Abstract
During meiosis, accurate separation of maternal and paternal chromosomes requires that they first be connected to one another through homologous recombination. Meiotic recombination has many intriguing but poorly understood features that distinguish it from recombination in mitotically dividing cells, and several of these features depend on the meiosis-specific DNA strand exchange protein Dmc1. Many questions about this protein have arisen since its discovery more than a decade ago, but recent genetic and biochemical breakthroughs promise to shed light on the unique behaviours and functions of this central player in the remarkable chromosome dynamics of meiosis.
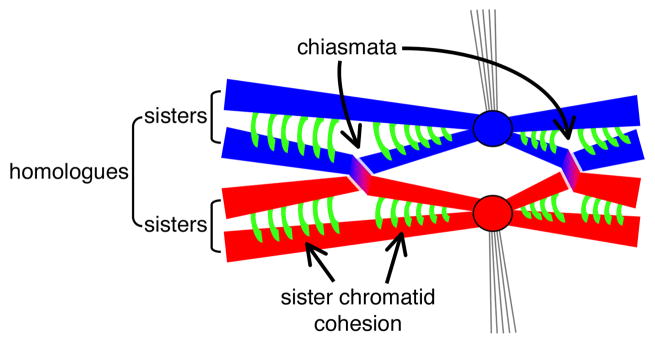
Sexual organisms must halve the chromosome number in gametes to maintain genome size with each generation. This goal is achieved through meiosis, in which two rounds of chromosome segregation follow a single round of DNA replication. The first meiotic division separates maternal and paternal copies of each chromosome, but in order for this segregation to occur properly, the chromosomes must first pair with their correct partner and then become physically connected so that they orient together on the meiotic spindle. Connection is established by the exchange of homologous chromosome arms (the point of crossover being called a “chiasma”) plus cohesion between sister chromatids1, 2 (Fig. 1). Exchange utilizes a specialized pathway of homologous recombination that repairs DNA double-strand breaks (DSBs)3—basically, the cell deliberately damages its own DNA and then uses the repair process to lock homologous chromosomes together. A central step in recombination involves proteins related to bacterial RecA, which catalyze the pairing and exchange of DNA strands between single-stranded DNA (ssDNA) formed at the break and intact, homologous double-stranded DNA (dsDNA)4. Most eukaryotes have two RecA homologues, the ubiquitous Rad51 and its meiosis-specific counterpart Dmc1 (ref. 5).
Figure 1. Connections formed between homologous chromosomes during meiosis.
Meiotic cells of most sexual organisms contain two copies of most chromosomes, one from each of the parents (red and blue). After DNA replication, each chromosome comprises a pair of sister chromatids held together by cohesion complexes (green). Sister centromeres (circles) attach as a single unit to microtubules (thin lines) from a spindle pole (not shown). Exchange of chromosome arms between non-sister chromatids yields a chiasma. Dissolution of sister chromatid cohesion along the arms allows the homologues to separate at the first meiotic division (not shown).
Meiotic recombination is subject to many layers of control that dictate, for example, the choice of DNA substrate for DSB repair, the distribution and timing of recombination events, and the integration of recombination with higher order chromosome structures2, 6. Executing these controls requires meiosis-specific factors (such as Dmc1) in addition to proteins (such as Rad51) that function during normal repair of DSBs in other cell types. The widely conserved Dmc1 plays a critical role in meiosis in most sexual organisms, but what its molecular functions are, and how and why it differs from its cousin Rad51 are not well understood5. Recent genetic studies have uncovered new information about roles of Dmc1 and accessory proteins that modulate Dmc1 function in vivo. These advances, coupled with breakthroughs in biochemical analysis of Dmc1 activities, promise to answer longstanding questions about the mechanism of meiotic recombination.
The challenges of meiotic recombination
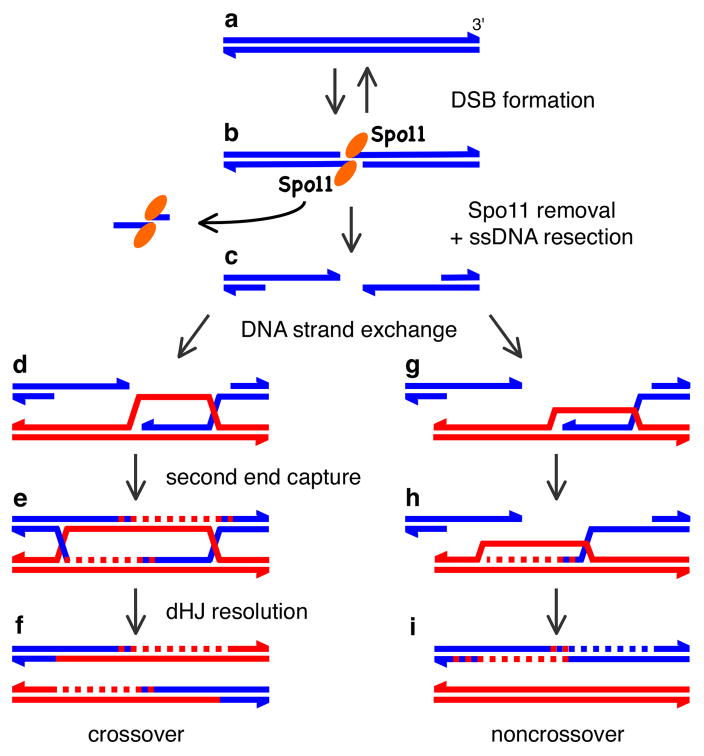
The molecular events of meiotic recombination have been most thoroughly described in the budding yeast Saccharomyces cerevisiae (Fig. 2). DSBs are formed by meiosis-specific Spo11 protein, which cuts DNA via a topoisomerase-like reaction to generate covalent protein-DNA linkages to the 5′ DNA ends on either side of the break3. After Spo11 is removed from the DNA ends, one or more exonucleases process the DSB to generate 3′ ssDNA tails. DNA strand exchange proteins bind these tails, forming helical nucleoprotein filaments which are the active structures that carry out the search for a homologous target and catalyze strand exchange. The detailed biochemical steps in this process are described below. Further processing of the strand exchange intermediates yields intact recombinant products that either have exchanged the flanking DNA arms (crossovers) or have not undergone exchange (noncrossovers) (Fig. 2).
Figure 2. DNA events in meiotic recombination.
a–c, Presynapsis. Spo11 (elipses) cleaves dsDNA, yielding a covalent Spo11-DNA complex. Endonuclease releases Spo11 bound to a short oligonucleotide, and 5′ DNA strands are degraded to yield 3′ ssDNA tails which are bound by Rad51 and Dmc1 (not shown). d–f, Crossover formation. d, Invasion of ssDNA from one end of the break forms an asymmetric strand exchange intermediate. e, DNA synthesis (dashed line) is primed from the invading 3′ end, the second DSB end is captured and primes DNA synthesis. Ligation yields a pair of Holliday junctions (dHJ). f, Resolution yields a mature product with exchanged flanking DNA. g–h, A noncrossover pathway. Strand invasion (g) and DNA synthesis (h) are inferred but have not been directly detected. A transient strand invasion complex may be dissociated, perhaps by DNA helicases, allowing newly synthesized DNA to anneal to complementary ssDNA on the other side of the break (i). Further DNA synthesis and ligation yield a mature noncrossover product.
The task of DNA strand exchange proteins in meiosis is remarkable. Each DSB generates about 1 kb of ssDNA, for which the homologous target duplex must be located and engaged. In yeast, this is the equivalent of searching through a 10-km-long string to find a particular 20-cm-long region. In human cells, that 20-cm stretch would need to be picked out of some 2500 km, enough to reach from London to Moscow. The problem is more complex, however, because within that 2500 km length would be three exact matches to the target (one on the sister chromatid, and one on each of the two chromatids of the homologous chromosome), but only the two targets on the homologue can be used. This strong preference for interhomologue recombination in meiosis is important because intersister recombination would not join homologues. This bias contrasts with mitotic cells, in which the sister chromatid is greatly preferred as the target for Rad51-mediated recombination7. The problem gets even worse because every meiotic cell makes many DSBs (hundreds per cell in yeast and mouse)8, 9. Thus, repair has to occur simultaneously at numerous places in each meiosis. Finally, the search for homology and the catalysis of strand exchange must be spatially and temporally coordinated with the development of higher order chromosome structures (e.g., synaptonemal complex, chromosome axes, and chromatin loops) that are required for the recombination event to successfully connect the homologues2, 6. All of this is accomplished in just a few hours in yeast and a few days in mammals. With all these issues to consider, it is clear why meiotic recombination is so tightly controlled, and why this process has fascinated chromosome biologists for decades.
The bacterial DNA strand exchange protein RecA had already been extensively characterized4 when the first eukaryotic homologues were identified in S. cerevisiae10–12. The widely conserved Rad51 protein gets its name from the radiation-sensitive phenotype caused by mutations in the gene. Not surprisingly, then, Rad51 is required for most homologous recombination reactions in both mitotic and meiotic cells13. In contrast, Dmc1 (“disrupted meiotic cDNA”) is meiosis-specific and, in many organisms, dmc1 mutants display reduced or absent meiotic recombination5. It was appreciated early that Dmc1 activities must be central to the unique aspects of meiotic (as opposed to mitotic) recombination10, 14. As such, several fundamental questions have stood since its discovery: Which aspects of meiotic recombination reflect specialized roles for Dmc1? Is it a DNA strand exchange protein, and what are the intrinsic biochemical properties that differentiate it from Rad51? What are the extrinsic factors that control and direct Dmc1, and to what extent do these dictate the unique properties of Dmc1?
Genetics and cytology of Dmc1
The physiological function and specialized roles of Dmc1 have been best characterized in S. cerevisiae5. In one commonly used laboratory strain (the “SK1” background), dmc1 mutants show a near complete block to recombination10. When the DNA intermediates of recombination were assayed, no stable strand invasion or double-Holliday junction formation was detected14, 15 (Fig. 2). Based in part on these observations, it was suggested that Dmc1 promotes an interhomologue-only recombination pathway that is unique to meiosis14; this role was also suggested by the observation that dmc1 mutants have increased recombination between ectopic sequence repeats (i.e., homologous sequences located at positions other than the equivalent position on the homologous chromosome)16. rad51 mutants are likewise profoundly defective for meiotic recombination, revealing that both DNA strand exchange proteins contribute critical, nonoverlapping functions5. In every organism examined, Rad51 and Dmc1 are components of multiprotein complexes, or nodules, that form on meiotic chromosomes at the sites where DSBs are being repaired8, 9.
A striking hallmark of meiotic recombination in most organisms is that crossovers are not randomly distributed along chromosomes. Instead, the presence of a crossover makes it less likely that another will form nearby17. This phenomenon, known as crossover interference, is as yet poorly understood, but Dmc1 may be involved since interference is lost or diminished when Dmc1 is absent18, 19.
Interestingly, the 5′ strands of DSBs are more extensively processed by exonuclease(s) when Dmc1 and/or Rad51 is missing10, 11, 14. This finding indicates that the nuclease(s) and DNA strand exchange proteins are coordinated in some fashion, for example through competition for the same substrate or by direct protein-protein interaction. This coordination likely serves to strike a balance between too little processing (and thus, not enough ssDNA to carry out the homology search and strand exchange) and too much processing (which may be deleterious for integrity of the genome). The idea of a hand-off from the DSB processing activity to the strand exchange proteins is attractive because functional coupling of sequential steps in the pathway might help meiotic DSB repair operate so efficiently that it can handle larger numbers of DSBs than would be tolerated by a mitotic cell.
Dmc1 is important for meiotic recombination in many organisms; for example, mice with targeted mutation of the Dmc1 gene are sterile and show hallmarks of poorly repaired DSBs20. However, Dmc1 is not absolutely essential in all circumstances. Some organisms, such as Drosophila melanogaster, Caenorhabditis elegans, and Neurospora crassa, have Rad51 but lack a Dmc1 orthologue17. In another frequently used S. cerevisiae laboratory strain (the “BR” background), and in Schizosaccharomyces pombe, dmc1 mutants still exhibit significant levels of meiotic recombination21, 22. Moreover, the recombination defect in S. cerevisiae dmc1 mutants can be largely suppressed by overexpression of Rad51 or Rad54 (a protein that stimulates Rad51; see below)19, 23. These findings may indicate that Rad51 and Dmc1 operate in multiple meiotic recombination pathways that only partially overlap in budding yeast19. However, it is also possible that under normal circumstances the two proteins function only together, and that only aberrant recombination occurs when one is missing.
Dmc1-mediated DNA strand exchange
While in vivo studies identify aspects of meiotic recombination that are influenced by Dmc1, they do not reveal the biochemical mechanism of Dmc1 action. Given its homology to RecA, it was long suspected that Dmc1 would have DNA strand exchange activity9, 24, but only relatively recently has this been demonstrated. Strand exchange by RecA-related proteins can be divided into three phases4 (Fig. 3a). During the presynaptic phase, the strand exchange protein assembles on ssDNA, creating a helical nucleoprotein filament in which the DNA is stretched and underwound relative to B-form DNA. Formation of a ternary complex of the ssDNA-protein filament with dsDNA initiates the synaptic phase. Juxtaposition of three DNA strands within the synaptic filament permits rapid homology sampling through transient Watson-Crick base pairing between the ssDNA and the complementary strand of the duplex partner. Sequential cycles of binding, sampling, and release of dsDNA are performed until homology is detected. A stable DNA joint is then formed by intertwining of the ssDNA with its complement from the homologous target. During the postsynaptic phase, the exchange of DNA strands is completed and the filament is dismantled. The generation of mature recombinant products (i.e., intact DNA duplexes with or without reciprocal exchange of flanking sequences) requires many other postsynaptic processes such as more extensive strand exchange via migration of branched DNA structures (D loops or Holliday junctions); resolution of branched DNA intermediates by helicases, nucleases, and/or topoisomerases; DNA synthesis; and resealing of DNA strand interruptions with DNA ligase13, 25.
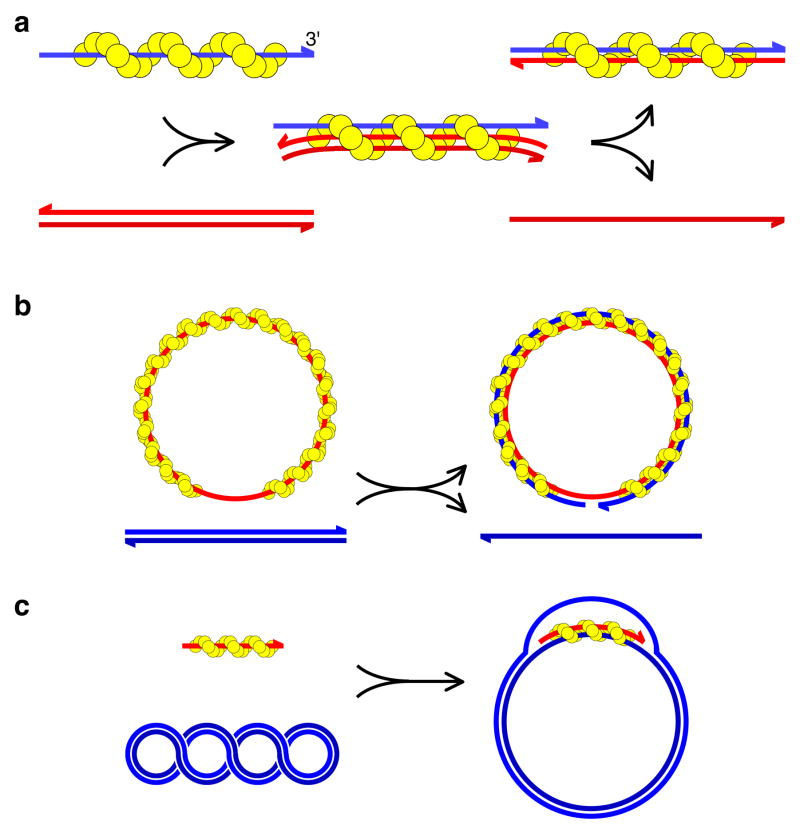
Figure 3. The DNA strand exchange reaction.
a, Three stages of strand exchange: presynaptic helical filament on ssDNA (left); synaptic complex with homologous dsDNA (middle); post-synaptic stage with the original ssDNA fully base-paired with the complementary strand from the dsDNA donor (right). b, c, Typical in vitro DNA strand exchange assay systems. b, Strand transfer from a linear duplex to a circular nucleoprotein filament. c, D-loop assay. Invasion of a short nucleoprotein filament into a negatively supercoiled circular duplex, creating a bubble, or D-loop.
Two types of in vitro DNA strand exchange assay are often used. The first detects transfer of one strand of a linear dsDNA onto a circular single-stranded nucleoprotein complex, creating a nicked duplex circle and a linear ssDNA (Fig. 3b). (Note that this situation contrasts with recombination in vivo, where a 3′ DNA end is provided by the initiating ssDNA.) The second assay detects invasion of a short ssDNA nucleoprotein complex into a negatively supercoiled duplex circle, displacing the noncomplementary strand into a D-loop structure (Fig. 3c). Because RecA has high specificity for binding ssDNA over dsDNA, strand exchange is efficient even when all reaction components are mixed simultaneously4. In contrast, Dmc1 is more sensitive to the order of addition because its ability to bind dsDNA can lead to non-productive complexes being formed26.
In early studies, Dmc1 displayed poor strand exchange activity relative to RecA or Rad51 (refs 27–30). Although RecA and its other homologues (eukaryotic Rad51, bacteriophage UvsX, and archaeal RadA) form hexameric to octameric rings in solution, the active oligomeric form for strand exchange is a helical nucleoprotein filament4. Electron microscopy of Dmc1, however, revealed nucleoprotein complexes that were exclusively rings, never helices29, 31 (Fig. 4a). The lack of helical filaments was surprising because Dmc1 has high sequence conservation with RecA-related proteins and because biophysical studies revealed that Dmc1 and Rad51 recognise homology in the same way32. It was suggested that rings were an artefact caused by lack of essential cofactors32, but models for ring-based DNA strand exchange were also proposed33.
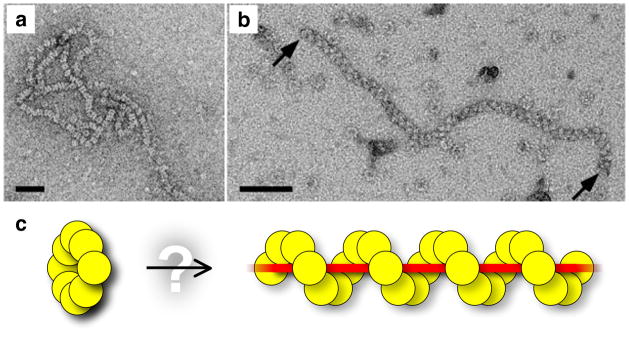
Figure 4. Oligomeric structure of Dmc1 nucleoprotein complexes.
a, b, Electron micrographs of Dmc1-DNA complexes (from refs 26, 31, with permission). Scale bars, 50 nm. a, Stacks of octameric Dmc1 rings on DNA. b, A helical Dmc1-ssDNA filament, similar to structures created by other RecA family members. c, A Dmc1 ring and a right-handed helical nucleoprotein filament. The dynamic relationship between rings and helical filaments is not known, but it is possible that filaments are formed by the opening and deposition of Dmc1 rings.
More recently, a robust Dmc1-mediated strand exchange reaction has been described, which differs from previous studies in using physiological pH and higher salt concentrations26, 34–36. Under these conditions, helical Dmc1-ssDNA filaments were formed26, 35, 37 (Fig. 4b), strongly indicating that the filament is the active oligomeric form for DNA strand exchange. Inefficient filament formation probably accounts for the weak strand exchange activity of earlier studies.
Yet, is there a function for the Dmc1 ring? It has been proposed that stacks of Dmc1 rings on DNA might be converted into an active helix, perhaps upon binding ATP38. As Dmc1 rings are oriented with alternating head-to-tail polarity31, 33, however, it is impossible to convert a stack of rings into a continuous helical filament without dismantling and reorienting at least half of the DNA-bound protein. A more likely possibility is that free, but not DNA-bound, rings are the form of Dmc1 (and of other RecA relatives) that assembles on DNA (Fig. 4c).
Ca2+ stimulates both filament formation on ssDNA and DNA strand exchange by bacterially expressed human and yeast Dmc1 proteins35, 36. Ca2+ also stimulates Rad51 (ref 39), so it has been suggested that Dmc1 and Rad51 activity might be modulated in vivo by variation in intracellular calcium levels35, 36. However, the free Ca2+ concentration required for optimal Dmc1 stimulation (~100 μM) is much higher than the free concentration in the cytosol (0.1–1 μM in yeast), and Ca2+ was not necessary for efficient DNA strand exchange by human Dmc1 expressed in insect cells and assayed in higher salt concentrations26. Therefore it is possible that Ca2+ acts as a nonphysiological stimulant, perhaps substituting for more physiological monovalent cations and/or mimicking the effects of accessory proteins.
DNA strand exchange proteins RecA, Rad51 and Dmc1 require nucleotide cofactor, but whether Dmc1 requires ATP hydrolysis remains controversial. Some studies indicate a strong requirement for ATP hydrolysis26, 28, 34, whereas others show strand exchange even when hydrolysis is hindered27, 35, 36, 40–42. Notably, neither RecA nor Rad51 requires ATP hydrolysis for strand invasion, but both require energy to turn over and release product DNA4.
The flurry of recent papers describing robust Dmc1-catalysed DNA strand exchange represent an important landmark because they open the door to a more detailed mechanistic understanding of Dmc1. The key question is, what are the intrinsic biochemical differences that functionally distinguish Dmc1 from Rad51? One apparent difference is the greater sensitivity of Dmc1 to reaction conditions in vitro, which seems to be related, at least in part, to the dynamics of ring vs. filament formation. At the moment, it is not obvious if or how these properties relate to the unique requirements of meiotic recombination. The challenge thus will be to sort out whether these are meaningful differences (as opposed to by-products of suboptimal in vitro conditions), and if so, to translate the differences into functional consequences in vivo.
Modulation of Dmc1 activity by extrinsic factors
RecA family members are remarkable enzymes, but do not function alone—RecA and Rad51 are profoundly affected by accessory proteins in vivo and in vitro13, 43. These accessory factors can function at one or more steps (presynaptic, synaptic, postsynaptic), and they serve to enhance (or sometimes restrain) catalytic efficiency and/or appropriately target strand exchange activities. Likewise, Dmc1 is also highly modulated5. Understanding how these factors affect Dmc1 is critical to understanding the mechanisms of the extremely regulated pathways of meiotic recombination.
RPA and recombination mediators
The ssDNA-binding protein RPA is required for efficient strand exchange by Rad51 but its effect is complex43. In positive terms, RPA minimises secondary structure in ssDNA, promoting Rad51 filament formation; it also sequesters both free ssDNA substrate and the displaced single-strand following strand exchange, preventing either from competing with dsDNA. However, RPA can also directly compete with Rad51 for binding to ssDNA, which is inhibitory. RPA is required for extensive Dmc1-catalysed strand exchange, presumably for the same reasons as for Rad51 (ref 26).
In vivo, RPA likely competes with Dmc1 for binding to ssDNA at meiotic DSBs. For Rad51-dependent reactions in vitro, inclusion of Rad52 or a complex of Rad55 and Rad57 overcomes this inhibitory effect and promotes assembly of Rad51 filaments on RPA-coated ssDNA13, 43. Proteins such as these are generally referred to as recombination ‘mediators’43. It is not yet known if they directly stimulate Dmc1 as well, but all are involved in meiotic recombination (at least in some organisms)13. The tumour suppressor protein Brca2 has also been implicated as a Rad51 mediator44. In many organisms, Brca2-deficient mutants have meiotic recombination defects45–48 and, in plants at least, Brca2 and Dmc1 interact physically46, 49.
Rdh54(Tid1)
Genetic studies in yeast indicate an important role in meiotic recombination for Rdh54 (also known as Tid1), a homologue of the DNA repair protein Rad54. Rdh54 is required for the timely conversion of DSBs into mature recombinant products50. In its absence, there is decreased colocalisation of Dmc1 and Rad51 in cytological nodules on chromosomes51, presumably reflecting a role for Rdh54 in dissociating Dmc1 from duplex DNA not associated with DSBs (D.K. Bishop, pers. comm.). Rad54 is less important in the meiotic context in yeast, so the role of Rdh54 is distinct50, 51. Rdh54 promotes D-loop formation by Rad51 in vitro52, 53, so it may function similarly with Dmc1. Fission and budding yeast Rdh54 and Dmc1 interact54, 55. Mammals have two known Rad54 family members, Rad54 and Rad54B. Human Rad54B interacts with and stimulates Dmc1 in vitro26, but the significance of this interaction is not yet clear because there is little or no meiotic recombination defect in mice lacking Rad54B, Rad54, or both56.
Insight into possible biochemical functions of Rdh54 with Dmc1 comes from consideration of its relative, Rad54, a member of the SWI2/SNF2 family of ATPases which appears to stimulate Rad51 activity through multiple mechanisms13, 43. Rad54 couples ATP hydrolysis with translocation along the DNA, thereby generating superhelical torsion in the DNA57, 58. Rad51 stimulates this activity, and the topological changes in the DNA in turn promote the ability of Rad51 to catalyse strand invasion and D-loop formation57, 58. Rad54 also possesses chromatin remodelling activity, stimulated by Rad51, indicating that another Rad54 function is to enhance the ability of Rad51 to target chromatinised dsDNA for strand exchange59, 60. Finally, Rad54 dismantles Rad51 complexes bound to dsDNA61; this activity may be useful post-synaptically to recycle limiting amounts of Rad51.
Hop2 and Mnd1
Genetic studies in yeast and mouse have identified a chromosome-associated, heterodimeric complex of Hop2 and Mnd1 proteins. Loss of these proteins causes chromosomes to synapse nonhomologously and meiotic DSBs to persist40, 62–67. Purified human, mouse, and budding yeast Hop2-Mnd1 complexes all stimulate D-loop formation and, where tested, DNA strand exchange by the Dmc1 proteins from the same organism40, 42, 68. The mammalian complexes also stimulate Rad5142, 68. Whether yeast Hop2-Mnd1 affects Rad51 activities has not been reported, but hop2 and mnd1 mutant phenotypes are much closer to those of dmc1 mutants than rad51, leading to the hypothesis that Hop2-Mnd1 functions primarily with Dmc1 (refs 40, 65, 66). Indeed, organisms that do not contain a Dmc1 orthologue also lack Hop2 and Mnd117. However, because Rad51 and Dmc1 are so closely tied together (see below), it is plausible that Hop2-Mnd1 could be specific for a Dmc1-dependent pathway and yet function biochemically to stimulate both Rad51 and Dmc1.
The molecular function of Hop2-Mnd1 remains unclear. The biochemical studies are consistent with direct effects on catalytic activities of Dmc1 and/or Rad51. A presynaptic role in aiding the binding of Dmc1 to ssDNA has been suggested42, but the genetics and biochemistry are currently most consistent with a synaptic and/or post-synaptic role40, 42, 64, 66, 67. Klein and colleagues have suggested a different model, in which Hop2-Mnd1 promotes Dmc1 activity indirectly by acting on the chromatin and/or higher order chromosome structures of the homologous target66. This model is motivated in part by the observation that Hop2-Mnd1 associates with chromatin independent of DSB formation, and does not co-localize with DSB sites or with Rad51 and Dmc1 complexes66.
Mei5 and Sae3
The evolutionarily conserved Mei5 and Sae3 proteins of budding yeast form DSB-dependent chromosomal complexes that colocalise with each other and with Dmc1 in a mutually dependent manner69, 70. Dmc1, Mei5, and Sae3 physically interact, and Mei5 and Sae3 co-purify as a complex69. Unrepaired meiotic DSBs accumulate in mei5 and sae3 mutants, indicating a defect in strand exchange69, 70. Rad51 foci form in the absence of Mei5, Sae3, or Dmc1, suggesting that the role of Mei5–Sae3 is Dmc1-specific69, 70. S. pombe contains two Mei5 homologues, Swi2 and Sfr1, and one Sae3 homologue, Swi5. Unlike in budding yeast, these proteins interact with Rhp51 (the S. pombe Rad51 orthologue), and function in recombination in both vegetative and meiotic cells71, 72.
The biochemical function(s) of Mei5-Sae3 is not yet known. The fact that Dmc1 is not loaded onto chromosomes in mei5 and sae3 mutants (unlike in hop2 and mnd1 mutants) suggests a direct presynaptic role promoting Dmc1 filament formation69, 70. Perhaps Mei5-Sae3 converts inactive Dmc1 rings into active helical filaments, or helps load Dmc1 onto RPA-coated ssDNA69 (as Rad52, Rad55, and Rad57 perform for Rad51). Indeed, genetic experiments in S. pombe support a mediator role for Swi5-Sfr1 in Rhp51-mediated recombination71. Now that Mei5 and Sae3 can be purified69, these ideas can be tested.
Rad51
The function of Dmc1 in meiotic recombination is closely coordinated with Rad51 (ref 5). As noted above, both proteins localize together to the sites where DSBs are being repaired8, 9. In the absence of Rad51, yeast Dmc1 localization is impaired, although Rad51 localization seems normal in the absence of Dmc1 (refs 9, 24). Precisely how the proteins are coordinated is not yet clear. Based in part on colocalization experiments, it has been proposed that they work together by forming mixed nucleoprotein filaments8. Homo-polymeric Rad51 and Dmc1 filaments arranged consecutively on the same ssDNA could also be envisioned. On the other hand, side-by-side immunostaining patterns have been observed for the two proteins under at least some circumstances on spread meiotic chromosomes8, 51, leading to the hypothesis that Rad51 and Dmc1 bind to opposite ends of each DSB15, 51. The idea of asymmetrical protein binding is attractive because the two ends of a DSB themselves behave differently—one end carries out the initial strand invasion, while the other end is captured in a separate reaction15 (see Fig. 2). If this idea is correct, it raises a couple of interesting questions: Which protein is responsible for initial strand invasion, and how is the asymmetric distribution of the proteins accomplished? It has been proposed that the Dmc1 nucleoprotein filament is the end which invades first15, although this remains to be verified. Recent studies of the early steps in DSB processing, when Spo11 is removed from its covalent attachment to DNA ends, have led to the hypothesis that asymmetry is set up very early, at or prior to DSB formation73. This also awaits confirmation.
Structural components of meiotic chromosomes
Meiotic recombination and large-scale chromosome structures are highly integrated, in part because a functional chiasma itself is a multicomponent structure comprising local exchange of homologous DNA duplexes (i.e., recombination) plus exchange of the proteinaceous axes of the homologues and local separation of sister chromatids2, 6 (Fig. 1). Achieving this integration requires crosstalk spanning size scales that differ by orders of magnitude, i.e., between the enzymes executing the DNA events of recombination and the structural proteins that make up μm-scale chromosome structures. The detailed molecular mechanisms involved are poorly understood. An extensive discussion is beyond the scope of this review (for further information see 1, 2, 6, 74), but a few points pertaining specifically to Dmc1 will be mentioned here.
The Red1 protein in S. cerevisiae provides one type of functional connection between Dmc1 and chromosome organization. Red1 is a major structural component of meiotic chromosomes75. In vivo, one function of Red1 is to promote, directly or indirectly, the loading of Dmc1 onto ssDNA at meiotic DSBs74. Another role of Red1 is to prevent stable DNA strand exchange between homologues when Dmc1 is absent14, 15. Whereas DSBs persist in dmc1 cells, cells lacking both Dmc1 and Red1 are able to repair meiotic DSBs by Rad51-mediated recombination between sister chromatids instead of between homologues. Thus, it appears that interplay between Dmc1 and Red1 is integral to the interhomologue bias of meiotic recombination.
Another connection between Dmc1 and chromosome structure is through Rec8, an evolutionarily conserved, meiosis-specific subunit of cohesin, the multiprotein complex that holds sister chromatids together in both mitosis and meiosis1, 76. In addition to defects in sister chromatid cohesion, rec8 mutants in S. cerevisiae have recombination phenotypes similar in some respects to dmc1 mutants, including inefficient repair and over-digestion by exonucleases76. Recombination defects are also apparent in a Rec8-defective mouse, suggesting that at least some of the roles of this protein in recombination are conserved77.
Outlook
This is an exciting time for students of meiotic chromosome dynamics in general, and of meiotic recombination in particular. The field faces several challenges. On the biological front, there is a need for more information about the specific processes influenced by Dmc1 (e.g., interhomologue bias, crossover interference), particularly in organisms other than budding yeast. Which aspects of Dmc1 function are conserved, and which are not? How does recombination differ in organisms that have Dmc1 versus those that do not? On the biochemical front, there is a need for deeper mechanistic understanding of mediators and other accessory proteins that influence Dmc1 activities. How do they modify Dmc1 activity? What is the interplay between Rad51 and Dmc1, and how do other factors impinge?
The biggest challenge, however, will be to connect the dots between the biochemical properties of Dmc1 and the in vivo functions revealed by genetics. One route to this goal will be to develop in vitro reactions that recapitulate increasingly complex aspects of chromosome behaviour, such as the connection between Dmc1 and chromosome structure proteins. Another route will be to apply the powerful genetic and cytological methods available in many organisms to test specific predictions from biochemical experiments, for example by examining phenotypes of mutants with specific biochemical defects in Dmc1 or other proteins. At such a point, we will be able to say that these too often disparate fields—biochemistry and genetics—have finally (re)combined.
Acknowledgments
Work from the authors’ laboratory is supported by a grant from the U.S. National Institutes of Health (to S.K.). M.J.N. is supported in part by a fellowship from the Human Frontiers Science Program. S.K. is a Leukemia and Lymphoma Society Scholar.
Footnotes
The authors declare no competing financial interests.
References
- 1.Petronczki M, Siomos MF, Nasmyth K. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell. 2003;112:423–440. doi: 10.1016/s0092-8674(03)00083-7. [DOI] [PubMed] [Google Scholar]
- 2.Kleckner N. Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma. 2006;115:175–194. doi: 10.1007/s00412-006-0055-7. [DOI] [PubMed] [Google Scholar]
- 3.Keeney S. Mechanism and control of meiotic recombination initiation. Curr Top Dev Biol. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- 4.Bianco PR, Tracy RB, Kowalczykowski SC. DNA strand exchange proteins: a biochemical and physical comparison. Front Biosci. 1998;3:D570–603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- 5.Shinohara A, Shinohara M. Roles of RecA homologues Rad51 and Dmc1 during meiotic recombination. Cytogenet Genome Res. 2004;107:201–207. doi: 10.1159/000080598. [DOI] [PubMed] [Google Scholar]
- 6.Zickler D, Kleckner N. Meiotic chromosomes: integrating structure and function. Annu Rev Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
- 7.Johnson RD, Jasin M. Double-strand-break-induced homologous recombination in mammalian cells. Biochem Soc Trans. 2001;29:196–201. doi: 10.1042/0300-5127:0290196. [DOI] [PubMed] [Google Scholar]
- 8.Tarsounas M, Morita T, Pearlman RE, Moens PB. RAD51 and DMC1 form mixed complexes associated with mouse meiotic chromosome cores and synaptonemal complexes. J Cell Biol. 1999;147:207–220. doi: 10.1083/jcb.147.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bishop DK. RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell. 1994;79:1081–1092. doi: 10.1016/0092-8674(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 10.Bishop DK, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 11.Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 12.Aboussekhra A, Chanet R, Adjiri A, Fabre F. Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol Cell Biol. 1992;12:3224–3234. doi: 10.1128/mcb.12.7.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 14.Schwacha A, Kleckner N. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell. 1997;90:1123–1135. doi: 10.1016/s0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- 15.Hunter N, Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- 16.Grushcow JM, et al. Saccharomyces cerevisiae checkpoint genes MEC1, RAD17 and RAD24 are required for normal meiotic recombination partner choice. Genetics. 1999;153:607–620. doi: 10.1093/genetics/153.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stahl FW, et al. Does crossover interference count in Saccharomyces cerevisiae? Genetics. 2004;168:35–48. doi: 10.1534/genetics.104.027789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shinohara M, Sakai K, Shinohara A, Bishop DK. Crossover interference in Saccharomyces cerevisiae requires a TID1/RDH54- and DMC1-dependent pathway. Genetics. 2003;163:1273–1286. doi: 10.1093/genetics/163.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsubouchi H, Roeder GS. The importance of genetic recombination for fidelity of chromosome pairing in meiosis. Dev Cell. 2003;5:915–925. doi: 10.1016/s1534-5807(03)00357-5. [DOI] [PubMed] [Google Scholar]
- 20.Di Giacomo M, et al. Distinct DNA damage-dependent and independent responses drive the loss of oocytes in recombination-defective mouse mutants. Proc Natl Acad Sci USA. 2005;102:737–742. doi: 10.1073/pnas.0406212102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rockmill B, Sym M, Scherthan H, Roeder GS. Roles for two RecA homologs in promoting meiotic chromosome synapsis. Genes Dev. 1995;9:2684–2695. doi: 10.1101/gad.9.21.2684. [DOI] [PubMed] [Google Scholar]
- 22.Young JA, Hyppa RW, Smith GR. Conserved and nonconserved proteins for meiotic DNA breakage and repair in yeasts. Genetics. 2004;167:593–605. doi: 10.1534/genetics.103.023762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bishop DK, et al. High copy number suppression of the meiotic arrest caused by a dmc1 mutation: REC114 imposes an early recombination block and RAD54 promotes a DMC1-independent DSB repair pathway. Genes Cells. 1999;4:425–444. doi: 10.1046/j.1365-2443.1999.00273.x. [DOI] [PubMed] [Google Scholar]
- 24.Shinohara A, Gasior S, Ogawa T, Kleckner N, Bishop DK. Saccharomyces cerevisiae recA homologues RAD51 and DMC1 have both distinct and overlapping roles in meiotic recombination. Genes Cells. 1997;2:615–629. doi: 10.1046/j.1365-2443.1997.1480347.x. [DOI] [PubMed] [Google Scholar]
- 25.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sehorn MG, Sigurdsson S, Bussen W, Unger VM, Sung P. Human meiotic recombinase Dmc1 promotes ATP-dependent homologous DNA strand exchange. Nature. 2004;429:433–437. doi: 10.1038/nature02563. [DOI] [PubMed] [Google Scholar]
- 27.Hong EL, Shinohara A, Bishop DK. Saccharomyces cerevisiae Dmc1 protein promotes renaturation of single-strand DNA (ssDNA) and assimilation of ssDNA into homologous super-coiled duplex DNA. J Biol Chem. 2001;276:41906–41912. doi: 10.1074/jbc.M105563200. [DOI] [PubMed] [Google Scholar]
- 28.Li Z, Golub EI, Gupta R, Radding CM. Recombination activities of HsDmc1 protein, the meiotic human homolog of RecA protein. Proc Natl Acad Sci USA. 1997;94:11221–11226. doi: 10.1073/pnas.94.21.11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masson JY, et al. The meiosis-specific recombinase hDmc1 forms ring structures and interacts with hRad51. EMBO J. 1999;18:6552–6560. doi: 10.1093/emboj/18.22.6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nara T, Hamada F, Namekawa S, Sakaguchi K. Strand exchange reaction in vitro and DNA-dependent ATPase activity of recombinant LIM15/DMC1 and RAD51 proteins from Coprinus cinereus. Biochem Biophys Res Commun. 2001;285:92–97. doi: 10.1006/bbrc.2001.5095. [DOI] [PubMed] [Google Scholar]
- 31.Passy SI, et al. Human Dmc1 protein binds DNA as an octameric ring. Proc Natl Acad Sci USA. 1999;96:10684–10688. doi: 10.1073/pnas.96.19.10684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta RC, Golub E, Bi B, Radding CM. The synaptic activity of HsDmc1, a human recombination protein specific to meiosis. Proc Natl Acad Sci USA. 2001;98:8433–8439. doi: 10.1073/pnas.121005298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kinebuchi T, et al. Structural basis for octameric ring formation and DNA interaction of the human homologous-pairing protein Dmc1. Mol Cell. 2004;14:363–374. doi: 10.1016/s1097-2765(04)00218-7. [DOI] [PubMed] [Google Scholar]
- 34.Sauvageau S, et al. Fission yeast Rad51 and Dmc1, two efficient DNA recombinases forming helical nucleoprotein filaments. Mol Cell Biol. 2005;25:4377–4387. doi: 10.1128/MCB.25.11.4377-4387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee MH, et al. Calcium ion promotes yeast Dmc1 activity via formation of long and fine helical filaments with single-stranded DNA. J Biol Chem. 2005;280:40980–40984. doi: 10.1074/jbc.M505896200. [DOI] [PubMed] [Google Scholar]
- 36.Bugreev DV, Golub EI, Stasiak AZ, Stasiak A, Mazin AV. Activation of human meiosis-specific recombinase Dmc1 by Ca2+ J Biol Chem. 2005;280:26886–26895. doi: 10.1074/jbc.M502248200. [DOI] [PubMed] [Google Scholar]
- 37.Chang YC, et al. Molecular visualization of the yeast Dmc1 protein ring and Dmc1-ssDNA nucleoprotein complex. Biochemistry. 2005;44:6052–6058. doi: 10.1021/bi048897q. [DOI] [PubMed] [Google Scholar]
- 38.Kinebuchi T, Kagawa W, Kurumizaka H, Yokoyama S. Role of the N-terminal domain of the human DMC1 protein in octamer formation and DNA binding. J Biol Chem. 2005;280:28382–28387. doi: 10.1074/jbc.M503372200. [DOI] [PubMed] [Google Scholar]
- 39.Bugreev DV, Mazin AV. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc Natl Acad Sci USA. 2004;101:9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen YK, et al. Heterodimeric complexes of Hop2 and Mnd1 function with Dmc1 to promote meiotic homolog juxtaposition and strand assimilation. Proc Natl Acad Sci USA. 2004;101:10572–10577. doi: 10.1073/pnas.0404195101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Enomoto R, et al. Positive role of the mammalian TBPIP/HOP2 protein in DMC1-mediated homologous pairing. J Biol Chem. 2004;279:35263–35272. doi: 10.1074/jbc.M402481200. [DOI] [PubMed] [Google Scholar]
- 42.Petukhova GV, et al. The Hop2 and Mnd1 proteins act in concert with Rad51 and Dmc1 in meiotic recombination. Nat Struct Mol Biol. 2005;12:449–453. doi: 10.1038/nsmb923. [DOI] [PubMed] [Google Scholar]
- 43.Sung P, Krejci L, Van Komen S, Sehorn MG. Rad51 recombinase and recombination mediators. J Biol Chem. 2003;278:42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- 44.Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- 45.Sharan SK, et al. BRCA2 deficiency in mice leads to meiotic impairment and infertility. Development. 2004;131:131–142. doi: 10.1242/dev.00888. [DOI] [PubMed] [Google Scholar]
- 46.Siaud N, et al. Brca2 is involved in meiosis in Arabidopsis thaliana as suggested by its interaction with Dmc1. EMBO J. 2004;23:1392–1401. doi: 10.1038/sj.emboj.7600146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kojic M, Kostrub CF, Buchman AR, Holloman WK. BRCA2 homolog required for proficiency in DNA repair, recombination, and genome stability in Ustilago maydis. Mol Cell. 2002;10:683–691. doi: 10.1016/s1097-2765(02)00632-9. [DOI] [PubMed] [Google Scholar]
- 48.Martin JS, Winkelmann N, Petalcorin MI, McIlwraith MJ, Boulton SJ. RAD-51-dependent and -independent roles of a Caenorhabditis elegans BRCA2-related protein during DNA double-strand break repair. Mol Cell Biol. 2005;25:3127–3139. doi: 10.1128/MCB.25.8.3127-3139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dray E, Siaud N, Dubois E, Doutriaux MP. Interaction between Arabidopsis Brca2 and its partners Rad51, Dmc1, and Dss1. Plant Physiol. 2006;140:1059–1069. doi: 10.1104/pp.105.075838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shinohara M, et al. Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homologue of RAD54, RDH54/TID1, in mitosis and meiosis. Genetics. 1997;147:1545–1556. doi: 10.1093/genetics/147.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shinohara M, Gasior SL, Bishop DK, Shinohara A. Tid1/Rdh54 promotes colocalization of Rad51 and Dmc1 during meiotic recombination. Proc Natl Acad Sci USA. 2000;97:10814–10819. doi: 10.1073/pnas.97.20.10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petukhova G, Stratton S, Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- 53.Petukhova G, Sung P, Klein H. Promotion of Rad51-dependent D-loop formation by yeast recombination factor Rdh54/Tid1. Genes Dev. 2000;14:2206–2215. doi: 10.1101/gad.826100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dresser ME, et al. DMC1 functions in a Saccharomyces cerevisiae meiotic pathway that is largely independent of the RAD51 pathway. Genetics. 1997;147:533–544. doi: 10.1093/genetics/147.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Catlett MG, Forsburg SL. Schizosaccharomyces pombe Rdh54 (TID1) acts with Rhp54 (RAD54) to repair meiotic double-strand breaks. Mol Biol Cell. 2003;14:4707–4720. doi: 10.1091/mbc.E03-05-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wesoly J, et al. Differential contributions of mammalian Rad54 paralogs to recombination, DNA damage repair, and meiosis. Mol Cell Biol. 2006;26:976–989. doi: 10.1128/MCB.26.3.976-989.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Komen S, Petukhova G, Sigurdsson S, Stratton S, Sung P. Superhelicity-driven homologous DNA pairing by yeast recombination factors Rad51 and Rad54. Mol Cell. 2000;6:563–572. doi: 10.1016/s1097-2765(00)00055-1. [DOI] [PubMed] [Google Scholar]
- 58.Ristic D, Wyman C, Paulusma C, Kanaar R. The architecture of the human Rad54-DNA complex provides evidence for protein translocation along DNA. Proc Natl Acad Sci USA. 2001;98:8454–8460. doi: 10.1073/pnas.151056798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jaskelioff M, Van Komen S, Krebs JE, Sung P, Peterson CL. Rad54p is a chromatin remodeling enzyme required for heteroduplex DNA joint formation with chromatin. J Biol Chem. 2003;278:9212–9218. doi: 10.1074/jbc.M211545200. [DOI] [PubMed] [Google Scholar]
- 60.Alexeev A, Mazin A, Kowalczykowski SC. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat Struct Biol. 2003;10:182–186. doi: 10.1038/nsb901. [DOI] [PubMed] [Google Scholar]
- 61.Solinger JA, Kiianitsa K, Heyer WD. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol Cell. 2002;10:1175–1188. doi: 10.1016/s1097-2765(02)00743-8. [DOI] [PubMed] [Google Scholar]
- 62.Leu JY, Chua PR, Roeder GS. The meiosis-specific Hop2 protein of S. cerevisiae ensures synapsis between homologous chromosomes. Cell. 1998;94:375–386. doi: 10.1016/s0092-8674(00)81480-4. [DOI] [PubMed] [Google Scholar]
- 63.Gerton JL, DeRisi JL. Mnd1p: an evolutionarily conserved protein required for meiotic recombination. Proc Natl Acad Sci USA. 2002;99:6895–6900. doi: 10.1073/pnas.102167899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Petukhova GV, Romanienko PJ, Camerini-Otero RD. The Hop2 protein has a direct role in promoting interhomolog interactions during mouse meiosis. Dev Cell. 2003;5:927–936. doi: 10.1016/s1534-5807(03)00369-1. [DOI] [PubMed] [Google Scholar]
- 65.Tsubouchi H, Roeder GS. The Mnd1 protein forms a complex with Hop2 to promote homologous chromosome pairing and meiotic double-strand break repair. Mol Cell Biol. 2002;22:3078–3088. doi: 10.1128/MCB.22.9.3078-3088.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zierhut C, Berlinger M, Rupp C, Shinohara A, Klein F. Mnd1 is required for meiotic interhomolog repair. Curr Biol. 2004;14:752–762. doi: 10.1016/j.cub.2004.04.030. [DOI] [PubMed] [Google Scholar]
- 67.Henry JM, et al. Mnd1/Hop2 facilitates Dmc1-dependent interhomolog crossover formation in meiosis of budding yeast. Mol Cell Biol. 2006;26:2913–2923. doi: 10.1128/MCB.26.8.2913-2923.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Enomoto R, et al. Stimulation of DNA strand exchange by the human TBPIP/Hop2-Mnd1 complex. J Biol Chem. 2006;281:5575–5581. doi: 10.1074/jbc.M506506200. [DOI] [PubMed] [Google Scholar]
- 69.Hayase A, et al. A protein complex containing Mei5 and Sae3 promotes the assembly of the meiosis-specific RecA homolog Dmc1. Cell. 2004;119:927–940. doi: 10.1016/j.cell.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 70.Tsubouchi H, Roeder GS. The budding yeast Mei5 and Sae3 proteins act together with Dmc1 during meiotic recombination. Genetics. 2004;168:1219–1230. doi: 10.1534/genetics.103.025700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Akamatsu Y, Dziadkowiec D, Ikeguchi M, Shinagawa H, Iwasaki H. Two different Swi5-containing protein complexes are involved in mating-type switching and recombination repair in fission yeast. Proc Natl Acad Sci USA. 2003;100:15770–15775. doi: 10.1073/pnas.2632890100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ellermeier C, Schmidt H, Smith GR. Swi5 acts in meiotic DNA joint molecule formation in Schizosaccharomyces pombe. Genetics. 2004;168:1891–1898. doi: 10.1534/genetics.104.034280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Blat Y, Protacio RU, Hunter N, Kleckner N. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell. 2002;111:791–802. doi: 10.1016/s0092-8674(02)01167-4. [DOI] [PubMed] [Google Scholar]
- 75.Smith AV, Roeder GS. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J Cell Biol. 1997;136:957–967. doi: 10.1083/jcb.136.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Klein F, et al. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999;98:91–103. doi: 10.1016/S0092-8674(00)80609-1. [DOI] [PubMed] [Google Scholar]
- 77.Xu H, Beasley MD, Warren WD, van der Horst GT, McKay MJ. Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev Cell. 2005;8:949–961. doi: 10.1016/j.devcel.2005.03.018. [DOI] [PubMed] [Google Scholar]