Abstract
This study aimed to investigate how long non-coding RNA (lncRNA) maternally expressed gene 3 (MEG3) inhibits the growth and metastasis of oral squamous cell carcinoma (OSCC) by regulating WNT/β-catenin signaling pathway in order to explore the antitumor effect of MEG3 and to provide a potential molecular target for the treatment of OSCC. The RT-qPCR technique was used to quantitatively analyze the expression of MEG3 in cancer and adjacent tissues collected from the patients after surgery. Using the Lipofectamine method, the MEG3 overexpression vector and the siRNA interference vector were constructed and transfected into SCC15 and Cal27 cells, respectively, followed by cell proliferation, apoptosis and metastasis analyses. The semi-quantitative analysis of the expression of the β-catenin protein in transfected cells was performed by the western blot analysis, and the activity of the WNT/β-catenin signaling pathway was analyzed using the TOP/FOP flash reporters. In addition, the cells were treated with decitabine to investigate the correlation between the MEG3 expression and the DNA methylation. Results showed that the expression level of MEG3 was significantly decreased in OSCC (p<0.05) and overexpression of MEG3 inhibited the proliferation and metastasis of cancer cells and promoted apoptosis. Importantly, MEG3 played a role as a tumor suppressor by inhibiting the WNT/β-catenin signaling pathway. In addition, the expression of the MEG3 was significantly affected by the degree of DNA methylation. It was concluded that the lncRNA MEG3 can inhibit the growth and metastasis of OSCC by negatively regulating the WNT/β-catenin signaling pathway.
Keywords: long non-coding RNA, maternally expressed gene 3, WNT/β-catenin pathway, oral squamous cell carcinoma, proliferation, metastasis
Introduction
Oral cancer is the 3rd most common cancer type in developing countries and the 6th most common worldwide (1). Oral squamous cell carcinoma (OSCC), which is the representative form of oral cancer, shows the highest incidence among all the head and neck cancers, and account for ~3% of the newly diagnosed cancer cases. Despite the corresponding diagnostic and clinical treatment strategies to prevent and treat OSCC, the survival rate of OSCC patients has not been significantly improved and is still below 50% (2). In the past, clinical studies on OSCC have generally restricted at the physiological and biochemical level, such as detecting the activity of basal cells in OSCC, computing the rate and frequency of proliferation and metastasis. However, the underlying molecular mechanism of OSCC tumor formation remained unclear. In order to improve the prognosis of OSCC patients and the treatment efficacy, the molecular mechanism of OSCC tumor formation and the identification of effective biological indicators and therapeutic targets are urgently needed.
Long non-coding RNA (lncRNA) is a class of newly discovered non-protein coding RNA transcripts >200 nt. Studies have shown that lncRNAs have important biological functions in a variety of physiological processes, such as carcinogenesis, cell proliferation, differentiation, apoptosis and cancer cell metastasis. Therefore, lncRNAs are often used as a biological indicator of cancer to study the molecular mechanism of tumorigenesis (3,4). In addition, lncRNAs have diverse functions, that is, lncRNAs can regulate various cellular processes at transcriptional, post-transcription, translational and epigenetic level in the forms of signal molecules, scaffolds and instructor (5,6). Maternally expressed gene 3 (MEG3), located in human chromosome 14q32.3 with a length of about 1.6 kb (7), is a lncRNA encoded by a maternally imprinted gene, MEG3 and the paternally imprinted gene DLK form the footprint. Studies have shown that the expression of the MEG3 was downregulated in a variety of human tumor cells, such as neuroblastoma, meningioma, bladder cancer, gastric cancer and glioma (8). In addition, due to the methylation of the promoter and differentially methylated DNA fragments, the expression of MEG3 can not be detected in some tumor tissues, such as human functionless adenomas (9). Moreover, MEG3 is a tumor suppressor gene that can inhibit the development of a tumor in a variety of cell lines, such as HCT116, HeLa and U87MG cells (10,11). However, the expression pattern and biological function of MEG3 in OSCC have not been reported yet.
The WNT/β-catenin signaling pathway is one of the classical pathways in the process of cell signal transduction, and the phosphorylation of β-catenin plays a key role in this signal transduction. The interaction of WNT protein and its receptor on the cell membrane can inhibit the phosphorylation of β-catenin induced by GSK-3β, which in turn leads to the accumulation of β-catenin in cytoplasm, and thus the accumulated β-catenin will enter the nucleus to interacts with TCF/LEF transcription factor to activate the expression of downstream target gene, so as to affect cell proliferation, differentiation and tumorigenesis (12). Studies have shown that small RNA molecules can regulate carcinogenesis by regulating WNT/β-catenin signaling pathway (4). This study investigates how lncRNA MEG3 inhibits the growth and metastasis of OSCC by regulating WNT/β-catenin signaling pathway and to provide potential molecular target for the treatment of OSCC.
Materials and methods
Subjects of study
Eighty-three OSCC tumor samples, as well as the corresponding normal tissues (from the unaffected area of 2 cm from the tumor) were collected during the period January 2011 to January 2016, and the patients signed an informed consent form. Additionally, the study was approved by the Ethics Committee of Xuzhou Stomatological Hospital. The sample consisted of 51 males and 32 females with an average age of 55.5±6.76 years. High tumor differentiation was found in 43 cases and middle to low differentiation was found in 40 cases. Furthermore, 72 cases had the tumor in tongue and 11 cases in gums, or other parts. None of the patients received chemotherapy and radiation therapy before surgery. All the samples were checked and the tumor and normal tissues were confirmed by three pathologists and samples were stored at −80°C.
Cells and reagents
Human OSCC cell lines SCC15 and Cal27 were generously presented by the Department of Stomatology, Wuhan University (Wuhan, China). Dulbecco's modified Eagle's medium (DMEM) and fetal bovine serum (FBS) (both from Gibco, Grand Island, NY, USA); TRIzol reagent, PrimeScript® RT reagent kit with gDNA Eraser and SYBR® Premix Ex Taq™ II (Takara Biotechnology Co., Ltd., Dalian, China); RIPA lysate (Thermo Fisher Scientific, Inc., Carlsbad, CA, USA); protease inhibitors and PVDF membranes (Roche, Basel, Switzerland); rabbit polyclonal TCF-4 antibody (dilution, 1:500; cat. no. ab185736), rabbit monoclonal cyclin D1 antibody (dilution, 1:500; cat. no. ab16663), rabbit polyclonal GAPDH antiboody (dilution, 1:500; cat. no. ab37168) and secondary goat anti-rabbit (HRP) IgG antibody (dilution, 1:5,000; cat. no. ab6721) were all purchased from Abcam (Cambridge, MA, USA). Cell Counting Kit-8 (CCK-8) (Dojindo, Kumamoto, Japan); Lipofectamine 3000 (Invitrogen Life Technologies, New York, NY, USA); Annexin V-FITC cell apoptosis detection kit (eBioscience, San Diego, CA, USA); 5-aza-2′-deoxycytidine (5-aza-CdR; Sigma, New York, NY, USA); TOP/FOP-Flash luciferase reporter vector (Millipore, Billerica, MA, USA); Dual-Luciferase assay kit (Promega, Madison, WI, USA); primers (Sangon, Shanghai, China).
Cell culture and transfection
SCC15 and Cal27 cells were cultured in DMEM (10% FBS) until the cells adhered; pcDNA3.1-MEG3 overexpression vector and the si-MEG3 interference vector and its corresponding control vector pcDNA3.1-Ctrl were constructed, si-Ctrl were transfected into cells using liposomes. After 48 h, the cells were collected for subsequent experiments and siRNA was synthesized by GenePharma (Shanghai, China). The sequences are listed in Table I.
Table I.
The sequences of siRNA and primers used in RT-qPCR.
| Primers/siRNA | Sequences |
|---|---|
| si-MEG3-3 | 5′-CCCUCUUGCUUGUCUUACUTT-3′ |
| MEG3-F | 5′-GCCCTAGGGGAGTGACTACA-3′ |
| MEG3-R | 5′-ACTCGGGACATACCTGCTCT-3′ |
| ACTB-F | 5′-CAGGGCGTGATGGTGGGCA-3′ |
| ACTB-R | 5′-CAAACATCATCTGGGTCATCTTCTC-3′ |
F, forward; R, reverse. MEG3, maternally expressed gene 3.
Real-time fluorescence quantitative PCR (RT-qPCR) analysis
RT-qPCR was used to quantify the relative expression levels of MEG3 in tissues and cells. Total RNA was extracted using TRIzol reagent according to the instructions of manuals and cDNA was synthesized by reverse transcription using PrimeScript® RT reagent kit with gDNA Eraser. Moreover, the PCR reaction system was prepared using SYBR® Premix Ex Taq™ II and PCR reaction was performed using CFX-96 Real-Time PCR (Bio-Rad, New York, NY, USA). Finally, the data were analyzed by 2−ΔΔCt method with β-actin used as endogenous control. All primers are listed in Table I.
Western blot analysis
The transfected cells were washed twice with phosphate-buffered saline (PBS) and lysed on ice in RIPA lysates containing protease inhibitors for 30 min. After that, the protein was quantified by using BCA method. The protein (50 µg) was subjected to SDS-PAGE electrophoresis to separate the protein. Then the protein was transferred to SDS-PAGE by an electric method. The membrane was blocked by TBST buffer containing 5% skimmed milk for 1 h at room temperature. Then primary rabbit polyclonal TCF-4 antibody (dilution, 1:500; cat. no. ab185736) and rabbit monoclonal cyclin D1 antibody (dilution, 1:500; cat. no. ab16663) were added and incubated overnight at 4°C. After washing, secondary goat anti-rabbit (HRP) IgG antibody (dilution, 1:5,000; cat. no. ab6721) was added and incubated for 2 h, and after incubation, the signal was detected by chemiluminescence method.
Methylation treatment
SCC15 and Cal27 (2.5×105) cells were seeded in a 6-well culture plate with fresh DMEM (containing 0, 5 and 10 µM of 5-aza-CdR) that was changed daily. After incubation for 5 days, RT-qPCR was used to detect the relative expression of MEG3.
Cell proliferation and apoptosis analysis
According to the kit manual instructions, cell proliferation assay was performed using the CCK-8. The transfected cells (5×103) were seeded in 96-well plates with 90 µl medium containing 10% FBS in each well. Then the CCK-8 solution (10 µl/well) was added at 0, 1, 2, 3 and 4 days, after incubation at 37°C for 2 h, and the number of cells in each well was calculated by measuring the absorbance at 450 nm. Apoptosis analysis was performed at 48 h after transfection. Cells were harvested from each well, and Annexin V-FITC Apoptosis Detection kit and BD FACSCalibur Flow Cell Analyzer (BD Biosciences, New York, NY, USA) were used to calculate the rate of apoptosis.
Cell migration assay
The artificial basement membrane was added to 24-well plates and DMEM containing transfected SCC15 and Cal27 cells (3×104) was added to the upper chamber. Furthermore, the lower chamber was filled with DMEM containing 20% FBS. After incubation for 24 h, the membrane was fixed with 4% methanol and rinsed with 0.1% crystal violet three times. Finally, the membrane with the colored cells on the surface was randomly divided into 5 regions for cell migration analysis.
Analysis of the activity of WNT signal pathway
TOP-Flash and FOP-Flash luciferase reporter vector and pcDNA3.1-MEG3 or si-MEG3 vector were co-transfected into OSCC cells. After incubation for 48 h, luciferase activity was measured by dual-luciferase assay kit.
Statistical analysis
Statistical analysis was performed using SPSS 17.0 statistical software (SPSS, Inc., Chicago, IL, USA). All experiments were repeated three times. The data were expressed as mean ± SD. The two-tailed t-test was applied. P<0.05 was considered to indicate a statistically significant difference.
Results
The expression of MEG3 is downregulated in OSCC cells
As shown in Fig. 1, the expression levels of MEG3 in 83 OSCC tissues and the corresponding normal tissues were detected by RT-qPCR technique. It was found that the expression level of MEG3 in OSCC tissue was significantly downregulated compared with normal oral tissue (p<0.05).
Figure 1.
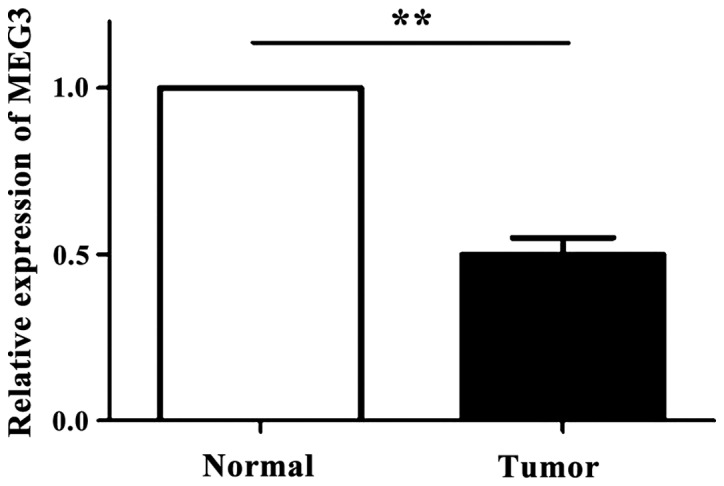
The relative expression of MEG3 in OSCC tissues and normal tissues (RT-qPCR showed that MEG3 expression level in OSCC tissues was significantly lower than that in normal tissues, **p<0.01). MEG3, maternally expressed gene 3; OSCC, oral squamous cell carcinoma.
The expression of MEG3 is regulated by DNA methylation
SCC15 and Cal27 cells were treated with different concentrations of 5-aza-CdR. The cells without 5-aza-CdR treatment were used as the control group. The results showed that compared with the control group, the expression of MEG3 in OSCC cells was significantly increased with the increase in the concentration of 5-aza-CdR (p<0.05) (Fig. 2).
Figure 2.
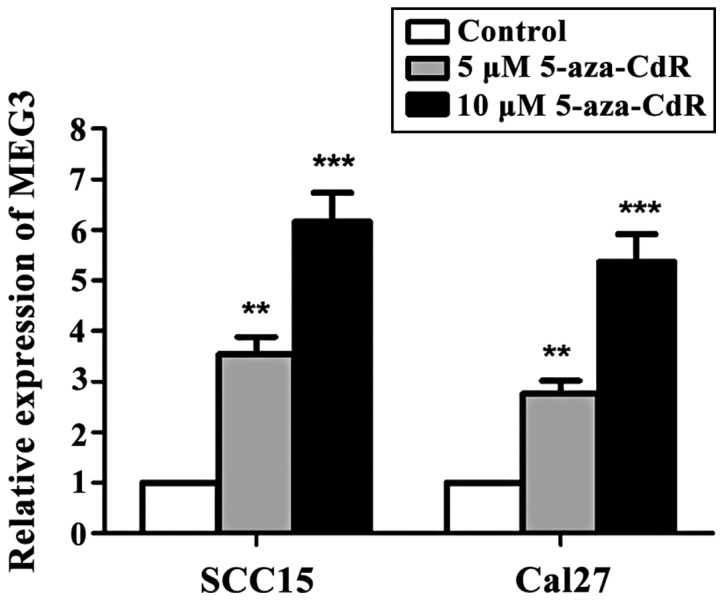
The relationship between the relative expression of MEG3 and the degree of DNA methylation. RT-qPCR showed that the expression of MEG3 was significantly incresed after 5-aza-CdR treatment compared with the control group (**p<0.01, ***p<0.001). MEG3, maternally expressed gene 3; 5-aza-CdR, 5-aza-2′-deoxycytidine.
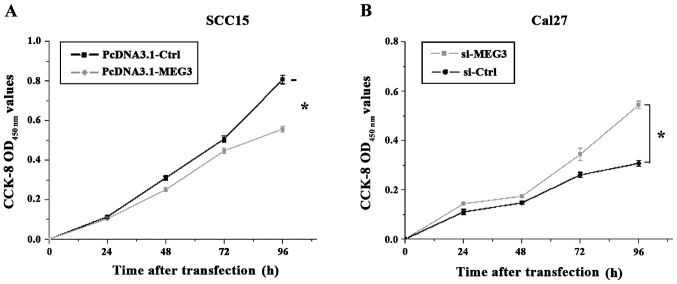
MEG3 inhibits OSCC cell proliferation
MEG3 overexpression vector pcDNA-MEG3 and the interfering vector si-MEG3 in the observation group, and the corresponding control vectors pcDNA-Ctrl and si-Ctrl were transfected into SCC15 and Cal27 cells, respectively. Then the CCK-8 was used to analyze the cell proliferation at 1, 2, 3 and 4 days after transfection. As shown in Fig. 3, the OD value measured at OD450 nm showed that the proliferation of the SCC15 cells with MEG3 overexpression was significantly inhibited compared to the control group (p<0.05). Furthermore, the proliferation rate of the Cal27 cells with reduced MEG3 expression was significantly increased compared to the control group (p<0.05).
Figure 3.
Cell proliferation of SCC15 and Cal27 transfected with different expression vectors. (A) The OD value measured at OD450 nm showed that the proliferation of the SCC15 cells was inhibited by MEG3 overexpression and (B) show that the proliferation rate of the Cal27 cells was increased by the reduced MEG3 expression, *p<0.05. MEG3, maternally expressed gene 3.
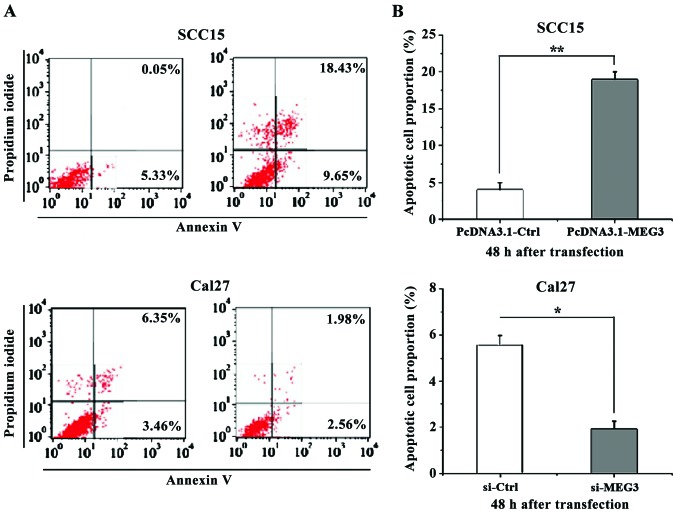
MEG3 promotes apoptosis of OSCC cells
For this, the apoptotic assay was performed at 48 h after transfection. Then Annexin V-FITC Apoptosis Detection kit and the Flow Cell Analyzer were used to detect the apoptotic cells, and the proportion of the apoptotic cells was calculated. The results showed that MEG3 overexpression in Cal27 cells had significantly increased the apoptosis rate of OSCC cells compared to the control group, while the reduced expression level of MEG3 in Cal27 cells had significantly decreased the apoptosis rate of OSCC cells compared to the control group (p<0.05) (Fig. 4).
Figure 4.
Apoptosis of SCC15 and Cal27 cells at 48 h after transfection. (A) The results of Annexin V-FITC cell apoptosis assay and flow cytometry analysis showed that the percentage of apoptotic cells was significantly higher in SCC15 cells with MEG3 overexpression compared with the control group at 48 h after transfection and (B) the percentage of apoptotic cells was significantly lower in Cal27 cells with MEG3 silencing compared with the control group. *p<0.05, **p<0.01. MEG3, maternally expressed gene 3.
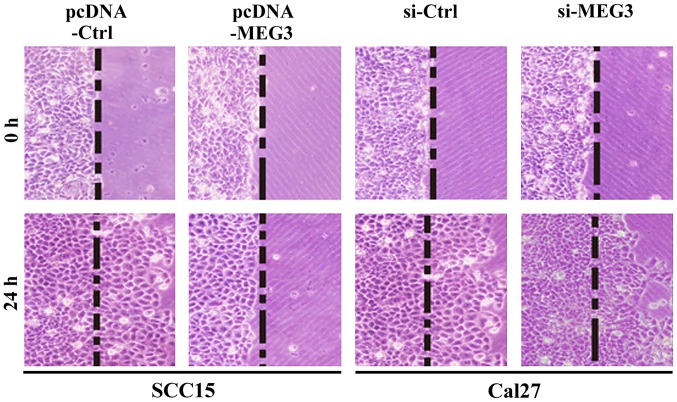
MEG3 inhibits OSCC cell migration
As seen in Fig. 5 the transfected OSCC cells were stained after incubation on the artificial basement membrane for 24 h. The results further showed that the cell migration of SCC15 cells with MEG3 overexpression was inhibited, whereas the cells in control group showed a certain degree of migration. The MEG3 silencing in Cal27 cells did not affect the cell migration, whereas the cells in control group showed a certain degree of migration. The results indicated that the expression of MEG3 can inhibit OSCC cell migration.
Figure 5.
Migration of SCC15 and Cal27 cells after transfecting with different expression vectors. The results showed that, compared with the control group, overexpression of MEG3 inhibited the migration of OSCC cells, while MEG3 silencing showed effect on OSCC cell migration. MEG3, maternally expressed gene 3; OSCC, oral squamous cell carcinoma.
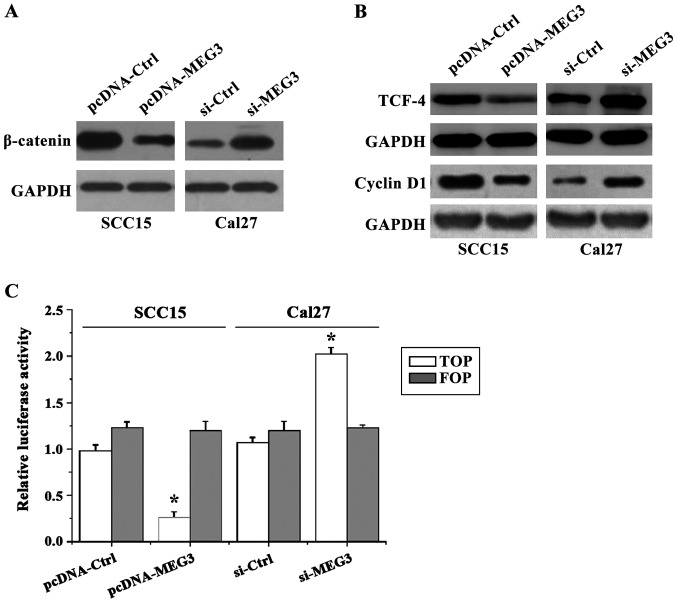
MEG3 regulates the activity of WNT/β-catenin signaling pathway in OSCC cells
In order to analyze the expression of related protein and the activity of WNT signal pathway in SCC15 and Cal27 cells after co-transfection of pcDNA3.1-MEG3 or si-MEG3, western blot analysis and transfection of TOP/FOP flash reporter vector have been used. The results showed (Fig. 6) that the expression level of WNT pathway core protein β-catenin in SCC15 cells with MEG3 overexpression was significantly lower than that in control group, whereas the expression level of β-catenin in Cal27 cells transfected with si-MEG3 was significantly higher than that in control group. Since the expression of β-catenin is directly related to the expression of T-cell factor/lymphoid enhancer factor (TCF/LEF) superfamily and the downstream target gene cyclin D1 (cyclin D1), therefore, the western blot analysis was used to detect the expression levels of TCF-4 and cyclin D1 protein. The results showed that the expression patterns of TCF-4 and cyclin D1 protein were similar to that of β-catenin, which supported our hypothesis. In addition, the results of TOP/FOP flash showed that the activity of TOP luciferase in SCC15 cells with MEG3 overexpression was significantly decreased, that is, the transcription activity of TCF/LEF was decreased compared to the control group. On the other hand, the activity of TOP luciferase in Cal27 cells with MEG3 silencing was significantly increased. The results were consistent with the results of western blot analysis (FOP-Flash with the mutation on the TCF binding site was used as a negative control).
Figure 6.
The activity of WNT/β-catenin pathway detected by western blot analysis and TOP/FOP flash experiments. The western blot results showed that overexpression of MEG3 resulted in decreased expression of β-catenin protein compared with the control group, so as to decrease the levels of downstream TCF-4 and cyclin D1 protein, while opposite results were found in cells with MEG3 silencing. The results of TOP/FOP flash experiments showed that the activity of luciferase was decreased by the transfection of MEG3 overexpression vector compared with the control group, while the activity of luciferase was increased after transfecting with MEG3 interference vector (*p<0.05). MEG3, maternally expressed gene 3.
Discussion
In the past few decades, studies on the regulatory role of MEG3 in specific tumorigenesis have shown that MEG3 is highly expressed in the central nervous system and the expression level is abnormally low in cancerous tissues (13), therefore, we speculated that MEG3 may also play an important regulatory role in the tumorigenesis of OSCC. In our study, we observed a decrease in the expression of MEG3 in OSCC tissue, which was consistent with the results of previous studies related to non-functional pituitary adenoma, such as gliomas, and meningiomas (8). In addition, epigenetic modifications induced tumor suppressor silencing through methylation at transcriptional level has attracted considerable attention in recent years (14). Recent studies showed that hypermethylation of the MEG3 promoter can inhibit MEG3 expression (15,16) and this finding has been confirmed for a variety of tumors (9). Our results also showed that the expression of MEG3 was increased with the increase of 5-aza-CdR concentration, which was further confirmed that MEG3 hypermethylation plays an important role in inhibiting MEG3 expression in OSCC.
In order to investigate the effects of MEG3 expression on OSCC cell proliferation, metastasis and apoptosis, pcDNA-MEG3 and si-MEG3 and their control vectors were transfected into SCC15 and Cal27 cells, respectively. The results showed that overexpression of MEG3 inhibited OSCC cell proliferation and metastasis, but promoted apoptosis and vice versa (17). This suggests that MEG3 may act as a tumor suppressor gene to inhibit OSCC.
The WNT/β-catenin signaling pathway, which regulates a variety of cellular processes, such as cell proliferation, invasion, and differentiation, is one of the classical pathways for signal transduction. The function of WNT/β-catenin signaling pathway has been extensively studied and reported. Recent studies have shown that downregulation of MEG3 in lung cancer cells can activate the WNT pathway (18). Therefore, in order to investigate the molecular mechanism of MEG3 regulated WNT pathway in OSCC cells, we detected and calculated the level of WNT pathway's core protein β-catenin by semi-quantitative analyses, and the activity of WNT pathway was detected using TOP/FOP flash reporter vector. Interestingly, both experiments showed that MEG3 levels were negatively correlated with the WNT pathway, that is, MEG3 inhibited OSCC cell proliferation, metastasis and promotes apoptosis by negatively regulating the WNT pathway (19,20). In addition, our findings provide new insights for future studies to investigate the tumor suppressive effect of MEG3 through local regulation of WNT/β-catenin signaling in OSCC cells.
In conclusion, our results suggest that MEG3 can possibly be used as a new biomarker for the diagnosis and treatment of the OSCC. It has also been found that DNA hypermethylation can inhibit MEG3 expression. In addition, MEG3 can inhibit the activity of OSCC WNT/β-catenin signaling pathway to inhibit tumor development. Therefore, the full understanding of this newly discovered functional mechanism of the MEG3 will be helpful for the diagnosis and treatment of OSCC.
References
- 1.Rosebush MS, Rao SK, Samant S, Gu W, Handorf CR, Pfeffer LM, Nosrat CA. Oral cancer: Enduring characteristics and emerging trends. J Tenn Dent Assoc. 2011;91:24–29. [PubMed] [Google Scholar]
- 2.Brocklehurst PR, Baker SR, Speight PM. Oral cancer screening: What have we learnt and what is there still to achieve? Future Oncol. 2010;6:299–304. doi: 10.2217/fon.09.163. [DOI] [PubMed] [Google Scholar]
- 3.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gutschner T, Diederichs S. The hallmarks of cancer: A long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qiu MT, Hu JW, Yin R, Xu L. Long noncoding RNA: An emerging paradigm of cancer research. Tumour Biol. 2013;34:613–620. doi: 10.1007/s13277-013-0658-6. [DOI] [PubMed] [Google Scholar]
- 6.Eades G, Zhang YS, Li QL, Xia JX, Yao Y, Zhou Q. Long non-coding RNAs in stem cells and cancer. World J Clin Oncol. 2014;5:134–141. doi: 10.5306/wjco.v5.i2.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wylie AA, Murphy SK, Orton TC, Jirtle RL. Novel imprinted DLK1/GTL2 domain on human chromosome 14 contains motifs that mimic those implicated in IGF2/H19 regulation. Genome Res. 2000;10:1711–1718. doi: 10.1101/gr.161600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou Y, Zhang X, Klibanski A. MEG3 noncoding RNA: A tumor suppressor. J Mol Endocrinol. 2012;48:R45–R53. doi: 10.1530/JME-12-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balik V, Srovnal J, Sulla I, Kalita O, Foltanova T, Vaverka M, Hrabalek L, Hajduch M. MEG3: A novel long noncoding potentially tumour-suppressing RNA in meningiomas. J Neurooncol. 2013;112:1–8. doi: 10.1007/s11060-012-1038-6. [DOI] [PubMed] [Google Scholar]
- 10.Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, Ansell PJ, Zhao J, Weng C, Klibanski A. Activation of p53 by MEG3 non-coding RNA. J Biol Chem. 2007;282:24731–24742. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- 11.Wang P, Ren Z, Sun P. Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. J Cell Biochem. 2012;113:1868–1874. doi: 10.1002/jcb.24055. [DOI] [PubMed] [Google Scholar]
- 12.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: Components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt JV, Matteson PG, Jones BK, Guan XJ, Tilghman SM. The Dlk1 and Gtl2 genes are linked and reciprocally imprinted. Genes Dev. 2000;14:1997–2002. [PMC free article] [PubMed] [Google Scholar]
- 14.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 15.Modali SD, Parekh VI, Kebebew E, Agarwal SK. Epigenetic regulation of the lncRNA MEG3 and its target c-MET in pancreatic neuroendocrine tumors. Mol Endocrinol. 2015;29:224–237. doi: 10.1210/me.2014-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braconi C, Kogure T, Valeri N, Huang N, Nuovo G, Costinean S, Negrini M, Miotto E, Croce CM, Patel T. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30:4750–4756. doi: 10.1038/onc.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao W, Chen X, He M. Inhibition of the Jagged/Notch pathway inhibits retinoblastoma cell proliferation via suppressing the PI3K/Akt, Src, p38MAPK and Wnt/β catenin signaling pathways. Mol Med Rep. 2014;10:453–458. doi: 10.3892/mmr.2014.2213. [DOI] [PubMed] [Google Scholar]
- 18.Xia Y, He Z, Liu B, Wang P, Chen Y. Downregulation of Meg3 enhances cisplatin resistance of lung cancer cells through activation of the WNT/β-catenin signaling pathway. Mol Med Rep. 2015;12:4530–4537. doi: 10.3892/mmr.2015.3897. [DOI] [PubMed] [Google Scholar]
- 19.Rowe MK, Chuang DM. Lithium neuroprotection: Molecular mechanisms and clinical implications. Expert Rev Mol Med. 2004;6:1–18. doi: 10.1017/S1462399404008385. [DOI] [PubMed] [Google Scholar]
- 20.Binnerts ME, Kim KA, Bright JM, Patel SM, Tran K, Zhou M, Leung JM, Liu Y, Lomas WE, III, Dixon M, et al. R-Spondin1 regulates Wnt signaling by inhibiting internalization of LRP6. Proc Natl Acad Sci USA. 2007;104:14700–14705. doi: 10.1073/pnas.0702305104. [DOI] [PMC free article] [PubMed] [Google Scholar]