Abstract
Calgranulin proteins are important mediators of innate immunity and are members of the S100 class of the EF-hand family of calcium binding proteins. Some S100 proteins have the capacity to bind transition metals with high affinity and effectively sequester them away from invading microbial pathogens in a process that is termed "nutritional immunity". S100A12 (EN-RAGE) binds both zinc and copper and is highly abundant in innate immune cells such as macrophages and neutrophils. We report a refined method for the expression, enrichment and purification of S100A12 in its active, metal-binding configuration. Utilization of this protein in bacterial growth and viability analyses reveals that S100A12 has antimicrobial activity against the bacterial pathogen, Helicobacter pylori. The antimicrobial activity is predicated on the zinc-binding activity of S100A12, which chelates nutrient zinc, thereby starving H. pylori which requires zinc for growth and proliferation.
Keywords: Biochemistry, Issue 123, Helicobacter pylori, nutritional immunity, S100A12, calgranulin C, zinc
Introduction
S100 proteins are a class of the EF-hand family of calcium binding proteins with a diverse array of functions1. They are expressed in a tissue and cell specific manner, and regulate a broad spectrum of cellular functions2,3. Unique to calcium binding proteins, S100 proteins exhibit both intracellular and extracellular functions4,5. Within the cell, Ca2+ binding induces a conformational change that exposes a hydrophobic surface that specifically targets protein binding partners6. This intracellular mechanism regulates important processes such as cell proliferation, differentiation and energy metabolism. In the extracellular milieu, S100 proteins exhibit two functions7. In one, they act as damage associated molecular pattern (DAMP) proteins and initiate a pro-inflammatory immune response through interaction with pattern recognition receptors8,9. Additionally, several members of the S100 protein class sequester transition metals, a function that serves to starve microbial pathogens in a process termed nutritional immunity10,11.
S100A12 (also known as calgranulin C and EN-RAGE) is highly expressed in macrophages and neutrophils and has been identified as a potential biomarker for inflammatory diseases12,13. In addition to binding calcium at its EF-Hand sites, S100A12 has two high affinity transition metal binding sites located at opposite ends of the dimer interface14,15. Each binding site is comprised of three histidine residues and one aspartic acid residue and can chelate zinc or copper16,17. Recently, we reported that S100A12-dependent zinc starvation is important in regulating Helicobacter pylori growth and the activity of pro-inflammatory virulence factors18.
H. pylori infects the stomach of about half of the world's human population; making it arguably one of the most successful bacterial pathogens19. Infection with H. pylori can lead to significant gastric disease outcomes including gastritis, peptic and duodenal ulcer, mucosa associated lymphoid tissue (MALT) lymphoma, and invasive gastric adenocarcinoma (stomach cancer). Stomach cancer is the leading cause of non-cardia cancer-associated death in the world, and the single biggest associated risk factor for stomach cancer is infection with H. pylori.
H. pylori persists in the gastric niche despite a robust immune response to the pathogen, underscoring the need for a better understanding of immune mechanisms of controlling this bacterial infection20,21,22,23. H. pylori-associated inflammation is characterized by a profound infiltration of polymorphonuclear cells, or neutrophils, which deposit a repertoire of antimicrobial proteins, including S100A12, at the site of infection18,24,25. In an effort to understand the complex dialogue between host and pathogen, we sought to refine the technique to purify S100A12 and use it to study the antimicrobial affect it exerts upon this medically relevant pathogen. The protocol below outlines an improved technique for S100A12 purification in its biologically active state; capable of binding nutrient metals with high affinity and chelating them away from invading microorganisms. Furthermore, the methods below highlight the utility of this protein as a critical reagent to study the mechanism by which innate antimicrobial molecules restrict the growth of bacterial pathogens.
S100A-family proteins have gained appreciation as an important group of innate immune system molecules which participate in immune signaling as well as host defense27. The most well-studied of these is calprotectin (MRP-8/14, calgranulin A/B, S100A8/A9)28,29,30. Calprotectin is a neutrophil-associated protein which forms a heterodimer of the S100A8 and S100A9 subunits which binds transition metals at the dimer interface31. Calprotectin has been shown to possess two metal binding sites: Site 1 can bind Zn2+, Mn2+, or Fe2+, and Site 2 can bind Zn2+ 31,32. Numerous reports have demonstrated that calprotectin has antimicrobial activities against diverse pathogens including Staphylococcus aureus, Candida albicans, Acinetobacter baumannii, Klebsiella pneumoniae, Escherichia coli, and H. pylori, and that the inhibitory effects are due to the metal chelation activity of calprotectin25,28,29.
Previous work demonstrated calprotectin exerts numerous activities on H. pylori including altering the lipid A structure in the outer membrane, repressing the cag-Type IV secretion system (which is a major proinflammatory virulence factor within H. pylori), inducing biofilm formation, and repressing H. pylori growth and viability in a dose-dependent manner25,33. Furthermore, genetic and biochemical assays revealed the antibacterial activity of calprotectin against H. pylori was largely derived from its ability to bind nutrient zinc25. H. pylori requires zinc, as was determined by previous research which utilized a chemically defined medium to ascertain the micronutrient requirements for this pathogen to grow and proliferate34. Additionally, calprotectin was highly abundant within H. pylori-infected tissues, and associated with neutrophilic infiltrates, indicating the host may be employing calprotectin as an antimicrobial strategy during infection and subsequent inflammation25,35.
Recent evidence from unbiased proteomics screening techniques suggests that under conditions where reactive oxygen species are plentiful, calprotectin undergoes post-translational modifications which alter the hexa-histidine binding site, thereby inhibiting the metal-binding activity of the protein36. As such, we hypothesized that other S100A-family proteins among the wide repertoire of these molecules could potentially act as auxiliary metal-chelators. We selected S100A12 for further study because it was not identified in the aforementioned screen for post-translational modification, it has the capacity to bind zinc, and it is highly abundant in human tissues derived from H. pylori-infected individuals.
Our work indicates that S100A12 can inhibit H. pylori growth and viability in a dose-dependent manner in the G27 strain of H. pylori, and that the antimicrobial activity of this protein can be reversed by the addition of excess nutrient zinc. This work complements our previous work indicating S100A12 exerted antimicrobial activity against PMSS1 and 7.13 strains of H. pylori, demonstrating its broad antibacterial activity against numerous clinical isolates and laboratory-adapted strains of H. pylori18. Together, these results confirm the importance of S100A12 as a mechanism to control bacterial growth and proliferation via nutritional immunity. Future studies of this important host-bacterial interaction could include exploiting the activity of S100A12 to reduce bacterial burden within host tissues, or determining the contribution of this protein to immune signaling in the context of H. pylori infection.
Protocol
1. Expression of S100A12
Transform competent BL21 DE3 cells with a pGEMEX-S100a12 plasmid using a standard heat shock protocol18. Add 1 to 5 µL of plasmid to 50 µL of bacteria in a microcentrifuge tube on ice. Incubate for 20 min.
Heat shock the cells at 42 °C for 30 s.
Incubate the cells on ice for 2 min.
Add 500 µL of SOC media to the cells. Incubate at 37 °C with shaking at 250 rpm on an orbital shaker for 1 h.
Plate 150 µL of the transformation reaction on LB-agar medium (supplemented with 100 µg/mL ampicillin). Incubate for 12-16 h at 37 °C.
Pick one colony. Inoculate 2 mL of LB (supplemented with 100 µg/mL ampicillin).
Incubate for 4-6 h at 37 °C on an orbital shaker (300 rpm). The OD600 should read between 1-3 absorbance units.
Add 500 µL of starter culture to 50 mL of ZYM-5052 autoinduction media26 supplemented with 100 µg/mL of ampicillin. For best aeration and maximum expression, use a 250 mL baffled Erlenmeyer flask. Shake (300 rpm) for 24 h, at 37 °C.
Transfer bacterial suspension to a centrifuge tube. Pellet the cells by centrifugation (4,000 x g, 10 min) at 4 °C.
Decant the media, log sample, and store cell paste at -80 °C. Sample is stable for years.
2. Purification of S100A12 Using Low Pressure Chromatography
Resuspend cells in 30 mL of 20 mM Tris, pH 8.0.
Sonicate the suspension on ice to lyse cells. Use ~20 W output, 5 s on and 5 s off cycle for 5 min.
Transfer the solution to high-speed centrifuge tubes. Clarify the cell lysate by centrifugation, 20,000 x g for 30 min at 4 °C.
Decant the supernatant and transfer to a clean 100 mL polypropylene beaker. Cool the solution by placing the beaker on ice. Add a stir bar and slowly add 11.20 g of ammonium sulfate. Allow the solution to stir on ice for an additional 1 h. This will create a 60% solution of ammonium sulfate and precipitate most of the E. coli endogenous proteins. S100A12 will remain soluble.
Centrifuge the solution at 4 °C, 20,000 x g for 20 min to pellet the precipitated protein.
Decant the supernatant and transfer to dialysis tubing (MWCO 3,500 kDa). Dialyze against 1 L of 20 mM Tris, pH 8.0 at 4 °C. Change the dialysis buffer twice. Allow 4 h in between changes.
- Anion exchange chromatography
- Perform chromatography on a low pressure system. Typical flow rate is 1 mL/min.
- Equilibrate a 5 mL Sepharose column with 10 mL of 20 mM Tris, pH 8.0.
- Load the ~40 mL of S100A12 solution using the sample pump (collect the flow through).
- Wash the column with 10 mL of 20 mM Tris, pH 8.
- Develop the column with a 0-30% gradient (Buffer B is 20 mM Tris, pH 8.0, 1 M NaCl) over 19 column volumes (CV, 95 mL). Collect 5 mL fractions.
- Take a 10 µL aliquot of each fraction and analyze using MES SDS PAGE with Coomassie staining. Run gel using constant voltage (20 V/cm) for 30 min.
- Pool fractions containing S100A12. S100A12 runs at ~10 kDa protein on a denaturing gel.
- Concentrate fractions to 5 mL using an ultrafiltration device (MWCO 10 kDa). Centrifuge at 3,000 x g for ~8 min. Take the top fraction.
- Size-exclusion chromatography
- Equilibrate S75 column with 1 CV (120 mL) of 20 mM Tris pH 8, 100 mM NaCl.
- Inject < 5 mL of concentrated S100A12 fractions (from the ion exchange chromatography).
- Develop the column at a flow rate of 1 mL/m over 120 mL. Collect 5 mL fractions.
- Take a 10 µL aliquot of each fraction and analyze using SDS PAGE with Coomassie staining. Validate protein identity using western blot or mass spectrometry (calculated molecular mass of monomeric subunit 10,575.0 Da, measured 10575.4 Da).
- Pool fractions
- Measure the absorbance of the protein A280 using a spectrophotometer. Use the SEC buffer as a blank. The calculated extinction coefficient for S100A12 homodimer is 5960 M-1 cm-1. NOTE: Typical yields are 35-45 mg of S100A12 per 50 mL of culture.
- Aliquot S100A12 into 1.5 mL microcentrifuge tubes (1 mg/tube), flash freeze in liquid nitrogen and store at -80 °C.
3. Antimicrobial Activity Assays
Streak H. pylori strain G27 onto tryptic soy agar plates supplemented with 5% sheep blood (blood agar plates). Grow 2-3 days at 37 °C in room air supplemented with 5% carbon dioxide.
Inoculate H. pylori into Brucella broth supplemented with 1x cholesterol. Culture overnight shaking (250 rpm) at 37 °C in room air supplemented with 5% carbon dioxide.
Dilute H. pylori 1:10 into 50% Brucella broth, 50% calprotectin buffer plus 1x cholesterol and culture in medium alone or supplemented with 100 µM zinc chloride plus 0, 100 or 1,000 µg/mL of purified S100A12. Culture overnight shaking (250 rpm) at 37 °C in room air supplemented with 5% carbon dioxide.
The following day, perform serial dilutions and plate onto blood agar plates. Allow bacterial colonies to grow for 2-3 days at 37 °C in room air supplemented with 5% carbon dioxide. Enumerate the colony forming units to calculate bacterial growth in the presence or absence of S100A12 and/or exogenous zinc.
Representative Results
S100A12 expression and purification
A three-step purification produced ~40 mg of recombinant S100A12 from 50 mL of bacterial culture. The first step was an ammonium sulfate precipitation of endogenous E. coli proteins. This step was followed by anion-exchange chromatography (Figure1A). The protein is tracked by a SDS-PAGE stained with Coomassie Brilliant Blue (Figure 1B). The last step of the purification procedure involved pooling fractions containing S100A12 for size-exclusion chromatography which separates proteins by molecular weight and shape (Figure 2A). S100A12 is a homodimer (92 amino acids per subunit) and has a total molecular weight of about 21 kDa. Fractions collected from size-exclusion chromatography were analyzed by SDS-PAGE and visualized by Coomassie staining (Figure 2B).
S100A12 represses bacterial growth via zinc chelation activity
To investigate the antimicrobial activity of S100A12, bacterial viability analyses were performed via quantitative microbiological culture techniques (Figure 3). Enumeration of bacterial cells reveals that exposure to 100 µg/mL of S100A12 in the presence or absence of an exogenous source of nutrient zinc does not significantly inhibit bacterial viability compared to controls without S100A12 added (P >0.05, One way ANOVA). However, exposure to 1000 µg/mL of S100A12 in medium alone results in a 69-fold decrease in bacterial viability compared to medium alone (P = 0.0193, Student's t Test, P >0.05 One Way ANOVA); a result that was reversed by the addition of an exogenous source of nutrient zinc (P = 0.023, Student's t Test). These results demonstrate the antimicrobial activity of S100A12 is dependent upon its zinc sequestration activity.
 Figure 1: Cell lysis and S100A12 purification by anion exchange chromatography. (a) Chromatogram of ion exchange purification. Trace of UV absorbance at 280 nm shown in blue, salt gradient depicted in pink, collected fractions marked in red. (b) SDS PAGE gel of purification steps. Lanes: 1) molecular weight standard 2) soluble lysate fraction 3) ammonium sulfate pellet 4) ammonium sulfate supernatant 5-14) Q chromatography fractions 6-15. Please click here to view a larger version of this figure.
Figure 1: Cell lysis and S100A12 purification by anion exchange chromatography. (a) Chromatogram of ion exchange purification. Trace of UV absorbance at 280 nm shown in blue, salt gradient depicted in pink, collected fractions marked in red. (b) SDS PAGE gel of purification steps. Lanes: 1) molecular weight standard 2) soluble lysate fraction 3) ammonium sulfate pellet 4) ammonium sulfate supernatant 5-14) Q chromatography fractions 6-15. Please click here to view a larger version of this figure.
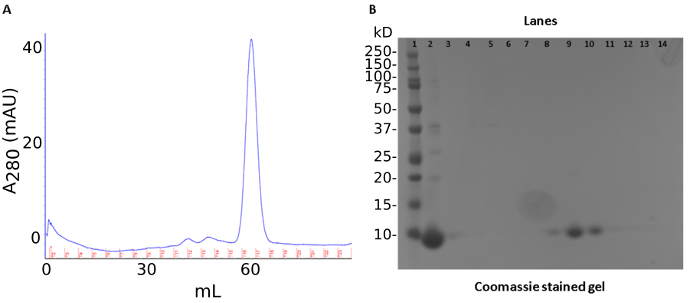 Figure 2: Size exclusion chromatography results of S100A12 purification. (a) Chromatogram of size-exclusion results. Trace of UV absorbance at 280 nm shown in blue, collected fractions marked in red. (b) SDS PAGE gel of size-exclusion chromatography. Lanes: 1) molecule weight marker 2) sample load 3-15) fractions 10-21. Please click here to view a larger version of this figure.
Figure 2: Size exclusion chromatography results of S100A12 purification. (a) Chromatogram of size-exclusion results. Trace of UV absorbance at 280 nm shown in blue, collected fractions marked in red. (b) SDS PAGE gel of size-exclusion chromatography. Lanes: 1) molecule weight marker 2) sample load 3-15) fractions 10-21. Please click here to view a larger version of this figure.
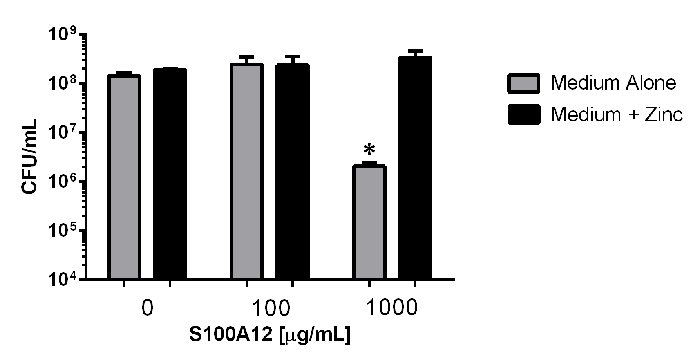 Figure 3: Quantitative culture analyses of bacterial viability in response to exposure to S100A12-dependent zinc chelation. Bacteria were exposed to 0, 100, or 100 µg/mL of S100A12 in medium alone (grey bars), or medium supplemented with 100 µM zinc chloride (black bars). Exposure to 1,000 µg/mL in the absence of an exogenous source of nutrient zinc results in significant inhibition of bacterial viability (*P <0.05, Student's t test compared to medium + zinc condition). Please click here to view a larger version of this figure.
Figure 3: Quantitative culture analyses of bacterial viability in response to exposure to S100A12-dependent zinc chelation. Bacteria were exposed to 0, 100, or 100 µg/mL of S100A12 in medium alone (grey bars), or medium supplemented with 100 µM zinc chloride (black bars). Exposure to 1,000 µg/mL in the absence of an exogenous source of nutrient zinc results in significant inhibition of bacterial viability (*P <0.05, Student's t test compared to medium + zinc condition). Please click here to view a larger version of this figure.
Discussion
An efficient protocol for both expression and purification of human S100A12 is presented. The E. coli expression system is the most common tool used for the production of recombinant proteins, particularly when mg quantities are required for biochemical and biophysical studies. A key enhancement of the procedure described here is the use of auto-induction media26 which increases the yield of purified protein by a factor of almost thirty as compared to expression with standard Luria-broth media18. Additionally, auto-induction media greatly simplifies the protein-expression workflow. Using conventional growth media, cultures must be continuously monitored so that an inducing agent can be added during the exponential growth phase. There is no need to monitor the doubling times when using auto-induction media. The cultures are allowed to grow to saturation. During the initial stages of growth with auto-induction media, E. coli uses glucose as a carbon source. Once the glucose is depleted, the bacteria switch to lactose as a carbon source which induces protein expression26. The composition of the auto-induction media is exquisitely tuned to allow for the growth of high density cultures and therefore increased expression of recombinant protein. In our experience with S100A12, auto-induction media greatly increases the amount of expressed protein, however we caution that this may be protein dependent and should be experimentally verified.
S100A12 is expressed in the cytoplasmic fraction of E. coli which suggests that it is soluble and well folded. The first step of the purification involves adding a high percentage of ammonium sulfate which precipitates many of the endogenous E. coli proteins. This step leverages the high stability and solubility of the S100 family of proteins. S100A12 is moderately acidic (pI 5.81). Thus S100A12 is purified further with a strong anion exchange resin. The last step of the purification process is a size-exclusion chromatography column which ensures that S100A12 is monodisperse and dimeric. This step of the protocol is crucial as S100A12 has been shown to form soluble oligomers15 and it is unknown what affect oligomerization may have on its antimicrobial activity. Since members of the S100 class share high sequence and structural homology, their physical properties are similar27. Hence, this method of purification of S100A12 may be broadly applicable to all S100 proteins that are expressed in the soluble fraction of E. coli.
Previous work has demonstrated that S100 proteins, such as calprotectin, have antimicrobial activity against bacterial pathogens such as H. pylori, and that this activity is dependent upon zinc-binding activity25. The antimicrobial activity of S100A12 is also zinc dependent, but the growth inhibition demonstrated in bacterial growth assays reveals that significantly more S100A12 is required to inhibit growth compared to other S100-family proteins such as calprotectin. Thus, it is critical to use higher concentrations of S100A12 than calprotectin to achieve similar phenotypes (growth inhibition or alterations in virulence). Future applications of this protocol could be applied to determine global changes in bacterial gene expression, metabolomic, or proteomic alterations in response to the metal sequestration imposed by S100A12.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
This research was supported by the Department of Veterans Affairs Career Development Award 1IK2BX001701, CTSA award UL1TR000445 from the National Center for Advancing Translational Sciences, the National Science Foundation Award Numbers 1547757 and 1400969, and NIH grant GM05551. Its contents are solely the responsibility of the authors and do not necessarily represent official views of the National Center for Advancing Translational Sciences or the National Institutes of Health.
References
- Schiopu A, Cotoi OS. S100A8 and S100A9: DAMPs at the crossroads between innate immunity, traditional risk factors, and cardiovascular disease. Mediators Inflamm. 2013;2013:828354. doi: 10.1155/2013/828354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, et al. S100A9 induced inflammatory responses are mediated by distinct damage associated molecular patterns (DAMP) receptors in vitro and in vivo. PLoS One. 2015;10(2):e0115828. doi: 10.1371/journal.pone.0115828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernov AV, et al. The calcium-binding proteins S100A8 and S100A9 initiate the early inflammatory program in injured peripheral nerves. J Biol Chem. 2015;290(18):11771–11784. doi: 10.1074/jbc.M114.622316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanjul M, et al. Presence of MRP8 and MRP14 in pancreatic cell lines: differential expression and localization in CFPAC-1 cells. Am J Physiol. 1995;268(5 Pt 1):C1241–C1251. doi: 10.1152/ajpcell.1995.268.5.C1241. [DOI] [PubMed] [Google Scholar]
- Vogl T, Gharibyan AL, Morozova-Roche LA. Pro-inflammatory S100A8 and S100A9 proteins: self-assembly into multifunctional native and amyloid complexes. Int J Mol Sci. 2012;13(3):2893–2917. doi: 10.3390/ijms13032893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SP, Shaw GS. A change-in-hand mechanism for S100 signalling. Biochem Cell Biol. 1998;76(2-3):324–333. doi: 10.1139/bcb-76-2-3-324. [DOI] [PubMed] [Google Scholar]
- Bresnick AR, Weber DJ, Zimmer DB. S100 proteins in cancer. Nat Rev Cancer. 2015;15(2):96–109. doi: 10.1038/nrc3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc E, Heizmann CW. The importance of Ca2+/Zn2+ signaling S100 proteins and RAGE in translational medicine. Front Biosci (Schol Ed) 2011;3:1232–1262. doi: 10.2741/223. [DOI] [PubMed] [Google Scholar]
- Goyette J, Geczy CL. Inflammation-associated S100 proteins: new mechanisms that regulate function. Amino Acids. 2011;41(4):821–842. doi: 10.1007/s00726-010-0528-0. [DOI] [PubMed] [Google Scholar]
- Kehl-Fie TE, Skaar EP. Nutritional immunity beyond iron: a role for manganese and zinc. Curr Opin Chem Biol. 2010;14(2):218–224. doi: 10.1016/j.cbpa.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zackular JP, Chazin WJ, Skaar EP. Nutritional Immunity: S100 Proteins at the Host-Pathogen Interface. J Biol Chem. 2015;290(31):18991–18998. doi: 10.1074/jbc.R115.645085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Realegeno S, et al. S100A12 Is Part of the Antimicrobial Network against Mycobacterium leprae in Human Macrophages. PLoS Pathog. 2016;12(6):e1005705. doi: 10.1371/journal.ppat.1005705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera C, McNeil HP, Geczy CL. S100 Calgranulins in inflammatory arthritis. Immunol Cell Biol. 2010;88(1):41–49. doi: 10.1038/icb.2009.88. [DOI] [PubMed] [Google Scholar]
- Moroz OV, et al. Structure of the human S100A12-copper complex: implications for host-parasite defence. Acta Crystallogr D Biol Crystallogr. 2003;59(Pt 5):859–867. doi: 10.1107/s0907444903004700. [DOI] [PubMed] [Google Scholar]
- Moroz OV, Blagova EV, Wilkinson AJ, Wilson KS, Bronstein IB. The crystal structures of human S100A12 in apo form and in complex with zinc: new insights into S100A12 oligomerisation. J Mol Biol. 2009;391(3):536–551. doi: 10.1016/j.jmb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Moroz OV, Dodson GG, Wilson KS, Lukanidin E, Bronstein IB. Multiple structural states of S100A12: A key to its functional diversity. Microsc Res Tech. 2003;60(6):581–592. doi: 10.1002/jemt.10300. [DOI] [PubMed] [Google Scholar]
- Cunden LS, Gaillard A, Nolan EM. Calcium Ions Tune the Zinc-Sequestering Properties and Antimicrobial Activity of Human S100A12. Chem Sci. 2016;7(2):1338–1348. doi: 10.1039/c5sc03655k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley KP, et al. The Human Antimicrobial Protein Calgranulin C Participates in Control of Helicobacter pylori Growth and Regulation of Virulence. Infect Immun. 2015;83(7):2944–2956. doi: 10.1128/IAI.00544-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cover TL, Blaser MJ. Helicobacter pylori in health and disease. Gastroenterology. 2009;136(6):1863–1873. doi: 10.1053/j.gastro.2009.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tham KT, et al. Helicobacter pylori genotypes, host factors, and gastric mucosal histopathology in peptic ulcer disease. Hum Pathol. 2001;32(3):264–273. doi: 10.1053/hupa.2001.21136. [DOI] [PubMed] [Google Scholar]
- Wotherspoon AC, Ortiz-Hidalgo C, Falzon MR, Isaacson PG. Helicobacter pylori-associated gastritis and primary B-cell gastric lymphoma. Lancet. 1991;338(8776):1175–1176. doi: 10.1016/0140-6736(91)92035-z. [DOI] [PubMed] [Google Scholar]
- Correa P, Piazuelo MB. The gastric precancerous cascade. J Dig Dis. 2012;13(1):2–9. doi: 10.1111/j.1751-2980.2011.00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Martel C, Forman D, Plummer M. Gastric cancer: epidemiology and risk factors. Gastroenterol Clin North Am. 2013;42(2):219–240. doi: 10.1016/j.gtc.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Algood HM, Gallo-Romero J, Wilson KT, Peek RM, Cover TL. Host response to Helicobacter pylori infection before initiation of the adaptive immune response. FEMS Immunol Med Microbiol. 2007;51(3):577–586. doi: 10.1111/j.1574-695X.2007.00338.x. [DOI] [PubMed] [Google Scholar]
- Gaddy JA, et al. The host protein calprotectin modulates the Helicobacter pylori cag type IV secretion system via zinc sequestration. PLoS Pathog. 2014;10(10):e1004450. doi: 10.1371/journal.ppat.1004450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW. Stable expression clones and auto-induction for protein production in E. coli. Methods Mol Biol. 2014;1091:17–32. doi: 10.1007/978-1-62703-691-7_2. [DOI] [PubMed] [Google Scholar]
- Gilston BA, Skaar EP, Chazin WJ. Binding of transition metals to S100 proteins. Sci China Life Sci. 2016;59(8):792–801. doi: 10.1007/s11427-016-5088-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin BD, et al. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science. 2008;319(5865):962–965. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- Kehl-Fie TE, et al. Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus. Cell Host Microbe. 2011;10(2):158–164. doi: 10.1016/j.chom.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, et al. Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell Host Microbe. 2012;11(3):227–239. doi: 10.1016/j.chom.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damo SM, et al. Molecular basis for manganese sequestration by calprotectin and roles in the innate immune response to invading bacterial pathogens. Proc Natl Acad Sci U S A. 2013;110(10):3841–3846. doi: 10.1073/pnas.1220341110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashige TG, Zhang B, Krebs C, Nolan EM. Human calprotectin is an iron-sequestering host-defense protein. Nat Chem Biol. 2015;11(10):765–771. doi: 10.1038/nchembio.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaddy JA, et al. Helicobacter pylori Resists the Antimicrobial Activity of Calprotectin via Lipid A Modification and Associated Biofilm Formation. MBio. 2015;6(6):e01349–e01315. doi: 10.1128/mBio.01349-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testerman TL, Conn PB, Mobley HL, McGee DJ. Nutritional requirements and antibiotic resistance patterns of Helicobacter species in chemically defined media. J Clin Microbiol. 2006;44(5):1650–1658. doi: 10.1128/JCM.44.5.1650-1658.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach ST, Mitchell HM, Geczy CL, Sherman PM, Day AS. S100 calgranulin proteins S100A8, S100A9 and S100A12 are expressed in the inflamed gastric mucosa of Helicobacter pylori-infected children. Can J Gastroenterol. 2008;22(5):461–464. doi: 10.1155/2008/308942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie-Grantham RP, et al. Myeloperoxidase-dependent lipid peroxidation promotes the oxidative modification of cytosolic proteins in phagocytic neutrophils. J Biol Chem. 2015;290(15):9896–9905. doi: 10.1074/jbc.M114.613422. [DOI] [PMC free article] [PubMed] [Google Scholar]


