Abstract
The objective of this article is to introduce preparation and characterization methods for nerve growth factor (NGF)-loaded, high-density, lipoprotein (HDL)-mimicking nanoparticles (NPs). HDLs are endogenous NPs and have been explored as vehicles for the delivery of therapeutic agents. Various methods have been developed to prepare HDL-mimicking NPs. However, they are generally complicated, time consuming, and difficult for industrial scale-up. In this study, one-step homogenization was used to mix the excipients and form the prototype NPs. NGF is a water-soluble protein of 26 kDa. To facilitate the encapsulation of NGF into the lipid environment of HDL-mimicking NPs, protamine USP was used to form an ion-pair complex with NGF to neutralize the charges on the NGF surface. The NGF/protamine complex was then introduced into the prototype NPs. Apolipoprotein A-I was finally coated on the surface of the NPs. NGF HDL-mimicking NPs showed preferable properties in terms of particle size, size distribution, entrapment efficiency, in vitro release, bioactivity, and biodistribution. With the careful design and exploration of homogenization in HDL-mimicking NPs, the procedure was greatly simplified, and the NPs were made scalable. Moreover, various challenges, such as separating unloaded NGF from the NPs, conducting reliable in vitro release studies, and measuring the bioactivity of the NPs, were overcome.
Keywords: Bioengineering, Issue 123, macromolecule, homogenization, gel filtration chromatography, in vitro release, neurite outgrowth, biodistribution
Introduction
Macromolecules, such as proteins, peptides, and nucleic acids, have been emerging as promising medications and have gained considerable attention in past decades1,2. Due to their high efficacy and specific action modes, they exhibit great therapeutic potential for the treatments of cancer, immune disease, HIV, and related conditions3,4. However, physiochemical properties, such as their large molecular size, three-dimensional structure, surface charges, and hydrophilic nature, make the in vivo delivery of these macromolecules very challenging. This considerably impedes their clinical use4. Recent advancements in drug delivery systems, such as microparticles, polymer nanoparticles (NPs), liposomes, and lipid NPs, overcame these challenges and significantly improved the in vivo delivery of macromolecules. However, some drawbacks regarding these delivery cargoes have been revealed, including low drug loading capacity, low entrapment efficiency, short half-life, loss of bioactivity, and undesirable side effects5,6,7,8. Effective carrier systems remain an area of research interest. Moreover, the development of analytical methods to characterize drug-loaded NPs is more challenging for macromolecules than for small molecules.
High-density lipoprotein (HDL) is a natural NP composed of a lipid core that is coated by apolipoproteins and a phospholipid monolayer. Endogenous HDL plays a critical role in the transport of lipids, proteins, and nucleic acids through its interaction with target receptors, such as SR-BI, ABCAI, and ABCG1. It has been explored as a vehicle for the delivery of different therapeutic agents9,10,11,12. Various methods have been developed to prepare HDL-mimicking NPs. Dialysis is a popular approach. In this method, NPs are formed by hydrating a lipid film using sodium cholate solution. The salt is then removed through a 2-day dialysis with three buffers13. Sonication methods fabricate NPs by sonicating a lipid mixture for 60 min under a heating condition; the NPs are further purified through gel chromatography14. Microfluidics generates NPs via a microfluidic device, which mixes phospholipids and apolipoprotein A-I (Apo A-I) solutions by creating microvortices in a focusing pattern15. Clearly, these methods can be time consuming, harsh, and difficult for industrial scale-up.
In this article, we introduce the preparation and characterization of novel HDL-mimicking NPs for nerve growth factor (NGF) encapsulation. NGF is a disulfide-linked polypeptide homodimer containing two 13.6-kDa polypeptide monomers. A novel procedure to prepare the NPs by homogenization, followed by the encapsulation of NGF into the NPs, was developed. The NGF HDL-mimicking NPs were characterized for particle size, size distribution, zeta potential, and in vitro release. Their bioactivity was evaluated for neurite outgrowth in PC12 cells. The biodistribution of NGF HDL-mimicking NPs was compared with that of free NGF after intravenous injection in mice.
Protocol
NOTE: The animal studies included in all procedures have been approved by the Institutional Animal Care and Use Committee at the University of North Texas Health Science Center.
1. Preparation of NGF HDL-mimicking Nanoparticles
Dissolve the excipients, phosphatidylcholine (PC), sphingomyelin (SM), phosphatidylserine (PS), cholesteryl oleate (CO), and D-α-tocopheryl polyethylene glycol succinate (TPGS), in ethanol to prepare stock solutions at 1 mg/mL. NOTE: The stock solutions were aliquoted and stored at -20 °C. The cholesteryl oleate was stored in dark bottles. The PC, SM, PS, and CO stock solutions were stable for up to 6, 3, 12, and 12 months, respectively, at -20 °C. The TPGS solution was stable for at least 12 months at -20 °C.
Mix 10 µL of NGF (1 mg/mL in water) with 10 µL of protamine USP (1 mg/mL in water) in a 1.5 mL microcentrifuge tube and let it stand for 10 min at room temperature to form the complex. NOTE: The NGF and protamine stock solutions were aliquoted and stored at -20 °C. Repeated freezing and thawing is not recommended for the NGF stock.
Add 59 µL of PC, 11 µL of SM, 4 µL of PS, 15 µL of CO, and 45 µL of TPGS to a glass vial. Mix and evaporate the ethanol under a gentle nitrogen stream for about 5 min; all excipients should form an oily, thin film at the bottom of the glass vial.
Add 1 mL of ultrapure (type 1) water to the vial and homogenize at 9,500 rpm (8,600 x g) for 5 min at room temperature to form the prototype NPs.
- Add the complex prepared in step 1.2 to the prototype NPs and incubate at 37 °C for 30 min with stirring by using a small stirring bar in the glass vial.
- Cool the NPs down by stirring at room temperature for another 30 min. After this cools, add 106 µL of Apo A-I (1.49 mg/mL) and stir at room temperature overnight to form the final NGF HDL-mimicking NPs.
2. Characterization of NGF HDL-mimicking Nanoparticles
Measure the particle size and zeta potential using a particle analyzer (see the Materials Table) as per the manufacturer's instructions.
- Use a cross-linked agarose gel filtration chromatography column to separate the unloaded NGF from the NGF HDL-mimicking NPs and determine the entrapment efficiency of the NGF.
- For the column preparation, transfer 15 mL of Sepharose 4B-CL suspension to a 50-mL beaker. Stir the bead suspension using a glass rod and pour some into a column (30 cm length × 1 cm diameter, with a glass frit at the bottom). Gently tap the column to get rid of bubbles. Allow the solvent to drain and the beads to settle for a few min.
- Continue to add the remaining suspension. Rinse the inside wall of column to clean the beads. Drain the solvent until the solvent level is slightly above the top of the stationary phase. Wash and condition the column with 20 mL of 1x phosphate-buffered saline (PBS, containing 137 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, and 2 mM KH2PO4).
- To determine the fractions containing unloaded NGF, load 200 µL of NGF solution (10 µg/mL) onto the gel filtration column (25 cm length x 1 cm diameter) and elute with 1x PBS.
- Collect a total of 12 fractions (1 mL for each fraction) and measure the concentration of NGF in each fraction using a Sandwich ELISA kit for NGF.
- Detect NGF from fractions 6 to 10 using the Sandwich ELISA kit. Obtain a chromatogram of unloaded NGF on the column. Wash the column with 20 mL of PBS.
- To determine the fractions containing NGF HDL-mimicking NPs, load 200 µL of the NPs onto the column and elute with 1x PBS. Collect a total of 12 fractions (1 mL for each fraction) and measure the intensity of particles in each fraction using the particle size analyzer. Detect the NGF NPs from fractions 2 to 4. NOTE: The intensity is a parameter measured by the particle analyzer, which tells how many nanoparticles may exist in the test solution. The more NPs exist in solution, the higher the intensity is. By this approach, we can confirm that the column can separate NGF NPs from unloaded NGF.
- To measure the entrapment efficiency of NGF, load 200 µL of NGF HDL-mimicking NPs onto the column and elute with 1x PBS. Collect a total of 12 fractions (1 mL for each fraction).
- Measure the unloaded NGF concentration from fractions 6 to 10 using the Sandwich ELISA kit.
- Add the NGF amounts measured from fractions 6 to 10 together as the unloaded NGF and calculate the entrapment efficiency of NGF using the following equation. %EE = (1 - unloaded NGF/total NGF added into NP) x 100% Equation (1)
3. In Vitro Release of NGF HDL-mimicking Nanoparticles
Study free NGF (10 µg/mL in water; n = 4) and NGF HDL-mimicking NPs (10 µg/mL; n = 4) in parallel.
Pre-treat 8 dialysis tubes (molecular weight cutoff: 300 kDa) by following the manufacturer's instructions.
Prepare 5% bovine serum albumin (BSA) in PBS as the release medium. Add 30 mL of release medium to a 50-mL centrifuge tube and warm it up to 37 °C in a shaker with a 135 rpm shaking speed. Place another 50 mL of release medium in the shaker for replacement.
Add 400 µL of release medium into the dialysis tube and quickly add 200 µL of the tested sample to make enough volume for dialysis.
Close the tube and quickly put the dialysis tube into the release medium (the centrifuge tube). Quickly withdraw 100 µL of the release medium from the outside dialysis tube as the sample for time 0. Immediately put the sample into -20 °C for later analysis. Add 100 µL of fresh release medium into the centrifuge tube to replace the withdrawn sample. Start the timer.
At 1, 2, 4, 6, 8, 24, 48, and 72 h, withdraw 100 µL of the release medium and replace it with 100 µL of fresh medium. Immediately put the withdrawn samples into -20 °C for later analysis.
After 72 h, take out all samples from -20 °C and thaw them at room temperature. Measure the NGF concentrations of the release samples using the Sandwich ELISA kit.
4. Bioactivity of NGF HDL-mimicking Nanoparticles (Neurite Outgrowth Study)
Culture PC12 cells in RPMI-1640 medium supplemented with 10% heat-inactivated horse serum, 5% fetal bovine serum, 100 µg/mL streptomycin, and 100 units/mL penicillin. Maintain the cells in a humidified incubator at 37 °C and with 5% CO2.
When the cells reach ~70-80% confluence, remove the culture medium and wash the cells with 1x PBS (approximately 2 mL per 10 cm2 culture surface area). Remove the wash solution. Trypsinize the cells by adding 0.25% Trypsin-EDTA solution (0.25% trypsin and 1 mM EDTA; 0.5 mL per 10 cm2 culture surface area) to the flask and incubate at 37 °C until most cells are detached.
Add two volumes of culture medium and spin down at 180 x g for 5 min. Remove the supernatant and resuspend the cell pellet in 5 mL of culture medium. Filter the cells through a 22 G, ½ inch needle to break cell clusters.
Split the cells at a ratio of 1:3 into a new cell culture flask. After incubating overnight, remove unattached PC12 cells. Allow the attached cells to remain for further growth.
Repeat this procedure for 3 passages to select the subculture of PC12 that has a strong adhesion property. Pre-coat a 6-well plate with 800 µL/well of rat tail collagen type I (100 µg/mL).
Trypsinize the selected PC12 cells as described in step 4.2 and filter them through a 22 G, ½ inch needle to break the cell clusters. Count the cells with a hemocytometer. Seed the cells overnight at a density of 10,000 cells/well in the pre-coated 6-well plate in step 4.5 to allow the cells to attach to the plate.
Dilute free NGF (10 µg/mL) and NGF HDL-mimicking NPs (10 µg/mL) with the culture medium (described in step 4.1) to prepare 0.5, 1, 5, 10, 50, and 100 ng/mL concentrations. Remove 2 mL of medium from each well and replace with 2 mL of the diluted free NGF or NGF HDL-mimicking NPs. Culture for 4 days.
On day 4, change the medium to fresh medium (described in step 4.1) containing the corresponding treatment and continue the treatment for another 3 days.
On day 7, visualize the cells with an inverted light microscope and image each well at random spots under 10X magnification.
5. Biodistribution of NGF HDL-mimicking Nanoparticles
Use adult BALB/c mice (male, 25-30 g) to test the tissue distribution of NGF NPs. Randomly divide the mice into three groups of 3 mice per group. Use a cone-shaped plastic restraint (e.g., decapicone) to restrain a mouse and wipe the tail with ethanol to promote vasodilation and the visibility of the vein.
Inject 100 µL of either saline, free NGF, or NGF HDL-mimicking NPs to each group of mice (3 groups) through the tail veins at a dose of 40 µg/kg of NGF. Use a 30½ G needle attached to a 1 mL syringe.
- At 30 min after the injection, anesthetize the mice using 3% inhaled isoflurane (in oxygen at flow rate of 2 L/min). Perform a tail and toe pinch to determine the depth of anesthesia. Collect blood by cardiac puncture.
- Withdraw approximately 1 mL of blood from the heart. Euthanize each mouse by cervical dislocation.
Place the mouse carcass in dorsal recumbency. Open the abdomen using surgical scissors and move fat and intestine aside using a cotton swab to expose the liver, spleen, and kidney. Harvest these tissues and rinse them in 1x PBS to clean the blood.
Immediately centrifuge the blood samples at 3,400 x g and 4 °C for 5 min to obtain the plasma. Store the plasma and tissues at -80 °C until the analyses.
To analyze the tissue samples, move the samples from -80 °C to 4 °C and suspend 100 mg of tissue sample in a 10x volume of extraction buffer (0.05 M sodium acetate, 1.0 M sodium chloride, 1% triton X-100, 1% BSA, 0.2 mM phenylmethanesulfonyl fluoride, and 0.2 mM benzethonium chloride). Homogenize at 10,000 rpm and 4 °C for 5 min.
Sacrifice two untreated mice to collect blank plasma and tissues, as described in steps 5.3 and 5.4. Prepare NGF standard solutions using blank plasma or blank tissue homogenates.
Determine the concentrations of NGF in the plasma and tissue homogenates using the Sandwich ELISA kit, as described above.
Representative Results
The engineering scheme of HDL-mimicking, α-tocopherol-coated NGF NPs prepared by an ion-pair strategy is shown in Figure 1. To neutralize the surface charges of NGF, protamine USP was used as an ion-pair agent to form a complex with NGF. To protect the bioactivity, prototype HDL-mimicking NPs were engineered, first using homogenization; then, the NGF/protamine complex was encapsulated into the prototype NPs. Homogenization provided sufficient energy and successfully promoted the mixing of the excipients. After a 3 min homogenization, consistent particle sizes (around 170 nm) were obtained for the prototype NPs (Figure 2). Apo A-I was incubated with the prototype NPs in different conditions, including 2 h of stirring at room temperature; 4 h of stirring at room temperature; 4 h of stirring at room temperature, followed by overnight incubation at 4 °C; and stirring at room temperature overnight. Over 26% of Apo A-I was incorporated in the NPs when stirred at room temperature overnight. To add the NGF-protamine complex, the complex was incubated with the prototype NPs for 30 min at 37 °C and then Apo A-I was added to the mixture to finish the final coating of Apo A-I on the surface of the NPs. Using the procedure described here, the final NGF HDL-mimicking NPs had particle sizes of 171.4 ±6.6 nm (n = 3), with 65.9% of NGF entrapment efficiency (Table 1). NGF HDL-mimicking NPs had a slight negative charge (Table 1). The NPs had a narrow size distribution, and incorporating NGF into the NPs did not affect the particle size (Figure 3).
To measure the entrapment efficiency of NGF, various methods were evaluated to separate unloaded NGF from NGF HLD-mimicking NPs. Unexpectedly, NGF cannot pass a separation filter (molecular weight cutoff: 100 kDa). Gel filtration columns, including Sephadex G-50, Sephadex G-100, and Sephacryl S-100, cannot separate unloaded NGF and NGF HDL-mimicking NPs, since both of them come out in the same fractions after elution. A Sepharose CL-4B column performed the separation with the optimized sample loading, elution buffer, and elution rate. As shown in Figure 4, unloaded NGF and NGF HDL-mimicking NPs were completely separated on a Sepharose CL-4B column.
A dialysis method was used to study the in vitro release of NGF HDL-mimicking NPs in PBS with 5% BSA, which was included to mimic the physiological conditions in blood. By increasing the size of the dialysis device (molecular weight cutoff: 300 kDa) and by adding PBS and BSA to the release medium, NGF did not bind with the dialysis membrane and freely passed through. As a result, the recovery of free NGF in this dialysis method was over 85% (Figure 5). NGF HDL-mimicking NPs showed a slow release profile, and about 10% of the NGF was released from the NPs over 72 h (Figure 5).
To test the bioactivity of NGF HDL-mimicking NPs, the subculture of PC12 cells that had a strong adhesion property was selected to conduct a neurite outgrowth assay. Figure 6 represents the imaging of neurite outgrowth when the cells were treated with 50 ng/mL free NGF (Figure 6A) and NGF HDL-mimicking NPs (Figure 6B). When the treatment concentration was higher than 10 ng/mL, neurite outgrowth was clearly observed by the microscope. At these high concentrations, free NGF and NGF HDL-mimicking NPs did not show a significant difference on the effect of neurite outgrowth. When the concentration of NGF was lower than 10 ng/mL, neurite outgrowth could not be observed clearly, both for free NGF and for NGF HDL-mimicking NPs. Biodistribution studies were performed to compare the in vivo behaviors of free NGF and NGF HDL-mimicking NPs. As shown in Figure 7, NGF HDL-mimicking NPs significantly increased the plasma concentration and decreased the uptake in the liver, kidney, and spleen.
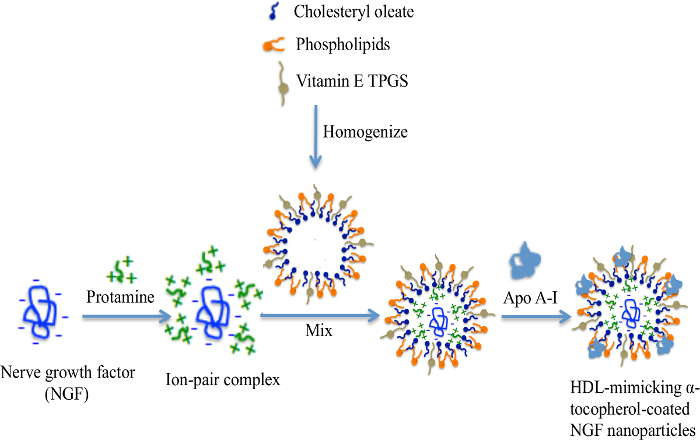 Figure 1:The engineering scheme of NGF HDL-mimicking nanoparticles prepared by an ion-pair strategy. NGF is a negatively charged hydrophilic molecule. A cationic peptide, protamine, was used to neutralize the charges and formed an ion-pair complex with NGF. Cholesteryl oleate, phospholipids, and TPGS formed self-assembly prototype NPs by homogenization. The NGF/protamine complex was incorporated into the prototype NPs. Finally, Apo A-I was coated on the NP surface after overnight incubation. This figure has been modified from Prathipati et al.16. Please click here to view a larger version of this figure.
Figure 1:The engineering scheme of NGF HDL-mimicking nanoparticles prepared by an ion-pair strategy. NGF is a negatively charged hydrophilic molecule. A cationic peptide, protamine, was used to neutralize the charges and formed an ion-pair complex with NGF. Cholesteryl oleate, phospholipids, and TPGS formed self-assembly prototype NPs by homogenization. The NGF/protamine complex was incorporated into the prototype NPs. Finally, Apo A-I was coated on the NP surface after overnight incubation. This figure has been modified from Prathipati et al.16. Please click here to view a larger version of this figure.
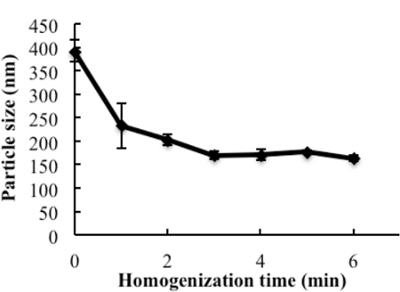 Figure 2:The influence of homogenization on the particle size of the prototype nanoparticles. The excipients, PC, SM, PS, CO, and TPGS, which were dissolved in ethanol, were added to glass vials, and the solvent was evaporated under N2 stream. 1 mL of water was added and homogenized at 9,500 rpm for different times. The particle sizes of subsequent nanoparticles were measured. Data are presented as the mean ± standard deviation (n = 4). This figure has been modified from Prathipati et al.16. Please click here to view a larger version of this figure.
Figure 2:The influence of homogenization on the particle size of the prototype nanoparticles. The excipients, PC, SM, PS, CO, and TPGS, which were dissolved in ethanol, were added to glass vials, and the solvent was evaporated under N2 stream. 1 mL of water was added and homogenized at 9,500 rpm for different times. The particle sizes of subsequent nanoparticles were measured. Data are presented as the mean ± standard deviation (n = 4). This figure has been modified from Prathipati et al.16. Please click here to view a larger version of this figure.
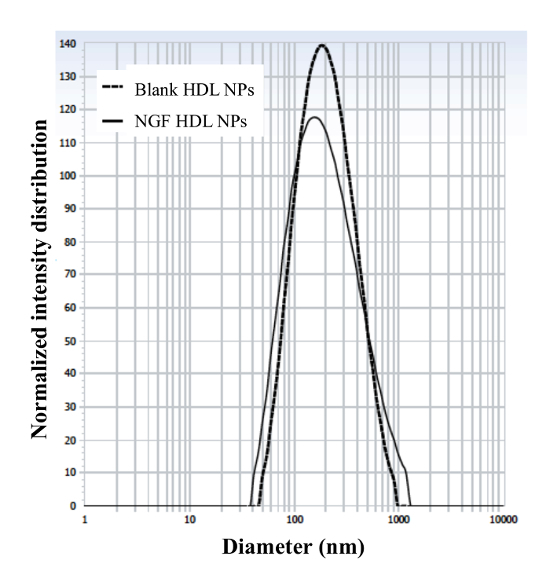 Figure 3: Particle size and size distribution of blank HDL-mimicking nanoparticles and NGF HDL-mimicking nanoparticles. The particle sizes and distributions were measured using a particle analyzer. This figure has been modified from Prathipati et al.16. Please click here to view a larger version of this figure.
Figure 3: Particle size and size distribution of blank HDL-mimicking nanoparticles and NGF HDL-mimicking nanoparticles. The particle sizes and distributions were measured using a particle analyzer. This figure has been modified from Prathipati et al.16. Please click here to view a larger version of this figure.
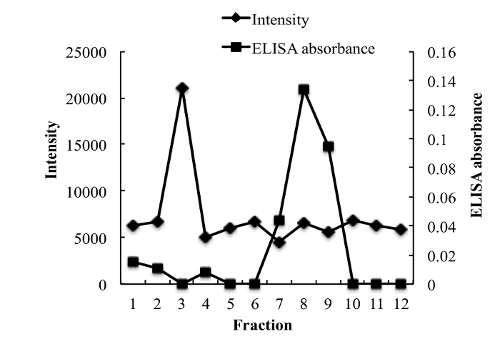 Figure 4: Chromatograms of free NGF and NGF HDL-mimicking nanoparticles on a Sepharose CL-4B column eluted by PBS. 200 µL of free NGF solution (10 µg/mL) and NGF NP solution were loaded onto the gel filtration column and eluted with 1x PBS. A total of twelve fractions (1 mL for each fraction) were collected for both samples. The NP intensity in each fraction was measured by a particle analyzer, and the concentration of NGF in each fraction was measured using a Sandwich ELISA kit. This figure has been modified from Prathipati et al.16.
Please click here to view a larger version of this figure.
Figure 4: Chromatograms of free NGF and NGF HDL-mimicking nanoparticles on a Sepharose CL-4B column eluted by PBS. 200 µL of free NGF solution (10 µg/mL) and NGF NP solution were loaded onto the gel filtration column and eluted with 1x PBS. A total of twelve fractions (1 mL for each fraction) were collected for both samples. The NP intensity in each fraction was measured by a particle analyzer, and the concentration of NGF in each fraction was measured using a Sandwich ELISA kit. This figure has been modified from Prathipati et al.16.
Please click here to view a larger version of this figure.
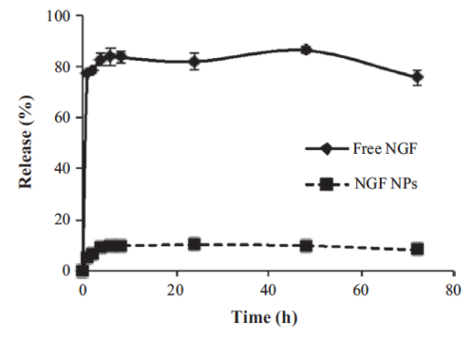 Figure 5: In vitro release of NGF HDL-mimicking nanoparticles measured by a dialysis method. PBS with 5% BSA was used as a release medium. 200 µL of free NGF solution (10 µg/mL) or NGF NPs were added to a dialysis tube supplemented with 400 µL of the release medium. The dialysis tube was put into 30 mL of pre-warmed release medium. The study was performed at 37 °C with 135 rpm shaking. At 1, 2, 4, 6, 8, 24, 48, and 72 h, 100 µL of the release medium was withdrawn and replaced with 100 µL of fresh medium. The NGF concentration in each sample was measured using a Sandwich ELISA kit. Data are presented as the mean ± standard deviation (n = 4). This figure has been modified from Prathipati et al.16. Please click here to view a larger version of this figure.
Figure 5: In vitro release of NGF HDL-mimicking nanoparticles measured by a dialysis method. PBS with 5% BSA was used as a release medium. 200 µL of free NGF solution (10 µg/mL) or NGF NPs were added to a dialysis tube supplemented with 400 µL of the release medium. The dialysis tube was put into 30 mL of pre-warmed release medium. The study was performed at 37 °C with 135 rpm shaking. At 1, 2, 4, 6, 8, 24, 48, and 72 h, 100 µL of the release medium was withdrawn and replaced with 100 µL of fresh medium. The NGF concentration in each sample was measured using a Sandwich ELISA kit. Data are presented as the mean ± standard deviation (n = 4). This figure has been modified from Prathipati et al.16. Please click here to view a larger version of this figure.
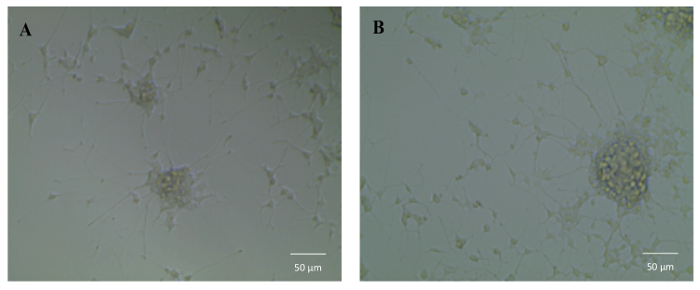 Figure 6:The influence of NGF HDL-mimicking nanoparticles on neurite outgrowth in PC12 cells. Cells were treated with 50 ng/mL free NGF (A) and NGF HDL-mimicking nanoparticles (B) for 7 days. The neurite was imaged using an inverted light microscope under 10X magnification. Please click here to view a larger version of this figure.
Figure 6:The influence of NGF HDL-mimicking nanoparticles on neurite outgrowth in PC12 cells. Cells were treated with 50 ng/mL free NGF (A) and NGF HDL-mimicking nanoparticles (B) for 7 days. The neurite was imaged using an inverted light microscope under 10X magnification. Please click here to view a larger version of this figure.
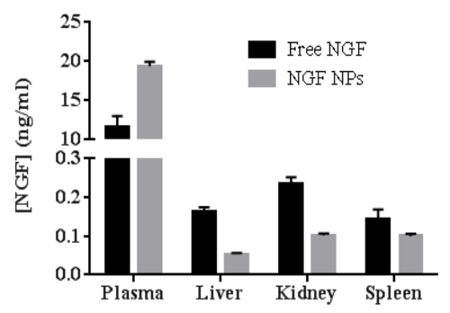 Figure 7:The comparison of biodistribution between free NGF and NGF HDL-mimicking nanoparticles in mice (n = 3). The mice were administered with 40 mg/kg of NGF by tail-vein injection and were sacrificed at 30 min after administration. The blood, liver, spleen, and kidney were collected, and the concentration of NGF in each sample was measured using a Sandwich ELISA kit. Data are shown as the mean ± standard deviation. This figure has been modified from Prathipati et al.16. Please click here to view a larger version of this figure.
Figure 7:The comparison of biodistribution between free NGF and NGF HDL-mimicking nanoparticles in mice (n = 3). The mice were administered with 40 mg/kg of NGF by tail-vein injection and were sacrificed at 30 min after administration. The blood, liver, spleen, and kidney were collected, and the concentration of NGF in each sample was measured using a Sandwich ELISA kit. Data are shown as the mean ± standard deviation. This figure has been modified from Prathipati et al.16. Please click here to view a larger version of this figure.
| Sample | Particle size (nm) | P.I. | EE% of NGF | Zeta potential (mV) |
| NGF HDL-mimicking NPs | 171.4 ±6.6 | 0.239 ±0.01 | 65.9 ±1.4 | -12.5 ±1.9 |
Table 1: Characterization of NGF HDL-mimicking nanoparticles (n = 3). Data are shown as the mean ± standard deviation. This table has been modified from Prathipati et al.16.
Discussion
In this study, we demonstrate a simple method to prepare HDL-mimicking NPs for NGF encapsulation. Various NP delivery systems have been studied to deliver proteins. Currently, many NP preparations involve dialysis, solvent precipitation, and film hydration. These processes are generally complicated and challenging upon scale-up. During this NP development, it was determined that the lipids had strong adhesion to the glass wall of the container, which led to the difficulties in hydrating the thin film and efficiently mixing the excipients. Given the multiple components in natural HDL NPs, mixing became very challenging and crucial for the preparation of HDL-mimicking NPs. According to the results, increasing the temperature and mixing time did not assist in NP formation. The homogenizer is a common instrument in industrial manufacturing and provides high energy and shear force for mixing. By applying this instrument to NP preparation, monodispersed NPs with the appropriate sizes were produced within 3 min (Figure 2 and Figure 3), which greatly reduced the manufacturing time and simplified the preparation procedure.
The great challenge to apply macromolecules to therapeutic administration is not only delivery, but also formulation. Proteins are normally water soluble and have large sizes and charges on their surfaces, which prevent the encapsulation of proteins into lipid-based NPs. Protamine USP, a polycation, was utilized to form an ion-pair complex with NGF. After mixing NGF and protamine, the white precipitate that indicated the formation of the insoluble complex was clearly observed. With the help of the complex, about 70% of the NGF was entrapped within the NPs, with a narrow size distribution (Table 1 and Figure 3). Moreover, the stability of proteins had to be considered during formulation development. To avoid the influence of homogenization on NGF stability, the procedure was adjusted to add an NGF complex after homogenization. Under a very mild incubation condition (30 min at 37 °C), the NGF complex was incorporated into the NPs. Different procedures were tested to add Apo A-I; however, overnight incubation at room temperature was still necessary to load enough Apo A-I on the surface of the NPs. Based on the results of the neurite outgrowth assay, the bioactivity of NGF after the NP preparation was successfully maintained with the procedure presented here (Figure 6). Moreover, encapsulating NGF into the HDL-mimicking NPs enhanced the blood circulation of NGF, as indicated by the biodistribution studies (Figure 7). These results demonstrate that NGF was successfully formulated with an NP delivery system. The HDL-mimicking NPs can assist NGF, allowing for a long half-life and in vivo stability.
Developing appropriate analytical methods is also challenging for protein-based nanomedicines. To measure the entrapment efficiency and in vitro release, it is mandatory to separate unloaded drug from drug-loaded nanoparticles. Membranes with certain molecular weight cutoffs are commonly used for this purpose and work well for small molecules. However, it is not easy to separate unloaded proteins and protein-loaded nanoparticles, mainly because of the similar sizes and binding properties of proteins. Free NGF and NGF HDL-loaded NPs cannot be separated using the membrane-based filtration device, although the molecule weight cutoff of the membrane is 100 kDa (the biggest pore size for commercial filtration devices). After several gel filtration columns were tested, free NGF and NGF HDL-mimicking NPs were finally separated using a Sepharose CL-4B column. However, the sample loading onto the column had to be kept at 200 µL in order to obtain reproducible chromatograms. Elution agents were also of importance. When water was used to elute NGF, a very broad chromatogram was obtained; NGF was detected from fractions 5 to 20. The broad chromatogram could have been caused by the strong interaction between NGF and the beads. After several elution agents, such as 5 mM NaCl and 50 mM NaCl, were tested, PBS was confirmed as a sufficient elution agent. Moreover, the separation challenge remained for the in vitro studies. Column separation is not suitable for in vitro release studies because of the number of released samples to be measured and the potential for the further release of NGF from the NPs when they pass the column. After various conditions were tested, the dialysis method described in the protocol was determined. According to the results (Figure 5), free NGF can freely pass the through membrane (molecule weight cutoff: 300 kDa) in PBS containing 5% BSA. With this positive control, the release result of NGF HDL-mimicking NPs became reliable. The increased pore size and decreased membrane binding caused by PBS and BSA contributed to the free penetration of NGF through the dialysis membrane.
In addition to the aforementioned issues, another challenge with the neurite outgrowth assay arose. There is a commercial neurite outgrowth assay kit that is based on a subclone of PC12 cells. However, the subclone of PC12 cells is no longer commercially available. Normal PC12 cells are floating cells that do not grow neurites very well and that are easy to lose during the experimental procedures. After several failures, a small population of PC12 cells that had a strong adhesion property was discovered. Thus, this population of cells was selected through several passages, as described in the protocol. Consequently, the assay was successfully conducted and demonstrates the comparable bioactivity of NGF HDL-mimicking NPs with free NGF (Figure 6).
The method reported here is a simple procedure for the preparation of HDL-mimicking NPs. We entrapped over 26% of Apo A-I on the NPs, which was over 3-fold higher than reported in the literature16. Over 65% of NGF was entrapped in the NPs, which was 2-fold higher than in the literature16. With these increases in the entrapment efficiency for both Apo A-I and NGF, it may not be necessary to purify the NPs. Consequently, the NP preparation was simplified. The unloaded Apo A-I and NGF in the NP preparation did not influence the NGF in vitro release (Figure 6) and the in vivo biodistribution (Figure 7), but it could influence the stability and in vitro cell studies. The studies here demonstrate that it is possible to further improve the entrapment efficiency of Apo A-I and NGF by modifying NP composition and preparation procedures. Moreover, the novel HDL-mimicking NPs and the strategies in these studies could be used to load other growth factors, although the ion-pair agent (protamine) could be changed to fit the specific growth factor. The long-term storage of protein formulations is challenging. The preferred way to keep protein-loaded NPs is to lyophilize them to dry powders. However, lyophilization could increase the particle size and decrease the entrapment efficiency of the protein after the reconstitution of lyophilized NPs. Few publications have discussed the lyophilization of HDL-mimicking NPs. We are studying the lyophilization of novel HDL-mimicking NPs. Eventually, HDL-mimicking NPs could become clinically applicable when they can be appropriately stored for a long period of time (at least 6 months).
Disclosures
The authors have nothing to disclose.
Acknowledgments
This work was supported by NIH R03 NS087322-01 to Dong, X.
References
- Bruno BJ, Miller GD, Lim CS. Basics and recent advances in peptide and protein drug delivery. Ther Deliv. 2013;4(11):1443–1467. doi: 10.4155/tde.13.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo ZC, Ren K, Liu X, Tang ZL, Yi GH. A high-density lipoprotein-mediated drug delivery system. Adv Drug Deliv Rev. 2016;106(Pt A):132–147. doi: 10.1016/j.addr.2016.04.030. [DOI] [PubMed] [Google Scholar]
- Lacko AG, Sabnis NA, Nagarajan B, McConathy WJ. HDL as a drug and nucleic acid delivery vehicle. Front Pharmacol. 2015;6:247–252. doi: 10.3389/fphar.2015.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishya R, Khurana V, Patel S, Mitra AK. Long-term delivery of protein therapeutics. Expert Opin Drug Deliv. 2015;12(3):415–440. doi: 10.1517/17425247.2015.961420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasic DD, Papahadjopoulos D. Medical application of liposomes. Elsevier; 1998. [Google Scholar]
- Samad A, Sultana Y, Aqil M. Liposomal drug delivery systems: an update review. Curr Drug Deliv. 2007;4(4):297–305. doi: 10.2174/156720107782151269. [DOI] [PubMed] [Google Scholar]
- Bezemer JM, Radersma R, Grijpma DW, Dijkstra PJ, van Blitterswijk CA, Feijen J. Microspheres for protein delivery prepared from amphiphilic multiblock copolymers: 2. Modulation of release rate. J Control Release. 2000;67(2-3):249–260. doi: 10.1016/s0168-3659(00)00212-1. [DOI] [PubMed] [Google Scholar]
- Patel A, Patel M, Yang X, Mitra AK. Recent advances in protein and peptide drug delivery: a special emphasis on polymeric nanoparticles. Protein Pept lett. 2014;21(11):1102–1120. doi: 10.2174/0929866521666140807114240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuai R, Li D, Chen YE, Moon JJ, Schwendeman A. High-density lipoproteins: nature's multifunctional nanoparticles. ACS Nano. 2016;10(3):3015–3041. doi: 10.1021/acsnano.5b07522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gursky O. Structural stability and functional remodeling of high-density lipoproteins. FEBS Lett. 2015;589(19 Pt A):2627–2639. doi: 10.1016/j.febslet.2015.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon KM, Thaxton CS. High-density lipoproteins for the systemic delivery of short interfering RNA. Expert Opin Drug Deliv. 2014;11(2):231–247. doi: 10.1517/17425247.2014.866089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon KM, Foit L, Angeloni NL, Giles FJ, Gordon LI, Thaxton CS. Synthetic high-density lipoprotein-like nanoparticles as cancer therapy. Cancer Treat Res. 2015;166:129–150. doi: 10.1007/978-3-319-16555-4_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch PG, Förtsch V, Hodler G, Bolli R. Production and characterization of a reconstituted high density lipoprotein for therapeutic applications. Vox Sang. 1996;71(3):155–164. doi: 10.1046/j.1423-0410.1996.7130155.x. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Chen J, Ding L, Jin H, Lovell JF, Corbin IR, Cao W, Lo PC, Yang M, Tsao MS, Luo Q, Zheng G. HDL-mimicking peptide-lipid nanoparticles with improved tumor targeting. Small. 2010;6(3):430–437. doi: 10.1002/smll.200901515. [DOI] [PubMed] [Google Scholar]
- Kim Y, Fay F, Cormode DP, Sanchez-Gaytan BL, Tang J, Hennessy EJ, Ma M, Moore K, Farokhzad OC, Fisher EA, Mulder WJ, Langer R, Fayad ZA. Single step reconstitution of multifunctional high-density lipoprotein-derived nanomaterials using microfluidics. ACS Nano. 2013;7(11):9975–9983. doi: 10.1021/nn4039063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prathipati P, Zhu J, Dong XD. Development of novel HDL-mimicking α-tocopherol-coated nanoparticles to encapsulate nerve growth factor and evaluation of biodistribution. Eur J Pharm and Biopharm. 2016;108:126–135. doi: 10.1016/j.ejpb.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]


