So the high throughput EcoTILLING approach was exploited in our research with minor modifications. On the one hand, gene cloning method was applied to get the plasmid DNA containing wild type genotype (Pina-D1a/Pinb-D1a) which minimized the use of control DNA templates and made the adjustment of PCR products concentration more effective than the wild type genomic DNA. On the other hand, nested PCR was used to improve the sensitivity and specificity.
The wide range of kernel hardness index varying from 1.4 to 102.7 was observed in overall collection of 1787 accessions. The high phenotypic variation in kernel hardness index is a reflection of the diverse nature of the scanned germplasm consisting of Chinese cultivars and landraces from 9 geographical regions and 141 foreign accessions from USA, Australia, Europe, and Japan. The distribution of hard and soft type wheats across nine geographical regions broadly divided China into two categories as North and South China. The first category included regions I, II, VIII, and X, where hard type accessions had higher frequency than soft type accession, and the second category consisted of regions III, IV, VI and IX, having more soft type accessions. This pattern of distribution of hard and soft type wheat accessions reflected the breeding preference. The hard type accessions are suitable for wheat noodles and steamed bread, which are preferential daily staple food for people living in North China. Conversely, people in South China prefer rice as staple food and use wheat as secondary food usually for making bakery products like biscuits and cookies etc. The similar distribution of hard and soft type accessions across North and South China was reported previously [10].
In total, six Pina and nine Pinb variants were detected in 1787 accessions, Pina-D1a and Pinb-D1b were the most abundant genotypes at Pina and Pinb, respectively (Table4). Corresponding to this, of 15 Pina and Pinb combinations, the most extensive distributed combinations were Pina-D1a/Pinb-D1b (43.1%), followed by Pina-D1a/Pinb-D1a (32.9%), Pina-D1a/Pinb-D1p (12.5%) and Pina-D1b/Pinb-D1a (6.7%) with corresponding average kernel hardness index of 67.7, 31.0, 68.0 and 79.3 (Table 5), which suggest that the order of kernel hardness index between these four combinations from high to low were Pina-D1b/Pinb-D1a, Pina-D1a/Pinb-D1p, Pina-D1a/Pinb-D1b and Pina-D1a/Pinb-D1a [24].
Regarding to the effects of these allelic variations on kernel hardness, the average kernel hardness index of each combinations are 60.0 or more, except Pina-D1a/Pinb-D1a, which supporting that wild type genotype (Pina-D1a/Pinb-D1a) was necessary for showing the soft kernel texture [9, 24]. In addition, the combination Pina-D1x/Pinb-D1ah had the highest average kernel hardness than all the remaining genotypes, indicating the null mutations of Pina or Pinb containing higher kernel hardness than others’ mutations [28].
The wild type Pina-D1a was detected as the most frequent allele of Pina with frequencies of 91.9, 80.4 and 82.3% in Chinese cultivars, landraces and foreign accessions, respectively (Table 5). Chinese cultivars are prone to have this wild type allele, which could be concluded from previous reports. In Chinese micro-core collection mainly consisting of landraces, Pina-D1a had a relatively low frequency (83.6%) [10], while this allele was found to be completely dominant (95.0%) in wheat cultivars released recently from the Yellow and Huai valley of China [11]. The phenomena suggested that this wild type Pina-D1a might preserve beneficial agronomic effects selected by breeders. The wild type of Pinb (Pinb-D1a) and its two mutants (Pinb-D1b and Pinb-D1p) were frequently detected in Chinese wheat germplasm [10, 11, 15, 19], and the frequency of mutated Pinb alleles was much higher than that of mutated Pina alleles, supporting the notion that hard wheats in China were mostly due to Pinb mutations rather than the ones arising from Pina [10, 15, 19]. Moreover, comparing to Chinese landraces and foreign accessions [10, 27], recently released varieties in this work (46.9%) and the Yellow and Huai valley (50.9%) [11] had a relatively high frequency in Pinb-D1b, which played a vital role on the improvement of kernel hardness in Chinese wheat breeding. These results are different from the gene pool investigations in North America, Europe, India, CIMMYT and Australia, where both Pina-D1b and Pinb-D1b alleles are the main sources of kernel hardness [13, 24, 29, 30].
Interestingly, the allele Pinb-D1c was completely missing in Chinese cultivars and landraces and was detected only in two foreign accessions. The reports of Pinb-D1c in genotypes from Ukraine, Portugal, Finland and European countries [24, 27, 29, 31] supported our finding. The allele Pinb-D1p was reported as restricted to Chinese wheat germplasm [10, 11, 15, 17, 19, 27, 28], but in this work we found nine foreign accessions with Pinb-D1p (Table 4). Similarly, though the allele Pina-D1r and Pina-D1s were found restricted to Chinese accessions in international collections of 803 landraces [24], 493 wheat cultivars [27], and 267 wheat cultivars and advanced lines [11], Pina-D1r and Pina-D1s were characterized in one and five foreign accessions, respectively. These data provided a broad representativeness of wheat accessions collected in this work. The stratified distribution of Pina and Pinb variants in different geographical regions might have been influenced by repeated use of the core germplasm in breeding and consumer’s preference in a particular region [31].
The simultaneous deletions at both Pina and Pinb loci are rarely reported so far [27, 28]. In 2005, Ikeda et al. reported the first double null mutation with the presence of D genome and absolute absence of Pina and Pinb proteins [28]. Later, three Chinese landraces and one Netherlands cultivar were detected absent of both Pina-D1 and Pinb-D1 genes, and this mutation was further deduced by primer walking strategy as an approximate 33-kb fragment deletion containing Pina and Pinb coding regions when compared with the BAC sequence of Chinese Spring on chromosome 5DS [27]. In this work, seven Chinese cultivars and three foreign accessions, designated as Pina-D1x/Pinb-D1ah, were observed lacking both Pina-D1 and Pinb-D1 genes, and these accessions were divided into three groups based on the presence or absence of amplicons surrounding the Ha locus on chromosome 5DS, which resulted in three deletion sizes of at least 25-kb, 63-kb and 90-kb, respectively. The accessions with Pina-D1x/Pinb-D1ah have the highest SKCS index among all 15 genotypes, and these grain textures similar with durum wheat were further observed high degree of adhesion between starch granules and protein matrix through SEM. However, these double null mutants are different from previously characterized double null mutants for the deletion size and kernel hardness [27]. Therefore, these double null mutants could be incorporated into quality improvement in bread wheat though further analysis is needed to clarify their exact deletion size.
Conclusion
The study herein verified many previous results regarding higher allelic diversity at Pinb locus; predominant prevalence of Pina-D1a and Pinb-D1b alleles; and association of various mutations at Pina and Pinb loci with SKCS index. The regions I, II, VIII, X foreign accessions were dominantly represented with hard type wheat while the soft type accessions were more frequent in regions III, IV, VI and IX. The Nei’s diversity, allelic and genotype frequency results together show the promising use of Chinese landraces and foreign accessions to broaden the genetic base of modern Chinese cultivars. The wild type Pina-D1a and Pinb-D1a, and one Pinb mutant (Pinb-D1b) are the most common alleles of six Pina and nine Pinb alleles, and a new double null genotype (Pina-D1x/Pinb-D1ah) possessed relatively high SKCS hardness index. More hard type varieties were released in recent years with different prevalence of genotypes in different regions. This work would benefit the understanding of the selection and molecular processes of kernel hardness across China and different breeding stages, and provide useful information for the improvement of wheat quality in China.
Methods
Wheat germplasm
A total of 1787 accessions were used for this study, including 1539 Chinese cultivars, 107 landraces and 141 accessions originated from USA, Australia, Europe and Japan and grouped together as foreign accessions (Table 1). This panel was obtained from the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences (IGDB, CAS), and the information about the origin and nature of each accession was collected from Chinese Crop Germplasm Resources Information System (http://www.cgris.net/cgris_english.html). All the accessions were grown at the experimental station of the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, in the 2009–2010 cropping season according to local crop management practices. Most accessions are from the winter wheat regions, but a few accessions are from the spring or winter-spring wheat regions, and to secure the survival of all the accessions, the seedlings were covered with a plastic film during the winter season. After harvesting, 300 physiologically mature seeds for each accession were subjected to measuring the kernel hardness using the Perten’s Single Kernel Characterization System (SKCS) 4100. In the successive 2010–2011 growing season, a subset of 623 accessions was grown and harvested for the kernel hardness measurement to assess the effect of years on kernel hardness. Each accession was planted on a 2-m row with inter row spacing of 0.25 cm and plant-to-plant distance of 5 cm within a row.
DNA isolation
Genomic DNA was extracted from single seedlings of each accessions growing in the greenhouse using CTAB procedure [32]. The isolated DNA was measured with the Nanodrop spectrophotometer (Thermo Fisher Scientific Inc., Wilmington, DE) and diluted to 100 ng/μl for further PCR analysis.
The modified EcoTILLING
High throughput EcoTILLING analysis was applied to identify Pina and Pinb alleles in the selected germplasm. The method was followed with Li et al. (2013) with minor modifications based on gene cloning to minimize the use of control DNA templates. Firstly, three dominant allelic variants of Pina (Pina-D1a from Chinese Spring) and Pinb (Pinb-D1a from Chinese Spring and Pinb-D1b from Neimai 11) were cloned into the pGEM-T Easy vector (Transgen, Beijing, China) as the control samples, respectively. The nested PCR was used to detect Pina and Pinb allelic variants (Fig. 1; Additional file 1: Table S3). The full length of Pina and Pinb were amplified individually from genomic DNA of tested samples with primers (Pina-Out-F/R, Pinb-Out-F/R) in the first step PCR reaction. Then, the PCR products and the plasmids (containing Pina-D1a, Pinb-D1a and Pinb-D1b, respectively) were diluted for 100-fold as templates for the second step PCR reaction individually. To improve the amplification efficiencies, the fluorescently labeled primers (LI-COR Biosciences, Lincoln, USA) were mixed with unlabeled primers (Pina-In-F/R, Pinb-In-F/R) in 1:1 ratio for the second step PCR reaction.
Screening the single nucleotide polymorphism of Pina and Pinb
For identifying the null alleles of Pina and Pinb, all of the products of nested PCR were detected with 1% agarose gels electrophoresis. Specific primers were further applied to identify the known null alleles, Pina-D1b [20], Pina-D1r and Pina-D1s [12] (Additional file 1: Table S3). For those samples whose nested PCR products of Pina and Pinb contained the target bands, different strategies were attempted to identify Pina and Pinb alleles [10]. The nested PCR products of four tested samples and one control were mixed at the same volumes, and the mixture were denatured and re-annealed to allow the formation of hetero-duplex between the wild type and mutant DNA molecules in a thermocycler as follows: 99 °C for 10 min, 70 cycles of 70 °C for 20 s decreasing 0.3 °C per cycle [33]. The resulted hetero-duplex mixtures were digested with CEL1 enzyme [34], and the cleavage reaction was stopped by adding 5 μl stop solution (2.5 μl 0.25 M EDTA and 2.5 μl formamide loading dye) [10]. To visualize the polymorphisms between the tested samples and the control, 1 μl of CEL1 enzyme digestion product was loaded into 6.5% polyacrylamide gels, and separated on the LI-COR 4300 DNA analyzer (LI-COR Biosciences, Lincoln, USA) at 1500 V/40 watts/45 °C for 4 h. The new allelic variants of Pina and Pinb were further confirmed by sequencing. The genetic diversity at each locus was calculated using Nei’s index [35] with formula H = 1–∑Pi 2, where H and Pi denote the genetic variation index and the frequency of the number of alleles at the locus, respectively.
For Pina and Pinb double null mutants, the BAC sequences (CT009735) flanking the Pina and Pinb genes were download from NCBI to design genome-specific primers surrounding these two genes. Four pairs of specific primers which located on different position of Pina and Pinb coding sequences were explored to investigate the deletion of Pina and Pinb in the wheat genomes (Additional file 1: Table S3), which could largely detect the Pina and Pinb genes in spite of chromosomal rearrangement. Moreover, Nine pairs of primer sets spanning an approximately 90-kb region were designed between −21,803 bp (reference to the ATG of the Pina gene) and +68,481 to clarify the molecular mechanism of the Pina and Pinb double null mutants (Additional file 1: Table S3). The size and position of deletions in these double null mutants were deduced based on the PCR amplification.
Kernel hardness measurement
The harvested seeds of 1787 accessions were cleaned and kept at dry indoor ventilation for 3 days to bring the moisture content to 11–13%. For each sample, kernel hardness index was measured with 300 seeds through the Perten’s Single Kernel Characterization System (SKCS) 4100 according to the manufacturer’s procedure (Perten Instruments North America Inc., Springfield, IL, USA). Chinese Spring was included as the reference for soft wheat with SKCS index of 25.0 ± 17.0. Regarding to kernel hardness based classification of the germplasm [24, 36], the categories normally include <40.0 (soft), 40.0–60.0 (medium), and >60.0 (hard) though different classification systems have been adopted in different countries [10, 24]. According to these categories, 1787 accessions with different SKCS index were classified into soft (<40.0), medium (40.0–60.0) and hard wheat (>60.0).
Assessment of grain texture by scanning electron microscopy (SEM)
The SEM images of physiologically mature grains from wheat accessions with double null mutants at Pina and Pinb loci were compared with those of wild type alleles at Pina and Pinb loci (Pina-D1a/Pinb-D1a) and of durum wheat lacking Pina and Pinb loci. Two grains from each accession were transversely sliced and placed onto glass microscope slides. The slides were fixed with double sided tape, and coated with gold in Dynavac CS300 coating unit [37]. Photographic images were captured at 2000-fold magnification using a ZEISS supra 10 vp field emission scanning electron microscope (Carl Zeiss Microscopy, NY, USA) at 10 kV.
Additional files
Year effect on SKCS value in 623 accessions. Table S2. Number of hard, soft and medium wheat in wheat cultivation regions when regarding to accession type. Table S3. Sequence, product size and annealing temperature of PCR primers used for Pina and Pinb amplification. Table S4. SKCS hardness index of Pina-D1x/Pinb-D1ah genotype. (DOCX 29 kb)
Identification of Pina and Pinb deletions in some accessions by a set of selected markers. (A) Primers AGPS-1 was used to check all of DNA quality. (B) Primers Pina-part amplified part of Pina coding sequence. (C) Primers Pina-cds amplified the Pina coding sequence. (D) Primers Pinb-part amplified part of Pinb coding sequence. (E) Primers Pinb-cds amplified the Pinb coding sequence. (F) Primers Pina-4 was used to check the deletion of Pina and Pinb downstream sequence. 1–7 show accessions Chinese Spring, NIL-Novos 67, Yunfengzao 21, Shan 150, 91G 149/Chang 128,865, Hedong TX-008, Xinong 8925–13 respectively. (TIFF 6824 kb)
Wheat seed protein analysis. (A) Western blot analysis of PINB protein. (B) SDS-PAGE gel of total proteins from wheat mature seeds. Lanes were loaded with 20 μg protein, 1–7 show accessions Chinese Spring, NIL-Novos 67, Yunfengzao 21, Shan 150, 91G 149/Chang 128,865, Hedong TX-008, Xinong 8925–13, respectively. (TIFF 7059 kb)
Sequence alignments of Pina (A) and Pinb (B) alleles. (A) Pina-D1a (DQ363911), Pina-D1l ([6, 15]), and Pina-D1y. (B) Pinb-D1u (EF620911), Pinb-D1a (DQ363913), Pinb-D1b (DQ363914), Pinb-D1c (KC585019), Pinb-D1d (KR259645), Pinb-D1p (AY581889), and Pinb-D1q (EF620909). (TIFF 7417 kb)
Acknowledgements
We thank two anonymous reviewers for their critical reviews and suggestions.
Funding
This work was supported by Ministry of Agriculture of China for transgenic research (2016ZX08009-003) and the National Science Foundation of China (31371610).
Availability of data and materials
Supporting data of Additional files 1, 2, 3, 4 are included as additional files in the manuscript.
Abbreviations
- ANOVA
Analysis of variance
- EcoTILLING
Ecotype targeting induced local lesions in genomes
- H
Nei’s index of allelic diversity index
- NCBI
National Center for Biotechnology Information
- nt
Nucleotide
- PINA
Puroindoline a
- PINB
Puroindoline b
- SEM
Scanning electron microscopy
- SKCS
Single kernel characterization system
- STS
Sequence tagged site
- WT
Wild type
Authors’ contributions
XM carried out most experiments and analyzed the data. XM, MS and DL wrote the manuscript. JW and XM analyzed the data. WY, XL and JS grew samples and measured the kernel hardness. AZ and DL conceptualized the experiments and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12870-017-1101-8) contains supplementary material, which is available to authorized users.
Contributor Information
Xiaoling Ma, Email: fanxiaoling@163.com.
Muhammad Sajjad, Email: sajjad@genetics.ac.cn.
Jing Wang, Email: sduwj@126.com.
Wenlong Yang, Email: wlyang@genetics.ac.cn.
Jiazhu Sun, Email: jzsun@genetics.ac.cn.
Xin Li, Email: lixin@genetics.ac.cn.
Aimin Zhang, Phone: +86 10 6480 6617, Email: amzhang@genetics.ac.cn.
Dongcheng Liu, Email: dcliu@genetics.ac.cn.
References
- 1.Pauly A, Pareyt B, Fierens E, Delcour JA. Wheat (Triticum Aestivum L. and T. Turgidum L. Ssp. Durum) kernel hardness: II. Implications for end-product quality and role of Puroindolines therein. Compr Rev Food Sci F. 2013;12(4):427–438. doi: 10.1111/1541-4337.12018. [DOI] [PubMed] [Google Scholar]
- 2.Jolly CJ, Rahman S, Kortt AA, Higgins TJV. Characterization of the wheat Mr 15000 “grain-softness protein” and analysis of the relationship between its accumulation in the whole seed and grain softness. Theor Appl Genet. 1993;86(5):589–597. doi: 10.1007/BF00838714. [DOI] [PubMed] [Google Scholar]
- 3.Morris CF, Greenblatt GA, Bettge AD, Malkawi HI. Isolation and characterization of multiple forms of Friabilin. J Cereal Sci. 1994;20(2):167–174. doi: 10.1006/jcrs.1994.1056. [DOI] [Google Scholar]
- 4.Gautier MF, Aleman ME, Guirao A, Marion D, Joudrier P. Triticum Aestivum puroindolines, two basic cystine-rich seed proteins: cDNA sequence analysis and developmental gene expression. Plant Mol Biol. 1994;25(1):43–57. doi: 10.1007/BF00024197. [DOI] [PubMed] [Google Scholar]
- 5.Bhave M, Morris CF. Molecular genetics of puroindolines and related genes: allelic diversity in wheat and other grasses. Plant Mol Biol. 2008;66(3):205–219. doi: 10.1007/s11103-007-9263-7. [DOI] [PubMed] [Google Scholar]
- 6.Morris CF, Bhave M. Reconciliation of D-genome puroindoline allele designations with current DNA sequence data. J Cereal Sci. 2008;48(2):277–287. doi: 10.1016/j.jcs.2007.09.012. [DOI] [Google Scholar]
- 7.Bhave M, Morris CF. Molecular genetics of puroindolines and related genes: regulation of expression, membrane binding properties and applications. Plant Mol Biol. 2008;66(3):221–231. doi: 10.1007/s11103-007-9264-6. [DOI] [PubMed] [Google Scholar]
- 8.Ali I, Sardar Z, Rasheed A, Mahmood T. Molecular characterization of the puroindoline-a and b alleles in synthetic hexaploid wheats and in silico functional and structural insights into Pina-D1. J Theor Biol. 2015;376:1–7. doi: 10.1016/j.jtbi.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Kumar R, Arora S, Singh K, Garg M. Puroindoline allelic diversity in Indian wheat germplasm and identification of new allelic variants. Breed Sci. 2015;65(4):319–326. doi: 10.1270/jsbbs.65.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, Sun JZ, Liu DC, Yang WL, Wang DW, Tong YP, Zhang AM. Analysis of Pina and Pinb alleles in the micro-core collections of Chinese wheat germplasm by Ecotilling and identification of a novel Pinb allele. J Cereal Sci. 2008;48(3):836–842. doi: 10.1016/j.jcs.2008.06.005. [DOI] [Google Scholar]
- 11.Chen F, Zhang FY, Xia XC, Dong ZD, Cui DQ. Distribution of puroindoline alleles in bread wheat cultivars of the yellow and Huai valley of China and discovery of a novel puroindoline a allele without PINA protein. Mol Breeding. 2011;29(2):371–378. doi: 10.1007/s11032-011-9553-2. [DOI] [Google Scholar]
- 12.Ikeda TM, Cong H, Suzuki T, Takata K. Identification of new Pina null mutations among Asian common wheat cultivars. J Cereal Sci. 2010;51(3):235–237. doi: 10.1016/j.jcs.2010.01.012. [DOI] [Google Scholar]
- 13.Morris CF, Lillemo M, Simeone MC, Giroux MJ, Babb SL, Kidwell KK. Prevalence of puroindoline grain hardness genotypes among historically significant north American spring and winter wheats. Crop Sci. 2001;41(1):218–228. doi: 10.2135/cropsci2001.411218x. [DOI] [Google Scholar]
- 14.Cane K, Spackman M, Eagles HA. Puroindoline genes and their effects on grain quality traits in southern Australian wheat cultivars. Aust J Agric Res. 2004;55(1):89–95. doi: 10.1071/AR03108. [DOI] [Google Scholar]
- 15.Chen F, He ZH, Xia XC, Xia LQ, Zhang XY, Lillemo M, Morris CF. Molecular and biochemical characterization of puroindoline a and b alleles in Chinese landraces and historical cultivars. Theor Appl Genet. 2006;112(3):400–409. doi: 10.1007/s00122-005-0095-z. [DOI] [PubMed] [Google Scholar]
- 16.Chen F, Yu YX, Xia XC, He ZH. Prevalence of a novel puroindoline b allele in Yunnan endemic wheats (Triticum Aestivum Ssp. Yunnanense king) Euphytica. 2007;156(1):39–46. doi: 10.1007/s10681-006-9347-5. [DOI] [Google Scholar]
- 17.Chang C, Zhang HP, Xu J, Li WH, Liu GT, You MS, Li BY. Identification of allelic variations of puroindoline genes controlling grain hardness in wheat using a modified denaturing PAGE. Euphytica. 2006;152(2):225–234. doi: 10.1007/s10681-006-9204-6. [DOI] [Google Scholar]
- 18.Martin JM, Frohberg RC, Morris CF, Talbert LE, Giroux MJ. Milling and bread baking traits associated with puroindoline sequence type in hard red spring wheat. Crop Sci. 2001;41(1):228–234. doi: 10.2135/cropsci2001.411228x. [DOI] [Google Scholar]
- 19.Xia LQ, Chen F, He ZH, Chen XM, Morris CF. Occurrence of puroindoline alleles in Chinese winter wheats. Cereal Chem. 2005;82(1):38–43. doi: 10.1094/CC-82-0038. [DOI] [Google Scholar]
- 20.Chen F, Zhang FY, Morris C, He ZH, Xia XC, Cui DQ. Molecular characterization of the Puroindoline a-D1b allele and development of an STS marker in wheat (Triticum Aestivum L.) J Cereal Sci. 2010;52(1):80–82. doi: 10.1016/j.jcs.2010.03.006. [DOI] [Google Scholar]
- 21.Wilkinson M, Wan YF, Tosi P, Leverington M, Snape J, Mitchell RAC, Shewry PR. Identification and genetic mapping of variant forms of puroindoline b expressed in developing wheat grain. J Cereal Sci. 2008;48(3):722–728. doi: 10.1016/j.jcs.2008.03.007. [DOI] [Google Scholar]
- 22.Chen F, Zhang FY, Cheng XY, Morris C, Xu HX, Dong ZD, Zhan KH, Cui DQ. Association of Puroindoline b-B2 variants with grain traits, yield components and flag leaf size in bread wheat (Triticum Aestivum L.) varieties of the yellow and Huai valleys of China. J Cereal Sci. 2010;52(2):247–253. doi: 10.1016/j.jcs.2010.06.001. [DOI] [Google Scholar]
- 23.Chen F, Xu HX, Zhang FY, Xia XC, He ZH, Wang DW, Dong ZD, Zhan KH, Cheng XY, Cui DQ. Physical mapping of puroindoline b-2 genes and molecular characterization of a novel variant in durum wheat (Triticum Turgidum L.) Mol Breeding. 2010;28(2):153–161. doi: 10.1007/s11032-010-9469-2. [DOI] [Google Scholar]
- 24.Qamar ZU, Bansal UK, Dong CM, Alfred RL, Bhave M, Bariana HS. Detection of puroindoline (Pina-D1 and Pinb-D1) allelic variation in wheat landraces. J Cereal Sci. 2014;60(3):610–616. doi: 10.1016/j.jcs.2014.07.007. [DOI] [Google Scholar]
- 25.Chen F, He ZH, Xia XC, Lillemo M, Morris C. A new puroindoline b mutation present in Chinese winter wheat cultivar Jingdong 11. J Cereal Sci. 2005;42(2):267–269. doi: 10.1016/j.jcs.2005.03.004. [DOI] [Google Scholar]
- 26.Morris C, Beecher B. The distal portion of the short arm of wheat (Triticum Aestivum L.) chromosome 5D controls endosperm vitreosity and grain hardness. Theor and Appl Genet. 2012;125(2):247–254. doi: 10.1007/s00122-012-1830-x. [DOI] [PubMed] [Google Scholar]
- 27.Chen F, Li HH, Cui DQ. Discovery, distribution and diversity of Puroindoline-D1 genes in bread wheat from five countries (Triticum Aestivum L.) BMC Plant Biol. 2013;13(1):125. doi: 10.1186/1471-2229-13-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ikeda TM, Ohnishi N, Nagamine T, Oda S, Hisatomi T, Yano H. Identification of new puroindoline genotypes and their relationship to flour texture among wheat cultivars. J Cereal Sci. 2005;41(1):1–6. doi: 10.1016/j.jcs.2004.10.002. [DOI] [Google Scholar]
- 29.Lillemo M, Chen F, Xia XC, William M, Pena RJ, Trethowan R, He ZH. Puroindoline grain hardness alleles in CIMMYT bread wheat germplasm. J Cereal Sci. 2006;44(1):86–92. doi: 10.1016/j.jcs.2006.03.004. [DOI] [Google Scholar]
- 30.Ram S, Boyko E, Giroux MJ, Gill BS. Null mutation in puroindoline a is prevalent in Indian wheats: Puroindoline genes are located in the distal part of 5DS. J Plant Biochem Biot. 2002;11(2):79–83. doi: 10.1007/BF03263140. [DOI] [Google Scholar]
- 31.Ma DY, Zhang Y, Xia XC, Morris CF, He ZH. Milling and Chinese raw white noodle qualities of common wheat near-isogenic lines differing in puroindoline b alleles. J Cereal Sci. 2009;50(1):126–130. doi: 10.1016/j.jcs.2009.03.006. [DOI] [Google Scholar]
- 32.Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1(4):19–21. doi: 10.1007/BF02712670. [DOI] [Google Scholar]
- 33.Li AX, Yang WL, Lou XY, Liu DC, Sun JZ, Guo XL, Wang J, Li YW, Zhan KH, Ling HQ, Zhang AM. Novel natural allelic variations at the Rht-1 loci in wheat. J Integr Plant Biol. 2013;55(11):1026–1037. doi: 10.1111/jipb.12103. [DOI] [PubMed] [Google Scholar]
- 34.Yang B, Wen X, Kodali NS, Oleykowski CA, Miller CG, Kulinski J, Besack D, Yeung JA, Kowalski D, Yeung AT. Purification, cloning, and characterization of the CEL I nuclease. Biochemistry. 2000;39(13):3533–3541. doi: 10.1021/bi992376z. [DOI] [PubMed] [Google Scholar]
- 35.Nei M. Analysis of gene diversity in subdivided populations. Proc Nat Acad Sci USA. 1973;70(12):3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaines CS, Finney PF, Fleege LM, Andrews LC. Predicting a hardness measurement using the single-kernel characterization system. Cereal Chem. 1996;73(2):278–283. [Google Scholar]
- 37.Brennan CS, Harris N, Smith D, Shewry PR. Structural differences in the mature endosperms of good and poor malting barley cultivars. J Cereal Sci. 1996;24(2):171–177. doi: 10.1006/jcrs.1996.0050. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Year effect on SKCS value in 623 accessions. Table S2. Number of hard, soft and medium wheat in wheat cultivation regions when regarding to accession type. Table S3. Sequence, product size and annealing temperature of PCR primers used for Pina and Pinb amplification. Table S4. SKCS hardness index of Pina-D1x/Pinb-D1ah genotype. (DOCX 29 kb)
Identification of Pina and Pinb deletions in some accessions by a set of selected markers. (A) Primers AGPS-1 was used to check all of DNA quality. (B) Primers Pina-part amplified part of Pina coding sequence. (C) Primers Pina-cds amplified the Pina coding sequence. (D) Primers Pinb-part amplified part of Pinb coding sequence. (E) Primers Pinb-cds amplified the Pinb coding sequence. (F) Primers Pina-4 was used to check the deletion of Pina and Pinb downstream sequence. 1–7 show accessions Chinese Spring, NIL-Novos 67, Yunfengzao 21, Shan 150, 91G 149/Chang 128,865, Hedong TX-008, Xinong 8925–13 respectively. (TIFF 6824 kb)
Wheat seed protein analysis. (A) Western blot analysis of PINB protein. (B) SDS-PAGE gel of total proteins from wheat mature seeds. Lanes were loaded with 20 μg protein, 1–7 show accessions Chinese Spring, NIL-Novos 67, Yunfengzao 21, Shan 150, 91G 149/Chang 128,865, Hedong TX-008, Xinong 8925–13, respectively. (TIFF 7059 kb)
Sequence alignments of Pina (A) and Pinb (B) alleles. (A) Pina-D1a (DQ363911), Pina-D1l ([6, 15]), and Pina-D1y. (B) Pinb-D1u (EF620911), Pinb-D1a (DQ363913), Pinb-D1b (DQ363914), Pinb-D1c (KC585019), Pinb-D1d (KR259645), Pinb-D1p (AY581889), and Pinb-D1q (EF620909). (TIFF 7417 kb)
Data Availability Statement
Supporting data of Additional files 1, 2, 3, 4 are included as additional files in the manuscript.


