Abstract
Single cell transcriptomics is becoming a common technique to unravel new biological phenomena whose functional significance can only be understood in the light of differences in gene expression between single cells. The technology is still in its early days and therefore suffers from many technical challenges. This review discusses the continuous effort to identify and systematically characterise various sources of technical variability in single cell expression data and the need to further develop experimental and computational tools and resources to help deal with it.
Keywords: Single cell, RNA-seq, Noise, Variability
1. Introduction
Next generation sequencing (NGS) technologies have revolutionized the way of approaching molecular biology to advance our understanding of the working principles of biological systems, including identification of the building blocks. Genome sequencing is now widely used across diverse fields in biology, ranging from medicine and population studies to animal breeding. However, the information encoded in the genome is static, an ensemble of the cell’s potentialities manifest once the process of transcription is triggered. Therefore, studying the transcriptome is essential to understand how genome information is decoded in a particular cell specific context, as the cells ultimately constitute as adaptable and dynamic entities.
NGS evolved from a range of laboratory techniques developed for expression analysis over the years. Initial experimental approaches include the early Northern blotting (Alwine et al., 1977), which targets a single gene and measures its expression levels through hybridization of a labelled probe. Advances in increasing throughput of transcriptome studies came with microarrays (Schena et al., 1995), a technology that used a similar probing approach, but increased the number of quantified transcripts by using tens of thousands of probes on a chip, onto which the RNA sample is hybridized. Both approaches described above are limited by the fact that probe design requires previous knowledge of the transcript sequences. To this end, the use of sequencing technologies such as Sanger sequencing and its derivatives, including expressed sequence tag (EST), improved access to the diversity of the transcriptomic landscape by overcoming the probe design constraint. Currently, however, the most widely used application of NGS technologies to transcriptomics is RNA sequencing (RNA-Seq) (Mortazavi et al., 2008), by which -potentially- all mRNA molecules in a cell can be sequenced, and hence characterized and quantified.
The importance of RNA-Seq is not only founded in its ability to access unknown transcripts and spliced variants, but also to increase microarray’s dynamic range (i.e. the lowly expressed transcripts could be successfully detected) and sensitivity (i.e. the expression level measurements show higher accuracy). RNA-Seq has therefore become the technology of choice to provide a high-throughput and fully quantitative approach to studying the transcriptome of a broad range of species, including the ones lacking full genome sequence availability. The technology has therefore been widely applied, replacing microarrays for the analysis of gene expression profile differences among cell populations, comparative transcriptomics and disease biomarker identifications (Wang et al., 2009). However, it has become apparent that not all cells within a population behave similarly when it comes to gene expression or splicing and, in this context, bulk RNA-Seq fails to address some important questions (Sandberg, 2014).
2. Single cell expression technologies and applications
Over the years, single-cell approaches have been developed in combination with microscopy to visualize gene expression patterns in individual cells. For example, single-molecule RNA fluorescence in situ hybridization (RNA FISH) technology combines probe hybridization with fluorescent labelling to resolve the location of a target transcript (Lubeck and Cai, 2012). The main disadvantage of RNA FISH is that, although parallelizable, it only allows access to a limited subset of genes. The implementation of single-cell microarrays (Iscove et al., 2002) presented itself again as a high-throughput alternative to RNA FISH, and although it helps overcome this main limitation, it suffers the drawbacks of bulk microarrays. Furthermore, the limited amount of starting material and the relatively low sensitivity of microarrays enforced high levels of pre-amplification, which can introduce significant biases.
In the light of these limitations, RNA Sequencing was implemented at the single-cell level, theoretically enabling access to the transcriptome of every individual cell in a population (Ramsköld et al., 2012; Tang et al., 2010). Essentially, single-cell RNA-Seq requires the following steps: single cell isolation, mRNA capture and reverse transcription to cDNA, cDNA amplification to improve the low transcript yields rendered by single cells, and sequencing (Picelli et al., 2014).
Over the last few years, single-cell RNAseq has been proven useful to unravel biological phenomena that can only be understood in the light of differences in gene expression between single cells, including:
-
•
Studying early embryonic development: In early stages of embryonic development, only a few cells contribute to activating the molecular machinery for cell differentiation. The characterisation of transcription changes in individual inner cell mass (ICM) cells of blastocysts was proven crucial to understand the complex transition from ICMs to embryonic stem cells (ESCs) (Tang et al., 2010). This approach set a precedent for subsequent studies of later and more complex stages in the process of cell commitment and differentiation into specific lineages. In this context, a spatial-temporal profiling of gene expression in embryonic development in Caenorhabditis elegans was used to study the evolution of the germ layers. The authors noted that the gene expression program of the mesoderm is induced after those of the ectoderm and endoderm and strikingly, the endoderm gene expression program activates earlier than ectoderm expression program, a phenomenon that is conserved across many species (Hashimshony et al., 2014).
-
•
Measuring diversity in cell populations: Single cell analysis is the most powerful tool to study the diversity between individual cells treated as homogenous in a typical bulk RNA-seq experiment. It has proven potential of providing valuable insights in some of the key problems in biomedical field e.g. tumour heterogeneity, which poses substantial challenges in cancer treatment. For example, single cell analysis can unravel intra- and inter-tumour differences (Patel et al., 2014) as well as distinguishing between malignant and non-malignant cells (Tirosh et al., 2016).
-
•
Identification of new rare cell types: Complex tissues often contain previously unidentified cell types that cannot be studied using bulk RNA-Seq, as it provides only an estimate of expression influenced by the abundance of the different cell types present. Single cell transcriptomics provides a promise to address this underlying diversity in order to assess meaningful differences in phenotype. Using this strategy, authors identified and characterised a rare population of dormant neural cells which were activated upon brain injury (Llorens-Bobadilla et al., 2015). Another example is the development of a computational approach (scLVM) to identify subpopulations of cells using latent variable models to account for hidden factors such as cell cycle. Namely, different sub-populations of cells corresponding to the differentiation stages during naive T cells to T helper 2 cells were identified (Buettner et al., 2015). Identification of rare cells is of high relevance, particularly characterisation of progenitor cells to understand vertebrate development. To this end, single cell RNA-Seq has been used to unravel transcription heterogeneity and lineage commitment in myeloid progenitors, in order to further demonstrate how Cebpe deletion results into diminishing of certain myeloid lineages (Paul et al., 2015).
-
•
Mapping developmental hierarchies: transcription dynamics during development and disease can be studied in much greater details using single cell studies, as bulk RNA-seq, by averaging out signal from multiple cells, misses out on the signal from rare developmentally relevant cells. However, single cell transcriptome profiling over time is not feasible. Taking advantage of the fact that an experiment characterising hundreds of unsynchronised cells from a population typically provides a snapshot of cells at various stages during differentiation, various methods for pseudo-time inference form single cell RNA-seq data have recently been developed (Haghverdi et al., 2016; Reid and Wernisch, 2016; Trapnell et al., 2014) and reviewed (Bacher and Kendziorski, 2016). As an example of this, single cell expression data has successfully been used to reconstruct the developmental progression of cells and identify transient and terminal states together with the branching decisions (Treutlein et al., 2014).
-
•
Understanding diverse features of transcription control: Single cell transcriptomics has facilitated unravelling mechanistic details of transcription control such as kinetics and bimodality, as well as studying other features such as allelic biases and transcription networks. Even though single cell transcriptomics does not measure expression changes in one gene over time, an overall rate of transcription between individual cells can be acquired and approximately represent the stochasticity of expression of a vast number of genes, facilitating estimation of kinetics of gene expression (Kim and Marioni, 2013). Recent studies have unravelled the stochastic modes of gene expression, which were not apparent at the population level. The functional implications of this stochasticity (i.e. changes on the phenotype of seemingly identical cells) can be explained by variation in gene regulation processes across individual cells (Munsky et al., 2012). Allelic biases in gene expression have also been investigated, including stochastic allelic expression in early embryogenesis (Tang et al., 2011) as a particularly relevant example. Finally, single cell transcriptome data is successfully used to reconstruct gene regulatory networks (Moignard et al., 2015).
In summary, single cell analysis has a huge potential to bring new insights into diverse fields of biological research. In the next sections, we will put this in context by discussing the technical challenges currently faced by single cell analysis to extract the ‘biological’ or functionally relevant variability from the data, which hinder its theoretical potential.
3. Technical variability in single cells
Despite the promise held by the approach, single-cell RNA-Seq is not free from biases. Quite contrarily, the low availability of starting material (i.e. RNA extracted from an individual cell) introduces high technical variability, making single-cell RNA-Seq data analysis especially challenging (Stegle et al., 2015). This typically results into many missing values (technical) or true absence of expression (biological) in typically lowly expressed transcripts, and discriminating both, although important, is not currently feasible. Furthermore, the necessary amplification of starting material introduces additional biases, such as 3′ end enrichment of signal and preferential amplification of some transcripts and/or mRNA fragments. Reassuringly, bulk RNA-Seq experiments can be recapitulated in silico by pooling 30 or more single cell transcriptomes in silico (Marinov et al., 2014), and used to estimate technical variability.
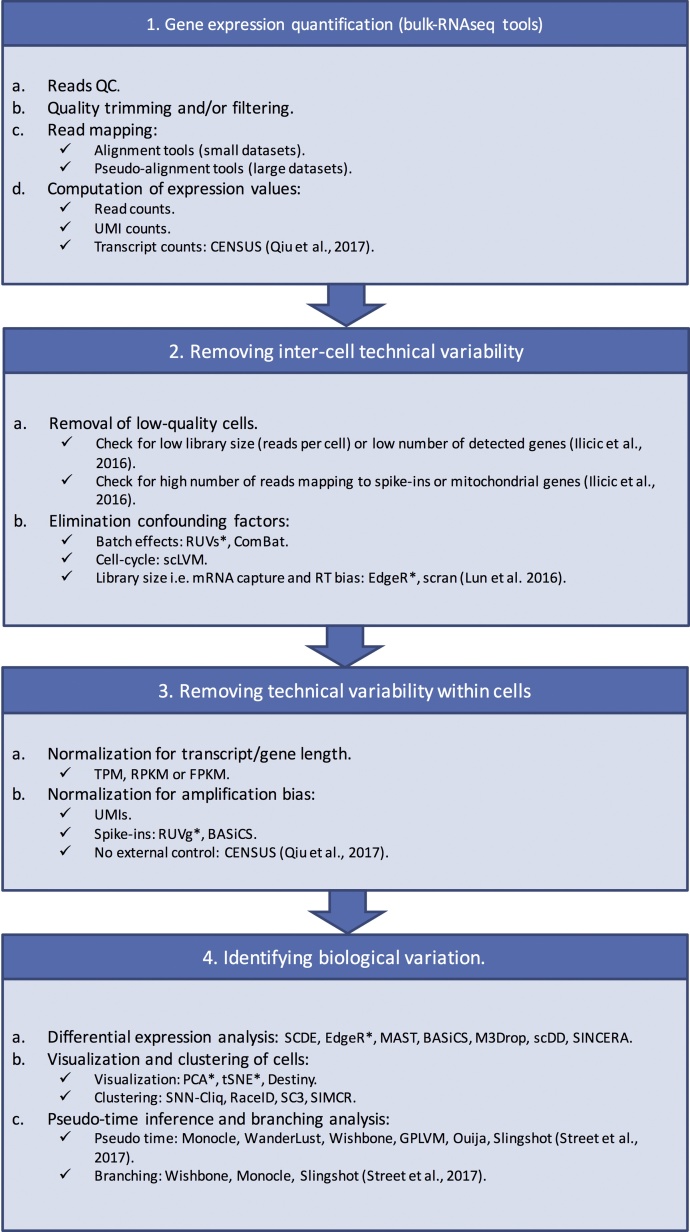
The technical variability in single-cell RNA-Seq can be divided into two categories: Inter-cell variability and within cell variability (Fig. 1).
Fig. 1.
Single-cell RNAseq data analysis workflow, including examples of computational methods available for each stage. Methods not developed specifically for single-cell RNAseq are marked with an asterisk. When reference is unspecified, see (Rostom et al., 2017) for full list.
3.1. Inter-cell variability
Inter-cell variability can appear as a result of the biological process under scrutiny, or can be due to unrelated phenomena, which can act as confounding factors. For example, the differences in cell cycle stage are responsible for a part of the gene expression variability between individual cells (Buettner et al., 2015), and differences in size affect the total amount of RNA extracted from individual cells (Stegle et al., 2015). In addition, cells can suffer stress or physical damage in cell capture systems used to isolate each single cell from the population, and capture sites can be occupied by more than one cell or by none. All of these events result in ‘low quality cells’, which have been estimated to be around 10–40% of the total number of cells in a dataset (Ilicic et al., 2016). The data from ‘low quality cells’ may be misleading and hinder meaningful biological interpretation. Hence, it is necessary to perform a cell filtering step in the data analysis pipeline.
Furthermore, RNA capture and amplification efficiency per cell is far from being uniform. This can be evaluated using external spike-ins as a control (Jiang et al., 2011, Stegle et al., 2015), a technique originally developed for bulk RNA-Seq. Simple measures include discarding cells with high proportion of reads mapping to mitochondrial genes or spike-in controls. As a complement to these, more complex statistical methods to eliminate confounding factors prior to data analysis have been developed. For example, variation due to differences in cell cycle stage was addressed by modelling cell-cycle variation as Gaussian processes, followed by linear regression, thus allowing the removal of noise caused by cell cycle (Buettner et al., 2015). Other quality control tools provide a model of the biological and technical features of low quality cells (Ilicic et al., 2016). The methods developed to identify sub-populations, including the popular principal component analysis (PCA), t-distributed stochastic neighbour embedding (t-SNE), or zero-inflated factor analysis (ZIFA) (Pierson and Yau, 2015) are also used to filter low quality cells. However, comprehensive quality controls with more than one metric are more effective for discarding true low quality samples, and are beginning to be implemented by R packages such as scone (McCarthy et al., 2016, Stegle et al., 2015). Even so, it should be noted that discarding multiple cells on the ground of technical quality can have a negative effect, since the number of singe cells studied in any experiment is generally limited due to high costs.
In addition to the biases listed above, batch effects are a source of technical variability that arises in the experimental stages required to generate sequencing data. Poor experimental design leads to completely confounded experiments in single-cell RNA-seq (that is, the observed variation cannot be separated into biological and technical), which can be avoided by introducing biological replicates of the different samples across all batches (Hicks et al., 2017).
3.2. Within cell variability
The main cause of technical noise across transcripts within a cell is the low amount of starting material available (mRNA) due to working with single cells (Brennecke et al., 2013). This noise component is dependent on the single-molecule capture efficiency, i.e. the fraction of mRNA molecules that are captured, amplified and subsequently sequenced from each cell (Marinov et al., 2014, Stegle et al., 2015). Notably, nano-litre volume sample preparation using microfluidics has been reported to improve capture efficiency (Wu et al., 2014) although more recent evaluations suggest that new library preparation methods, such as Smart-seq2, have outperformed these approaches when it comes to the number of molecules captured (Ziegenhain et al., 2017; Table 1).
Table 1.
Qualitative comparison of library preparation methods for single-cell RNAseq. Note that full-length methods are not compatible with UMIs, and that all UMI methods capture only the 3′ end of the transcript. In-vitro transcription methods also include UMIs. Precision defined as reproducibility of the gene expression quantification. More the (+) signs, higher the costs or amplification bias or number of genes or cells (Ziegenhain et al., 2017).
| Technology | No. of cells | No. of genes | Amplification bias | Cost | |
|---|---|---|---|---|---|
| Full-length cDNA methods | Smart-seq/C1 | + | ++ | +++ | ++ |
| Smart-seq2 | ++ | ++++ | +++ | +++ | |
| UMI methods | SCRB-seq | +++ | +++ | ++ | + |
| Drop-seq | ++++ | + | ++ | + | |
| In-vitro transcription methods | MARS-seq | +++ | + | + | + |
| CEL-seq2/C1 | ++ | ++ | + | ++ |
Apart from capture efficiency, single cell analysis entails other inconveniences, namely the impossibility to use technical replicates and the PCR amplification biases, such as overrepresentation of the 3′ end of transcripts due to poly-A priming (Wu et al., 2014). Regarding uneven transcript coverage and length normalization, FPKM units (fragments per kilobase per million reads), although largely used in RNA-seq, have proven unreliable for cell-to-cell comparison when the total amount of RNA per cell differs largely (Lin et al., 2012). Such limitations, however, can be partially overcome by an alternative normalization using TPM units (transcripts per million) (Wagner et al., 2012), although whether TPM is the best unit of transcription readout is still disputed.
Specific experimental designs including external controls have been implemented to account for technical variation within each cell, including spike-in quantification of molecules of known abundance and sequence, whole-transcriptome spikes from a distantly related organism or a set of artificial spike-in mix (ERRC) (Jiang et al., 2011). These spike-ins, although used to detect low-quality cells, also serve as estimators of uneven reverse transcription and PCR amplification within each cell. To detect this, pool/split experiments are yet another way. In this case, RNA pooled from multiple cells is subsequently divided in separate reactions consisting of equal material to be used for the library construction. Variation in the pool/split experiment, as in the spike-in read counts, will be solely attributed to technical noise (Marinov et al., 2014). Using these additional controls demands further experimental and computational resources, takes over a relatively high number of the reads in the sequencing library, and not all experimental platforms can accommodate them (Bacher and Kendziorski, 2016). Furthermore, it has been recently shown that the amplification bias is stronger in endogenous genes than in ERCC spike-ins (Tung et al., 2017).
Another design for noise control is the use of unique molecular identifiers (UMIs). UMIs are short random DNA sequences (typically 6-8bp) linked to cDNA before the amplification step in order to estimate the absolute number of molecules in the cell lysate, while correcting for the amplification bias (Islam et al., 2014). During data analysis, reads with the same UMI (and mapping site) are interpreted as PCR duplicates and collapsed, preventing them to be interpreted as coming from genuine gene expression. However, UMIs will be present only in reads originating at the 3′ end of transcripts, and therefore are not compatible with isoform or allele specific expression studies, as they require full-length sequencing of the transcript. Given the limitations of both UMIs and spike-ins, a computational method that does not rely on any external controls has recently been developed; instead, CENSUS uses read counts and a generative model to estimate lysate (i.e. before reverse-transcription) transcript counts for each gene (Qiu et al., 2017).
Importantly, the low abundance of starting material has other implications, notably, not all transcripts are similarly affected by technical noise. To this extent, it is expected that transcripts with low levels of expression (resulting in low read counts) will be detected with less accuracy, as the sensitivity of methods available is limited, whereas expression of high-read count genes can be more reliably measured (Ramsköld et al., 2012). Brennecke et al. (2013) performed a single-cell RNA-Seq analysis of plant cells to establish the relationship between transcript level and sensitivity to technical noise using spike-ins as a control. They noted that, for spike-ins with an expected read count of up to 100 per cell, technical noise was maximal, making it impossible to assess whether inter-sample variability was biologically meaningful or solely due to noise. A more recent study by Kim et al. (2015) also used spike-ins to model technical noise and stated that only about 12% of variability in lowly expressed genes can be attributed to biological factors in contrast to over 55% of variability for highly expressed genes.
While technical improvements in single-cell RNA-Seq protocols are still in development, new computational tools are necessary to help discriminate genes where technical variability obfuscates the relevant biological one. We recently introduced a correlation-based method to identify the genes where biological variation was higher than technical, and create a high-quality subset that would be suitable for further analysis (Mantsoki et al., 2016). In addition, we noted that highly expressed genes generally have low coefficients of variation (CV), whereas lowly expressed genes present a wide range of CV values.
4. Future challenges and opportunities
There is an ongoing effort to identify and systematically characterise various sources of technical variability in single cell expression data and further develop computational tools and resources to help deal with it. Following the steps of development of UMIs and spike-ins to take into account the technical variability in single cell experiments, there is a constant need for development of experimental and computational protocols, as current ones are not exempt of limitations (Table 1), as it has been recently reviewed (Rostom et al., 2017, Ziegenhain et al., 2017).
Since technical variation can be introduced at various steps, ranging from pipetting variations during sample processing steps to amplification or sequencing bias, it is important to develop strategies that estimate and remove batch effects. Eliminating such biases in bulk RNA-seq can be done by performing technical replicates, which is constrained by the low amount of material in the case of single-cell RNA-seq. Hence, more research is necessary to allow correct discrimination of technical and biological variability. Furthermore, many computational challenges remain, including determining whether TPM is the best measure for transcript length normalization, or to what extent bulk RNA-seq analysis tools can be applied to single cell RNA-seq.
As more and more single cell datasets are generated, data integration will be inevitable. This poses a great challenge, as it was recently proven that the conversion of reads to molecules using UMIs is impacted by both biological and technical variation, indicating that UMI counts are not an unbiased estimator of gene expression levels (Tung et al., 2017). Thus, an additional challenge will be to account for biases in a way that is standardised enough to make datasets from different studies, which will be differently affected by noise, comparable.
In summary, single-cell RNA-Seq holds tremendous potential to unravel gene expression heterogeneity at an individual cell level. To fulfil this promise, the most urgent concern is that biological variation needs to be efficiently separated from technical variation. Faced with such a challenging endeavour, the joint efforts of both experimental and computational biologists are needed, as the increase in multi-disciplinary teams will help tackle different aspects of this issue and design comprehensive solutions. Apart from delineating technical and biological variation, the computational biology field can contribute in various ways to the ever growing single-cell analysis field, namely performing a systematic assessment of the many existing quality control and normalization methods, and developing statistical and computational tools for effectively discovering the biology behind high dimensional data generated by single cell RNA-seq.
Competing interests
The authors declare no completing interests.
Acknowledgements
GD is a Cascade fellow (H2020, Marie Curie). AJ is a Chancellor’s fellow at the Roslin Institute, University of Edinburgh. A.J. lab is supported by from Biotechnology and Biological Sciences Research Council (BBSRC, BB/J004235/1).
References
- Alwine J.C., Kemp D.J., Stark G.R. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc. Natl. Acad. Sci. U. S. A. 1977;74:5350–5354. doi: 10.1073/pnas.74.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher R., Kendziorski C. Design and computational analysis of single-cell RNA-sequencing experiments. Genome Biol. 2016;17 doi: 10.1186/s13059-016-0927-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke P., Anders S., Kim J.K., Kołodziejczyk A.A., Zhang X., Proserpio V., Baying B., Benes V., Teichmann S.A., Marioni J.C., Heisler M.G. Accounting for technical noise in single-cell RNA-seq experiments. Nat. Methods. 2013;10:1093–1095. doi: 10.1038/nmeth.2645. [DOI] [PubMed] [Google Scholar]
- Buettner F., Natarajan K.N., Casale F.P., Proserpio V., Scialdone A., Theis F.J., Teichmann S.A., Marioni J.C., Stegle O. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat. Biotechnol. 2015;33:155–160. doi: 10.1038/nbt.3102. [DOI] [PubMed] [Google Scholar]
- Hashimshony T., Feder M., Levin M., Hall B.K., Yanai I. Spatiotemporal transcriptomics reveals the evolutionary history of the endoderm germ layer. Nature. 2014;519:219–222. doi: 10.1038/nature13996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks S.C., Townes F.W., Teng M., Irizarry R.A. 2017. Missing Data and Technical Variability in Single-Cell RNA- Sequencing Experiments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilicic T., Kim J.K., Kolodziejczyk A.A., Bagger F.O., McCarthy D.J., Marioni J.C., Teichmann S.A. Classification of low quality cells from single-cell RNA-seq data. Genome Biol. 2016;17:29. doi: 10.1186/s13059-016-0888-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iscove N.N., Barbara M., Gu M., Gibson M., Modi C., Winegarden N. Representation is faithfully preserved in global cDNA amplified exponentially from sub-picogram quantities of mRNA. Nat. Biotechnol. 2002;20:940–943. doi: 10.1038/nbt729. [DOI] [PubMed] [Google Scholar]
- Islam S., Zeisel A., Joost S., La Manno G., Zajac P., Kasper M., Lönnerberg P., Linnarsson S. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods. 2014;11:163–166. doi: 10.1038/nmeth.2772. [DOI] [PubMed] [Google Scholar]
- Jiang L., Schlesinger F., Davis C.A., Zhang Y., Li R., Salit M., Gingeras T.R., Oliver B. Synthetic spike-in standards for RNA-seq experiments. Genome Res. 2011;21:1543–1551. doi: 10.1101/gr.121095.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.K., Marioni J.C. Inferring the kinetics of stochastic gene expression from single-cell RNA-sequencing data. Genome Biol. 2013;14:R7. doi: 10.1186/gb-2013-14-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.K., Kolodziejczyk A.A., Ilicic T., Illicic T., Teichmann S.A., Marioni J.C. Characterizing noise structure in single-cell RNA-seq distinguishes genuine from technical stochastic allelic expression. Nat. Commun. 2015;6:8687. doi: 10.1038/ncomms9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.Y., Lovén J., Rahl P.B., Paranal R.M., Burge C.B., Bradner J.E., Lee T.I., Young R.A. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens-Bobadilla E., Zhao S., Baser A., Saiz-Castro G., Zwadlo K., Martin-Villalba A. Single-cell transcriptomics reveals a population of dormant neural stem cells that become activated upon brain injury. Cell Stem Cell. 2015;17:329–340. doi: 10.1016/j.stem.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Lubeck E., Cai L. Single-cell systems biology by super-resolution imaging and combinatorial labeling. Nat. Methods. 2012;9:743–748. doi: 10.1038/nmeth.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantsoki A., Devailly G., Joshi A. Gene expression variability in mammalian embryonic stem cells using single cell RNA-seq data. Comput. Biol. Chem. 2016;63:52–61. doi: 10.1016/j.compbiolchem.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinov G.K., Williams B.A., McCue K., Schroth G.P., Gertz J., Myers R.M., Wold B.J. From single-cell to cell-pool transcriptomes: stochasticity in gene expression and RNA splicing. Genome Res. 2014;24:496–510. doi: 10.1101/gr.161034.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy D., Wills Q., Campbell K. Bioconductor R Package Version 1.2.0. 2016. scater: single-cell analysis toolkit for gene expression data in R. [Google Scholar]
- Moignard V., Woodhouse S., Haghverdi L., Lilly A.J., Tanaka Y., Wilkinson A.C., Buettner F., Macaulay I.C., Jawaid W., Diamanti E., Nishikawa S.-I., Piterman N., Kouskoff V., Theis F.J., Fisher J., G?ttgens B. Decoding the regulatory network of early blood development from single-cell gene expression measurements. Nat. Biotechnol. 2015;33:269–276. doi: 10.1038/nbt.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Munsky B., Neuert G., van Oudenaarden A. Using gene expression noise to understand gene regulation. Science. 2012;336:183–187. doi: 10.1126/science.1216379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A.P., Tirosh I., Trombetta J.J., Shalek A.K., Gillespie S.M., Wakimoto H., Cahill D.P., Nahed B.V., Curry W.T., Martuza R.L., Louis D.N., Rozenblatt-Rosen O., Suvà M.L., Regev A., Bernstein B.E. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul F., Arkin Y., Giladi A., Jaitin D., Kenigsberg E., Keren-Shaul H., Winter D., Lara-Astiaso D., Gury M., Weiner A., David E., Cohen N., Lauridsen F., Haas S., Schlitzer A., Mildner A., Ginhoux F., Jung S., Trumpp A., Porse B., Tanay A., Amit I. Transcriptional heterogeneity and lineage commitment in myeloid progenitors. Cell. 2015;163:1663–1677. doi: 10.1016/j.cell.2015.11.013. [DOI] [PubMed] [Google Scholar]
- Picelli S., Faridani O.R., Björklund A.K., Winberg G., Sagasser S., Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- Pierson E., Yau C. ZIFA: dimensionality reduction for zero-inflated single-cell gene expression analysis. Genome Biol. 2015;16:241. doi: 10.1186/s13059-015-0805-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Hill A., Packer J., Lin D., Ma Y.-A., Trapnell C. Single-cell mRNA quantification and differential analysis with Census. Nat. Methods. 2017;14:309–315. doi: 10.1038/nmeth.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsköld D., Luo S., Wang Y.-C., Li R., Deng Q., Faridani O.R., Daniels G.A., Khrebtukova I., Loring J.F., Laurent L.C., Schroth G.P., Sandberg R. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostom R., Svensson V., Teichmann S.A., Kar G. Computational approaches for interpreting scRNA-seq data. FEBS Lett. 2017 doi: 10.1002/1873-3468.12684. http://onlinelibrary.wiley.com/doi/10.1002/1873-3468.12684/abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R. Entering the era of single-cell transcriptomics in biology and medicine. Nat. Methods. 2014;11:22–24. doi: 10.1038/nmeth.2764. [DOI] [PubMed] [Google Scholar]
- Schena M., Shalon D., Davis R.W., Brown P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Stegle O., Teichmann S.A., Marioni J.C. Computational and analytical challenges in single-cell transcriptomics. Nat. Rev. Genet. 2015;16:133–145. doi: 10.1038/nrg3833. [DOI] [PubMed] [Google Scholar]
- Tang F., Barbacioru C., Bao S., Lee C., Nordman E., Wang X., Lao K., Surani M.A. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell. 2010;6:468–478. doi: 10.1016/j.stem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F., Barbacioru C., Nordman E., Bao S., Lee C., Wang X., Tuch B.B., Heard E., Lao K., Surani M.A. Deterministic and stochastic allele specific gene expression in single mouse blastomeres. PLoS One. 2011;6:e21208. doi: 10.1371/journal.pone.0021208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I., Izar B., Prakadan S.M., Wadsworth M.H., Treacy D., Trombetta J.J., Rotem A., Rodman C., Lian C., Murphy G., Fallahi-Sichani M., Dutton-Regester K., Lin J.-R., Cohen O., Shah P., Lu D., Genshaft A.S., Hughes T.K., Ziegler C.G.K., Kazer S.W., Gaillard A., Kolb K.E., Villani A.-C., Johannessen C.M., Andreev A.Y., Van Allen E.M., Bertagnolli M., Sorger P.K., Sullivan R.J., Flaherty K.T., Frederick D.T., Jané-Valbuena J., Yoon C.H., Rozenblatt-Rosen O., Shalek A.K., Regev A., Garraway L.A. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein B., Brownfield D.G., Wu A.R., Neff N.F., Mantalas G.L., Espinoza F.H., Desai T.J., Krasnow M.A., Quake S.R. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung P.-Y., Blischak J.D., Hsiao C.J., Knowles D.A., Burnett J.E., Pritchard J.K., Gilad Y. Batch effects and the effective design of single-cell gene expression studies. Sci. Rep. 2017;7:39921. doi: 10.1038/srep39921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner G.P., Kin K., Lynch V.J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012;131:281–285. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A.R., Neff N.F., Kalisky T., Dalerba P., Treutlein B., Rothenberg M.E., Mburu F.M., Mantalas G.L., Sim S., Clarke M.F., Quake S.R. Quantitative assessment of single-cell RNA-sequencing methods. Nat. Methods. 2014;11:41–46. doi: 10.1038/nmeth.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegenhain C., Vieth B., Parekh S., Reinius B., Guillaumet-Adkins A., Smets M., Leonhardt H., Heyn H., Hellmann I., Enard W. Comparative analysis of single-cell RNA sequencing methods. Mol. Cell. 2017;65:631–643. doi: 10.1016/j.molcel.2017.01.023. (e4) [DOI] [PubMed] [Google Scholar]