Abstract
The pathogenesis of chronic rejection, Bronchiolitis Obliterans Syndrome (BOS) following lung transplantation (LT) is poorly understood. We hypothesized that development of antibodies to HLA (DSA) is associated with dysregulation of microRNA (miRNA) that predisposes BOS. Towards this, miRNA profiling of mononuclear cells from 10 stable LT (DSA−BOS−), 10 LT with DSA+BOS− (DSA group) and 10 LT with DSA+BOS+ (BOS group) were performed. Prediction by mirPath indicated that differential miRNAs in DSA+BOS− compared to stable are significantly up-regulated (relative fold >2, p < 0.05) for TGF-β and B cell receptor signal pathways. A total of seventy-four miRNAs were up-regulated and six miRNAs were down regulated in LT with DSA+BOS+ when compared to stable (relative fold >2, p < 0.05). There was also significant enrichment of cell cycle and gap junction pathways. An inverse correlation between expression of two key miRNAs and their target genes were observed: miR-369-5p and miR-548d were down regulated in DSA+ LT while their gene targets in TGF-β signal pathways were up-regulated. In addition, miR-628-5p and miR-134 were down regulated and their target genes (B cell development) were up-regulated. Therefore, we conclude that alloimmunity induced changes in miRNAs affecting the TGF-β and B cell receptor signal pathways play important roles in BOS development.
Introduction
Lung transplantation (LT) is a treatment option for end-stage lung diseases including idiopathic pulmonary fibrosis, chronic obstructive pulmonary disease, and cystic fibrosis (1,2). However, long-term survival remains a challenge due to development of Bronchiolitis Obliterans Syndrome (BOS) (3). Approximately 50% of LT recipients (LTx) develop BOS by five years after transplantation (3). Many etiologies for the pathogenesis of BOS have been proposed and it is accepted that development of BOS is due to immunological insults. Our study demonstrated a correlation between the development of antibodies (Abs) to donor mismatched HLA (DSA) during post-transplantation and the risk for BOS (4). Furthermore, Abs against HLA has been eluted from lungs that underwent chronic rejection (5). Further support for the possible role for anti-HLA in the immunopathogenesis of obliterative airway disease (OAD) has been provided by studies in which administration of anti-HLA class I intra-bronchially into produced OAD in the native lungs (6). However, the mechanisms leading to BOS following de novo development of anti-HLA still remains unknown.
MiRNAs are a class of non-coding small RNAs, 22–25 nucleotides in length, which bind to the 3′UTR of target genes and thereby repress translation and/or induce degradation of target gene mRNA (7). Aberrant expression of miRNAs is associated with initiation and progression of pathological processes including immune-system disorders like asthma and rheumatoid arthritis (8,9). Furthermore, miRNAs have been shown to influence toll-like receptor signaling, inflammatory gene expression, T cell and B cell differentiation, lineage specificity and development of fibrosis, cirrhosis, and graft loss (10). Studies also demonstrated that miRNA dysregulation may result in chronic antibody-mediated rejection following kidney transplantation (11–13). Therefore, to test the hypothesis that immune responses to alloantigens as evidenced by development of Abs to donor HLA will dysregulate miRNAs involved in regulating immune responses, we determined miRNA expression profiles following human LT, including the LTx who developed DSA to mismatched MHC and developed BOS.
Materials and Methods
Patient selection
Thirty LTx at Barnes Jewish Hospital/Washington University School of Medicine were enrolled between January 2009 and March 2013. This study was approved by the institutional review board and written informed consent was obtained. Among the 30 patients included, 10 developed DSA but had normal FEV1 (Forced Expiratory Volume in 1 s) and had no evidence of BOS two years after LT (DSA+BOS− group, 20 ± 10 months from LT); 10 developed DSA and had a clinical diagnosis of BOS (DSA+BOS+ group, 25 ± 9 months from LT), and 10 didn't develop DSA and did not have any evidence of BOS within a two year period (stable group, 22 ± 8 months from LT) (Table 1). Stable group and DSA+BOS− group were available for follow-up. There were no significant differences of mean fluorescent intensity (MFI) values of DSA and antibodies to MHC class I or class II in the patients between DSA+BOS− and DSA+BOS+ groups. There were also no significant differences (p > 0.05) in the development of antibodies to self-antigens (Ka1 T and Col V) among three groups. BOS was defined by the criteria set forth by the International Society for Heart & Lung Transplantation (ISHLT) (14). LTx from these three groups were similar in age, gender, race, underlying diseases, and had bilateral LT. The standard immunotherapy protocol consisted of tacrolimus, mycophenolate mofetil, and prednisone.
Table 1. Demographic data on three groups of lung transplant recipients.
| Groups | Stable (n = 10) | DSA+BOS− (n = 10) | DSA+BOS+ (n = 10) |
|---|---|---|---|
| Age | 53 ± 9 | 54 ± 8 | 53 ± 10 |
| Gender | 6M/4F | 6M/4F | 4M/6F |
| Race (C/AA/others) | 9C/1AA | 10C | 8C/2AA |
| Diagnosis (IPF/COP /CF/others) | 3/3/3/1 | 5/2/2/1 | 3/3/2/2 |
| Type of LTx (bilateral/single LTx) | 10BLT | 10BLT | 10BLT |
| Time of sampling from LTx (months) | 22 ± 8 | 20 ± 10 | 25 ± 9 |
| Time of DSA onset (days) | NA | 16 ± 10 | 24 ± 23 |
| Time of BOS onset (months) | NA | NA | 21 ± 9 |
M, male; F, female; C, Caucasian; AA, African American; CF, cystic fibrosis; IPF, idiopathic pulmonary fibrosis; COPD, chronic obstructive pulmonary disease; BLT, bilateral lung transplant; DSA+BOS−, LTx recipients with developed DSA but had no evidence of BOS two years after LT; DSA+BOS+, LTx recipients with developed DSA and had a clinical diagnosis of BOS.
Cell isolation from peripheral blood mononuclear cells (PBMCs), bronchoalveolar lavage (BAL), and miRNA extraction
PBMCs were separated from blood using the Ficoll-hypaque density separation (Amersham, Uppsala, Sweden) (15). BAL fluid from bronchoscopy was centrifuged at 300 g and 4°C for 10 min to separate cells. There were no statistically significant differences in total blood mononuclear cell (PBMC) counts and PBMC profiles among stable, DSA+BOS−, and DSA+BOS+ patient groups. PBMCs and BAL cells were suspended in TRIzol (Agilent Technologies, Morrisville, NC) and stored at −80°C until RNA extraction. Total RNA was extracted using the mirVana miRNA isolation kit according to the manufacturer's specification (Ambion, Austin, TX).
miRNA expression profiling
The TaqMan Low-Density Array Human miRNA Panel v1.0 (Applied Biosystems, Foster City, CA) was utilized for global miRNA profiling. The panel includes two 384-well microfluidic cards (human miRNA pool A and pool B) that contain primers and probes for 746 different human miRNAs in addition to six small nucleolar RNAs that function as endogenous controls for data normalization. RNA extracted from PBMC among stable, DSA+BOS− and DSA+BOS+ patients were pooled separately and used for miRNA profiling. Equal quantity of RNA (30 ng) from PBMCs (n = 10) from each of the groups were pooled and reverse transcribed for cDNA synthesis using the TaqMan Multiplex RT set (Applied Biosystems) for TaqMan Array Human miRNA Panels. Each RT reaction was diluted 62.5-fold with water and 55 μL of each diluted product was combined with 55 μL of TaqMan 2× Universal PCR Master Mix, No AmpErase UNG (Applied Biosystems). One-hundred microliters of the sample/master mix for each multiplex pool were loaded into fill reservoirs on the microfluidic card. The array was then centrifuged and mechanically sealed with the Applied Biosystems sealer device. Quantitative PCR was performed on an Applied BioSystems 7900HT thermocycler (Applied Biosystems) using the manufacturer's recommendations.
TaqMan miRNA assay for individual miRNAs
Independent sets of PBMC and BAL were used for qPCR confirmation. Total RNA was isolated, and six individual miRNAs were further quantitated by TaqMan miRNA assays (Applied Biosystems) and RNU44 was used as controls to normalize data.
Quantification of messenger RNAs
To determine the expression of target genes, cDNA from independent sets of PBMC was synthesized using Super-Script II reverse transcriptase with random hexamers (Life Technologies, Carlsbad, CA). Real-time PCR was performed using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA). GAPDH was detected as endogenous control.
miRNA target prediction and pathway analysis
DIANA-mirPath (16) was employed to perform the enrichment analysis of predicted target genes by one or more miRNAs in biological pathways. The software performs analysis of multiple miRNA target genes to all known KEGG pathways (Kyoto Encyclopedia of Genes and Genomes) based on two algorithms, microT-CDS (17,18) and miRTarBase (19). The input dataset enrichment was performed by a Pearson's Chi-squared test, and FDR correction was used for the number of pathways. The graphical output of the program provides an overview of the parts of the pathway modulated by selected miRNAs facilitating the interpretation and presentation of the results. The statistical significance associated with the identified biological pathways was calculated by the mirPath software (http://microrna.gr/mirpath).
Statistical analysis
The selection of 10 samples within each group was based upon sample availability and statistical test power. The efficiency of pooling total RNA in microarray experiments was based upon published methods (20,21) and the RNA pooling enabled us to reduce number of arrays without a loss of precision. Data analysis on mi RNA expression levels from TaqMan Low-Density Array was performed by the SDS software version 2.2.2 (Applied Biosystems). Assays that had Ct values >35 were removed from the analysis. The delta Ct values were calculated by using RNU44 as the endogenous control. We initially designed the experiments to minimize the impact of covariates by matching the samples for key confounding factors. Therefore, we used a univariate test to screen for differentially expressed miRNAs, and validated the result with a separate set of samples. A Student's t-test was performed to detect differentially expressed miRNAs. Bonferroni procedure was used to calculate adjusted p values to control false discovery rate (FDR) among stable, DSA+BOS− and DSAþBOS+ patients groups, respectively (22). Heatmap was performed by R software. The Mann–Whitney test was performed using GraphPad Prism 5 to determine the significance of miRNA levels of PBMC and BAL for independent validation. The association between expression levels of mRNAs and miRNAs was analyzed using the Pearson's correlation. All data were expressed as mean ± SE unless otherwise specified. p values less than 0.05 were considered significant.
Results
miRNAs are differentially expressed in DSA+BOS− LT recipients compared to stable LT recipients
To identify differentially expressed miRNAs in LT patients, we profiled the expression of 746 miRNAs using the TaqMan miRNA array. The study design is illustrated in Figure 1. The total RNA obtained from PBMCs from 10 individuals in each group (stable, DSA+BOS− and DSA+BOS+) was pooled with equal quantity (30 ng). The selection of pooled samples (10 per group) are approximately equivalent to 4–7 independent samples in each group, if samples were not pooled, and also assumes a wide range of values for the ratio of biological variations among the samples over the technical variations in the assays (21) (Figure S1). Global miRNA expression profiling was performed on the pooled RNA using TaqMan Low-density Human miRNA Array in combination with Megaplex RT and Megaplex pre-amplification techniques. Preliminary evaluation of candidate differentially expressed miRNAs was performed by Heatmap, potential target prediction and pathway enrichment and finally independent validation of differentially expressed miRNAs using the TaqMan miRNA assay.
Figure 1. Study design.
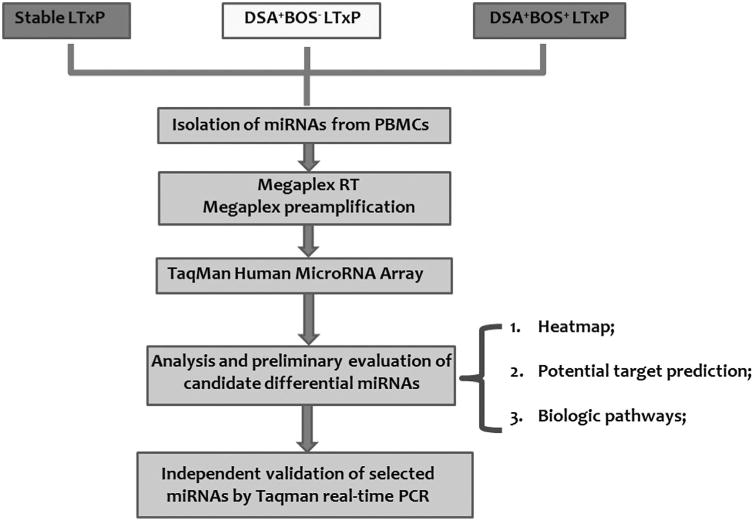
The workflow of this study was performed in three steps: (1) global expression profiling of PBMC miRNA pools extracted from three pools of LT recipients (stable, DSA+BOS– and DSA+BOS+) using TaqMan Low-Density Human miRNA Array, in combination with Megaplex RT and Megaplex pre-amplification techniques; (2) analysis and preliminary evaluation of candidate miRNAs by heatmap, potential target prediction and pathway enrichment; and (3) independent validation of differentially expressed miRNAs.
To investigate the relative abundance of detected miRNAs, they were normalized in each pool (stable, DSA+BOS− and DSA+BOS+) to RNU44 in the corresponding pool. The results identified 349 miRNAs (∼46.8%) (Ct <35 in at least one pool were classed as detectable), indicating that global miRNA expression profiling is a feasible method to identify aberrantly expressed miRNAs. Subsequently we investigated the miRNA expression profile after de novo development of DSA. Comparison of the miRNA expression profile in the DSA+BOS− to the stable patients demonstrated that 31 miRNAs were up regulated and 14 miRNAs were down regulated in the DSA+BOS− group (relative fold >2, p < 0.05) (Figure 2A and Table S1), suggesting that these miRNAs might be important in DSA mediated immune responses after LT. The heatmap, which represents the mean fold change of differential miRNA signature in DSA+BOS− patients, was generated by 45 differential miRNAs (31 over-expressed miRNAs and 14 under-expressed miRNAs in the DSA+BOS− group, Figure 2B). In order to examine the biologic pathways modulated by the differentially expressed miRNAs in DSA+BOS− patients, we applied DIANA-mirPath on the DSA-related deregulated miRNA signature. Thirty-two KEGG biological processes were significantly modulated by the differentially expressed miRNAs in DSA+BOS− patients (p < 0.05, FDR corrected). Among them, the TGF beta signal pathway (p < 0.0001) was the most prominent pathway enriched in quantities with DSA-related differentially expressed miRNA patterns (Table 2 and Figure S2). Furthermore, B cell receptor signal pathway (p = 0.0013, Figure S4), T cell receptor signal pathway (p < 0.0001), chemokine signaling pathway (p < 0.0001), cytokine-cytokine receptor interaction (p = 0.0075) were significantly modulated by the differentially expressed miRNAs in DSA+BOS− patients (Table 2). These results strongly suggest that immune response associated biologic pathways were affected following de novo development of DSA.
Figure 2. Differentially expressed miRNA patterns in LT patients.
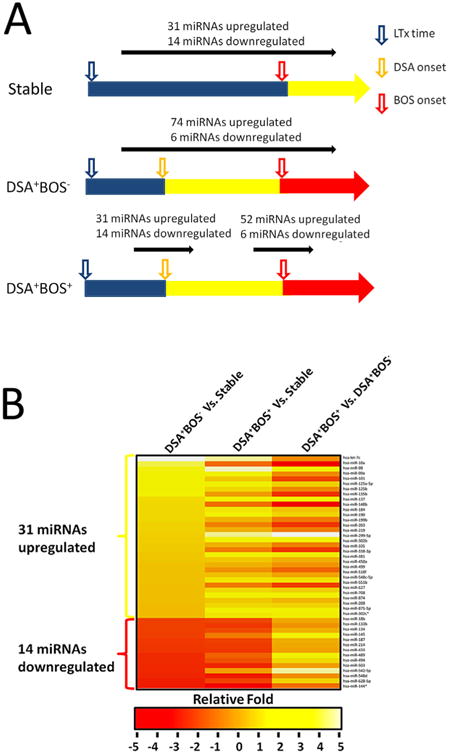
(A) Preliminary data analysis of miRNA expression levels from TaqMan Low-Density Array was performed by using the SDS software. Data were normalized and then analyzed to define miRNAs that are differentially expressed among stable, DSA+BOS− and DSA+BOS+ patients' pools. Assays that had Ct values >35 were removed from the analysis. The delta Ct values were calculated by using RNU44 as the endogenous control. A Student's t-test was performed to detect differentially expressed miRNAs among stable, DSA+BOS− and DSA+BOS+ patients' pools. miRNAs in DSA+BOS− and DSA+BOS+ pools with relative fold >2 and p-value less than 0.05 were considered as significantly differentially expressed when compared to those in stable transplants. (B) Heatmap representation of the mean fold change in DSA+BOS− related differential miRNA signature. Two-dimensional grid matrix displaying 45 differential miRNAs (31 over expressed miRNAs and 14 under expressed miRNAs from DSA+BOS− group) in stable, DSA+BOS− and DSA+BOS+ groups was obtained by the functional heatmap in R. Columns refer to three comparison: DSA+BOS− and DSA+BOS+ relative to stable as well as DSA+BOS+ compared to DSA+BOS− as the third column. Rows stand for the 45 differential miRNAs. Each entry of the grid refers to relative fold (log2) between the expression level of a given miRNA on DSA+BOS− or DSA+BOS+ pools relative to stable pools. The color of each entry is determined by the value of that fold difference, ranging from red (negative values) to yellow (positive values).
Table 2. Shared biologic pathways enriched by differentially expressed miRNAs in DSA+BOS– patients and DSA+BOS+ patients.
| Comparison | DSA+BOS− versus Stable | |
|---|---|---|
|
|
|
|
| KEGG pathway | p-value, FDR corrected | Selected miRNAs |
| TGF-beta signaling pathway | 8.05E-28 | miR-98, miR-125a-5p, miR-548c-5p, miR-133b, miR-134, miR-433,, miR-627, miR-874 |
| MAPK signaling pathway | 8.05E-28 | miR-125a-5p, miR-137, miR-184, miR-203, miR-134, miR-433, miR-489, miR-494 |
| PI3K-Akt signaling pathway | 8.05E-28 | let-7c, miR-299-5p, miR-338-3p, miR-381, miR-627, miR-874, miR-875-5p |
| Ubiquitin mediated proteolysis | 2.46E-20 | miR-125a-5p, miR-137, miR-203, miR-299-5p, miR-875-5p, miR-134, miR-433 |
| Adherens junction | 3.23E-19 | miR-137, miR-203, miR-299-5p, miR-338-3p, miR-381, miR-875-5p |
| Focal adhesion | 7.40E-18 | miR-299-5p, miR-338-3p, miR-381, miR-627, miR-874, miR-134, miR-433, miR-489 |
| Wnt signaling pathway | 2.77E-15 | let-7c, miR-98, miR-134, miR-433, miR-494, miR-503, miR-628-5p |
| mTOR signaling pathway | 3.57E-10 | let-7c, miR-299-5p, miR-338-3p, miR-381, miR-548c-5p, miR-874, miR-628-5p |
| B cell receptor signaling pathway | 9.00E-10 | miR-184, miR-203, miR-299-5p, miR-338-3p, miR-381, miR-134, miR-433, miR-628-5p |
| Regulation of actin cytoskeleton | 1.49E-09 | let-7c, miR-338-3p, miR-381, miR-548c-5p, miR-627, miR-874, miR-875-5p, miR-494 |
| Toll-like receptor signaling pathway | 1.52E-08 | miR-125a-5p, miR-137, miR-627, miR-875-5p, miR-133b, miR-433, miR-489, miR-503 |
| Jak-STAT signaling pathway | 2.68E-08 | let-7c, miR-98, miR-299-5p, miR-338-3p, miR-381, miR-548c-5p, miR-627 |
| Hedgehog signaling pathway | 5.39E-08 | miR-338-3p, miR-381, miR-548c-5p, miR-875-5p, miR-503, miR-628-5p |
| Bacterial invasion of epithelial cells | 1.19E-07 | let-7c, miR-98, miR-338-3p, miR-381, miR-548c-5p, miR-627, miR-874, miR-503, miR-628-5p |
| Notch signaling pathway | 6.70E-07 | miR-137, miR-299-5p, miR-338-3p, miR-433, miR-489, miR-494, miR-628-5p |
| Regulation of autophagy | 6.12E-06 | miR-299-5p, miR-338-3p, miR-381, miR-548c-5p, miR-627, miR-133b, miR-494, miR-628-5p |
| Tight junction | 1.35E-05 | let-7c, miR-98, miR-299-5p, miR-338-3p, miR-133b, miR-134, miR-433, miR-503, miR-628-5p |
| T cell receptor signaling pathway | 2.94E-05 | miR-184, miR-299-5p, miR-338-3p, miR-134, miR-433, miR-489, miR-542-5p, miR-628-5p |
| Fc gamma R-mediated phagocytosis | 4.00E-05 | miR-203, miR-299-5p, miR-338-3p, miR-381, miR-503, miR-628-5p |
| Chemokine signaling pathway | 4.00E-05 | miR-98, miR-125a-5p, miR-627, miR-874, miR-875-5p, miR-489, miR-542-5p, miR-628-5p |
| Apoptosis | 1.22E-04 | let-7c, miR-98, miR-299-5p, miR-338-3p, miR-381, miR-542-5p, miR-628-5p |
| HIF-1 signaling pathway | 2.28E-04 | miR-125a-5p, miR-137, miR-548c-5p, miR-875-5p, miR-489, miR-494, miR-503, miR-628-5p |
| Cytokine-cytokine receptor interaction | 7.50E-03 | let-7c, miR-203, miR-299-5p, miR-627, miR-874, miR-134, miR-433, miR-494, miR-503 |
|
| ||
| Comparison | DSA+BOS+ versus Stable | |
|
|
|
|
| KEGG pathway | p-value, FDR corrected | Differentially expressed miRNAs |
|
| ||
| TGF-beta signaling pathway | 1.13E-20 | miR-98, miR-125a-5p, miR-548c-5p, miR-548d-5p, miR-582-3p, miR-875-5p, miR-369-5p |
| MAPK signaling pathway | 4.46E-31 | miR-152, miR-193a-5p, miR-410, miR-422a, miR-450b-5p, miR-542-3p, miR-875-5p, miR-542-5p |
| PI3K-Akt signaling pathway | 5.26E-40 | miR-125a-5p, miR-329, miR-338-3p, miR-422a, miR-450b-5p, miR-582-3p, miR-627, miR-542-5p |
| Ubiquitin mediated proteolysis | 7.00E-27 | miR-193a-5p, miR-338-3p, miR-372, miR-381, miR-410, miR-450b-5p, miR-483-5p, miR-542-3p |
| Adherens junction | 9.48E-17 | let-7c, miR-1, miR-338-3p, miR-372, miR-548c-5p, miR-548d-5p, miR-582-3p, miR-511, miR-875-5p |
| Focal adhesion | 4.44E-33 | miR-137, miR-140-3p, miR-152, miR-381, miR-410, miR-548d-5p, miR-582-3p, miR-376c |
| Wnt signaling pathway | 3.20E-15 | miR-140-3p, miR-152, miR-193a-5p, miR-542-3p, miR-376c, miR-511, miR-941, miR-875-5p |
| mTOR signaling pathway | 1.24E-09 | miR-1, miR-98, miR-125a-5p, miR-137, miR-338-3p, miR-450b-5p, miR-542-3p, miR-511 |
| B cell receptor signaling pathway | 1.35E-03 | let-7c, miR-329, miR-338-3p, miR-372, miR-381, miR-376c, miR-511, miR-134, miR-433, miR-628-5p |
| Regulation of actin cytoskeleton | 6.10E-21 | miR-98, miR-125a-5p, miR-137, miR-450b-5p, miR-483-5p, miR-627, miR-376c, miR-511, miR-875-5p |
| Toll-like receptor signaling pathway | 3.58E-03 | let-7c, miR-1, miR-98, miR-125a-5p, miR-193a-5p, miR-329, miR-338-3p, miR-875-5p, miR-542-5p |
| Jak-STAT signaling pathway | 6.21E-14 | miR-98, miR-203, miR-329, miR-338-3p, miR-422a, miR-450b-5p, miR-542-3p, miR-548d-5p, miR-582-3p |
| Hedgehog signaling pathway | 1.93E-08 | let-7c, miR-152, miR-203, miR-329, miR-338-3p, miR-450b-5p, miR-548c-5p, miR-548d-5p, miR-582-3p |
| Bacterial invasion of epithelial cells | 2.81E-07 | let-7c, miR-1, miR-140-3p, miR-152, miR-203, miR-422a, miR-548d-5p, miR-582-3p, miR-627, miR-875-5p |
| Notch signaling pathway | 1.85E-09 | let-7c, miR-125a-5p, miR-137, miR-338-3p, miR-381, miR-410, miR-582-3p, miR-511 |
| Regulation of autophagy | 3.88E-07 | let-7c, miR-203, miR-338-3p, miR-372, miR-381, miR-548d-5p, miR-627, miR-376c, miR-511 |
| Tight junction | 9.11E-04 | miR-125a-5p, miR-372, miR-381, miR-410, miR-548d-5p, miR-582-3p, miR-511, miR-875-5p |
| T cell receptor signaling pathway | 8.70E-07 | miR-98, miR-125a-5p, miR-137, miR-338-3p, miR-372, miR-381, miR-450b-5p, miR-941, miR-542-5p |
| Fc gamma R-mediated phagocytosis | 4.92E-02 | miR-137, miR-140-3p, miR-381, miR-410, miR-450b-5p, miR-671-3p, miR-376c, miR-511, miR-875-5p |
| Chemokine signaling pathway | 4.29E-04 | miR-125a-5p, miR-137, miR-140-3p, miR-548d-5p, miR-582-3p, miR-627, miR-219-1-3p, miR-542-5p |
| Apoptosis | 4.98E-02 | miR-140-3p, miR-152, miR-329, miR-422a, miR-450b-5p, miR-511, miR-941, miR-875-5p, miR-542-5p |
| HIF-1 signaling pathway | 9.48E-17 | miR-193a-5p, miR-203, miR-329, miR-542-3p, miR-548c-5p, miR-219-1-3p, miR-376c, miR-875-5p |
| Cytokine-cytokine receptor interaction | 1.91E-03 | miR-329, miR-338-3p, miR-542-3p, miR-219-1-3p, miR-376c, miR-511, miR-875-5p |
miRNAs are differentially expressed in DSA+BOS+ LT recipients compared to stable LT recipients
In order to identify the differentially expressed miRNAs in the DSA+BOS+ group, we compared miRNA expression levels between the DSA+BOS+ and the stable group. There were 74 miRNAs up regulated and 6 miRNAs down regulated in the DSA+BOS+ group, compared to those of stable (relative fold >2, p < 0.05) (Figure 2A and Table S2). In addition, 26 miRNAs were up regulated and 3 miRNAs were down regulated in both DSA+BOS+ versus stable and DSA+BOS− versus stable comparisons (relative fold >2, p < 0.05, FDR corrected) (Figure 3A), indicating that these differentially expressed miRNAs are associated with de novo development of DSA in LTx with BOS.
Figure 3. Analysis of the differentially expressed miRNAs in LT patients.
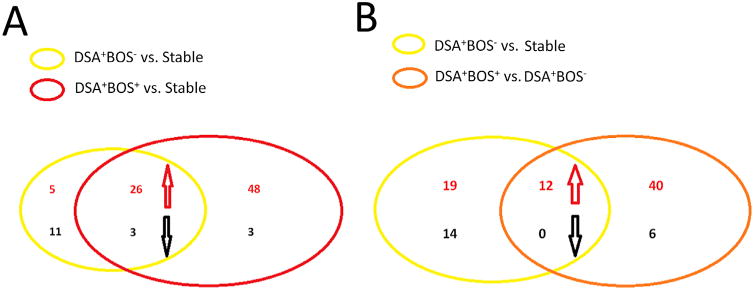
(A) Venn diagram showing a total of 26 miRNAs that were up regulated and three miRNAs that were down regulated in both DSA+BOS+ versus stable and DSA+BOS− versus stable comparisons. (B) Venn diagram showing a total of 12 miRNAs that were up regulated in both comparisons of DSA+BOS− versus stable and DSA+BOS+ versus DSA+BOS−.
We next performed comparative pathway analysis using DIANA-mirPath on differentially expressed miRNAs in DSA+BOS+ patients. There were 35 biologic pathways enriched in differentially expressed miRNAs in DSA+BOS+ patients (p < 0.05, FDR corrected). Compared with the pathways enriched by differential miRNAs in DSA+BOS− patients, there are 25 dysregulated biologic pathways enriched in DSA- and BOS-related dysregulated miRNA signatures in DSA+BOS− and DSA+BOS+ patients (Table 2). In addition, the TGF beta signal pathway (DSA+BOS−: p < 0.0001, Figure S2 and DSA+BOS+: p < 0.0001, Figure S3) was also enriched prominently (Table 2). Furthermore, several other pathways involved in the dysregulation of immune-response including B cell receptor (Figure S5) and T cell receptor signal pathways, chemokine signaling pathway and cytokine-cytokine receptor interaction pathway were also affected (Table 2). These results demonstrate that the dysregulation of TGF beta signaling and immune response pathways persisted in LTx who had persistent DSA leading to BOS.
Finally, we found 10 additional signal pathways that were dysregulated only in the DSA+BOS+ patients. They were inositol phosphate metabolism (p < 0.0001), gap junction (p < 0.0001), cell cycle (p < 0.0001), pertussis (p = 0.0002), phosphatidylinositol signaling system (p = 0.0072), basal transcription factors (p = 0.0074), pentose phosphate pathway (p = 0.037), N-glycan biosynthesis (p = 0.037), Toxoplasmosis (p = 0.037), and epithelial cell signaling in Helicobacter pylori infection (p = 0.049) (Table 3).
Table 3. Additional biological cellular pathways enriched by differentially expressed miRNAs in DSA+BOS+ patients.
| DSA+BOS+ | ||
|---|---|---|
|
| ||
| KEGG pathway | p-value, FDR corrected | Selected miRNAs |
| Inositol phosphate metabolism | 8.62E-06 | miR-193a-5p, miR-203, miR-329, miR-338-3p, miR-372, miR-381, miR-410, miR-582-3p, miR-627 |
| Gap junction | 1.02E-05 | miR-152, miR-193a-5p, miR-203, miR-329, miR-338-3p, miR-372, miR-381, miR-548d-5p, miR-627 |
| Cell cycle | 4.23E-50 | let-7c, miR-1, miR-98, miR-125a-5p, miR-137, miR-140-3p, miR-152, miR-203, miR-329, miR-338-3p, mkiR-372 |
| Pertussis | 2.44E-04 | let-7c, miR-1, miR-98, miR-125a-5p, miR-137, miR-405b-5p, miR-542-3p, miR-548c-5p, miR-219-1-3p, miR-875-5p |
| Phosphatidylinositol signaling system | 7.21E-03 | miR-1, miR-125a-5p, miR-372, miR-381, miR-410, miR-548d-5p, miR-582-3p, miR-627, miR-376c, miR-511 |
| Basal transcription factors | 7.44E-03 | miR-98, miR-125a-5p, miR-137, miR-152, miR-329, miR-338-3p, miR-548c-5p, miR-548d-5p, miR-376c, miR-511 |
| Pentose phosphate pathway | 3.78E-02 | miR-1, miR-125a-5p, miR-137, miR-329, miR-338-3p, miR-372, miR-542-3p |
| N-Glycan biosynthesis | 3.78E-02 | let-7c, miR-98, miR-125a-5p, miR-338-3p, miR-372, miR-381, miR-410, miR-627, miR-671-3p, miR-376c |
| Toxoplasmosis | 3.79E-02 | miR-329, miR-338-3p, miR-372, miR-410, miR-450b-5p, miR-542-3p, miR-548c-5p, miR-875-5p, miR-5542-5p |
| Epithelial cell signaling in Helicobacter pylori infection | 4.98E-02 | miR-140-3p, miR-152, miR-329, miR-338-3p, miR-372, miR-381, miR-582-3p, miR-875-5p, miR-542-5p |
miRNAs are differentially expressed in DSA+BOS+ LT recipients compared to DSA+BO− LT recipients
By comparing the miRNA profiles between DSA+BOS+ and DSA+BOS− groups, we identified 52 candidate miRNAs up regulated and 6 miRNAs down regulated (relative fold >2, p < 0.05, Figure 2A and Table S3). We then compared two differentially expressed miRNA patterns from DSA+BOS− versus stable and DSA+BOS+ versus DSA+BOS−. Twelve miRNAs were up regulated in both comparisons (Figure 3B), indicating that these differentially expressed miRNAs may play important roles in DSA mediated BOS development.
Comparative pathway analysis by DIANA-mirPath demonstrated that 39 dysregulated biologic pathways were enriched (p < 0.05, FDR corrected, Table 4) in differentially expressed miRNAs in DSA+BOS+ LTx compared to DSA+BOS− LTx. Among them, Wnt signaling (p < 0.0001), TGF-beta signal (p < 0.0001), and B cell receptor signal pathways (p < 0.0001) were ranked as the most enriched by comparing miRNA profiles between the DSA+BOS+ and DSA+BOS− groups. In addition, inositol phosphate metabolism (p < 0.0001), gap junction (p < 0.0001), and cell cycle (p < 0.0001) were also enriched when comparing the DSA+BOS+ and DSA+BOS− groups (Table 4).
Table 4. Biologic pathways enriched by differentially expressed miRNAs in DSA+BOS− versus DSA+BOS+ patients.
| Comparison | DSA+BOS+ versus DSA+BOS | |
|---|---|---|
|
|
|
|
| KEGG pathway | p-value, FDR corrected | Differentially expressed miRNAs |
| PI3K-Akt signaling pathway | 4.87E-52 | miR-133a, miR-133b, miR-137, miR-152, miR-193a-5p, miR-450b-5p, miR-483–5p, miR-489, miR-618 |
| Focal adhesion | 8.55E-45 | let-7c, miR-301b, miR-338–3p, miR-381, miR-483–5p, miR-542–3p, miR-582–3p, miR-618, miR-627 |
| Wnt signaling pathway | 1.20E-39 | miR-133b, miR-134, miR-137, miR-152, miR-193a-5p, miR-495, miR-542–3p, miR-582–3p, miR-941, miR-184 |
| Regulation of actin cytoskeleton | 5.40E-36 | let-7c, miR-134, miR-137, miR-152, miR-193a-5p, miR-338–3p, miR-450b-5p, miR-483–5p, miR-489, miR-495 |
| RNA transport | 1.74E-29 | miR-98, miR-152, miR-193a-5p, miR-301b, miR-450b-5p, miR-483–5p, miR-542–3p, miR-618, miR-184 |
| Endocytosis | 1.25E-25 | miR-134, miR-137, miR-152, miR-193a-5p, miR-301b, miR-450b-5p, miR-483–5p, miR-582–3p, miR-618 |
| Ubiquitin mediated proteolysis | 2.08E-24 | miR-152, miR-193a-5p, miR-301b, miR-450b-5p, miR-483–5p, miR-542–3p, miR-582–3p, miR-618 |
| MAPK signaling pathway | 2.08E-24 | miR-193a-5p, miR-301b, miR-331–5p, miR-338–3p, miR-381, miR-450b-5p, miR-627, miR-941, miR-184 |
| TGF-beta signaling pathway | 2.17E-23 | miR-152, miR-193a-5p, miR-301b, miR-338–3p, miR-381, miR-542–3p, miR-582–3p, miR-618 |
| ErbB signaling pathway | 2.17E-23 | let-7c, miR-1, miR-193a-5p, miR-301b, miR-338–3p, miR-542–3p, miR-582–3p, miR-627 |
| Adherens junction | 1.55E-19 | miR-98, miR-133a, miR-193a-5p, miR-301b, miR-338–3p, miR-495, miR-582–3p, miR-618 |
| mTOR signaling pathway | 8.24E-15 | miR-134, miR-137, miR-152, miR-193a-5p, miR-301b, miR-495, miR-542–3p, miR-582–3p |
| mRNA surveillance pathway | 2.11E-14 | miR-193a-5p, miR-301b, miR-338–3p, miR-381, miR-450b-5p, miR-495, miR-542–3p, miR-582–3p, miR-618 |
| Insulin signaling pathway | 3.90E-13 | let-7c, miR-152, miR-193a-5p, miR-338–3p, miR-381, miR-450b-5p, miR-489, miR-582–3p |
| Chemokine signaling pathway | 3.03E-10 | miR-152, miR-193a-5p, miR-301b, miR-338–3p, miR-381, miR-582–3p, miR-618, miR-627, miR-184 |
| Tight junction | 1.19E-09 | miR-133b, miR-134, miR-137, miR-152, miR-301b, miR-495, miR-542–3p, miR-582–3p, miR-618 |
| Bacterial invasion of epithelial cells | 1.26E-09 | miR-133a, miR-133b, miR-134, miR-381, miR-450b-5p, miR-495, miR-542–3p, miR-582–3p, miR-618, miR-627 |
| HIF-1 signaling pathway | 1.89E-09 | let-7c, miR-134, miR-137, miR-152, miR-381, miR-450b-5p, miR-489, miR-495, miR-542–3p, miR-582–3p |
| Gap junction | 2.87E-09 | let-7c, miR-1, miR-193a-5p, miR-301b, miR-338–3p, miR-381, miR-495, miR-618, miR-627 |
| Inositol phosphate metabolism | 9.12E-08 | miR-1, miR-301b, miR-338–3p, miR-381, miR-450b-5p, miR-582–3p, miR-618, miR-627 |
| RNA degradation | 6.14E-07 | let-7c, miR-98, miR-338–3p, miR-381, miR-450b-5p, miR-495, miR-542–3p, miR-582–3p |
| Hedgehog signaling pathway | 2.64E-06 | miR-133a, miR-133b, miR-137, miR-152, miR-301b, miR-338–3p, miR-495, miR-582–3p |
| Protein processing in endoplasmic reticulum | 8.65E-06 | miR-152, miR-193a-5p, miR-301b, miR-338–3p, miR-489, miR-495, miR-542–3p, miR-582–3p, miR-618 |
| Fc gamma R-mediated phagocytosis | 1.23E-05 | miR-301b, miR-338–3p, miR-381, miR-450b-5p, miR-489, miR-495, miR-542–3p, miR-582–3p, miR-184 |
| Jak-STAT signaling pathway | 2.72E-05 | let-7c, miR-193a-5p, miR-301b, miR-338–3p, miR-381, miR-542–3p, miR-582–3p, miR-627 |
| B cell receptor signaling pathway | 3.24E-05 | let-7c, miR-1, miR-137, miR-152, miR-301b, miR-495, miR-542–3p, miR-582–3p, miR-941, miR-184 |
| Cell cycle | 7.94E-05 | miR-98, miR-133a, miR-133b, miR-338–3p, miR-381, miR-450b-5p, miR-489, miR-582–3p, miR-618, miR-184 |
| Regulation of autophagy | 8.51E-05 | let-7c, miR-133b, miR-137, miR-152, miR-301b, miR-338–3p, miR-381, miR-495, miR-542–3p, miR-627 |
| Phosphatidylinositol signaling system | 8.99E-05 | miR-1, miR-133a, miR-137, miR-338–3p, miR-381, miR-450b-5p, miR-495, miR-582–3p, miR-618, miR-627 |
| Pathogenic Escherichia coli infection | 2.79E-04 | miR-133b, miR-134, miR-137, miR-152, miR-301b, miR-495, miR-542–3p, miR-582–3p, miR-618 |
| Calcium signaling pathway | 4.38E-04 | miR-134, miR-137, miR-152, miR-193a-5p, miR-450b-5p, miR-495, miR-542–3p, miR-582–3p |
| VEGF signaling pathway | 4.77E-04 | let-7c, miR-338–3p, miR-381, miR-450b-5p, miR-489, miR-582–3p, miR-941, miR-184 |
| T cell receptor signaling pathway | 5.94E-04 | let-7c, miR-1, miR-152, miR-193a-5p, miR-301b, miR-338–3p, miR-495, miR-542–3p, miR-184 |
| Epithelial cell signaling in Helicobacter pylori infection | 1.82E-03 | miR-137, miR-152, miR-301b, miR-338–3p, miR-381, miR-542–3p, miR-582–3p, miR-618, miR-627 |
| Apoptosis | 1.93E-03 | let-7c, miR-1, miR-98, miR-381, miR-450b-5p, miR-495, miR-542–3p, miR-941 |
| Pentose phosphate pathway | 6.62E-03 | miR-1, miR-133a, miR-133b, miR-137, miR-338–3p, miR-495, miR-542–3p |
| Leukocyte transendothelial migration | 1.13E-02 | let-7c, miR-1, miR-134, miR-137, miR-381, miR-450b-5p, miR-495, miR-542–3p, miR-582–3p, miR-618 |
| SNARE interactions in vesicular transport | 1.42E-02 | let-7c, miR-1, miR-98, miR-133a, miR-338–3p, miR-483–5p, miR-489, miR-495, miR-618 |
| Notch signaling pathway | 2.35E-02 | let-7c, miR-98, miR-338–3p, miR-381, miR-450b-5p, miR-489, miR-495, miR-542–3p, miR-582–3p |
Validation of miRNA array expression using independent cohorts
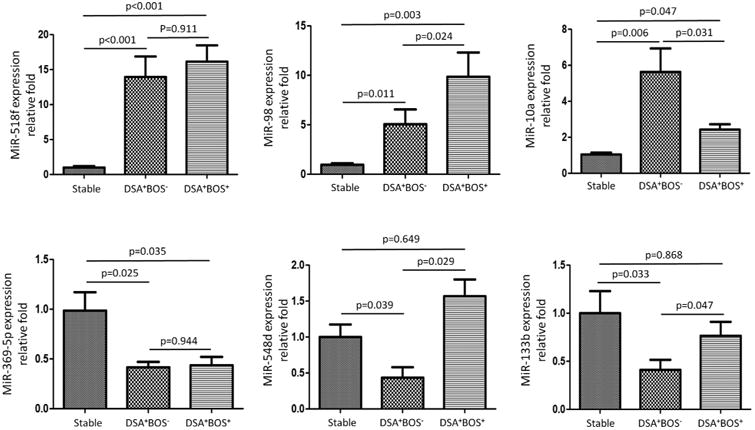
We employed TaqMan RT-PCR to validate the expression level of differentially expressed miRNAs. Six miRNAs (miR-518f, miR-98, miR-10a, miR-369-5p, miR-548d, and miR-133b) were selected for validation based on published association with T and B cell differentiation, lineage specificity and development of post-transplant fibrosis, cirrhosis, and graft loss (10). They were validated by TaqMan RT-qPCR using individual PBMC and BAL samples from three independent groups of 9 stable, 7 DSA+BOS−, and 10 DSA+BOS+ LTx. There were no significant differences in age, gender, race, underlying diseases, and medical treatment among the three groups analyzed (Table S4). In agreement with the miRNA array data, miR-518f, miR-98 and miR-10a were significantly increased in PBMCs from DSA+BOS− and DSA+BOS+ groups when compared to the stable group. MiR-369-5p, miR-548d, and miR-133b were significantly under expressed in the DSA+BOS− compared to the stable (Figure 4). Furthermore, four of them (miR-518f, miR-10a, miR-369-5p, and miR-133b) were found to be differentially expressed in BAL cells. Expression of miR-10a and miR-518f was up regulated in DSA+BOS− and DSA+BOS+ patients compared to in the stable. Expression of miR-369-5p and miR-133b was under expressed in DSA+BOS− and DSA+BOS+ patients relative to the stable (Figure 5). Taken together, independent validation by TaqMan RT-qPCR confirmed the validity of previous finding of differential expression of miRNAs using TaqMan Low-Density Array. In addition, the results demonstrate that miR-518f and miR-98 were up regulated in the DSA+BOS− group and continue to have higher levels in DSA+BOS+ group (Figures 4 and 5).
Figure 4. Validation of the miRNA array on independent patients by qPCR.
Quantitative expression of miR-518f, miR-98, miR-10a, miR-369-5p, miR-548d, and miR-133b was assessed byTaqMan real-time RT-PCR on RNA samples of PBMCs from 9 stable LT recipients, 7 LT recipients with DSA+BOS−, and 10 DSA+BOS+ LT recipients. Relative expression of these miRNAs was calculated and observed statistically differentially expressed compared to the stable group (p < 0.05, Mann–Whitney U-test). Bonferroni procedure was used to calculate adjusted p values to control FDR among stable, DSA+BOS− and DSA+BOS+ patient groups, respectively. The delta Ct values were calculated by using RNU44 as the endogenous control. All qRT-PCR reactions were performed in triplicate and the Ct values greater than 35 from the RT-PCR assays were treated as 35.
Figure 5. Validation of the differentially expressed miRNAs in BAL cells by individual qPCR.
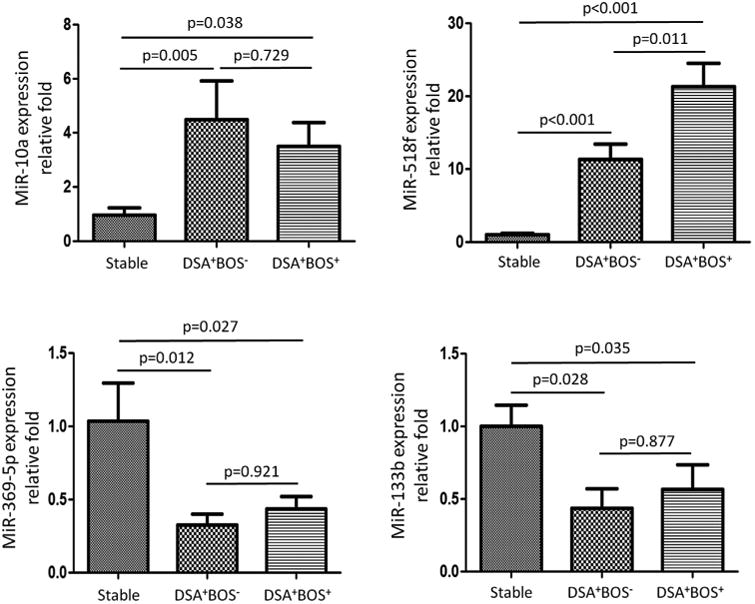
Quantitative expression of miR-518f, miR-10a, miR-369-5p, and miR-133b was evaluated by TaqMan RT-PCR in RNA samples of BAL cells from 9 stable LT recipients, 7 LT recipients with DSA+BOS−, and 10 DSA+BOS+ LT recipients. The delta Ct values were calculated by using RNU44 as the endogenous control. The Mann-Whitney test was performed to determine the significance of miRNA levels of BAL cells. Bonferroni procedure was used to calculate adjusted p values to control FDR among stable, DSA+BOS− and DSA+BOS+ patient groups, respectively.
Correlation of differentially expressed miRNAs in TGF beta signal pathway with the target genes
We further determined the potential target genes for differentially expressed miRNAs based on pathway analysis via the mirPath algorithm. We focused on the TGF beta signaling pathway for further investigation since up regulation of the TGF-beta signaling pathway has been known to be a risk factor for BOS after LT (23). Three down-regulated miRNAs (miR-133b, miR-369-3p, and miR-548d) were associated with up regulation of the TGF beta signaling pathway by regulating their gene targets (Figure 6A). Further RT-qPCR confirmed that miR-369-3p was down-regulated while its gene target, latent TGF-beta binding proteins (LTBP1) (encoding latent TGF-beta binding proteins), mRNA expression was up regulated in DSA+BOS+ versus stable patients (Pearson's correlation p-value: p < 0.001, R2 = 0.981, Figure 6B); Similarly, there was an inverse correlation between expression of miR-369-3p and LTBP1 mRNA in DSA+BOS− versus stable patients (p < 0.001, R2 = 0.969, Figure 6B). We further observed that miR-548d was down regulated, consistent with up regulation of its potential target, TGFRB1 (encoding transforming growth factor, beta receptor 1) mRNA expression in DSA+BOS+ versus stable patients (p < 0.001, R2 = 0.431, Figure 6C). An inverse correlation between expression of miR-548d and TGFBR1 mRNA were also found in DSA+BOS− patients compared to those in the stable group (p < 0.001, R2 = 0.672, Figure 6C). These results clearly demonstrate that differentially expressed miRNAs regulate the TGF beta signaling pathway which in turn leads to deregulation of their target genes, de novo DSA development and BOS.
Figure 6. Differentially expressed miRNAs in TGF beta signal pathway.
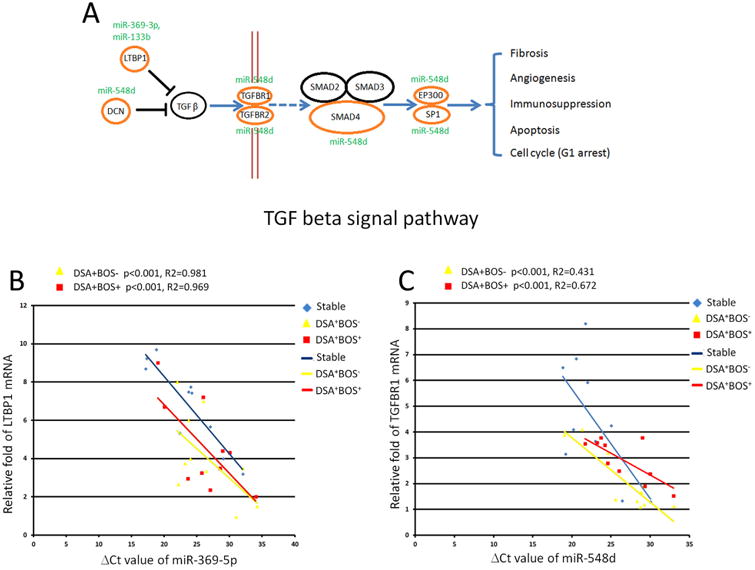
(A) Diagram of TGF beta signal pathway. Three underexpressed miRNAs (miR-369-3p, miRNA-133b, and miR-548d) were predicted to participate in TGF beta signal pathway through regulating their potential gene targets. (B) miRNA-369-5p was underexpressed while its potential target, LTBP1, was up-regulated in the DSA+BOS− or DSA+BOS+ patients. The p values were from Pearson's correlation tests. Blue dots represent the stable samples. Yellow dots represent DSA+BOS− patients' samples, and red dots represent the DSA+BOS+ patient samples. X-axis: miRNA expression; Y-axis: mRNA expression. (C) miR-548d was down regulated with inversely up-regulation of TGFBR1 mRNA levels in the DSA+BOS− or DSA+BOS+ patients.
Correlation of differentially expressed miRNAs in B cell receptor signal pathway with the target genes
B cell receptor signaling pathways play a crucial role in humoral responses and have been implicated in chronic rejection (24, 25). We therefore selected two differentially expressed miRNAs (miR-628–5p and miR-134) to determine their roles in B cell receptor signaling and development of DSA and BOS (Figure 7A). MiR-628–5p was down-regulated while the mRNA for its potential target gene, BTK (Bruton agammaglobulinemia tyrosine kinase), was up regulated in DSA+BOS− patients compared to stable (p < 0.001, R2 = 0.623, Figure 7B). There was an inverse correlation between expression of miR-628–5p and BTK mRNA in DSA+BOS+ versus stable patients (p < 0.001, R2 = 0.752, Figure 7B). Similarly, miR-134 which was reduced and its target gene, BLNK (B cell linker) mRNA expression was increased in DSA+BOS- subjects relative to that of the stable (p < 0.001, R2 = 0.992, Figure 7C). We also noted an inverse correlation between expression of miR-134 and BLNK mRNA found in DSA+BOS+ patients compared to those in the stable (p < 0.001, R2 = 0.863, Figure 7C). These data indicate that differentially expressed miRNAs regulating B cell receptor signal pathway can dysregulate their target genes in LTx with de novo development of DSA leading to BOS.
Figure 7. Differentially expressed miRNAs in B cell receptor signal pathway.
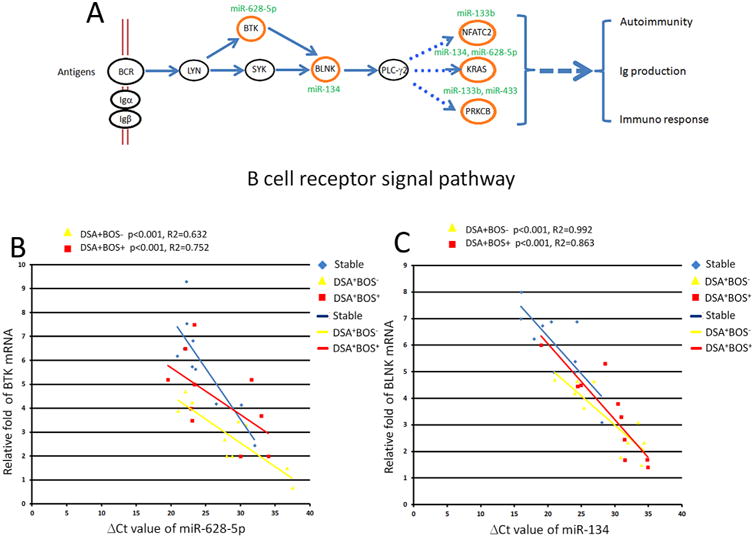
(A) Diagram of B cell receptor signal pathway. Four differentially expressed miRNAs (including miR-133b, miR-134, miR-433, and miR-628-5p) were selected as examples for examining miRNA-mRNA relationships in B cell receptor signal pathway. (B) miRNA-628-5p was down-regulated in the DSA+BOS− patients with over expression of its potential target, BTK. (C) miRNA-134 was down-regulated in the DSA+BOS− or DSA+BOS+ patients, while its potential target, BLNK, was up-regulated.
Discussion
The goal of the present study was to investigate whether miRNAs are aberrantly expressed in LT recipients who develop DSA leading to BOS. In order to determine differential expression of miRNAs, we generated three pools of PBMCs from stable, DSA+BOS− and DSA+BOS+ LT patients. Using Taqman Low-Density miRNA Array method we evaluated the miRNA expression profiles and a relative change of at least twofold was deemed significant. Comprehensive pathway analyses indicated that dysregulated biologic processes including B cell, Toll-like, and T cell receptor pathways, TGF-β and chemokine signal pathways, and cytokine-cytokine receptor interactions were significantly altered in DSA+BOS− and DSA+BOS+ LT patients. Chronic allograft rejection has been attributed to immune responses to donor HLA (26,27). A strong correlation between de novo development of DSA and BOS following human LT has been reported (28,29). Results from the current study provide evidence that development of DSA after LT, can induce differential expression of miRNA that influences the overall immunity. We further demonstrate a differential expression of four miRNAs (miR-10a, miR-518f, miR-369-5p, and miR-133b) at the local site, that is, in the cells from BAL fluid. MiR-10a is known to be involved in the commitment and differentiation of hematopoietic cells (30,31), and has a crucial role in the stability and functioning of regulatory T cells (Tregs) (31). In this study, miR-10a expression was significantly increased in PBMC from DSA+BOS− and DSA+BOS+ patients. The miR-133b located in the IL17A/F locus is known to be co-regulated with IL-17 production in αβ and γδ T cells (32). We found that miR-133b was significantly under expressed in the DSA+BOS− group compared to that of stable. Therefore, dysregulation of these two miRNAs: miR-10a and miR-133b in LT patients possibly lead to Treg cells dysfunction and aberrant IL-17 production, which have been proposed as the key mediators in the development of BOS. Analysis of Treg and IL-17 dependent pathways following DSA development after LT and its correlation to BOS need further investigation.
Of particular importance is our finding of significant up regulation of the TGF beta signal pathway both in the DSA+BOS− as well as DSA+BOS+ groups. Persistent dysregulation in LT patients who developed BOS is of great interest as it suggests that development of alloimmunity induces this change early and persists during BOS development. TGF-β expression has been reported to be increased in patients with BOS even before the onset of airflow obstruction (33,34). TGF-β itself was not up regulated in the sample set of miRNAs chosen, but downstream mediators including the TGF beta receptors and LTBP1, were markedly up regulated (Figure 6B and Figure 6C). Additionally, the Wnt signaling pathway is also of interest since Wnts have diverse roles in governing cell fate, proliferation, migration, polarity, and death (35). The significance of the Wnt pathway in allograft rejection and in BOS following human LT is currently unknown. Moreover, DSA+BOS+ patients also demonstrated dysregulation of miRNAs, which are involved in the cell cycle and gap junction signaling pathways. It is of interest that the development of Abs to lung associated self-antigen K-alpha 1 tubulin, an epithelial gap junction, significantly correlates with the development of BOS (36).
There are limitations in the present study. First, using miRNA array methods to analyze miRNA expression patterns is an evolving area. The standard methods of statistical analysis and analysis of significance have not yet been firmly established. In particular, choosing the appropriate threshold for determining statistical significance is, in part, dependent on the purpose of the analysis. We evaluated the miRNA expression profiles with greater than twofold increase because of the optimization of numbers of candidate miRNAs and high specificity of results afforded by this level of statistical stringency. Secondly, we failed to validate the elevated expression of miR-875-5p, miR-548c-5p, and miR-548d-5p in the DSA+BOS+ patients. A probable reason for this discrepancy is that although the TaqMan miRNA array is promising for simultaneously measuring miRNA expression in a high-throughput manner (37,38) and the introduction of an miRNA cDNA pre-amplification step significantly reduces the amount of RNA needed (39). It has a potential for pre-amplification bias that may be responsible for variations in the Ct values. This may diminish the reproducibility with miRNAs of low abundance. Future studies should include stratification analysis of a larger cohort to eliminate sample selection biases. Finally, a larger sample size may be needed to more accurately determine how anti-HLA can dysregulate the transcriptional activation and regulation of miRNA expression patterns in LT patients.
In summary, our study, for the first time, demonstrated that the alloimmune response as evidenced by development of DSA following human LT leads to changes in miRNA expression affecting primarily the TGF-beta and B cell receptor signal pathways. This is noted early following LT when there is no evidence of BOS. These miRNAs changes persist and expand to other cell cycle signal pathways in LT patients during development of BOS. The clinical significance of these findings include potential use of miRNAs as early markers for identifying risk factors in development of BOS which needs further investigation.
Supplementary Material
Figure S1: Method employed for estimating the number of patients by pooled and non-pooled designs. The efficiency of pooling ten patients into one sample was determined based on biological and technical variations within the assay as described earlier (21). The plot shows equivalent number of patients when ten samples were not pooled for independent microRNA arrays. The varying lambda (λ = σb2/σe2) is the ratio of underlying biological variations among the ten samples over the technical variations among the assays.
Figure S2: The KEGG pathway “TGF-beta signaling pathway” was significantly altered between DSAþBOS-LTx patients and Stable patients subjects at baseline, with 17 miRNAs (hsa-let-7c, hsa-miR-98, hsa-miR-125a-5p, hsa-miR-137, hsa-miR-203, hsa-miR-299-5p, hsa-miR-338-3p, hsa-miR-381, hsa-miR-548c-5p, hsa-miR-874, hsa-miR-875-5p, hsa-miR-133b, hsa-miR-134, hsa-miR-433, hsa-miR-494, hsa-miR-503, hsa-miR-628-5p) targeting 46 genes. Targeted genes are indicated by yellow (one miRNA) and brown (multiple miRNAs) colors in the pathway map.
Figure S3: The KEGG pathway “TGF-beta signaling pathway” was significantly altered between DSA+BOS+LTx patients and Stable patients subjects at baseline, with 29 miRNAs (hsa-let-7c, hsa-miR-1, hsa-miR-98, hsa-miR-125a-5p, hsa-miR-137, hsa-miR-140-3p, hsa-miR-152, hsa-miR-193a-5p, hsa-miR-203, hsa-miR-329, hsa-miR-331-5p, hsa-miR-338-3p, hsa-miR-372, hsa-miR-381, hsa-miR-410, hsa-miR-422a, hsa-miR-450b-5p, hsa-miR-542-3p, hsa-miR-548c-5p, hsa-miR-548d-5p, hsa-miR-582-3p, hsa-miR-627, hsa-miR-671-3p, hsa-miR-219-1-3p, hsa-miR-376c, hsa-miR-511, hsa-miR-941, hsa-miR-875-5p, hsa-miR-542-5p) targeting 126 genes. Targeted genes are indicated by yellow (one miRNA) and brown (multiple miRNAs) colors in the pathway map.
Figure S4: The KEGG pathway “B cell receptor signaling pathway” was significantly altered between DSA+BOS-LTx patients and Stable patients subjects at baseline, with 20 miRNAs (hsa-let-7c, hsa-miR-98, hsa-miR-125a-5p, hsa-miR-137, hsa-miR-184, hsa-miR-203, hsa-miR-299-5p, hsa-miR-338-3p, hsa-miR-381, hsa-miR-548c-5p, hsa-miR-627, hsa-miR-874, hsa-miR-133b, hsa-miR-134, hsa-miR-433, hsa-miR-489, hsa-miR-494, hsa-miR-503, hsa-miR-542-5p, hsa-miR-628-5p) targeting 34 genes. Targeted genes are indicated by yellow (one miRNA) and brown (multiple miRNAs) colors in the pathway map.
Figure S5: The KEGG pathway “B cell receptor signaling pathway” was significantly altered between DSA+BOS+LTx patients and Stable patients subjects at baseline, with 24 miRNAs (hsa-let-7c, hsa-miR-1, hsa-miR-98, hsa-miR-125a-5p, hsa-miR-137, hsa-miR-140-3p, hsa-miR-152, hsa-miR-203, hsa-miR-329, hsa-miR-338-3p, hsa-miR-372, hsa-miR-381, hsa-miR-410, hsa-miR-450b-5p, hsa-miR-542-3p, hsa-miR-548c-5p, hsa-miR-548d-5p, hsa-miR-582-3p, hsa-miR-627, hsa-miR-219-1-3p, hsa-miR-376c, hsa-miR-511, hsa-miR-941, hsa-miR-542-5p) targeting 34 genes. Targeted genes are indicated by yellow (one miRNA) and brown (multiple miRNAs) colors in the pathway map.
Table S1: Differential miRNAs when DSA+BOS- versus stable.
Table S2: Differential miRNAs when DSA+BOS+ versus stable.
Table S3: Differential microRNAs when DSA+BOS+ versus DSA+BOS-.
Table S4: Demographic data on the lung transplant recipients in the replicative independent cohort.
Acknowledgments
We thank Billie Glasscock for her assistance in preparing this manuscript. This work is supported by NIH RO1 HL056643-13 and the BJC foundation (TM).
Abbreviations
- Abs
antibodies
- BAL
bronchoalveolar lavage
- BOS
bronchiolitis obliterans syndrome
- BLNK
B cell linker
- BTK
Bruton agammaglobulinemia tyrosine kinase
- DSA
antibodies to mismatched donor HLA
- HLA
human leukocyte antigens
- LT
lung transplantation
- LTBP1
latent TGF-beta binding proteins
- miRNA
microRNA
- OAD
obliterative airway disease
- PBMCs
peripheral blood mononuclear cells
- RT-qPCR
Real Time quantitative PCR
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- TGFRB1
transforming growth factor beta receptor 1
- Tregs
regulatory T cells
Footnotes
Disclosure: The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
Supporting Information: Additional supporting information may be found in the online version of this article.
References
- 1.Arcasoy SM, Kotloff RM. Lung transplantation. N Engl JMed. 1999;340:1081–1091. doi: 10.1056/NEJM199904083401406. [DOI] [PubMed] [Google Scholar]
- 2.Trulock EP, Edwards LB, Taylor DO, Boucek MM, Keck BM, Hertz MI. Registry of the International Society for Heart and Lung Transplantation: Twenty-third official adult lung and heart-lung transplantation report–2006. J Heart Lung Transplant. 2006;25:880–892. doi: 10.1016/j.healun.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Estenne M, Hertz MI. Bronchiolitis obliterans after human lung transplantation. Am J Respir Crit Care Med. 2002;166:440–444. doi: 10.1164/rccm.200201-003pp. [DOI] [PubMed] [Google Scholar]
- 4.Jaramillo A, Smith MA, Phelan D, et al. Development of ELISA-detected anti-HLA antibodies precedes the development of bronchiolitis obliterans syndrome and correlates with progressive decline in pulmonary function after lung transplantation. Transplantation. 1999;67:1155–1161. doi: 10.1097/00007890-199904270-00012. [DOI] [PubMed] [Google Scholar]
- 5.Moschl P, Lubec G, Keiler A, Salem G, Glockler M. Donor- and organ-specific evaluation of antibodies eluted from canine lung allografts rejected by immunosuppressively treated and untreated recipients. Respiration. 1979;38:12–17. doi: 10.1159/000194053. [DOI] [PubMed] [Google Scholar]
- 6.FukamiN, Ramachandran S, Saini D, et al. Antibodies to MHC class I induce autoimmunity: role in the pathogenesis of chronic rejection. J Immunol. 2009;182:309–318. doi: 10.4049/jimmunol.182.1.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 8.Wang JW, Li K, Hellermann G, Lockey RF, Mohapatra S, Mohapatra S. Regulating the Regulators: microRNA and asthma. World Allergy Organ J. 2011;4:94–103. doi: 10.1186/1939-4551-4-6-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furer V, Greenberg JD, Attur M, Abramson SB, Pillinger MH. The role of microRNA in rheumatoid arthritis and other autoimmune diseases. Clin Immunol. 2010;136:1–15. doi: 10.1016/j.clim.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Sarma NJ, Tiriveedhi V, Ramachandran S, Crippin J, Chapman W, Mohanakumar T. Modulation of immune responses following solid organ transplantation by microRNA. Exp Mol Pathol. 2012;93:378–385. doi: 10.1016/j.yexmp.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scian MJ, Maluf DG, Mas VR. MiRNAs in kidney transplantation: potential role as new biomarkers. Expert Rev Mol Diagn. 2013;13:93–104. doi: 10.1586/erm.12.131. [DOI] [PubMed] [Google Scholar]
- 12.Sui W, Lin H, Peng W, et al. Molecular dysfunctions in acute rejection after renal transplantation revealed by integrated analysis of transcription factor, microRNA and long noncoding RNA. Genomics. 2013;102:310–322. doi: 10.1016/j.ygeno.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 13.Sui W, Dai Y, Huang Y, Lan H, Yan Q, Huang H. Microarray analysis of microRNA expression in acute rejection after renal transplantation. Transpl Immunol. 2008;19:81–85. doi: 10.1016/j.trim.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 14.Estenne M, Maurer JR, Boehler A, et al. Bronchiolitis obliterans syndrome 2001: An update of the diagnostic criteria. J Heart Lung Transplant. 2002;21:297–310. doi: 10.1016/s1053-2498(02)00398-4. [DOI] [PubMed] [Google Scholar]
- 15.Bach MK, Brashler JR. Isolation of subpopulations of lymphocytic cells by the use of isotonically balanced solutions of Ficoll. I. Development of methods and demonstration of the existence of a large but finite number of subpopulations. Exp Cell Res. 1970;61:387–396. doi: 10.1016/0014-4827(70)90462-3. [DOI] [PubMed] [Google Scholar]
- 16.Papadopoulos GL, Alexiou P, Maragkakis M, Reczko M, Hatzi-georgiou AG. DIANA-mirPath: Integrating human and mouse microRNAs in pathways. Bioinformatics. 2009;25:1991–1993. doi: 10.1093/bioinformatics/btp299. [DOI] [PubMed] [Google Scholar]
- 17.Reczko M, Maragkakis M, Alexiou P, Grosse I, Hatzigeorgiou AG. Functional microRNA targets in protein coding sequences. Bioinformatics. 2012;28:771–776. doi: 10.1093/bioinformatics/bts043. [DOI] [PubMed] [Google Scholar]
- 18.Paraskevopoulou MD, Georgakilas G, Kostoulas N, et al. DIANA-microT web server v5.0: Service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41:W169–W173. doi: 10.1093/nar/gkt393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu SD, Lin FM, Wu WY, et al. MiRTarBase: A database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011;39:D163–D169. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kainkaryam RM, Bruex A, Woolf PJ, Schiefelbein J. Smart pooling of mRNA samples for efficient transcript profiling. Methods Mol Biol. 2012;876:189–194. doi: 10.1007/978-1-61779-809-2_15. [DOI] [PubMed] [Google Scholar]
- 21.Kendziorski CM, Zhang Y, Lan H, Attie AD. The efficiency of pooling mRNA in microarray experiments. Biostatistics. 2003;4:465–477. doi: 10.1093/biostatistics/4.3.465. [DOI] [PubMed] [Google Scholar]
- 22.Kowalski A, Enck P. Statistical methods: multiple significance tests and the Bonferroni procedure. Psychother Psychosom Med Psychol. 1995;60:286–287. doi: 10.1055/s-0030-1248493. [DOI] [PubMed] [Google Scholar]
- 23.El-Gamel A, Sim E, Hasleton P, et al. Transforming growth factor beta (TGF-beta) and obliterative bronchiolitis following pulmonary transplantation. J Heart Lung Transplant. 1999;18:828–837. doi: 10.1016/s1053-2498(99)00047-9. [DOI] [PubMed] [Google Scholar]
- 24.Morscio J, Dierickx D, Tousseyn T. Molecular pathogenesis of B-cell posttransplant lymphoproliferative disorder: What do we know so far. Clin Dev Immunol. 2013;150835 doi: 10.1155/2013/150835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukami N, Ramachandran S, Takenaka M, Weber J, Subramanian V, Mohanakumar T. An obligatory role for lung infiltrating B cells in the immunopathogenesis of obliterative airway disease induced by antibodies to MHC class I molecules. Am J Transplant. 2012;12:867–876. doi: 10.1111/j.1600-6143.2011.03917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lachmann N, Terasaki PI, Budde K, et al. Anti-human leukocyte antigen and donor-specific antibodies detected by luminex posttransplant serve as biomarkers for chronic rejection of renal allografts. Transplantation. 2009;87:1505–1513. doi: 10.1097/TP.0b013e3181a44206. [DOI] [PubMed] [Google Scholar]
- 27.Terasaki PI, Cai J. Human leukocyte antigen antibodies and chronic rejection: From association to causation. Transplantation. 2008;86:377–383. doi: 10.1097/TP.0b013e31817c4cb8. [DOI] [PubMed] [Google Scholar]
- 28.Jaramillo A, Smith MA, Phelan D, et al. Temporal relationship between the development of anti-HLA antibodies and the development of bronchiolitis obliterans syndrome after lung transplantation. Transplant Proc. 1999;31:185–186. doi: 10.1016/s0041-1345(98)01495-x. [DOI] [PubMed] [Google Scholar]
- 29.Palmer SM, Davis RD, Hadjiliadis D, et al. Development of an antibody specific to major histocompatibility antigens detectable by flow cytometry after lung transplant is associated with bronchiolitis obliterans syndrome. Transplantation. 2002;74:799–804. doi: 10.1097/00007890-200209270-00011. [DOI] [PubMed] [Google Scholar]
- 30.Undi RB, Kandi R, Gutti RK. MicroRNAs as Haematopoiesis Regulators. Adv Hematol. 2013;695754 doi: 10.1155/2013/695754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeker LT, Zhou X, Gershberg K, et al. MicroRNA 10a marks regulatory T cells. PLoS ONE. 2012;7:e36684. doi: 10.1371/journal.pone.0036684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haas JD, Nistala K, Petermann F, et al. Expressionof miRNAs miR-133b and miR-206 in the Il17a/f locus is co-regulated with IL-17 production in alphabeta and gammadelta T cells. PLoS ONE. 2011;6:e20171. doi: 10.1371/journal.pone.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charpin JM, Valcke J, Kettaneh L, Epardeau B, Stern M, Israel-Biet D. Peaks of transforming growth factor-beta mRNA inalveolar cells of lung transplant recipients as an early marker ofchronic rejection. Transplantation. 1998;65:752–755. doi: 10.1097/00007890-199803150-00027. [DOI] [PubMed] [Google Scholar]
- 34.Magnan A, Mege JL, Escallier JC, et al. Balance between alveolar macrophage IL-6 and TGF-beta in lung-transplant recipients. Marseille and Montreal Lung Transplantation Group. Am J Respir Crit Care Med. 1996;153:1431–1436. doi: 10.1164/ajrccm.153.4.8616577. [DOI] [PubMed] [Google Scholar]
- 35.Huelsken J, Behrens J. The Wnt signalling pathway. J Cell Sci. 2002;115:3977–3978. doi: 10.1242/jcs.00089. [DOI] [PubMed] [Google Scholar]
- 36.Goers TA, Ramachandran S, Aloush A, Trulock E, Patterson GA, Mohanakumar T. De novo production of K-alpha1 tubulin-specific antibodies: Role inchronic lung allograft rejection. J Immunol. 2008;180:4487–4494. doi: 10.4049/jimmunol.180.7.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hui AB, Shi W, Boutros PC, et al. Robust global micro-RNA profiling with formalin-fixed paraffin-embedded breast cancer tissues. Lab Invest. 2009;89:597–606. doi: 10.1038/labinvest.2009.12. [DOI] [PubMed] [Google Scholar]
- 38.Lu B, Xu J, Chen J, Yu J, Xu E, Lai M. TaqMan low density array is roughly right for gene expression quantification in colorectal cancer. Clin Chim Acta. 2008;389:146–151. doi: 10.1016/j.cca.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 39.Mestdagh P, Feys T, Bernard N, et al. High-throughput stem-loop RT-qPCR miRNA expression profiling using minute amounts of input RNA. Nucleic Acids Res. 2008;36:e143. doi: 10.1093/nar/gkn725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Method employed for estimating the number of patients by pooled and non-pooled designs. The efficiency of pooling ten patients into one sample was determined based on biological and technical variations within the assay as described earlier (21). The plot shows equivalent number of patients when ten samples were not pooled for independent microRNA arrays. The varying lambda (λ = σb2/σe2) is the ratio of underlying biological variations among the ten samples over the technical variations among the assays.
Figure S2: The KEGG pathway “TGF-beta signaling pathway” was significantly altered between DSAþBOS-LTx patients and Stable patients subjects at baseline, with 17 miRNAs (hsa-let-7c, hsa-miR-98, hsa-miR-125a-5p, hsa-miR-137, hsa-miR-203, hsa-miR-299-5p, hsa-miR-338-3p, hsa-miR-381, hsa-miR-548c-5p, hsa-miR-874, hsa-miR-875-5p, hsa-miR-133b, hsa-miR-134, hsa-miR-433, hsa-miR-494, hsa-miR-503, hsa-miR-628-5p) targeting 46 genes. Targeted genes are indicated by yellow (one miRNA) and brown (multiple miRNAs) colors in the pathway map.
Figure S3: The KEGG pathway “TGF-beta signaling pathway” was significantly altered between DSA+BOS+LTx patients and Stable patients subjects at baseline, with 29 miRNAs (hsa-let-7c, hsa-miR-1, hsa-miR-98, hsa-miR-125a-5p, hsa-miR-137, hsa-miR-140-3p, hsa-miR-152, hsa-miR-193a-5p, hsa-miR-203, hsa-miR-329, hsa-miR-331-5p, hsa-miR-338-3p, hsa-miR-372, hsa-miR-381, hsa-miR-410, hsa-miR-422a, hsa-miR-450b-5p, hsa-miR-542-3p, hsa-miR-548c-5p, hsa-miR-548d-5p, hsa-miR-582-3p, hsa-miR-627, hsa-miR-671-3p, hsa-miR-219-1-3p, hsa-miR-376c, hsa-miR-511, hsa-miR-941, hsa-miR-875-5p, hsa-miR-542-5p) targeting 126 genes. Targeted genes are indicated by yellow (one miRNA) and brown (multiple miRNAs) colors in the pathway map.
Figure S4: The KEGG pathway “B cell receptor signaling pathway” was significantly altered between DSA+BOS-LTx patients and Stable patients subjects at baseline, with 20 miRNAs (hsa-let-7c, hsa-miR-98, hsa-miR-125a-5p, hsa-miR-137, hsa-miR-184, hsa-miR-203, hsa-miR-299-5p, hsa-miR-338-3p, hsa-miR-381, hsa-miR-548c-5p, hsa-miR-627, hsa-miR-874, hsa-miR-133b, hsa-miR-134, hsa-miR-433, hsa-miR-489, hsa-miR-494, hsa-miR-503, hsa-miR-542-5p, hsa-miR-628-5p) targeting 34 genes. Targeted genes are indicated by yellow (one miRNA) and brown (multiple miRNAs) colors in the pathway map.
Figure S5: The KEGG pathway “B cell receptor signaling pathway” was significantly altered between DSA+BOS+LTx patients and Stable patients subjects at baseline, with 24 miRNAs (hsa-let-7c, hsa-miR-1, hsa-miR-98, hsa-miR-125a-5p, hsa-miR-137, hsa-miR-140-3p, hsa-miR-152, hsa-miR-203, hsa-miR-329, hsa-miR-338-3p, hsa-miR-372, hsa-miR-381, hsa-miR-410, hsa-miR-450b-5p, hsa-miR-542-3p, hsa-miR-548c-5p, hsa-miR-548d-5p, hsa-miR-582-3p, hsa-miR-627, hsa-miR-219-1-3p, hsa-miR-376c, hsa-miR-511, hsa-miR-941, hsa-miR-542-5p) targeting 34 genes. Targeted genes are indicated by yellow (one miRNA) and brown (multiple miRNAs) colors in the pathway map.
Table S1: Differential miRNAs when DSA+BOS- versus stable.
Table S2: Differential miRNAs when DSA+BOS+ versus stable.
Table S3: Differential microRNAs when DSA+BOS+ versus DSA+BOS-.
Table S4: Demographic data on the lung transplant recipients in the replicative independent cohort.


