Abstract
Sensing and responding to our environment requires functional neurons that act in concert. Neuronal cell loss resulting from degenerative diseases cannot be replaced in humans, causing a functional impairment to integrate and/or respond to sensory cues. In contrast, zebrafish (Danio rerio) possess an endogenous capacity to regenerate lost neurons. Here, we will focus on the processes that lead to neuronal regeneration in the zebrafish retina. Dying retinal neurons release a damage signal, tumor necrosis factor α, which induces the resident radial glia, the Müller glia, to reprogram and re-enter the cell cycle. The Müller glia divide asymmetrically to produce a Müller glia that exits the cell cycle and a neuronal progenitor cell. The arising neuronal progenitor cells undergo several rounds of cell divisions before they migrate to the site of damage to differentiate into the neuronal cell types that were lost. Molecular and immunohistochemical studies have predominantly provided insight into the mechanisms that regulate retinal regeneration. However, many processes during retinal regeneration are dynamic and require live-cell imaging to fully discern the underlying mechanisms. Recently, a multiphoton imaging approach of adult zebrafish retinal cultures was developed. We will discuss the use of live-cell imaging, the currently available tools and those that need to be developed to advance our knowledge on major open questions in the field of retinal regeneration.
Keywords: multiphoton microscopy, live-cell imaging, zebrafish, interkinetic nuclear migration, tissue culture, retinal regeneration, Müller glia, neuronal progenitor cell, differentiation, phagocytosis
Introduction
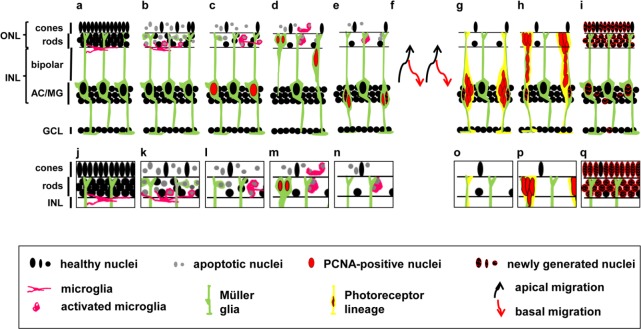
Neurogenesis in the adult human central nervous system (CNS) is restricted to limited regions of the brain i.e., the subventricular zone and the dentate gyrus (Ming and Song, 2011; Goncalves et al., 2016). Consequently, neuronal damage and death in most of the CNS cannot be repaired leading to functional impairment of the nervous system (Dimou and Gotz, 2014). In contrast, lower vertebrates such as fish, reptiles and amphibians possess an endogenous capacity to regenerate lost neurons in the brain, spinal cord and sensory organs, such as the retina and ear (Hitchcock et al., 2004; Kaslin et al., 2008; Burns and Corwin, 2013). Research in goldfish (Carassiusauratus) provided the first insight that neurons within the adult retina can be regenerated in response to a variety of damage stimuli including surgical, laser and chemical lesions (Maier and Wolburg, 1979; Braisted and Raymond, 1992; Hitchcock et al., 1992; Braisted et al., 1994; Hitchcock et al., 2004). Recently, zebrafish (Danio rerio) has become the preferred teleost species to investigate retinal regeneration due to the availability of tools to manipulate gene function (Wyatt et al., 2015). In the adult zebrafish retina, radial glial cells, the Müller glia, are induced to reprogram and re-enter the cell cycle in response to a variety of damage stimuli (Figure 1; Fausett and Goldman, 2006; Bernardos et al., 2007; Fimbel et al., 2007; Kassen et al., 2007; Powell et al., 2016). Damaged retinal neurons release tumor necrosis factor alpha, which acts as a damage signal to initiate Müller glia reprogramming and induces their re-entry into the cell cycle (Nelson et al., 2013). Proliferating Müller glia nuclei replicate their DNA in the basal region of the inner nuclear layer (INL), where they are typically located, and migrate to the apical surface of the retina, the outer nuclear layer (ONL), to divide asymmetrically to yield a Müller glia and a neuronal progenitor cell (NPC; Figure 1c–e; Nagashima et al., 2013; Lahne et al., 2015). Both the Müller glia and neuronal progenitor cells (NPC) return to the INL, where the Müller glia has been suggested to exit the cell cycle and the NPCs continue to proliferate (Figure 1e–h; Nagashima et al., 2013; Lahne et al., 2015). This migratory pattern in phase with the cell cycle, termed interkinetic nuclear migration (INM), was originally observed in developing neuro-epithelia (Sauer and Walker, 1959; Pearson et al., 2005; Baye and Link, 2007). Like NPC nuclei during development, those in the regenerating retina also undergo INM before they locate to the site of damage and differentiate into the lost neurons (Figure 1f–i; Vihtelic and Hyde, 2000; Lahne et al., 2015).
Figure 1.
Schematic of the time course of the regenerating zebrafish retina.
Simplified diagram of a healthy retina (a). Exposure of zebrafish to constant intense light induces photoreceptor cell death (b). The dying photoreceptors are engulfed by microglia (pink, a–e, k–n) or Müller glia (green, b, c, k, l). At subsequent time points, a subset of Müller glia re-enters the cell cycle, expressing the proliferating cell nuclear antigen, PCNA (c). Proliferating Müller glia nuclei undergo interkinetic nuclear migration (INM) to the outer nuclear layer (ONL) to divide, producing a neuronal progenitor cell (NPC) and a Müller glia (d). Both nuclei return to the inner nuclear layer (INL) where the Müller glia exits the cell cycle (checkered nucleus), while the NPC continues to proliferate (e). NPCs also undergo INM (f, black and red arrows represent apical and basal migration, respectively), producing clusters of NPCs that increase expression of commitment factors that drive NPCs towards the photoreceptor lineage in the light-damaged retina (g). Committed NPCs migrate to the ONL (h), differentiating into the lost neurons to repair the damaged retina (i, checkered nuclei). In addition, a few inner retinal neurons are also produced during the regeneration time course (i, checkered nuclei in INL and ganglion cell layer (GCL)). A subregion of the ONL was magnified to display events in the ONL in detail (j–q). AC: Amacrine cell; MG: Müller glia.
Research has predominantly focused on identifying signaling mechanisms that regulate Müller glia and NPC proliferation (Gorsuch and Hyde, 2014; Lenkowski and Raymond, 2014). In contrast, less attention has been given to other cellular processes that occur during retinal regeneration, such as phagocytosis of dying neurons and their role in inducing proliferation, the migration of NPCs to the site of damage, and the integration of newly differentiated neurons into the existing neuronal network. Our current knowledge of the mechanisms governing retinal regeneration predominantly stems from research that employs molecular and immunohistochemical approaches, while live-cell imaging techniques of the adult zebrafish retina have not been available until recently (Bailey et al., 2012; Weber et al., 2013; Lahne et al., 2015, 2017). Spectral domain optical coherence tomography (SD-OCT) was previously used to monitor damage and the recovery of the retinal structure in live adult zebrafish following light- or ouabain-induced damage (Bailey et al., 2012; Weber et al., 2013). However, this technique has not yet been employed to study mechanisms regulating retinal regeneration in zebrafish. Moreover, SD-OCT does not allow visualizing cellular details or processes such as INM. Recently, we developed a multiphoton imaging approach to monitor INM in live retinal cultures from adult light-damaged zebrafish (Lahne et al., 2015, 2017). This approach is not only applicable to investigating INM, but also other dynamic processes that occur during retinal regeneration. Here, we will summarize research areas during zebrafish retinal regeneration that would benefit from performing live-cell imaging and give an overview of tools that are available or that have to be developed to advance our understanding of retinal regeneration.
Phagocytosis of Dying Neurons
Müller glia phagocytosis
Müller glia were proposed to phagocytose dying cells in the regenerating zebrafish retina based on the presence of cytoplasmic TUNEL (Terminal deoxynucleotidyl transferase dUTP nick end labeling) signal or rod photoreceptor-driven GFP in Tg(XlRho:EGFP)fl1 zebrafish Figure 1a–c; Bailey et al., 2010). Live-cell imaging will be a powerful tool to verify that Müller glia engulf dying retinal neurons in the adult zebrafish retina. Elegant live-cell imaging studies of the chick and mouse utricle, a balance organ, showed that the glial cells (support cells) in these sensory epithelia phagocytose dying hair cells (sensory cells; Bird et al., 2010; Monzack et al., 2015). RCAS-driven (retroviral vector) mosaic expression of β-actin-enhanced green fluorescent protein (EGFP) in both the glial cells and hair cells of utricles revealed a two-step process: initially the actin rich hair bundles were excised, followed by the phagocytosis of the dying hair cells by surrounding glial support cells (Bird et al., 2010). During this process, a reorganization of the actin cytoskeleton occurred, which indicates that the actin cytoskeleton plays an active role in hair cell phagocytosis (Bird et al., 2010; Monzack et al., 2015). Hair cells and photoreceptors are similarly structured in that both consist of apical membrane protrusions that form the sensory units, the stereocilial hair bundles or the rod and cone photoreceptor inner and outer segments, respectively. Moreover, similar proteins such as the Usher syndrome family of proteins are present in both stereocilial hair bundles and photoreceptor inner and outer segments (Mathur and Yang, 2015). As such, it could be envisioned that Müller glia, similar to the removal of dying hair cells by glial supporting cells, initially excise the inner/outer segment of photoreceptors before they engulf the photoreceptor cell bodies. In support of this model, uptake of the rod inner segment marker zs-4 was not observed in Müller glia (Bailey et al., 2010). The recent establishment of live-cell imaging of zebrafish retinal explants by multiphoton microscopy (Lahne et al., 2015, 2017) will reveal whether and how Müller glia phagocytose dying photoreceptors in the damaged zebrafish retina. Transgenic lines that express fluorescent proteins with different excitation spectra in rod photoreceptor cells and Müller glia, such as the Tg[rho:nsfB. Eco-EGFP] and Tg[gfap-2A-tdtomato], respectively, will be useful in determining whether Müller glia phagocytose dying rod photoreceptors. Interestingly, Bailey et al. (2010) did not observe uptake of fluorescent protein that stemmed from a cone-photoreceptor-specific transgene. Thus, it will be important to examine whether phagocytosis of cellular debris by Müller glia is specific to rod photoreceptors in the zebrafish retina or whether other retinal cell types are also phagocytosed. Live-cell imaging of double transgenic lines that identify Müller glia and other retinal cell types, such as ganglion and amacrine cells in the Tg[elavl3:EGFP] transgenic zebrafish, in combination with a damage paradigm that predominantly ablates these neurons, will answer whether each neuronal cell type is phagocytosed by Müller glia in the regenerating zebrafish retina.
The use of cell death markers that can only enter cells with compromised cell membranes and subsequently bind to DNA, such as propidium iodide or ethidium homodimer will help distinguish healthy from dying neurons (Pinheiro et al., 2009; Bird et al., 2010; Zhao et al., 2015a). Furthermore, this approach will correlate the onset of membrane permeabilization and phagocytosis, as was previously shown in chick and mouse hair cell epithelia (Bird et al., 2010; Monzack et al., 2015). To address the mechanisms by which phagocytosis is mediated, transgenic lines that express the fluorescently-conjugated actin binding protein, life-act, are available for visualizing the remodeling of the actin cytoskeleton (Behrndt et al., 2012). Ideally, fluorescently-conjugated life-act would be expressed in a cell-type specific manner, e.g., to observe the contribution of actin cytoskeletal reorganization in Müller glia versus the dying neurons. The availability of cell-type specific GAL4-transgenic driver lines that can be crossed with Tg[UAS:lifeact-GFP] zebrafish will reveal actin cytoskeletal changes during photoreceptor cell death/Müller glia-mediated phagocytosis (Helker et al., 2013; Kawakami et al., 2016).
The presence of cytoplasmic TUNEL signal in proliferating Müller glia at time points subsequent to maximal photoreceptor cell death in the light-damaged zebrafish retina indicated that uptake of cell debris might instruct Müller glia to re-enter the cell cycle (Bailey et al., 2010). Following the fate of Müller glia by longer-term live-cell imaging would correlate whether only Müller glia that phagocytosed dying photoreceptors re-entered the cell cycle or whether those that did not phagocytose dying cells are also recruited into the cell cycle. Photo-conversion of Kaede, a fluorescent protein that changes its spectral properties from green to red fluorescing following exposure to UV light, could be used to specifically mark the Müller glia that phagocytosed dying neurons and track their fate (Hatta et al., 2006; Wilson et al., 2016). A challenge however might be the ability to perform long-term imaging. While it was reported that cultures are viable for up to 6 hours of multiphoton imaging, longer imaging periods were not assessed (Lahne et al., 2015). Culture conditions and imaging frequencies might have to be adjusted to accommodate longer-term imaging to examine the fate of cells.
Microglia phagocytosis of dying neurons
Microglia possess a variety of functions (Brawek and Garaschuk, 2017). Importantly, they are responsible for engulfing cell debris and dying cells, thereby maintaining a physiologically viable environment for healthy cells (Sousa et al., 2017; Brawek and Garaschuk, 2017). Additionally, microglia can induce apoptosis in neurons in the inflamed brain and in neuronal precursor cells during nervous system development (Cunningham et al., 2013; Neher et al., 2013; Brown and Neher, 2014). In retinitis pigmentosa models, microglia have been suggested to engulf both living and dead photoreceptors and disruption of the microglial function delayed the degeneration phenotype (Zhao et al., 2015a). Recently, microglial behavior was investigated by live-cell imaging of larval Tg[rho:YFP-ntr] zebrafish in response to rod photoreceptor cell death (White et al., 2017). Microglia that were resident in the nerve fiber layer of the larval retina migrated to the site of injury, the ONL, to engulf dying photoreceptors (White et al., 2017). As it was previously shown that dying cells release fluorescent proteins upon membrane disintegration, this sophisticated study would have benefited from the use of dyes that enter dying cells to convincingly demonstrate phagocytosis of dying neurons by microglia (Monzack et al., 2015). This approach would also establish whether microglial engulfment of dying cells induced neuronal apoptosis in zebrafish similar to their effect in retinitis pigmentosa models (Zhao et al., 2015a). In the future, it will be necessary to investigate whether microglia in the adult regenerating retina behave similar to those in larval zebrafish (Figure 1a–e). In the adult retina, microglia reside in the nerve fiber layer, the inner plexiform/basal inner nuclear layer, and the outer plexiform layer (Bailey et al., 2010). As such, the kinetics of microglial activation might differ as they are distributed closer to the site of injury.
As both microglia and Müller glia were suggested to phagocytose dying retinal cells, it will be necessary to examine their contribution to the removal of cell debris. Interestingly, if Müller glia engulf photoreceptors in a two-step process, initially excising the photoreceptor inner and outer segment, similar to glial cells in hair cell epithelia (Bird et al., 2010; Monzack et al., 2015), then potentially microglia phagocytose the released inner/outer segment structures. Additionally, Müller glia were not observed to take up cone photoreceptor components/fluorescence (Bailey et al., 2010). Thus, microglia might play the predominant role in the removal of dying cone photoreceptors. To distinguish the contributions of these two cell types, the mechanisms that regulate the initiation, recognition, and engulfment of cellular debris will have to be determined. While it is possible that both cell types employ similar mechanisms, cell-type specific knockdown strategies could be used to manipulate expression levels of genes important for the recognition and engulfment of debris. Subsequent monitoring of the phagocytic activity of the two cell types would reveal their individual involvement. Interestingly, ablation of microglia resulted in reduced Müller glia proliferation and a decreased regenerative capacity in larval zebrafish (White et al., 2017). While it still has to be established that a similar effect is observed in the adult zebrafish retina, it will be crucial to understand the cross-talk between microglia and Müller glia that instruct Müller glia to re-enter the cell cycle.
Mechanisms Governing INM of Müller Glia and NPCs
In both the developing and regenerating zebrafish retina, nuclei of proliferating cells (i.e., Müller glia and NPCs) undergo INM (Pearson et al., 2005; Baye and Link, 2007; Norden et al., 2009; Nagashima et al., 2013; Lahne et al., 2015). Live-cell imaging studies have predominantly provided insight into the mechanisms governing INM in the developing retina (Pearson et al., 2005; Del Bene et al., 2008; Norden et al., 2009). In contrast, live-cell imaging was only used to confirm that INM occurs in the regenerating retina, but was not employed to examine the regulatory processes that facilitate INM (Lahne et al., 2015, 2017). For example, Tg[gfap:nGFP]mi2004 zebrafish that express GFP in nuclei under the control of the Müller glia-specific promoter, glial fibrillary acidic protein (gfap), were used to determine the velocities before nuclear envelope breakdown and after cell division. However, the velocity after nuclear envelop breakdown could not be determined in Tg[gfap:nGFP]mi2004 zebrafish due to the redistribution of GFP throughout the cell following nuclear envelop breakdown. Studying INM in retinal explants from Tg[h2afva:h2afva-GFP]kca6 zebrafish, which express a fusion protein consisting of histone 2 and GFP that remains attached to the DNA throughout the cell cycle, would overcome this limitation (Pauls et al., 2001).
During neuroepithelial development, both actin-myosin mediated contraction, as well as the action of microtubules, has been suggested to facilitate INM in various tissues (Del Bene et al., 2008; Norden et al., 2009; Schenk et al., 2009; Tsai et al., 2010; Yu et al., 2011; Spear and Erickson, 2012). Inhibiting either actin filament formation or Rho-associated coiled-coil kinases implicated actin-myosin contraction as a mediator of INM in the regenerating retina. However, the contribution of microtubules in regulating INM in the regenerating retina was not investigated. In the chick neural tube, actin-myosin and microtubules both play a role at different stages of INM (Spear and Erickson, 2012). Tools are available to monitor the re-arrangement of the actin cytoskeleton (e.g., Tg(actc1b:LIFEACT-EGFP)) or the microtubule apparatus (e.g., Tg(UAS:EGFP-tuba2)) in zebrafish (Asakawa and Kawakami, 2010; Behrndt et al., 2012). Live-cell imaging of retinal cultures that express transgenes that can detect cytoskeletal changes, in combination with pharmacological or genetic approaches to disrupt the function of these cellular components, will reveal the contribution of distinct cytoskeletal networks in facilitating INM in the regenerating retina. Interestingly, based on the velocity, basal migration of nuclei in the regenerating retina occurs in an actively-driven manner, which is in contrast to retinal development where basal migration was suggested to be a passive stochastic process (Norden et al., 2009; Lahne et al., 2015). It is possible that Müller glia and NPC INM is regulated by different mechanisms and that NPCs in the regenerating retina might behave more similar to NPCs in the developing retina. Thus, NPC basal migration might occur in a passive manner, while Müller glia basal migration is driven by active forces. This hypothesis will have to be examined by assessing migration kinetics at different stages of the regeneration response using stage-specific transgenic zebrafish lines (Fausett and Goldman, 2006; Thummel et al., 2008; Lahne et al., 2015).
Deciphering the mechanisms that regulate basal migration might be challenging as pharmacological inhibition or genetic disruption of signaling pathways/cytoskeletal components might affect both apical and basal migration. The recent development of photo-activatable drugs, such as caged rho-associated coiled-coil kinase inhibitor Rockout, might help elucidate the mechanisms regulating basal nuclear migration, specifically, if photo-activation of these pharmacological agents was induced immediately after mitosis was observed by live-cell imaging (Morckel et al., 2012). Additionally, generating transgenic zebrafish that express optically-controllable tools, such as those that activate extracellularly regulated kinases or inhibit calcium/calmodulin-dependent protein kinase II (Johnson et al., 2017; Murakoshi et al., 2017) will offer a variety of means to examine the processes governing basal INM. Alternatively, plasmids that encode photoactivatable proteins could be electroporated into retinal explants; however, low electroporation efficiencies have been reported previously, especially in the central retina (Kustermann et al., 2008).
The signaling mechanisms that regulate the re-organization of the actin network or other cytoskeletal components during INM have not been investigated. A variety of signaling pathways, including changes in calcium levels, can regulate the remodeling of the actin cytoskeleton and/or activate actin-myosin mediated contraction (Okamoto et al., 2007; Chazeau and Giannone, 2016). During retinal development, increases in intracellular calcium levels were observed prior to the movement of nuclei and chelation of intracellular calcium resulted in reduced migration speed (Pearson et al., 2005). In the adult regenerating retina, it has not been established whether calcium levels change in relation to nuclear movement. Performing calcium imaging experiments in whole retinal explants while monitoring INM might be challenging, as the entire thickness of the retina would have to be imaged which requires several minutes. In contrast, changes in intracellular calcium levels could occur in a shorter time frame and events would be missed. Previously, changes in intracellular calcium concentrations have been monitored in cone photoreceptors in retinal slices from adult zebrafish using the genetically encoded calcium indicator GCaMP3 (Giarmarco et al., 2017). Culturing retinal slices might offer an alternative approach to retinal whole mounts, thereby allowing imaging of INM and calcium changes in a single plane; however, it would have to be first established whether retinal slice cultures are viable for prolonged periods. A number of other fluorescent-based sensors have been developed to measure activity changes of a variety of signaling pathways (e.g., protein kinases, cAMP) in real-time in vitro (Takao et al., 2005; Gesellchen et al., 2011). Recently, some of these sensors were successfully tested for their applicability to investigate zebrafish embryonic development (Fritz et al., 2013; Zhao et al., 2015b). In the future, employing such fluorescent-based signaling sensors along with live-cell imaging will gain insight into the spatio-temporal changes of signaling events that regulate INM during retinal regeneration. Additionally, such approaches will reveal the effectiveness of pharmacological or genetic manipulations of the respective signaling pathway.
Regulation of Cell Fate Choices
The role of INM in cell fate choices
Disrupting INM in the developing retina resulted in early cell cycle exit and increased production of early born neurons at the expense of those generated later during retinal development, potentially due to the exposure of basally located nuclei to aberrant Notch signaling levels, as overexpression of the Notch intracellular domain rescued the cell cycle exit and differentiation defects (Del Bene et al., 2008). In agreement, nuclei that migrated more basally following mitosis were more likely to produce neurons following the next round of mitosis rather than remaining in the cell cycle (Baye and Link, 2007). Thus, INM likely plays a role in regulating cell cycle exit decisions. Similarly, increased numbers of early born neurons were observed when INM was blocked in the regenerating zebrafish retina (Lahne et al., 2015). Interestingly, in the adult regenerating zebrafish retina, the two nuclei that formed following cell division were observed to migrate different distances basally into the INL or further into the inner plexiform layer (Figure 1e, g; Lahne et al., 2015). Monitoring the fate of nuclei that had migrated to different basal locations using longer-term live-cell imaging would reveal whether nuclear migration distances have similar functional implications as in retinal development. Photoconversion of Kaede-expressing transgenic lines could trace the nuclei that had migrated most basally and subsequently determine their cell fate (Hatta et al., 2006; Wilson et al., 2016). However, it must be established whether NPCs in retinal explant cultures behave similarly to those in vivo, i.e., whether they are able to commit to a neuronal lineage and subsequently mature into neurons (Thummel et al., 2008; Thomas et al., 2012; Conner et al., 2014; Lahne et al., 2015).
Recently, it was demonstrated that photoreceptor cell death results in the generation of other neuronal cell types such as ganglion, amacrine and bipolar cells, that were not lost in response to intense light (Figure 1i, checkered nuclei in the INL and GCL; Vihtelic et al., 2006; Lahne et al., 2015; Powell et al., 2016). In the developing retina, the different cell types are produced in a sequential, but overlapping order (Carter-Dawson and LaVail, 1979; Young, 1985; He et al., 2012; Suzuki et al., 2013). Thus, the different retinal cell types that are produced in the regenerating retina might be generated in a similar sequential fashion (Figure 2). To use live-cell imaging to investigate whether NPCs follow a similar developmental pattern of cell fate specification, markers/transgenic lines have to be identified that distinguish between the different cell types. During zebrafish retinal development, a number of transgenic lines were generated to investigate the pattern and mechanisms governing cell type specification (Poggi et al., 2005; Baye and Link, 2007; Jusuf et al., 2011; Suzuki et al., 2013; Weber et al., 2014). Currently, only the Tg[atoh7:GFP] and Tg[nrd:EGFP] transgenic zebrafish upregulate GFP expression in NPCs in the light-damaged zebrafish retina (Thomas et al., 2012; Conner et al., 2014; Lahne et al., 2015).
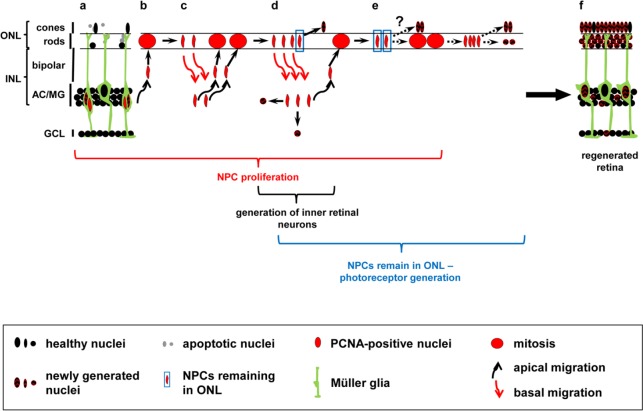
Figure 2.
Hypothetical model of NPC-mediated generation of neuronal subtypes in the regenerating zebrafish retina.
Diagram of a light-damaged retina after the initial Müller glia cell division that produced a cell cycle exiting Müller glia (a, checkered nucleus) and an NPC that continues to proliferate (a, red nucleus). NPCs undergo several rounds of cell divisions (b–e), migrating repetitively in phase with the cell cycle between the INL where they replicate their DNA and the ONL where mitosis occurs (b–d, round red nucleus). Black and red arrows indicate apical and basal nuclear migration, respectively. Mitosis potentially produces cells with neurogenic and proliferative fates allowing a subset of cells to sequentially exit the cell cycle (d, e), thereby generating cells that differentiate into the different retinal subtypes. A subset of ONL cell divisions produce nuclei that remain in the ONL (d, e, blue boxed nuclei). These ONL remaining cells could either directly exit the cell cycle and differentiate into the lost photoreceptors (d, e; checkered ONL based nuclei) or undergo an additional cell division before giving rise to photoreceptors (e) resulting in the regeneration of the damaged retina (f). This model is based on the following research: 1) Müller glia divide asymmetrically yielding a cell-cycle exiting Müller glia and an NPC that continues proliferating (Nagashima et al., 2013), 2) only 61 percent of nuclei return from the ONL to the INL after cell division (Lahne et al., 2015), and 3) both photoreceptors and inner retinal neurons are produced following light damage (Vihtelic and Hyde, 2000; Lahne et al., 2015; Powell et al., 2016). The exact sequence of events leading to the differentiation of the different neuronal subtypes in the regenerating zebrafish retina is currently unknown. AC: Amacrine cell; GCL: ganglion cell layer; INL: inner nuclear layer; MG: Müller glia; NPC: neuronal progenitor cell; ONL: outer nuclear layer; PCNA: proliferating cell nuclear antigen.
To understand the processes regulating neuronal differentiation in the regenerating retina, it also has to be established how the newly arising photoreceptors locate to the ONL. Based on the position of proliferating nuclei clusters in the INL of fixed tissue sections at 72 hours of light treatment (Figure 1g) and their accumulation in the ONL at later time points (Figure 1h), it was suggested that NPCs migrate from the INL to the ONL along the radial Müller glia processes (Figure 1g, h; Vihtelic and Hyde, 2000; Gorsuch and Hyde, 2014). However, it is possible that the distribution of NPCs to the ONL is mediated by INM e.g., either one nucleus remains in the ONL while the second returns to the INL or alternatively both nuclei stay in the ONL (Figure 2). Live-cell imaging of retinal cultures showed that only 61 percent of Müller glia/NPC nuclei in Tg[gfap:nGFP] zebrafish returned to the INL, supporting that many of the cells remain in the ONL to potentially differentiate into photoreceptors (Lahne et al., 2015). In the developing retina, red cone precursor cells can either arise from an asymmetric division at the apical surface that also produces a ganglion cell or a horizontal cell precursor (Suzuki et al., 2013). The arising precursor cell nuclei subsequently migrated basally before the red cone precursor cell nuclei migrated apically to divide symmetrically to give rise to two red cone photoreceptors (Suzuki et al., 2013). During retinal regeneration, a detailed analysis of the location of both daughter cells after cell division and their fate at different stages of the proliferative response is necessary. It is also unclear, whether all the nuclei that remain in the ONL exit the cell cycle (Figure 2d, e). Live-cell imaging of retinal explants from Fucci zebrafish (e.g., Tg[-3.5ubb:Cerulean-gmnn-2A-mCherry-cdt1]), which express different fluorescent proteins in G0/G1 versus S/G2/M-phases, could elucidate whether nuclei that stay in the ONL exit the cell cycle (Sugiyama et al., 2009; Bouldin and Kimelman, 2014a, b). Furthermore, it is unknown whether those nuclei that remain in the ONL differentiate into specific cone photoreceptor subtypes or rod photoreceptors. Tg[gfap:nGFP] zebrafish in combination with e.g., Tg[trβ2:tdTomato] transgenics could give insight whether a subset of those ONL-remaining nuclei differentiate into red cone photoreceptors.
Influence of division plane orientation on cell fate choices
The orientation of the cleavage plane during cell division has been correlated with the specification of retinal cell types in the developing retina (Cayouette and Raff, 2003; Weber et al., 2014). Specifically, the majority of NPCs in the mouse retina divide with a cleavage plane that is oriented horizontally to the apical surface of the retina; however, the number of cell divisions with a vertical orientation of the cleavage plane increases during postnatal retinal development (Cayouette and Raff, 2003). Cell divisions with vertical cleavage planes were associated with the generation of rod photoreceptors (Cayouette and Raff, 2003). In the regenerating zebrafish retina, approximately ten percent of divisions are oriented in a vertical manner, while 90 percent are horizontally oriented during early Müller glia/NPC divisions (Lahne et al., 2015). The significance of these vertical cell divisions is currently unknown. One possibility is that the cells resulting from a vertical division exit the cell cycle and differentiate. This could be examined by live-cell imaging of retinal explants from Fucci zebrafish (e.g., Tg[-3.5ubb:Cerulean-gmnn-2A-mCherry-cdt1]; (Sugiyama et al., 2009; Bouldin and Kimelman, 2014a, b). However, to determine the cell fates of vertically dividing cells, transgenic lines have to be identified that can faithfully represent the differentiation of each retinal cell type specifically in the regenerating zebrafish retina. The orientation of NPC division planes at later stages of lineage commitment still have to be determined. Transgenic zebrafish, such as Tg[atoh7:GFP] and Tg[nrd:EGFP], will enable correlating division planes of NPCs during stages of their commitment to either a neuronal or a photoreceptor lineage, respectively (Poggi et al., 2005; Thomas et al., 2012; Conner et al., 2014; Lahne et al., 2015). Understanding how the division plane orientation influences the generation of specific retinal cell types and the mechanisms that govern this process, might ultimately provide us tools to manipulate the division plane to obtain increased numbers of a desired cell type to regenerate those lost in the diseased mammalian retina.
Functional Integration of Regenerated Neurons into the Existing Neuronal Circuit
Reforming synaptic contacts was previously studied in response to either photoreceptor or bipolar cell regeneration in larval zebrafish (D’Orazi et al., 2016; Yoshimatsu et al., 2016). H3 horizontal cells re-established contacts with regenerated UV-cones, maintaining their preferential connectivity, while regenerated bipolar cells changed their connectivity pattern, forming less frequently contacts with red cones (D’Orazi et al., 2016; Yoshimatsu et al., 2016). In larval zebrafish, developmental signaling networks may still be active, thus regulating the re-establishment of neuronal networks. In contrast, integrating newly regenerated photoreceptors into the existing neuronal network and the mechanisms that drive this process have not been investigated in the light-damaged adult zebrafish retina. In addition to the production of photoreceptors, other retinal neurons including amacrine, ganglion and bipolar cells were also recently shown to be produced following light-damage (Lahne et al., 2015; Powell et al., 2016). It will be interesting to investigate whether these neuronal cell types also integrate into the existing neuronal circuit. Live-cell imaging could establish whether regenerated neurons generate processes that are appropriate for the specific cell type as established during retinal development (Godinho et al., 2005; Mumm et al., 2006; Schroeter et al., 2006). However, as the majority of inner retinal neurons are maintained, transgenic lines will have to be identified that allow tracking of the newly generated ganglion, amacrine and bipolar cells and distinguish them from those that were maintained during the damaging insult. Lineage-tracing experiments using Müller glia or NPC-specific Cre-recombinase expressing lines in conjunction with a floxed fluorescent reporter zebrafish line would enable the identification of newly generated neurons and would allow monitoring the outgrowth of axons and dendritic arbors by live-cell imaging (Ramachandran et al., 2012; Briona et al., 2015). Alternatively, transgenic zebrafish lines that transiently upregulate the expression of fluorescent reporter proteins in NPCs might be suitable to detect newly generated neurons if these fluorescent reporter proteins are stable for extended periods. In the light-damaged retina, GFP under the control of the atoh7 promotor is specifically expressed in a subset of NPCs (Conner et al., 2014; Lahne et al., 2015). Moreover, during zebrafish retinal development atoh7:GFP-positive cells give rise to ganglion, amacrine and photoreceptor cells (Poggi et al., 2005) and as such, the Tg[atoh7:EGFP] transgenic line could be tested for its potential to identify new-born ganglion, amacrine and photoreceptor cells during the regenerative response. Ideally, suitable transgenic zebrafish would be crossed to transgenic lines expressing fluorophores of a different spectrum that identifies the various neuronal cell types that were maintained while the damaging insult occurred. Live-cell imaging of retinal cultures of such double transgenic lines during the regenerative phase would reveal whether newly generated neurons contact those that were maintained throughout the damaging period.
Monitoring the establishment of contacts between newly generated neurons and those maintained in the regenerated retina does not reveal whether these cells functionally interact. Recently, zebrafish lines were developed that drive the Gal/UAS-regulated expression of optogenetic tools that allow the manipulation of cellular ion concentrations and thus membrane potential and cell excitability. Transgenic lines available that express light-gated ion channels or pumps include channelrhodopsin (non-selective cation channel), archearhodopsin 3 (proton pump) and Halo (chloride pump, Kimura et al., 2013). Live-cell imaging of retinal cultures from these transgenic lines would offer the possibility to manipulate the membrane potential and thereby firing probabilities of neurons in a spatially restricted manner, potentially allowing only the activation of single cells by laser light of a specific wavelength. However, to employ these tools to investigate the functional interaction between existing and newly formed neurons, they would have to be combined with an approach such as calcium- or voltage-sensitive imaging that permits detecting a functional output in a network of cells. Transgenic lines that express genetically encoded calcium-sensitive reporters such as GCaMP or commercially available synthetic calcium or voltage-sensing probes could be employed (Friedrich and Korsching, 1998; Renninger and Orger, 2013). While this sophisticated approach would reveal the extent newly generated neurons interact with existing neurons, it might harbor several challenges as newly/existing cells would have to be imaged while calcium/membrane potentials of neighboring cells are measured and optogenetic tools are simultaneously stimulated.
Alternatively, neurons produced in excess such as ganglion, amacrine and bipolar cells that are not lost during light-damage or photoreceptors that are generated following N-methyl-D-aspartate (NMDA)-induced inner retinal neuronal loss might undergo apoptosis (Vihtelic et al., 2006; Powell et al., 2016). In support, following poke-induced injury, a decline in the number of newly generated cells that were pulsed with bromodeoxyuridine (BrdU) during the proliferative phase was observed after the cessation of proliferation, suggesting that overproduced neurons might undergo apoptosis (Fausett and Goldman, 2006). During nervous system development, neurons that were produced in excess are removed by microglia (Marin-Teva et al., 2004; Cunningham et al., 2013). Moreover, microglia do not only remove apoptotic cells, but they can also induce apoptosis (e.g., of neuronal precursor cells in the developing brain), thus playing an active role in the control of organ size and circuit formation (Cunningham et al., 2013). Currently, it remains unknown whether neurons born in excess in the regenerating retina are removed by microglial activity similar to nervous system development (Marin-Teva et al., 1999; Marin-Teva et al., 2004). Live-cell imaging approaches using retinal cultures from double-transgenic zebrafish lines that identify both newly generated neurons and microglia (e.g., Tg[mpeg:EGFP]) would reveal whether microglia engulf newly generated neurons (Ellett et al., 2011; Morsch et al., 2015). Additionally, the viability status of the engulfed cell can be assessed by exposing retinal cultures during the imaging period to dyes that are excluded from healthy cells and that are only incorporated into the DNA following cell membrane breakdown (Zhao et al., 2015a). Uptake of such a dye following microglial phagosome formation would indicate that microglia induce neuronal cell death rather than clearing apoptotic cells. It will be interesting to compare the percentages of newly generated neuronal cell types that are induced to die relatively to those integrated into the neuronal circuit in various damage models that ablate different cell types. Understanding the mechanisms that regulate/maintain the balance between integration and cell death might help direct the integration of the correct number of cells following cell transplantation of induced pluripotent stem cells or precursor cells in the future.
Advantages and Limitations of Two-Photon Imaging of Retinal Cultures
The development of the multiphoton imaging technique to monitor dynamic processes in retinal explant cultures during retinal regeneration such as phagocytosis of dying neurons by Müller glia or interkinetic nuclear migration is a major technical advancement (Lahne et al., 2015, 2017). Culture systems offer the advantage that the tissue of interest can be exposed to pharmacological agents in an easy and controlled manner, permitting flexibility to maintain constant concentrations without affecting other tissues/organs. However, in regard to live-cell imaging, it has to be kept in mind when designing the experiment that some drugs are light-sensitive. This limiting factor can be overcome by continuously replacing the drugs utilizing perfusion systems (Scudder et al., 1993; Tiruchinapalli et al., 2003). While cell culture systems offer great means to study cellular processes by live-cell imaging the components included in the culture media might positively or negatively affect the behavior of cells, e.g., it was previously shown that microglia become activated in vitro (Stence et al., 2001). Additionally, continuous exposure of cells to laser light can result in the generation of radicals that reduce the viability of cells within the tissue (Coutu and Schroeder, 2013). In the future, techniques will need to be developed to perform in vivo imaging of the adult zebrafish retina that allow monitoring of cells in their physiological environment, thereby overcoming limitations associated with tissue culture systems.
Depending on the multiphoton system available, the user might experience technical restrictions. For instance, the choice of cell death markers/transgenic lines might be limited depending on the range of excitation wavelengths that the system available permits. Additionally, the effective wavelength for the different fluorescent proteins/marker might not overlap and therefore, sequential imaging at different excitation wavelength might have to be executed (Bestvater et al., 2002; Spiess et al., 2005). Sequential imaging would potentially be a disadvantage: 1) as slow image acquisition might hinder or reduce the sensitivity to detect the events of interest, and 2) the events imaged using distinct fluorescent probes might be temporally offset. The latter might not be problematic for correlating events imaged with distinct fluorescent probes if small z-stacks are acquired. However, if imaging throughout the thickness of the retina is required, this can result in an inability to obtain conclusive temporal information. Hence, the investigator will have to weigh the advantages and disadvantages to identify the best viable approach to investigate their research question.
Conclusion
The development of live-cell imaging approaches will expand our understanding of the dynamic processes that occur during retinal regeneration and the mechanisms that regulate these. Live-cell imaging will also allow tracking of proliferating cells and reveal fate decisions that could otherwise not be discerned. However, in order to use the technique to its full potential, the method will have to be further developed and adjusted according to the research question. Additionally, establishing transgenic zebrafish lines that identify cells at specific stages of the regeneration time course and those that express tools to manipulate signaling pathway will open new avenues to expand our knowledge of neuronal regeneration. A deeper understanding of the mechanisms facilitating retinal regeneration in zebrafish will ultimately aid the design of regenerative therapies in human.
Footnotes
Funding: This work was supported by NIH-NEI grants to DRH (R01-EY018417, R01-EY024519) and the Center for Zebrafish Research, University of Notre Dame, USA.
Conflicts of interest: None declared.
Plagiarism check: Checked twice by iThenticate.
Peer review: Externally peer reviewed.
Open peer review report:
Reviewer: Colin Barnstable, Pennsylvania State University College of Medicine, USA.
Comments to authors: An authoritative and deep review of a large body of work in zebrafish. This will stand as the key source for this information and should be publicized as much as possible when published. This is a superb review of retinal regeneration in zebrafish. It covers almost all of the recent data that have elucidated concepts of regeneration and the role of Muller glial cells in providing a source of retinal progenitors.
References
- Asakawa K, Kawakami K. A transgenic zebrafish for monitoring in vivo microtubule structures. Dev Dyn. 2010;239:2695–2699. doi: 10.1002/dvdy.22400. [DOI] [PubMed] [Google Scholar]
- Bailey TJ, Davis DH, Vance JE, Hyde DR. Spectral-domain optical coherence tomography as a noninvasive method to assess damaged and regenerating adult zebrafish retinas. Invest Ophthalmol Vis Sci. 2012;53:3126–3138. doi: 10.1167/iovs.11-8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TJ, Fossum SL, Fimbel SM, Montgomery JE, Hyde DR. The inhibitor of phagocytosis, O-phospho-L-serine, suppresses muller glia proliferation and cone cell regeneration in the light-damaged zebrafish retina. Exp Eye Res. 2010;91:601–612. doi: 10.1016/j.exer.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baye LM, Link BA. Interkinetic nuclear migration and the selection of neurogenic cell divisions during vertebrate retinogenesis. J Neurosci. 2007;27:10143–10152. doi: 10.1523/JNEUROSCI.2754-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrndt M, Salbreux G, Campinho P, Hauschild R, Oswald F, Roensch J, Grill SW, Heisenberg CP. Forces driving epithelial spreading in zebrafish gastrulation. Science. 2012;338:257–260. doi: 10.1126/science.1224143. [DOI] [PubMed] [Google Scholar]
- Bernardos RL, Barthel LK, Meyers JR, Raymond PA. Late-stage neuronal progenitors in the retina are radial muller glia that function as retinal stem cells. J Neurosci. 2007;27:7028–7040. doi: 10.1523/JNEUROSCI.1624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestvater F, Spiess E, Stobrawa G, Hacker M, Feurer T, Porwol T, Berchner-Pfannschmidt U, Wotzlaw C, Acker H. Two-photon fluorescence absorption and emission spectra of dyes relevant for cell imaging. J Microsc. 2002;208:108–115. doi: 10.1046/j.1365-2818.2002.01074.x. [DOI] [PubMed] [Google Scholar]
- Bird JE, Daudet N, Warchol ME, Gale JE. Supporting cells eliminate dying sensory hair cells to maintain epithelial integrity in the avian inner ear. J Neurosci. 2010;30:12545–12556. doi: 10.1523/JNEUROSCI.3042-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouldin CM, Kimelman D. Cdc25 and the importance of G2 control: Insights from developmental biology. Cell Cycle. 2014a;13:2165–2171. doi: 10.4161/cc.29537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouldin CM, Kimelman D. Dual fucci: A new transgenic line for studying the cell cycle from embryos to adults. Zebrafish. 2014b;11:182–183. doi: 10.1089/zeb.2014.0986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braisted JE, Raymond PA. Regeneration of dopaminergic neurons in goldfish retina. Development. 1992;114:913–919. doi: 10.1242/dev.114.4.913. [DOI] [PubMed] [Google Scholar]
- Braisted JE, Essman TF, Raymond PA. Selective regeneration of photoreceptors in goldfish retina. Development. 1994;120:2409–2419. doi: 10.1242/dev.120.9.2409. [DOI] [PubMed] [Google Scholar]
- Brawek B, Garaschuk O. Monitoring in vivo function of cortical microglia. Cell Calcium. 2017;64:109–117. doi: 10.1016/j.ceca.2017.02.011. [DOI] [PubMed] [Google Scholar]
- Briona LK, Poulain FE, Mosimann C, Dorsky RI. Wnt/ss-catenin signaling is required for radial glial neurogenesis following spinal cord injury. Dev Biol. 2015;403:15–21. doi: 10.1016/j.ydbio.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GC, Neher JJ. Microglial phagocytosis of live neurons. Nat Rev Neurosci. 2014;15:209–216. doi: 10.1038/nrn3710. [DOI] [PubMed] [Google Scholar]
- Burns JC, Corwin JT. A historical to present-day account of efforts to answer the question: “What puts the brakes on mammalian hair cell regeneration?”. Hear Res. 2013;297:52–67. doi: 10.1016/j.heares.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter-Dawson LD, LaVail MM. Rods and cones in the mouse retina. II. autoradiographic analysis of cell generation using tritiated thymidine. J Comp Neurol. 1979;188:263–272. doi: 10.1002/cne.901880205. [DOI] [PubMed] [Google Scholar]
- Cayouette M, Raff M. The orientation of cell division influences cell-fate choice in the developing mammalian retina. Development. 2003;130:2329–2339. doi: 10.1242/dev.00446. [DOI] [PubMed] [Google Scholar]
- Chazeau A, Giannone G. Organization and dynamics of the actin cytoskeleton during dendritic spine morphological remodeling. Cell Mol Life Sci. 2016;73:3053–3073. doi: 10.1007/s00018-016-2214-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner C, Ackerman KM, Lahne M, Hobgood JS, Hyde DR. Repressing notch signaling and expressing TNFalpha are sufficient to mimic retinal regeneration by inducing muller glial proliferation to generate committed progenitor cells. J Neurosci. 2014;34:14403–14419. doi: 10.1523/JNEUROSCI.0498-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutu DL, Schroeder T. Probing cellular processes by long-term live imaging--historic problems and current solutions. J Cell Sci. 2013;126:3805–3815. doi: 10.1242/jcs.118349. [DOI] [PubMed] [Google Scholar]
- Cunningham CL, Martinez-Cerdeno V, Noctor SC. Microglia regulate the number of neural precursor cells in the developing cerebral cortex. J Neurosci. 2013;33:4216–4233. doi: 10.1523/JNEUROSCI.3441-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bene F, Wehman AM, Link BA, Baier H. Regulation of neurogenesis by interkinetic nuclear migration through an apical-basal notch gradient. Cell. 2008;134:1055–1065. doi: 10.1016/j.cell.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimou L, Gotz M. Glial cells as progenitors and stem cells: New roles in the healthy and diseased brain. Physiol Rev. 2014;94:709–737. doi: 10.1152/physrev.00036.2013. [DOI] [PubMed] [Google Scholar]
- D’Orazi FD, Zhao XF, Wong RO, Yoshimatsu T. Mismatch of synaptic patterns between neurons produced in regeneration and during development of the vertebrate retina. Curr Biol. 2016;26:2268–2279. doi: 10.1016/j.cub.2016.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellett F, Pase L, Hayman JW, Andrianopoulos A, Lieschke GJ. Mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood. 2011;117:e49–56. doi: 10.1182/blood-2010-10-314120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fausett BV, Goldman D. A role for alpha1 tubulin-expressing muller glia in regeneration of the injured zebrafish retina. J Neurosci. 2006;26:6303–6313. doi: 10.1523/JNEUROSCI.0332-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimbel SM, Montgomery JE, Burket CT, Hyde DR. Regeneration of inner retinal neurons after intravitreal injection of ouabain in zebrafish. J Neurosci. 2007;27:1712–1724. doi: 10.1523/JNEUROSCI.5317-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich RW, Korsching SI. Chemotopic, combinatorial, and noncombinatorial odorant representations in the olfactory bulb revealed using a voltage-sensitive axon tracer. J Neurosci. 1998;18:9977–9988. doi: 10.1523/JNEUROSCI.18-23-09977.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz RD, Letzelter M, Reimann A, Martin K, Fusco L, Ritsma L, Ponsioen B, Fluri E, Schulte-Merker S, van Rheenen J, Pertz O. A versatile toolkit to produce sensitive FRET biosensors to visualize signaling in time and space. Sci Signal. 2013;6:rs12. doi: 10.1126/scisignal.2004135. [DOI] [PubMed] [Google Scholar]
- Gesellchen F, Stangherlin A, Surdo N, Terrin A, Zoccarato A, Zaccolo M. Measuring spatiotemporal dynamics of cyclic AMP signaling in real-time using FRET-based biosensors. Methods Mol Biol. 2011;746:297–316. doi: 10.1007/978-1-61779-126-0_16. [DOI] [PubMed] [Google Scholar]
- Giarmarco MM, Cleghorn WM, Sloat SR, Hurley JB, Brockerhoff SE. Mitochondria maintain distinct Ca2+ pools in cone photoreceptors. J Neurosci. 2017;37:2061–2072. doi: 10.1523/JNEUROSCI.2689-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho L, Mumm JS, Williams PR, Schroeter EH, Koerber A, Park SW, Leach SD, Wong RO. Targeting of amacrine cell neurites to appropriate synaptic laminae in the developing zebrafish retina. Development. 2005;132:5069–5079. doi: 10.1242/dev.02075. [DOI] [PubMed] [Google Scholar]
- Goncalves JT, Schafer ST, Gage FH. Adult neurogenesis in the hippocampus: From stem cells to behavior. Cell. 2016;167:897–914. doi: 10.1016/j.cell.2016.10.021. [DOI] [PubMed] [Google Scholar]
- Gorsuch RA, Hyde DR. Regulation of muller glial dependent neuronal regeneration in the damaged adult zebrafish retina. Exp Eye Res. 2014;123:131–140. doi: 10.1016/j.exer.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta K, Tsujii H, Omura T. Cell tracking using a photoconvertible fluorescent protein. Nat Protoc. 2006;1:960–967. doi: 10.1038/nprot.2006.96. [DOI] [PubMed] [Google Scholar]
- He J, Zhang G, Almeida AD, Cayouette M, Simons BD, Harris WA. How variable clones build an invariant retina. Neuron. 2012;75:786–798. doi: 10.1016/j.neuron.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helker CS, Schuermann A, Karpanen T, Zeuschner D, Belting HG, Affolter M, Schulte-Merker S, Herzog W. The zebrafish common cardinal veins develop by a novel mechanism: Lumen ensheathment. Development. 2013;140:2776–2786. doi: 10.1242/dev.091876. [DOI] [PubMed] [Google Scholar]
- Hitchcock P, Ochocinska M, Sieh A, Otteson D. Persistent and injury-induced neurogenesis in the vertebrate retina. ProgRetin Eye Res. 2004;23:183–194. doi: 10.1016/j.preteyeres.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Hitchcock PF, Lindsey Myhr KJ, Easter SS, Jr, Mangione-Smith R, Jones DD. Local regeneration in the retina of the goldfish. J Neurobiol. 1992;23:187–203. doi: 10.1002/neu.480230209. [DOI] [PubMed] [Google Scholar]
- Johnson HE, Goyal Y, Pannucci NL, Schupbach T, Shvartsman SY, Toettcher JE. The spatiotemporal limits of developmental ERK signaling. Dev Cell. 2017;40:185–192. doi: 10.1016/j.devcel.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jusuf PR, Almeida AD, Randlett O, Joubin K, Poggi L, Harris WA. Origin and determination of inhibitory cell lineages in the vertebrate retina. J Neurosci. 2011;31:2549–2562. doi: 10.1523/JNEUROSCI.4713-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaslin J, Ganz J, Brand M. Proliferation, neurogenesis and regeneration in the non-mammalian vertebrate brain. Philos Trans R SocLond B Biol Sci. 2008;363:101–122. doi: 10.1098/rstb.2006.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassen SC, Ramanan V, Montgomery JE, T Burket C, Liu CG, Vihtelic TS, Hyde DR. Time course analysis of gene expression during light-induced photoreceptor cell death and regeneration in albino zebrafish. Dev Neurobiol. 2007;67:1009–1031. doi: 10.1002/dneu.20362. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Asakawa K, Hibi M, Itoh M, Muto A, Wada H. Gal4 driver transgenic zebrafish: Powerful tools to study developmental biology, organogenesis, and neuroscience. Adv Genet. 2016;95:65–87. doi: 10.1016/bs.adgen.2016.04.002. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Satou C, Fujioka S, Shoji W, Umeda K, Ishizuka T, Yawo H, Higashijima S. Hindbrain V2a neurons in the excitation of spinal locomotor circuits during zebrafish swimming. Curr Biol. 2013;23:843–849. doi: 10.1016/j.cub.2013.03.066. [DOI] [PubMed] [Google Scholar]
- Kustermann S, Schmid S, Biehlmaier O, Kohler K. Survival, excitability, and transfection of retinal neurons in an organotypic culture of mature zebrafish retina. Cell Tissue Res. 2008;332:195–209. doi: 10.1007/s00441-008-0589-5. [DOI] [PubMed] [Google Scholar]
- Lahne M, Gorsuch RA, Nelson CM, Hyde DR. Culture of adult transgenic zebrafish retinal explants for live-cell imaging by multiphoton microscopy. J Vis Exp. 2017 doi: 10.3791/55335. doi:10.3791/55335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahne M, Li J, Marton RM, Hyde DR. Actin-cytoskeleton- and rock-mediated INM are required for photoreceptor regeneration in the adult zebrafish retina. J Neurosci. 2015;35:15612–15634. doi: 10.1523/JNEUROSCI.5005-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenkowski JR, Raymond PA. Muller glia: Stem cells for generation and regeneration of retinal neurons in teleost fish. Prog Retin Eye Res. 2014;40:94–123. doi: 10.1016/j.preteyeres.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier W, Wolburg H. Regeneration of the goldfish retina after exposure to different doses of ouabain. Cell Tissue Res. 1979;202:99–118. doi: 10.1007/BF00239223. [DOI] [PubMed] [Google Scholar]
- Marin-Teva JL, Cuadros MA, Calvente R, Almendros A, Navascues J. Naturally occurring cell death and migration of microglial precursors in the quail retina during normal development. J Comp Neurol. 1999;412:255–275. [PubMed] [Google Scholar]
- Marin-Teva JL, Dusart I, Colin C, Gervais A, van Rooijen N, Mallat M. Microglia promote the death of developing purkinje cells. Neuron. 2004;41:535–547. doi: 10.1016/s0896-6273(04)00069-8. [DOI] [PubMed] [Google Scholar]
- Mathur P, Yang J. Usher syndrome: Hearing loss, retinal degeneration and associated abnormalities. Biochim Biophys Acta. 2015;1852:406–420. doi: 10.1016/j.bbadis.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming GL, Song H. Adult neurogenesis in the mammalian brain: Significant answers and significant questions. Neuron. 2011;70:687–702. doi: 10.1016/j.neuron.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monzack EL, May LA, Roy S, Gale JE, Cunningham LL. Live imaging the phagocytic activity of inner ear supporting cells in response to hair cell death. Cell Death Differ. 2015;22:1995–2005. doi: 10.1038/cdd.2015.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morckel AR, Lusic H, Farzana L, Yoder JA, Deiters A, Nascone-Yoder NM. A photoactivatable small-molecule inhibitor for light-controlled spatiotemporal regulation of rho kinase in live embryos. Development. 2012;139:437–442. doi: 10.1242/dev.072165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morsch M, Radford R, Lee A, Don EK, Badrock AP, Hall TE, Cole NJ, Chung R. In vivo characterization of microglial engulfment of dying neurons in the zebrafish spinal cord. Front Cell Neurosci. 2015;9:321. doi: 10.3389/fncel.2015.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumm JS, Williams PR, Godinho L, Koerber A, Pittman AJ, Roeser T, Chien CB, Baier H, Wong RO. In vivo imaging reveals dendritic targeting of laminated afferents by zebrafish retinal ganglion cells. Neuron. 2006;52:609–621. doi: 10.1016/j.neuron.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakoshi H, Shin ME, Parra-Bueno P, Szatmari EM, Shibata AC, Yasuda R. Kinetics of endogenous CaMKII required for synaptic plasticity revealed by optogenetic kinase inhibitor. Neuron. 2017;94:37–47.e5. doi: 10.1016/j.neuron.2017.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima M, Barthel LK, Raymond PA. A self-renewing division of zebrafish muller glial cells generates neuronal progenitors that require N-cadherin to regenerate retinal neurons. Development. 2013;140:4510–4521. doi: 10.1242/dev.090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher JJ, Emmrich JV, Fricker M, Mander PK, Thery C, Brown GC. Phagocytosis executes delayed neuronal death after focal brain ischemia. Proc Natl Acad Sci U S A. 2013;110:E4098–107. doi: 10.1073/pnas.1308679110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CM, Ackerman KM, O’Hayer P, Bailey TJ, Gorsuch RA, Hyde DR. Tumor necrosis factor-alpha is produced by dying retinal neurons and is required for muller glia proliferation during zebrafish retinal regeneration. J Neurosci. 2013;33:6524–6539. doi: 10.1523/JNEUROSCI.3838-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norden C, Young S, Link BA, Harris WA. Actomyosin is the main driver of interkinetic nuclear migration in the retina. Cell. 2009;138:1195–1208. doi: 10.1016/j.cell.2009.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Narayanan R, Lee SH, Murata K, Hayashi Y. The role of CaMKII as an F-actin-bundling protein crucial for maintenance of dendritic spine structure. Proc Natl Acad Sci U S A. 2007;104:6418–6423. doi: 10.1073/pnas.0701656104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauls S, Geldmacher-Voss B, Campos-Ortega JA. A zebrafish histone variant H2A. F/Z and a transgenic H2A.F/Z:GFP fusion protein for in vivo studies of embryonic development. Dev Genes Evol. 2001;211:603–610. doi: 10.1007/s00427-001-0196-x. [DOI] [PubMed] [Google Scholar]
- Pearson RA, Luneborg NL, Becker DL, Mobbs P. Gap junctions modulate interkinetic nuclear movement in retinal progenitor cells. J Neurosci. 2005;25:10803–10814. doi: 10.1523/JNEUROSCI.2312-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro AC, da Silva AJ, Prado MA, CordeiroMdo N, Richardson M, Batista MC, de Castro Junior CJ, Massensini AR, Guatimosim C, Romano-Silva MA, Kushmerick C, Gomez MV. Phoneutria spider toxins block ischemia-induced glutamate release, neuronal death, and loss of neurotransmission in hippocampus. Hippocampus. 2009;19:1123–1129. doi: 10.1002/hipo.20580. [DOI] [PubMed] [Google Scholar]
- Poggi L, Vitorino M, Masai I, Harris WA. Influences on neural lineage and mode of division in the zebrafish retina in vivo. J Cell Biol. 2005;171:991–999. doi: 10.1083/jcb.200509098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell C, Cornblath E, Elsaeidi F, Wan J, Goldman D. Zebrafish muller glia-derived progenitors are multipotent, exhibit proliferative biases and regenerate excess neurons. Sci Rep. 2016;6:24851. doi: 10.1038/srep24851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Reifler A, Wan J, Goldman D. Application of cre-loxP recombination for lineage tracing of adult zebrafish retinal stem cells. Methods Mol Biol. 2012;884:129–140. doi: 10.1007/978-1-61779-848-1_8. [DOI] [PubMed] [Google Scholar]
- Renninger SL, Orger MB. Two-photon imaging of neural population activity in zebrafish. Methods. 2013;62:255–267. doi: 10.1016/j.ymeth.2013.05.016. [DOI] [PubMed] [Google Scholar]
- Sauer ME, Walker BE. Radioautographic study of interkinetic nuclear migration in the neural tube. Proc Soc Exp Biol Med. 1959;101:557–560. doi: 10.3181/00379727-101-25014. [DOI] [PubMed] [Google Scholar]
- Schenk J, Wilsch-Brauninger M, Calegari F, Huttner WB. Myosin II is required for interkinetic nuclear migration of neural progenitors. Proc Natl Acad Sci U S A. 2009;106:16487–16492. doi: 10.1073/pnas.0908928106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeter EH, Wong RO, Gregg RG. In vivo development of retinal ON-bipolar cell axonal terminals visualized in nyx::MYFP transgenic zebrafish. Vis Neurosci. 2006;23:833–843. doi: 10.1017/S0952523806230219. [DOI] [PubMed] [Google Scholar]
- Scudder KM, Christian GD, Ruzicka J. Flow injection fluorescence microscopy: A novel tool for the study of cells through controlled perfusion. Exp Cell Res. 1993;205:197–204. doi: 10.1006/excr.1993.1077. [DOI] [PubMed] [Google Scholar]
- Sousa C, Biber K, Michelucci A. Cellular and molecular characterization of microglia: A unique immune cell population. Front Immunol. 2017;8:198. doi: 10.3389/fimmu.2017.00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear PC, Erickson CA. Apical movement during interkinetic nuclear migration is a two-step process. Dev Biol. 2012;370:33–41. doi: 10.1016/j.ydbio.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess E, Bestvater F, Heckel-Pompey A, Toth K, Hacker M, Stobrawa G, Feurer T, Wotzlaw C, Berchner-Pfannschmidt U, Porwol T, Acker H. Two-photon excitation and emission spectra of the green fluorescent protein variants ECFP, EGFP and EYFP. J Microsc. 2005;217:200–204. doi: 10.1111/j.1365-2818.2005.01437.x. [DOI] [PubMed] [Google Scholar]
- Stence N, Waite M, Dailey ME. Dynamics of microglial activation: A confocal time-lapse analysis in hippocampal slices. Glia. 2001;33:256–266. [PubMed] [Google Scholar]
- Sugiyama M, Sakaue-Sawano A, Iimura T, Fukami K, Kitaguchi T, Kawakami K, Okamoto H, Higashijima S, Miyawaki A. Illuminating cell-cycle progression in the developing zebrafish embryo. Proc Natl Acad Sci U S A. 2009;106:20812–20817. doi: 10.1073/pnas.0906464106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki SC, Bleckert A, Williams PR, Takechi M, Kawamura S, Wong RO. Cone photoreceptor types in zebrafish are generated by symmetric terminal divisions of dedicated precursors. Proc Natl Acad Sci U S A. 2013;110:15109–15114. doi: 10.1073/pnas.1303551110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao K, Okamoto K, Nakagawa T, Neve RL, Nagai T, Miyawaki A, Hashikawa T, Kobayashi S, Hayashi Y. Visualization of synaptic Ca2+/calmodulin-dependent protein kinase II activity in living neurons. J Neurosci. 2005;25:3107–3112. doi: 10.1523/JNEUROSCI.0085-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JL, Ochocinska MJ, Hitchcock PF, Thummel R. Using the tg(nrd:Egfp)/albino zebrafish line to characterize in vivo expression of neurod. PLoS One. 2012;7:e29128. doi: 10.1371/journal.pone.0029128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel R, Kassen SC, Enright JM, Nelson CM, Montgomery JE, Hyde DR. Characterization of muller glia and neuronal progenitors during adult zebrafish retinal regeneration. Exp Eye Res. 2008;87:433–444. doi: 10.1016/j.exer.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiruchinapalli DM, Oleynikov Y, Kelic S, Shenoy SM, Hartley A, Stanton PK, Singer RH, Bassell GJ. Activity-dependent trafficking and dynamic localization of zipcode binding protein 1 and beta-actin mRNA in dendrites and spines of hippocampal neurons. J Neurosci. 2003;23:3251–3261. doi: 10.1523/JNEUROSCI.23-08-03251.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JW, Lian WN, Kemal S, Kriegstein AR, Vallee RB. Kinesin 3 and cytoplasmic dynein mediate interkinetic nuclear migration in neural stem cells. Nat Neurosci. 2010;13:1463–1471. doi: 10.1038/nn.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihtelic TS, Hyde DR. Light-induced rod and cone cell death and regeneration in the adult albino zebrafish (danio rerio) retina. J Neurobiol. 2000;44:289–307. doi: 10.1002/1097-4695(20000905)44:3<289::aid-neu1>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Vihtelic TS, Soverly JE, Kassen SC, Hyde DR. Retinal regional differences in photoreceptor cell death and regeneration in light-lesioned albino zebrafish. Exp Eye Res. 2006;82:558–575. doi: 10.1016/j.exer.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Weber A, Hochmann S, Cimalla P, Gartner M, Kuscha V, Hans S, Geffarth M, Kaslin J, Koch E, Brand M. Characterization of light lesion paradigms and optical coherence tomography as tools to study adult retina regeneration in zebrafish. PLoS One. 2013;8:e80483. doi: 10.1371/journal.pone.0080483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber IP, Ramos AP, Strzyz PJ, Leung LC, Young S, Norden C. Mitotic position and morphology of committed precursor cells in the zebrafish retina adapt to architectural changes upon tissue maturation. Cell Rep. 2014;7:386–397. doi: 10.1016/j.celrep.2014.03.014. [DOI] [PubMed] [Google Scholar]
- White DT, Sengupta S, Saxena MT, Xu Q, Hanes J, Ding D, Ji H, Mumm JS. Immunomodulation-accelerated neuronal regeneration following selective rod photoreceptor cell ablation in the zebrafish retina. Proc Natl Acad Sci U S A. 2017;114:E3719–E3728. doi: 10.1073/pnas.1617721114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SG, Wen W, Pillai-Kastoori L, Morris AC. Tracking the fate of her4 expressing cells in the regenerating retina using her4:Kaede zebrafish. Exp Eye Res. 2016;145:75–87. doi: 10.1016/j.exer.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt C, Bartoszek EM, Yaksi E. Methods for studying the zebrafish brain: Past, present and future. Eur J Neurosci. 2015;42:1746–1763. doi: 10.1111/ejn.12932. [DOI] [PubMed] [Google Scholar]
- Yoshimatsu T, D’Orazi FD, Gamlin CR, Suzuki SC, Suli A, Kimelman D, Raible DW, Wong RO. Presynaptic partner selection during retinal circuit reassembly varies with timing of neuronal regeneration in vivo. Nat Commun. 2016;7:10590. doi: 10.1038/ncomms10590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RW. Cell differentiation in the retina of the mouse. Anat Rec. 1985;212:199–205. doi: 10.1002/ar.1092120215. [DOI] [PubMed] [Google Scholar]
- Yu J, Lei K, Zhou M, Craft CM, Xu G, Xu T, Zhuang Y, Xu R, Han M. KASH protein syne-2/nesprin-2 and SUN proteins SUN1/2 mediate nuclear migration during mammalian retinal development. Hum Mol Genet. 2011;20:1061–1073. doi: 10.1093/hmg/ddq549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Zabel MK, Wang X, Ma W, Shah P, Fariss RN, Qian H, Parkhurst CN, Gan WB, Wong WT. Microglial phagocytosis of living photoreceptors contributes to inherited retinal degeneration. EMBO Mol Med. 2015a;7:1179–1197. doi: 10.15252/emmm.201505298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Wan X, Li Y, Zhou W, Peng L. Multiplexed 3D FRET imaging in deep tissue of live embryos. Sci Rep. 2015b;5:13991. doi: 10.1038/srep13991. [DOI] [PMC free article] [PubMed] [Google Scholar]