Abstract
Anabolic metabolism in lymphocytes promotes plasmablast and cytotoxic T cell differentiation at the expense of self-renewal. Heightened expression and function of the transcription factor IFN regulatory factor 4 (IRF4) accompany enhanced anabolic induction and full commitment to functional differentiation in B cells and CD8+ T cells. In this study, we used a genetic approach to determine whether IRF4 plays an analogous role in Th1 cell induction. Our findings indicate that IRF4 promotes determined Th1 cell differentiation in tandem with anabolic metabolism of CD4+ T cells. IRF4-deficient CD4+ T cells stimulated in vitro exhibit impaired induction of Th1 gene expression and defective silencing of T cell factor 1 expression. IRF4-deficient CD4+ T cells also undergo a shift toward catabolic metabolism, with reduced mammalian target of rapamycin activation, cell size, and nutrient uptake, as well as increased mitochondrial clearance. These findings suggest that the ability to remodel metabolic states can be an essential gateway for altering cell fate.
INTRODUCTION
Emerging evidence has implicated cellular metabolism as a defining feature, if not a frank determinant of lymphocyte fate and function (1). Naive and quiescent memory lymphocytes rely on catabolic metabolism, including mitochondrial oxidative phosphorylation, fatty acid β-oxidation, and autophagy, for successful homeostasis and renewal. In contrast, activation, proliferation, and induction of effector T cells and plasmablasts require anabolic metabolism, including inducible glucose uptake and aerobic glycolysis (reviewed in Ref. 1).
Activated progenitors that have undergone several divisions are capable of giving rise to asymmetric daughter cells, owing to unequal anabolic PI3K/mammalian target of rapamycin (mTOR) activation (2–4). A differentiated sibling cell undergoes inactivation of FoxO1 and irreversible silencing of Pax5 or T cell factor 1 (TCF1), whereas a self-renewing sibling cell with lesser PI3K activation maintains expression of Pax5 or TCF1 and the ability to give rise to self-renewing and differentiated progeny (2–4). Determined CD4+ Th1 cell differentiation in vivo is also characterized by asymmetric outcomes wherein production of a Th1 cell that silences TCF1 is accompanied by the production of a self-renewing sibling cell that maintains expression of TCF1 (3, 5). Th1 cell determination is also driven by anabolic PI3K/mTOR signaling and aerobic glycolysis (3, 5–11).
The transcription factor IFN regulatory factor 4 (IRF4) is critical for the differentiation of germinal center B cells and, at high concentrations, plasma cells(3, 4, 12–14).IRF4 has been implicated in dose-dependent anabolic gene induction of CD8+ effector T cells that silence TCF1 and plasmablasts that silence Pax5, at least in part because of its induction by PI3K/mTOR signaling and its governance of numerous genes of the glucose uptake and metabolism pathway (2, 4, 15–19). The roles of IRF4 in CD4+ effector differentiation have been partly elucidated (20–26). In view of the recent suggestion that IRF4 governs oxidative and glycolytic metabolism in CD4+ T cells (20), we deployed an in vitro Th1-induction system to discern whether IRF4 governs the anabolic switch required for Th1 differentiation.
MATERIALS AND METHODS
Mice and Th1 culture
Wild-type (WT) C57BL/6 and IRF4-knockout (KO) (14) littermate mice served as sources of CD4+ T cells. Both male and female mice were used at age 8–10 wk, and mice were housed in specific pathogen–free conditions. All experiments were conducted in accordance with National Institutes of Health and Columbia University Institutional Animal Care and Use Committee guidelines. All efforts were made to minimize animal suffering and the number of animals used. Naive CD4+ T cells were purified from spleens by magnetic cell separation (Miltenyi Biotec) and subsequently labeled with the cell proliferation dye (CPD) CellTrace Violet (Thermo Fisher). A total of 5 ×105 cells was seeded in 48-well tissue culture plates precoated with anti-CD3 (1 μg/ml) and anti-CD28(1 μg/ml) inIscove’s Modified EagleMedium (Corning) supplemented with 10% FBS, IL-2 (20 IU/ml), and IL-12 (10 ng/ ml). For experiments with retroviral expression of T-bet–GFP, cells were activated for ≥36 h before spinfection, as previously described (3). Following spinfection, cells were returned to the original culture media and cultured for an additional 48 h before flow cytometric analysis.
Flow cytometry
Staining for flow cytometry analysis was performed as described (3, 4). For phospho-flow cytometry, samples were fixed with 4% paraformaldehyde for 15 min, followed by permeabilization with ice-cold methanol for 5 min before Ab staining. Cytokine production was assayed following PMA (50 ng/ml) and ionomycin (1 μg/ml) restimulation for 4 h. Abs used in this study include CD25 (clone PC61; BioLegend), CD62L (clone MEL-14; BD Biosciences), CD98 (clone RL388; BioLegend), anti-Glut1 (clone EPR3915; Abcam), T-bet (clone 4B10; BioLegend), TCF1 (clone C63D9; Cell Signaling Technology), phospho-S6 (clone S235/236; Cell Signaling Technology), granzyme B (clone GB11; BioLegend), IFN-γ(cloneXMG1.2;BD Biosciences), TNF-α(clone MP6-XT22; BD Biosciences), and goat anti-rabbit Alexa Fluor 647 secondary Ab (Thermo Fisher). Flow cytometry samples were acquired on a BD LSR II or BD Fortessa, and analysis was performed with FlowJo software (TreeStar, San Carlos, CA).
Glucose uptake following TCR stimulation was measured by incubating cells in 2-NBDG (100 μM; Cayman Chemical) for 45 min at 37°C in glucose-free RPMI 1640 supplemented with 10% dialyzed FBS. Mitoclearance was assessed by labeling naive CD4+ T cells with MitoTracker Green (200 nM; Thermo Fisher), followed by two washes with complete media. Cells were then labeled with CellTrace Violet and activated in Th1-inducing conditions. Clearance of mitochondrial fluorescence represents the sum of passive dilution during cell division plus active destruction through autophagy (3). Total mitochondrial mass and mitochondrial membrane potential were determined by labeling cells with MitoTracker Red or TMRE (Thermo Fisher), respectively, according to the manufacturer’s instructions.
Statistical analyses
Statistical analyses were performed using GraphPad Prism (version 6). The p values and significance cutoffs are specified in each figure legend.
RESULTS
Impaired Th1 differentiation in IRF4-KO CD4+ T cells
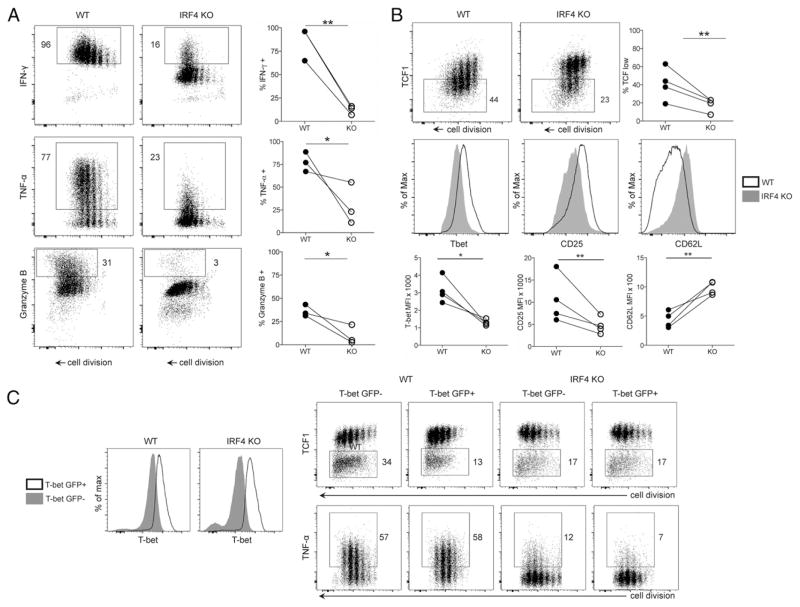
Naive WT and IRF4-KO (14) CD4+ T cells were activated in Th1-inducing conditions in vitro for 4 d prior to flow cytometry. Compared with WT cells, proliferating IRF4-deficient cells expressed substantially less Th1 cytokines (IFN-γ, TNF-α) and effector molecules (granzyme B) (Fig. 1A). Consistent with a defect in functional Th1 gene expression, IRF4-deficient cells also failed to extinguish expression of TCF1 (Fig. 1B), which is characteristically silenced during terminal Th1 differentiation (5, 7, 27, 28). IRF4-deficient cells also exhibited reduced induction of the Th1 markers T-bet and CD25, as well as increased maintenance of CD62 ligand expression (Fig. 1B). Transduction of IRF4-deficient cells with T-bet–GFP retrovirus failed to rescue defects in TCF1 silencing and cytokine induction (Fig. 1C). The present results, together with recent observations using Listeria infection (20), suggest that IRF4 is required for the induction of optimal Th1 differentiation but not simply as an inducer of Th1-specific genes.
FIGURE 1. IRF4 is required for efficient Th1 CD4+ effector cell differentiation.
(A) CD4+ T cells were labeled with CPD, stimulated with anti-CD3/CD28 + IL-2/IL-12, and restimulated with PMA/ionomycin on day 4, followed by FACS analysis. Representative FACS plots of indicated protein versus CPD (left panels). Quantification of the percent positive population in WT and IRF4-KO CD4+ T cells (right panels). (B) Representative FACS plots of TCF1 expression versus CPD after 4 d of activation in Th1-inducing conditions (top left and middle panels). Quantification of TCF1-low population in WT and IRF4-KO CD4+ T cells (top right panel). Representative line graphs of T-bet, CD25, and CD62L staining of CD4+ T cells (middle row). Quantification of protein expression in WT and IRF4-KO CD4+ T cells (bottom row). (C) Increased T-bet protein expression in WT and IRF4-KO CD4+ T cells following transduction with T-bet–GFP retrovirus (open graph) compared with untransduced cells (shaded graph) at day 4 postactivation (left panels). Expression of TCF1 (upper right panels) and TNF-α (lower right panels) by CD4+ T cells transduced (GFP+) or untransduced (GFP−) with T-bet–GFP retrovirus, at day 4 postactivation. Plots are representative of three independent experiments. *p < 0.05, **p < 0.01, paired t test.
Impaired anabolism in IRF4-KO CD4+ T cells
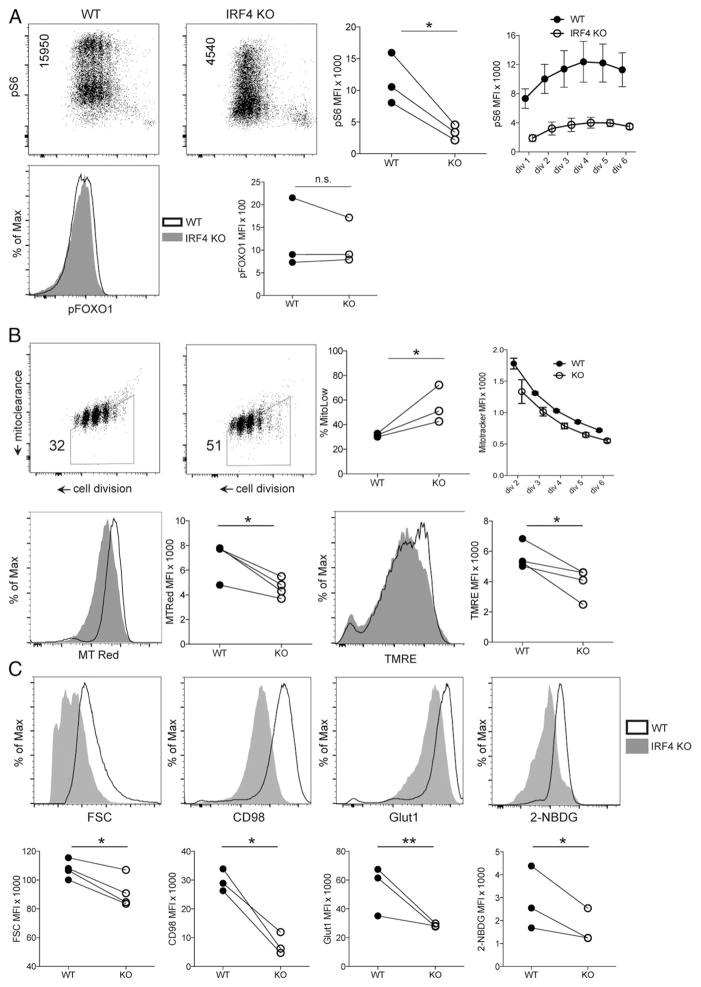
Like plasmablast and CD8+ effector differentiation, CD4+ Th1 differentiation requires sufficient anabolic activation, including aerobic glycolysis (Warburg effect) (3, 5–11). Consistent with the previously described role for IRF4 in the anabolic activation of B cells and CD8+ T cells (3, 4, 16), we found that IRF4-deficient CD4+ T cells had impaired activation of the mTOR pathway, as assessed by phosphorylation of ribosome subunit S6 (Fig. 2A). Deficient mTOR activation was evident in early and late division cells and was maintained throughout the duration of the experiment, suggesting that IRF4 plays a primary role in supporting PI3K/mTOR activation. Despite the impairment in phosphorylation of S6, levels of FoxO1 phosphorylation (another target of PI3K signaling) appeared intact in IRF4-deficient cells (Fig. 2A), which is consistent with the ability of IRF4-deficient B cells to undergo inducible nuclear displacement of FoxO1 (4).
FIGURE 2. IRF4 is required for anabolic induction of CD4+ T cells.
(A) CD4+ T cells activated under Th1-inducing conditions were analyzed by phospho-flow cytometry for mTOR activity by ribosomal S6 phosphorylation levels at day 4 post–TCR stimulation (upper left and middle left panels). FACS plot statistic indicates mean fluorescence intensity (MFI) of phospho-S6. Quantification of phospho-S6 levels (upper middle right panel). MFI of phospho-S6 at the indicated cell divisions (upper right panel). Error bars denote SEM. Representative line graph of phospho-FoxO1 levels in WT and IRF4-KO CD4+ T cells at day 4 post–TCR stimulation (lower left panel). Quantification of phospho-FoxO1 MFI (lower right panel). (B) WT and IRF4-KO CD4+ T cells were pulse-labeled with CPD and MitoTracker Green, followed by TCR stimulation and flow cytometry analysis after 66 h of culture (upper left and middle left panels). MitoTracker fluorescence (y-axis) decreases with each cell division (x-axis) as the pulse-labeled mitochondria age and are passively apportioned, as well as actively cleared by mitophagy (hence, “mitoclearance”). Frequency statistic indicates the percentage of cells in the MitoTracker-low trapezoidal gate. Quantification of the frequency of cells in the MitoTracker-low gate (upper middle right panel). MFI of pulsed MitoTracker fluorescence at the indicated cell divisions (upper right panel). Error bars denote SEM. Representative line graph of total mitochondrial content measured by MitoTracker Red staining immediately prior to flow cytometry, at day 4 of culture (lower left panel). Quantification of MitoTracker Red MFI (lower middle left panel). Representative line graph of mitochondrial membrane potential measured by TMRE staining on day 4 of culture (lower middle right panel). Quantification of TMRE MFI (lower right panel). (C) Representative line graphs of CD4+ T cells labeled with CPD, stimulated in Th1 conditions, and analyzed by flow cytometry on day 4 of culture for forward scatter (FSC; cell size), expression of the indicated nutrient transporters (CD98 and Glut1), and glucose uptake (2-NBDG) (upper panels). Quantification of the indicated flow cytometry parameters for WT and IRF4-KO CD4+ T cells (lower panels). *p < 0.05, **p < 0.01, paired t test. n.s., not significant.
Consistent with a model wherein IRF4 sits in a critical node of anabolic metabolism, and whose deficiency results in catabolism (3, 4, 16), we found that the cellular descendants of IRF4-deficient cells exhibited enhanced clearance of aged mitochondria (Fig. 2B), an indicator of catabolic mitochondrial autophagy, which appears to predict a diminished likelihood of differentiation (3). Differences in the levels of aged mitochondria were evident beginning at the second division and continuing on to later divisions (Fig. 2B), further suggesting a primary role for IRF4 in sustaining nutritive signaling. Despite enhanced mitochondrial elimination, dividing IRF4-deficient CD4+ T cells had lower total mitochondrial content and lower mitochondrial membrane potential (Fig. 2B).
Consistent with a switch toward more catabolic metabolism, IRF4-deficient cells exhibited reduced blasting cell size, along with defective expression of CD98, a shared chain of some amino acid transporters, and Glut1 (8), the predominant inducible glucose transporter of CD4+ T cells (Fig. 2C). Reduced Glut1 induction was also associated with diminished glucose uptake in IRF4-deficient cells (Fig. 2C). These results are consistent with recent analyses showing defective glycolytic flux in IRF4-deficient CD4+ T cells (20).
DISCUSSION
IRF4 appears to function as a factor that permits anabolic metabolism to be interwoven with terminal lymphocyte differentiation. It had been previously suggested that IRF4 acts downstream of mTOR signaling in T cells (15). The present finding that mTOR activation also requires IRF4 function is consistent with a feed-forward feed-back relationship that characterizes binary or bistable systems (3). As such, perturbations of upstream signals or downstream processes of anabolism cause abortive anabolic induction, with default to catabolism (3). By licensing upstream nutritive signaling, nutrient uptake and utilization, and rewiring the transcriptional circuitry of cell fate, IRF4 couples metabolic and lineage choice.
Mitochondrial biogenesis and fusion, which accompany the autophagy of older mitochondria in physiological memory T cells, are essential for optimal oxidative metabolism (3, 29). The present findings that IRF4-deficient cells exhibit a reduction in total mitochondrial content and mitochondrial membrane potential suggest that the severity of nutrient deprivation in the absence of IRF4 prevents basal replacement/biogenesis of new mitochondria to offset the elimination of aged organelles. Therefore, the resultant defects in mitochondrial function would be consistent with recent findings that oxidative metabolism is reduced in IRF4-deficient CD4+ T cells (20). Defective oxygen consumption of IRF4-deficient CD4+ T cells might then be regarded as a reflection of the severity of the catabolic state rather than an inability to engage in catabolic self-digestion per se.
The present results suggest that IRF4 is not simply directing two parallel, but unrelated, processes (Th1 differentiation and anabolism). Instead, defective anabolism in IRF4-deficient cells is probably analogous to perturbation of glucose availability, both being causal to defective Th1 differentiation and function (8, 10, 11). Presumably, anabolic metabolism, which apparently requires IRF4, drives Th1 differentiation, in part through its ability to support silencing of TCF1, a negative regulator of T-bet induction (7, 27, 28). Whether IRF4 and anabolism promote silencing of TCF1 solely by inactivating FoxO1, a guardian of lymphocyte self-renewal (3, 4), or through other gene regulatory mechanisms will require further investigation. Nonetheless, the binary nature of cellular anabolism versus catabolism is a seemingly robust framework to balance the opposing cell fates of differentiation and self-renewal and an attractive target to dampen or augment immune responses.
Acknowledgments
This work was supported by National Institutes of Health Grants AI113365 and AI076458 (to S.L.R.) and by the Charles Revson Foundation.
Abbreviations used in this article
- CPD
cell proliferation dye
- IRF4
IFN regulatory factor 4
- KO
knockout
- mTOR
mammalian target of rapamycin
- TCF1
T cell factor 1
- WT
wild-type
Footnotes
DISCLOSURES
The authors have no financial conflicts of interest.
References
- 1.Pearce EL, Pearce EJ. Metabolic pathways in immune cell activation and quiescence. Immunity. 2013;38:633–643. doi: 10.1016/j.immuni.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin WW, Nish SA, Yen B, Chen YH, Adams WC, Kratchmarov R, Rothman NJ, Bhandoola A, Xue HH, Reiner SL. CD8(+) T lymphocyte self-renewal during effector cell determination. Cell Reports. 2016;17:1773–1782. doi: 10.1016/j.celrep.2016.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams WC, Chen YH, Kratchmarov R, Yen B, Nish SA, Lin WW, Rothman NJ, Luchsinger LL, Klein U, Busslinger M, et al. Anabolism-associated mitochondrial stasis driving lymphocyte differentiation over self-renewal. Cell Reports. 2016;17:3142–3152. doi: 10.1016/j.celrep.2016.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin WH, Adams WC, Nish SA, Chen YH, Yen B, Rothman NJ, Kratchmarov R, Okada T, Klein U, Reiner SL. Asymmetric PI3K signaling driving developmental and regenerative cell fate bifurcation. Cell Reports. 2015;13:2203–2218. doi: 10.1016/j.celrep.2015.10.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nish SA, Zens KD, Kratchmarov R, Lin WW, Adams WC, Chen YH, Yen B, Rothman NJ, Bhandoola A, Xue HH, et al. CD4+ T cell effector commitment coupled to self-renewal by asymmetric cell divisions. J Exp Med. 2017;214:39–47. doi: 10.1084/jem.20161046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ray JP, Staron MM, Shyer JA, Ho PC, Marshall HD, Gray SM, Laidlaw BJ, Araki K, Ahmed R, Kaech SM, Craft J. The interleukin-2–mTORc1 kinase axis defines the signaling, differentiation, and metabolism of T helper 1 and follicular B helper T cells. Immunity. 2015;43:690–702. doi: 10.1016/j.immuni.2015.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu T, Shin HM, Moseman EA, Ji Y, Huang B, Harly C, Sen JM, Berg LJ, Gattinoni L, McGavern DB, Schwartzberg PL. TCF1 is required for the T follicular helper cell response to viral infection. Cell Reports. 2015;12:2099–2110. doi: 10.1016/j.celrep.2015.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Macintyre AN, V, Gerriets A, Nichols AG, Michalek RD, Rudolph MC, Deoliveira D, Anderson SM, Abel ED, Chen BJ, Hale LP, Rathmell JC. The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metab. 2014;20:61–72. doi: 10.1016/j.cmet.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng M, Yin N, Chhangawala S, Xu K, Leslie CS, Li MO. Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science. 2016;354:481–484. doi: 10.1126/science.aaf6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blagih J, Coulombe F, Vincent EE, Dupuy F, Galicia-Vázquez G, Yurchenko E, Raissi TC, van der Windt GJ, Viollet B, Pearce EL, et al. The energy sensor AMPK regulates T cell metabolic adaptation and effector responses in vivo. Immunity. 2015;42:41–54. doi: 10.1016/j.immuni.2014.12.030. [DOI] [PubMed] [Google Scholar]
- 11.Chang CH, Curtis JD, Maggi LB, Jr, Faubert B, Villarino AV, O’Sullivan D, Huang SC, van der Windt GJ, Blagih J, Qiu J, et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willis SN, Good-Jacobson KL, Curtis J, Light A, Tellier J, Shi W, Smyth GK, Tarlinton DM, Belz GT, Corcoran LM, et al. Transcription factor IRF4 regulates germinal center cell formation through a B cell-intrinsic mechanism. J Immunol. 2014;192:3200–3206. doi: 10.4049/jimmunol.1303216. [DOI] [PubMed] [Google Scholar]
- 13.Sciammas R, Shaffer AL, Schatz JH, Zhao H, Staudt LM, Singh H. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 2006;25:225–236. doi: 10.1016/j.immuni.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 14.Klein U, Casola S, Cattoretti G, Shen Q, Lia M, Mo T, Ludwig T, Rajewsky K, Dalla-Favera R. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- 15.Yao S, Buzo BF, Pham D, Jiang L, Taparowsky EJ, Kaplan MH, Sun J. Interferon regulatory factor 4 sustains CD8(+) T cell expansion and effector differentiation. Immunity. 2013;39:833–845. doi: 10.1016/j.immuni.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Man K, Miasari M, Shi W, Xin A, Henstridge DC, Preston S, Pellegrini M, Belz GT, Smyth GK, Febbraio MA, et al. The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat Immunol. 2013;14:1155–1165. doi: 10.1038/ni.2710. [DOI] [PubMed] [Google Scholar]
- 17.Raczkowski F, Ritter J, Heesch K, Schumacher V, Guralnik A, Höcker L, Raifer H, Klein M, Bopp T, Harb H, et al. The transcription factor interferon regulatory factor 4 is required for the generation of protective effector CD8+ T cells. Proc Natl Acad Sci USA. 2013;110:15019–15024. doi: 10.1073/pnas.1309378110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nayar R, Schutten E, Bautista B, Daniels K, Prince AL, Enos M, Brehm MA, Swain SL, Welsh RM, Berg LJ. Graded levels of IRF4 regulate CD8+ T cell differentiation and expansion, but not attrition, in response to acute virus infection. J Immunol. 2014;192:5881–5893. doi: 10.4049/jimmunol.1303187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nayar R, Enos M, Prince A, Shin H, Hemmers S, Jiang JK, Klein U, Thomas CJ, Berg LJ. TCR signaling via Tec kinase ITK and interferon regulatory factor 4 (IRF4) regulates CD8+ T-cell differentiation. Proc Natl Acad Sci USA. 2012;109:E2794–E2802. doi: 10.1073/pnas.1205742109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahnke J, Schumacher V, Ahrens S, Käding N, Feldhoff LM, Huber M, Rupp J, Raczkowski F, Mittrücker HW. Interferon regulatory factor 4 controls TH1 cell effector function and metabolism. Sci Rep. 2016;6:35521. doi: 10.1038/srep35521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brüstle A, Heink S, Huber M, Rosenplänter C, Stadelmann C, Yu P, Arpaia E, Mak TW, Kamradt T, Lohoff M. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat Immunol. 2007;8:958–966. doi: 10.1038/ni1500. [DOI] [PubMed] [Google Scholar]
- 22.Lohoff M, Mittrücker HW, Prechtl S, Bischof S, Sommer F, Kock S, Ferrick DA, Duncan GS, Gessner A, Mak TW. Dysregulated T helper cell differentiation in the absence of interferon regulatory factor 4. Proc Natl Acad Sci USA. 2002;99:11808–11812. doi: 10.1073/pnas.182425099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mittrücker HW, Matsuyama T, Grossman A, Kündig TM, Potter J, Shahinian A, Wakeham A, Patterson B, Ohashi PS, Mak TW. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. 1997;275:540–543. doi: 10.1126/science.275.5299.540. [DOI] [PubMed] [Google Scholar]
- 24.Staudt V, Bothur E, Klein M, Lingnau K, Reuter S, Grebe N, Gerlitzki B, Hoffmann M, Ulges A, Taube C, et al. Interferon-regulatory factor 4 is essential for the developmental program of T helper 9 cells. Immunity. 2010;33:192–202. doi: 10.1016/j.immuni.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 25.Rengarajan J, Mowen KA, McBride KD, Smith ED, Singh H, Glimcher LH. Interferon regulatory factor 4 (IRF4) interacts with NFATc2 to modulate interleukin 4 gene expression. J Exp Med. 2002;195:1003–1012. doi: 10.1084/jem.20011128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tominaga N, Ohkusu-Tsukada K, Udono H, Abe R, Matsuyama T, Yui K. Development of Th1 and not Th2 immune responses in mice lacking IFN-regulatory factor-4. Int Immunol. 2003;15:1–10. doi: 10.1093/intimm/dxg001. [DOI] [PubMed] [Google Scholar]
- 27.Choi YS, Gullicksrud JA, Xing S, Zeng Z, Shan Q, Li F, Love PE, Peng W, Xue HH, Crotty S. LEF-1 and TCF-1 orchestrate T(FH) differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nat Immunol. 2015;16:980–990. doi: 10.1038/ni.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu L, Cao Y, Xie Z, Huang Q, Bai Q, Yang X, He R, Hao Y, Wang H, Zhao T, et al. The transcription factor TCF-1 initiates the differentiation of T(FH) cells during acute viral infection. Nat Immunol. 2015;16:991–999. doi: 10.1038/ni.3229. [DOI] [PubMed] [Google Scholar]
- 29.Buck MD, O’Sullivan D, Klein Geltink RI, Curtis JD, Chang CH, Sanin DE, Qiu J, Kretz O, Braas D, van der Windt GJ, et al. Mitochondrial dynamics controls T cell fate through metabolic programming. Cell. 2016;166:63–76. doi: 10.1016/j.cell.2016.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]