Abstract
C′ dots are fluorescent inorganic-organic hybrid nanoparticles synthesized in water comprised of a silica core with a covalently embedded near infrared dye, and a polyethylene glycol (PEG) outer layer. C′ dots containing the integrin specific ligand, cycloRGDyC, are the first of their kind particles approved for human clinical trials. In the continued clinical development of these nanoparticles, high-resolution analytical approaches are needed. Here we investigate the use of reversed phase high performance liquid chromatography (RP-HPLC) to analyze cycloRGDyC-Cy5-C′ dots. Given the stability and protein-like size, we reasoned that these nanoparticles would be compatible under RP-HPLC conditions typically used to characterize peptides and proteins. Our results show that RP-HPLC provides excellent resolution, showing significant heterogeneity of these nanoparticles. C′ dots also exhibit unusual peak profiles where RP-HPLC chromatogram peak shapes change from run to run, possibly due to the conformational heterogeneity or charge distribution of the particle surface due to the PEG groups. In addition we describe a novel thiol-mediated release of C′ dot ligands to directly estimate cycloRGDyC by exposing the particles to organic thiols. Ligand release is presumably afforded by a reverse Michael reaction mechanism.
Advances in nanotechnology have largely been driven by the development of novel materials with interesting properties emerging at the nanoscale. In biomedical research, nanoparticles in particular, have garnered attention largely due to their physical attributes – discrete and relatively stable particles in the nanometer size regime with at least one novel/superior property.1, 2 Their potential utility in biomedical applications, such as disease diagnosis, imaging, and drug delivery, are only now being realized as a number of products have reached the clinical stage.1–5
One of the major challenges with the development of nanoparticles as a viable clinical product is the arduous and complex process to rigorously assess their stability, toxicity, and activity at the preclinical stage.1–3,6,7 Given the wide range of particle morphologies, preclinical characterization of nanoparticles has largely relied on techniques used in the field of materials and polymer research, for example, UV-vis absorption spectroscopy, dynamic light scattering (DLS), zeta-potential, thermogravimetric analysis (TGA), or transmission electron microscopy (TEM), etc. These tools have provided necessary information regarding the size, shape, composition, and spectral properties of particles, respectively.8,9 More recently, chromatographic approaches, such as gel permeation/filtration or size exclusion chromatography (GPC or SEC), have also become integrated into the workflow of nanoparticle characterization and purification.9–11
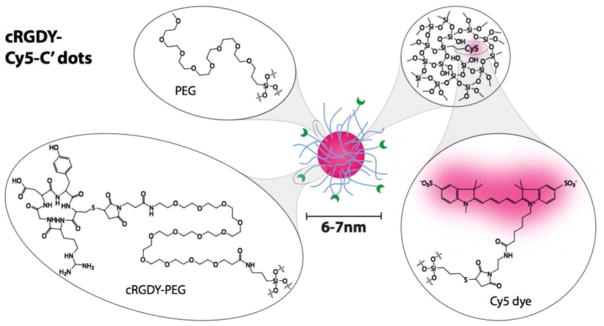
Our research has been focused on the clinical development of specific types of ultrasmall (i.e. sizes below 10 nm) fluorescent silica nanoparticles, known as C′ dots (Figure 1). C′ dots are synthesized in water as opposed to alcoholic solutions in earlier efforts leading to C dots.12 C′ dots are inorganic-organic hybrids comprised of a silica core and a polyethylene glycol (PEG) shell.10,13 For medical applications, embedded within the core are near infrared dyes (e.g., Cy5 or Cy5.5), which results in dramatically enhanced optical properties over the free dye as a result of the rigid silica matrix,10,14,15 while the PEG surface is partially functionalized with targeting peptides that can further be labeled with radioisotopes.16 Consequently, C′ dots have been shown to be clinically viable for dual-modality PET-optical imaging, and are a first-in-kind particle probe approved for human use.5 Additionally the stability, flexible surface chemistry, and particularly the small size of these particles – which affords good tissue penetration and predictable clearance profiles17 – make them ideal candidates as drug delivery vehicles, an application which is currently being explored.18
Figure 1.
CycloRGDyC-Cy5-C′ dots. These particles are comprised of four components: 1) the cycloRGDyC ligand; 2) PEG oligomers; 3) a silica core; 4) embedded NIR dye (Cy5).
Characterization of C′ dots have included the standard regiment of approaches as listed above: UV-vis absorption spectroscopy, DLS, zeta-potential, TGA, TEM, and GPC.10 Additionally, the embedded dyes of C′ dots have enabled the use of fluorescence correlation spectroscopy (FCS) to simultaneously obtain information on hydrodynamic size, particle concentration, and relative particle brightness,12,14 which has helped optimizing synthesis parameters by assessing batch-to-batch variations.
In this report, we introduce the use of RP-HPLC as a tool for C′ dot analysis. RP-HPLC has been used to a limited capacity for nanoparticle characterization,19–21 as most nanoparticle systems are incompatible due to their large size or relative instability (particles formed by self-assembly or aggregation, e.g. liposomes). We reasoned that C′ dots – hybrid particles that exhibit good stability, UV-vis (as well as fluorescence) activity, and are in the size regime of proteins – may be compatible with analytical HPLC approaches. Three detection systems are used for particle analysis: a diode array detector (DAD), an evaporative light scattering detector (ELSD), and an electrospray ionization-single quadropole mass detector (ESI-SQD). In addition to UV-vis absorption afforded by an embedded Cy5 dye (Figure 1), the ELSD will provide detection of non-UV active material, such as free PEG or silica-based components. While we do not anticipate mass detection of particles by the ESI-SQD, it was used to monitor the presence of lower molar mass impurities. In addition we describe a new approach to directly estimate the number of surface ligands (cycloRGDyC) on the particle through the use of organic thiols.
Experimental
Analytical RP-HPLC analysis of C′ dots
CycloRGDyC-Cy5-C′ dots were synthesized as described elsewhere10. Samples were analyzed on a Waters Alliance HPLC System or Autopure LCMS System (2767 Sample Manager, 2996 Photodiode Array Detector, 2420 ELS Detector, Micromass ZQ, 2525 Binary Gradient Module, Column Fluidics Organizer, 515 HPLC Pump, Pump Control Module II) using a linear gradient of 5–95% acetonitrile (ACN) in water with 0.05% trifluroacetic acid (TFA) or formic acid (FA) for 10 minutes at 1.2 mL/min on a C18 4.6×50mm, 300 Å, 5 μm reversed phase XBridge analytical column (Waters). Slower gradients runs were performed using a linear gradient of 5–95% ACN for 90 min. Samples were analyzed at 280 nm or 650 nm. 15 μM C′ dot stocks (in water) were diluted as needed for RP-HPLC analyses, and the injection volume used was 20–50 μL.
Thiol-mediated release assay
For a typical assay, 1 μL of 1 M DTT was added to 50 μL of 15 μM C′ dots and incubated at room temperature or 37°C overnight. To test glutathione, 25 μL of glutathione stock (100 mM) was added to 50 μL of 15 μM C′ dot and incubated overnight. The reaction was analysed by analytical HPLC. The released cycloRGDyC concentrations were determined by comparing peak integrations with a standard curve generated using unconjugated cycloRGDyC peptide.
Results and Discussion
HPLC analysis of C′ dots
For our initial studies, we sought to assess the overall compatibility of C′ dots in an analytical RP-HPLC system. We reasoned that the ~6 nm diameter of the particle is analogous to a globular protein, so a reversed phase C18 column with a 300 Å pore size was selected along with a standard linear gradient of 5–95% ACN in water (with 0.05% TFA). As shown in Figure 2, the particle elutes as a broad signal containing multiple peaks when monitored at 650 nm (C′ dots contain the near-infrared-dye Cy5, which exhibits a characteristic absorption maximum at 650 nm) and by the ELSD. Interestingly, peak shapes changed from run-to-run.
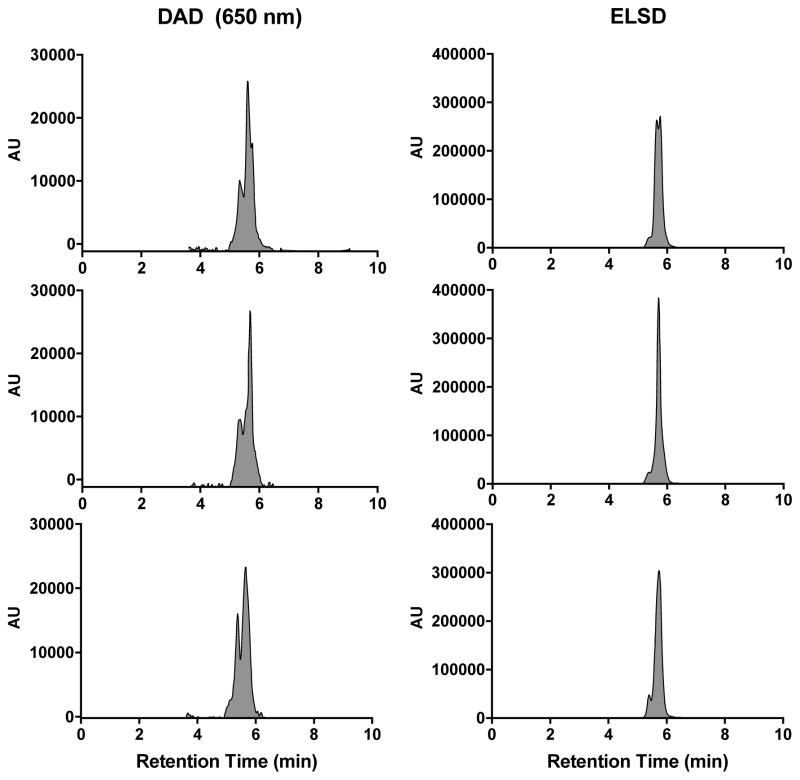
Figure 2.
RP-HPLC chromatograms of cyclo(RGDyC)-Cy5-C′ dots, 1. (a) DAD at 650 nm; (b) ELSD. Three consecutive analyses of the same sample (triplicate). 1.5 μM particle concentration; 20 μL injection volume; chromatographic conditions were 5–95% ACN (0.05% TFA) in 10 min.
When samples were analyzed in triplicate, the overall retention times were similar, but peak shapes differed significantly between each run (Figure 2). Remarkably, peak integrations proved to be very consistent, with relative standard deviations (RSD) of 7%, suggesting excellent run-to-run reproducibility (Table 1). Similar results were observed when monitored by the ELSD – peak profiles varied significantly from run-to-run, whereas peak integrations showed excellent consistency (RSD = 5%) (Figure 3b, Table 1).
Table 1.
Peak Integrations for Three Identical RP-HPLC Runs.
| Run | ||||
|---|---|---|---|---|
| 1 | 2 | 3 | +/− RSD (%) | |
| DAD | 11698 | 10218 | 11426 | 7 |
| ELSD | 85467 | 78326 | 86755 | 5 |
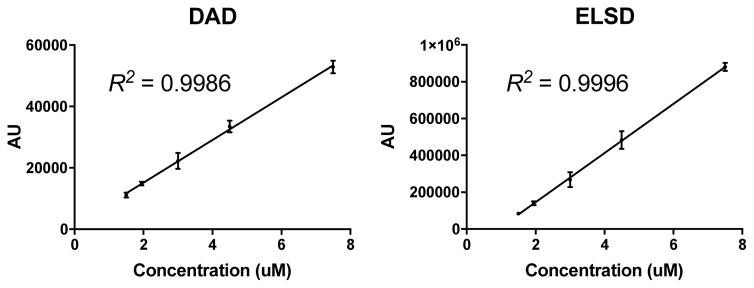
Figure 3.
Peak areas versus particle concentration. Five particle concentrations (1.5, 2.0, 3.0, 4.5, 8.0 μM) were analyzed by RP-HPLC (triplicate). 20 μL injection volume; chromatographic conditions were 5–95% ACN (0.05% TFA) in 10 min. Chromatogram peaks from the DAD (650 nm) and ELSD were integrated and plotted verses concentration.
To further highlight the consistency of the RP-HPLC for C′ dot analysis, a nanoparticle concentration curve was generated. Particle samples of varying concentrations ranging from 1.5 μM to 8 μM were analyzed and peak integrations at 650 nm where determined and plotted against concentrations. As shown in Figure 3A, an excellent linear response was observed with the DAD (R2 = 0.9986). Even concentrations down to 0.5 μM and up to 60 μM (not shown) were also in the linear range of the detector. Unexpectedly, the ELSD response also exhibited excellent linearity in the 1.5–8 μM range (R2 = 0.9996). In summary, both detectors provide consistent linearity in the ~1–10 μM concentration range.
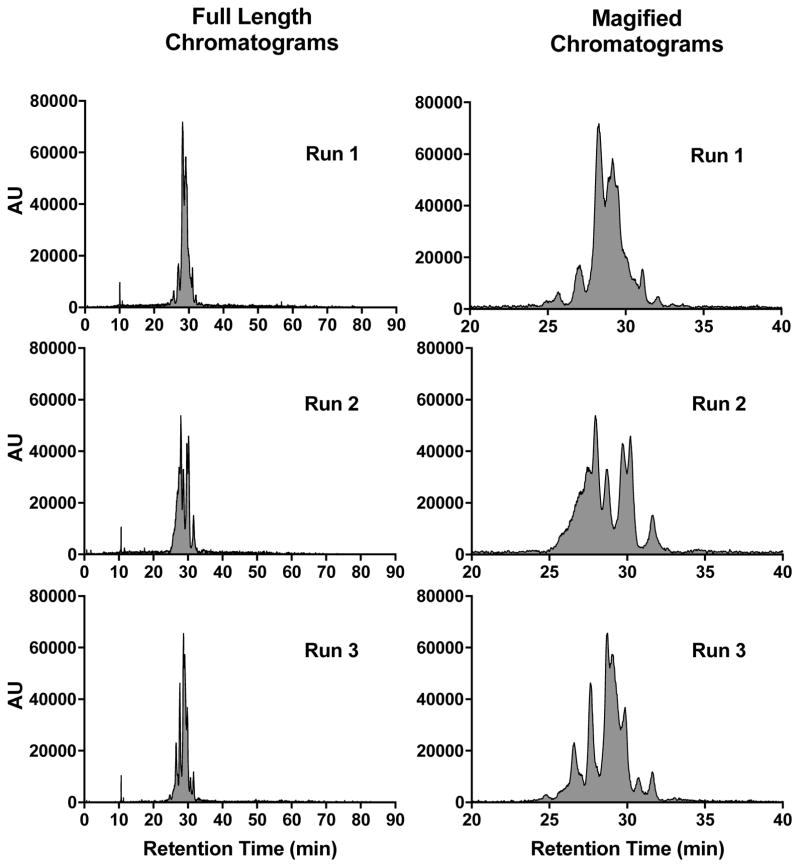
The peak shapes seen in the HPLC chromatograms indicate a heterogeneous mixture of compounds. To further investigate the nature of the observed peaks, nanoparticle samples were analyzed using a slower or more shallow gradient. While initial analyses (Figure 2) were performed at 5–95% ACN in 10 min (gradient of 9% ACN/min), Figure 4 shows chromatograms run at 5–95% ACN in water for 90 min, which translates into a gradient of 1% ACN/min. Under these conditions, the broad peaks as evident in Figure 2, are better resolved, revealing numerous peaks. However, once again, run-to-run variations in peak profiles are observed. Yet remarkably, the peak integrations at 650 nm demonstrate excellent consistency (RSD = 5.6%).
Figure 4.
RP-HPLC chromatograms of cycloRGDyC-C′ dot using a slower gradient. Three consecutive analyses of the same sample (triplicate) were performed. Panels on the left show the full-length chromatograms (0–90 min); panels on the right show a magnified portion of the chromatograms (20–40 min). RP-HPLC conditions were 5–95% ACN (0.05% TFA) in 90 min; DAD at 650 nm.
Possible explanations for the changing peak shapes are that the particle is chemically degrading or undergoing dynamic changes in surface charge or PEG conformations. Interestingly, C′ dots containing no cycloRGDyC peptide also exhibit the same changing peak profiles (data not shown). This suggests that the peptide ligands are not the main contributors to the observed peak features. The HPLC analyses are run under acidic conditions, so one possibility would be that the particles are being slowly hydrolyzed. However, cycloRGDyC-Cy5-C′ dots subjected to acidic RP-HPLC conditions (0.05% TFA in water) over time (1–16 hours), showed no distinction with regard to peak shape, retention time, or peak integrations (data not shown). Also, when these particles are analyzed by GPC, elution volumes were identical to non-treated particles. Weaker acid conditions (0.05% FA in water) showed no effect, as RP-HPLC chromatograms were consistent with TFA runs.
The run-to-run variations in peak profiles may be explained by the particle undergoing dynamic changes in surface charge and PEG conformations. This is consistent with our observations that the zeta-potential of C′ dots is never exactly zero, but always slightly negative.10 Additionally, the remaining free silanol groups on the C′ dot surfaces can undergo intramolecular interactions with the PEG groups. For example, the formation of hydrogen bonds between the ether oxygen of PEGs and silicic acid has been described.13,22 Taken together, this suggests that the particles are stable under HPLC conditions, and the changing peak profiles may be due to the dynamic nature of the PEG surface layer.
Thiol-mediated release of ligands
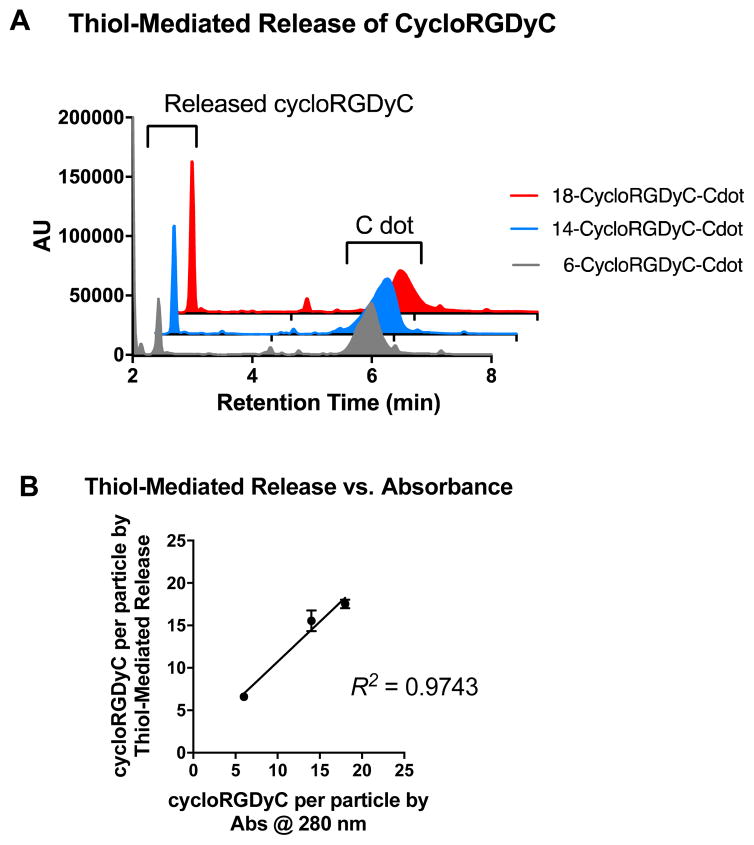
While exposing the cycloRGDyC-Cy5-C′ dots to various in vitro assay conditions and monitoring by RP-HPLC, we observed an unexpected phenomenon where the cycloRGDyC could be released in the presence of commonly used small organic thiols such as dithiothreitol (DTT). In the synthesis of the nanoparticle, cycloRGDyC is attached to heterobifunctional PEGs through a thiol-maleimide linkage (Figure 5).10 Recently it has been shown in the context of antibody drug conjugates that thiol-maleimides can be susceptible to reverse Michael reactions in the presence of other intermolecular thiols. These studies have shown this reaction to be highly sequence dependent on antibodies. To assess the stability of the thiol-maleimide linkage on the C′ dots, the particles were subjected to thiol-containing compounds DTT or glutathione for 24 hrs at 37 °C, then analyzed by LCMS (glutathione is ubiquitous in vivo tripeptide containing cysteine). Under these conditions we observed efficient release of the cycloRGDyC when DTT was present. However minimal release (<5%) was observed with glutathione (data not shown) at up to 48 hours. Furthermore, C′ dots incorporating different amounts of the ligand could be assessed using this method. Typically, ligand quantitation is estimated by UV absorption spectroscopy.10 Three C′ dots, estimated to contain on average 6, 14, or 18 cycloRGDyC peptides by UV, were treated with DTT and analyzed by RP-HPLC (Figure 6A). A standard curve with the unconjugated cycloRGDyC peptide was also generated. Peak integrations of the released peptides from the 6, 14 and 18 cycloRGDyC-Cy5-C′ dot samples were obtained; and their corresponding concentrations and ligand per particle values were determined from the standard curve. Figure 6B shows a plot comparing the levels of cycloRGDyC per particles as estimated by UV spectroscopy verses this thiol-mediated release assay, and good correlation is observed between these two methods. In addition to estimating ligand number or concentration, thiol-mediated release may provide insight into the spatial location of the ligands. Given the accessibility of the DTT to the maleimides, and subsequent release of the peptides, this supports the notion that the peptide is likely near or on the surface of the particle, and not embedded within, consistent with the schematic in Figure 1.
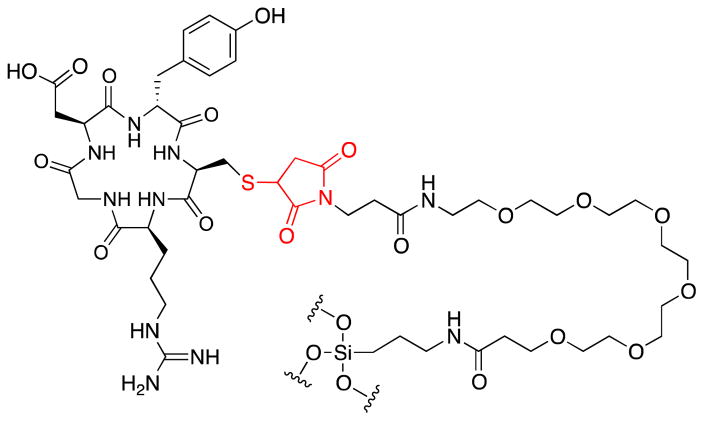
Figure 5.
Cyclo(RGDyC) on C′ dots. The cyclo(RGDyC) peptide is covalently attached to the particle through a cysteine-maleimido-PEG-silyl group. The thiol-maleimide group (red) may be susceptible to a reverse Michael reaction.
Figure 6.
Thiol-mediated release of cycloRGDyC from C′ dots. (A) C′ dots containing different numbers of cycloRDyC (6, grey; 14, blue; and 18, red) are treated with DTT then analyzed by RP-HPLC. (B) Plot comparing cycloRGDyC per particle estimates from UV absorbance verses thiol-mediated release
Conclusions
In this report, we expand the use of traditional analytical tools for the characterization of inorganic-organic hybrid nanoparticles to high resolution HPLC. In particular, we employ two RP-HPLC based analytical approaches for a specific type of ultrasmall silica based nanoparticles called C′ dots. As the clinical applications of these nanoparticles expand, such new approaches are needed to refine characterization and assessment of their exact composition and structure. While detailed peak shapes were inconsistent from run-to-run, which could represent different charged or conformational states of the PEGylated particle surface; other features were found to be highly reproducible, such as retention time and peak integrations, providing an overall predictable and linear response. Further optimization with solvent conditions may be needed (i.e. the use of different solvent additives such as buffers, salts or ion pairing reagents) and is currently ongoing. However, the data suggests that RP-HPLC may provide a high resolution means of separating distinct subpopulations of nanoparticles. An unexpected phenomenon was observed where cysteine-maleimides incorporated onto the particles were susceptible to low molar mass thiols, but not glutathione. Excess DTT was used to release cycloRGDyC from the C′ dot, and released peptide could be measured to obtain ligand to particle estimates.
Acknowledgments
The authors gratefully acknowledge Dr. Feng Chen for his thoughtful insights and suggestions; and support by the National Cancer Institute of the National Institutes of Health under Award Number U54CA199081. We would like to further acknowledge the NMR Core Facility at MSKCC and Hunter Mass Spectrometry for use of their instrumentation. This work made use of the Cornell Center for Materials Research shared facilities supported through the NSF MRSEC program (DMR-1120296), and the Nanobiotechnology Center shared research facilities at Cornell.
References
- 1.Dawidczyk CM, Kim C, Park JH, Russell LM, Lee KH, Pomper MG, Searson PC. J Controlled Release. 2014;187:133–144. doi: 10.1016/j.jconrel.2014.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ediriwickrema A, Saltzman WM. ACS Biomater Sci Eng. 2015;1:64–78. doi: 10.1021/ab500084g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eifler A, Thaxton CS. In: Biomedical Nanotechnology. Hurst SJ, editor. Vol. 726. Humana Press; 2011. pp. 325–338. ch. 21. [Google Scholar]
- 4.Sun T, Zhang YS, Pang B, Hyun DC, Yang M, Xia Y. Angew Chem, Int Ed. 2014;53:12320–12364. doi: 10.1002/anie.201403036. [DOI] [PubMed] [Google Scholar]
- 5.Phillips E, Penate-Medina O, Zanzonico PB, Carvajal RD, Mohan P, Ye Y, Humm J, Gönen M, Kalaigian H, Schöder H, Strauss HW, Larson SM, Wiesner U, Bradbury MS. Sci Transl Med. 2014;6:260ra149. doi: 10.1126/scitranslmed.3009524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bobo D, Robinson KJ, Islam J, Thurecht KJ, Corrie SR. Pharm Res. 2016;33:2373–2387. doi: 10.1007/s11095-016-1958-5. [DOI] [PubMed] [Google Scholar]
- 7.Cho EJ, Holback H, Liu KC, Abouelmagd SA, Park J, Yeo Y. Mol Pharm. 2013;10:2093–2110. doi: 10.1021/mp300697h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piao Y, Burns A, Kim J, Wiesner U, Hyeon T. Adv Funct Mater. 2008;18:3745–3758. [Google Scholar]
- 9.Lin PC, Lin S, Wang PC, Sridhar R. Biotechnol Adv. 2014;32:711–726. doi: 10.1016/j.biotechadv.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma K, Mendoza C, Hanson M, Werner-Zwanziger U, Zwanziger J, Wiesner U. Chem Mater. 2015;27:4119–4133. [Google Scholar]
- 11.Lopez-Serrano A, Olivas RM, Landaluze JS, Camara C. Anal Methods. 2014;6:38–56. [Google Scholar]
- 12.Ow H, Larson DR, Srivastava M, Baird BA, Webb WW, Wiesner U. Nano Lett. 2005;5:113–117. doi: 10.1021/nl0482478. [DOI] [PubMed] [Google Scholar]
- 13.Ma K, Zhang D, Cong Y, Wiesner U. Chem Mater. 2016;28:1537–1545. [Google Scholar]
- 14.Larson DR, Ow H, Vishwasrao HD, Heikal AA, Wiesner U, Webb WW. Chem Mater. 2008;20:2677–2684. [Google Scholar]
- 15.Herz E, Ow H, Bonner D, Burns A, Wiesner U. J Mater Chem. 2009;19:6341–6347. [Google Scholar]
- 16.Benezra M, Penate-Medina O, Zanzonico PB, Schaer D, Ow H, Burns A, DeStanchina E, Longo V, Herz E, Iyer S, Wolchok J, Larson SM, Wiesner U, Bradbury MS. J Clin Invest. 2011;121:2768–2780. doi: 10.1172/JCI45600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burns AA, Vider J, Ow H, Herz E, Penate-Medina O, Baumgart M, Larson SM, Wiesner U, Bradbury M. Nano Lett. 2008;9:442–448. doi: 10.1021/nl803405h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoo B, Ma K, Zhang L, Burns A, Sequeira S, Mellinghoff I, Brennan C, Wiesner U, Bradbury MS. Bioorg Med Chem. 2015;23:7119–7130. doi: 10.1016/j.bmc.2015.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.González-Fuenzalida RA, Moliner-Martínez Y, Molins-Legua C, Parada-Artigues V, Verdú-Andrés J, Campins-Falcó P. Anal Chem. 2016;88:1485–1493. doi: 10.1021/acs.analchem.5b04751. [DOI] [PubMed] [Google Scholar]
- 20.Xie S, Paau MC, Zhang Y, Shuang S, Chan W, Choi MMF. Nanoscale. 2012;4:5325–5332. doi: 10.1039/c2nr30984j. [DOI] [PubMed] [Google Scholar]
- 21.Truillet C, Lux F, Tillement O, Dugourd P, Antoine R. Anal Chem. 2013;85:10440–10447. doi: 10.1021/ac402429p. [DOI] [PubMed] [Google Scholar]
- 22.Preari M, Spinde K, Lazic J, Brunner E, Demadis KD. J Am Chem Soc. 2014;136:4236–4244. doi: 10.1021/ja411822s. [DOI] [PubMed] [Google Scholar]