Abstract
Assessment of antibiotic action with new drug development directed towards anaerobic bacteria is difficult and technically demanding. To gain insight into possible MOA, morphologic changes associated with antibiotic exposure can be visualized using scanning electron microscopy (SEM). Integrating SEM imaging with traditional kill curves may improve our insight into drug action and advance the drug development process. To test this premise, kill curves and SEM studies were conducted using drugs with known but different MOA (vancomycin and metronidazole). C. difficile cells (R20291) were grown with or without the presence of antibiotic for up to 48 h. Throughout the 48 h interval, cells were collected at multiple time points to determine antibiotic efficacy and for imaging on the SEM. Consistent with previous reports, vancomycin and metronidazole had significant bactericidal activity following 24 h of treatment as measured by colony-forming unit (CFU) counting. Using SEM imaging we determined that metronidazole had significant effects on cell length (> 50% reduction in cell length for each antibiotic; P< 0.05) compared to controls and vancomycin. While the phenotypic response to drug treatment has not been documented previously in this manner, they are consistent with the drug's MOA demonstrating the versatility and reliability of the imaging and measurements and the application of this technique for other experimental compounds.
Keywords: Immunology, Issue 123, Clostridium difficile, scanning electron microscopy, antibiotics, vancomycin, metronidazole, ridinilazole, fidaxomicin
Introduction
Clostridium difficile is a gram positive, spore-forming bacterium, causing approximately 500,000 infections annually in the US and is considered a threat level urgent pathogen by the Centers for Disease Control and Prevention (CDC), the highest level of risk.1 The past decade has seen considerable drug development in antimicrobials with activity against C. difficile.2,3In vitro studies are a necessary component of the drug development process.4 Traditionally, in vitro susceptibility and time kill studies are used to validate future animal and other in vivo studies.
While these methods serve an important role for evaluating killing action, they do not capture the cells' phenotypic response to pharmacological treatment. By incorporating scanning electron microscopy (SEM) with standard killing kinetic studies, a more thorough characterization of the antibiotic direct effects is possible.5,6,7 Here, we present a method where SEM is used as a means to profile the efficacy of antibiotic treatment.
Protocol
1. Isolating C. difficile from Different Environmental or Clinical Sources
Environmental isolates: Using a pre-sterilized cotton gauge (lightly wetted with 0.85% NaCl), swab the surface of any area of interest (floor, door, handle, shelf, etc.).8 Make sure to wear sterile gloves and place the swab in a sterilized tube after completed.
Clinical isolates (stool): Plate 10 to 100 mg of clinical stool samples onto cefoxitin-cycloserine-fructose agar (CCFA) using an inoculation loop and incubate under strict anaerobic conditions for 48 - 72 h. Store isolated colonies of C. difficile stock in cryovials at -80 °C for further analyses.7,9,10
Enrich the environmental swab samples in brain heart infusion (BHI) broth with 0.05% sodium taurocholate and place in an anaerobic chamber at 37 °C for 5 days. Centrifuge 1 mL of the culture at 10,000 x g and resuspend the pellet in 100 µL of ethanol.
Plate the resuspended cells (50 µL) onto cycloserinecefoxitin fructose agar (CCFA) plates and incubate in an anaerobic chamber at 37 °C for 40 - 48 h. Store isolated colonies of C. difficile stock in cryovials at -80 °C for further analyses.
Test suspected C. difficile colonies using the latex agglutination reagent or PCR.
2. Culturing C. difficile and Killing Kinetic Procedures
Grow purified environmental or clinical C. difficile strains on blood agar plates in an anaerobic chamber at 37 °C for 48 h.
Take one isolated colony with an inoculation loop, transfer it into 5 mL BHI medium in a 15 mL tube, and grow for 24 h in an anaerobic chamber at 37 °C .
Dilute the pre-cultures 1:100 to approximately 106 colony-forming units (CFU)/mL in fresh pre-reduced BHIS (BHI plus 5 g/L yeast extract and 1% L-cysteine) supplemented with 0.1% sodium taurocholate and the appropriate concentration of antibiotic (T0).
Collect a 1 mL sample with a pipette at each time point (T0, T6, T24, T48) and plate/spread a small aliquot (100 µL in serial dilutions) onto a blood agar plate. Let the cells grow on the blood agar plate for 48 h in an anaerobic chamber at 37 °C and count the resulting number of colonies to determine CFU.
3. Preparing Samples for Scanning Electron Microscopy
Collect 1 mL of cells from each time point in microcentrifuge tubes and centrifuge at 10,000 x g for 10 min. Discard the supernatant and wash cells in PBS.
Centrifuge the samples at 10,000 x g for 10 min again and discard the supernatant. Re-dilute cells in 1 mL of 4% paraformaldehyde and incubate for 1 h at room temperature.
Centrifuge samples again for 10 min at 10,000 x g and discard supernatant. Wash cells twice with distilled water and redilute in 100 µL of distilled water. Adjust volume depending on the turbidity of the solution. NOTE: Making multiple serial dilutions is a good idea.
Label coverslips and add 40 µL of the sample on it. Incubate for 15 min in a flow hood to evaporate the liquid and allow the cells to adhere to the coverslip. If liquid is still present, use a blower to remove the liquid.
Place labeled coverslips with cells in a desk sputtering machine and tape down. Secure pure gold in the sputtering machine. Turn machine on and commence sputtering at low pressure (50 mTorr). Coat cells for 30 s at 80 mA, which translates to 20 nm of gold coating.
Transfer coated cells to the scanning electron microscope.
4. Imaging C. difficile Cells on a Scanning Electron Microscope
Vent the scanning electron microscope (SEM) properly by pressing the vent button on the computer software.
Using carbon tape, secure the coated coverslips onto the metal stage. Once the SEM is vented, the door should easily open. Lock the metal stage into the SEM chamber by screwing it in.
Click the PUMP button on the computer software. The SEM will be useable when the system reads "Vac OK".
Click on the SE detector under the detectors tab. Turn on the beam by clicking on the button that displays the voltage. Start imaging at a lower voltage (5 kV) before increasing voltage (up to 15 kV). An image will appear after the beam is turned on.
Using the tracking function, find an area on the coated coverslips to image. Zoom into the region and find rod-shaped structures; these are clostridium difficile.
Zoom in and focus the image to calibrate the system. This should be done at multiple working distances: 15, 9 and 5 mm. Image at a working distance of 5 mm.
Switch on the ultra high resolution imaging mode and begin to focus and optimize astigmatism.
- Begin to focus at high magnification. Use the coarse and fine focus toggles for this. Adjust astigmatism as well to obtain a clearer image.
- Adjust astigmatism at high magnification. To do this, adjust the astigmatism toggles (they look similar to the fine/coarse focus toggles) and check the image for clarity by digitally zooming in on the computer software.
se the slow scan function to collect a high quality image. Save the collected image as a .TIFF file, which will be used for analysis. Make sure the data bar is selected if measurements will be made during analysis.
Collect images at different angles (up to 52° by manually turning the angle directly on the SEM. Angled images tend to reveal more depth information.
Change beam voltage depending on the cells that are being imaged. Keep this mostly between 5 - 15 kV for all experiments.
After imaging is completed, turn the beam off and raise the working distance to 20 mm. The chamber can then be vented and the stage can be removed. Copy all of the images onto a drive for further analysis.
5. Image Processing and Analysis
Download and install the freely available software, FIJI (http://fiji.sc), on to the computer.
Open the image file in FIJI.
Using the line function, precisely trace the scale bar.
Click on the Analyze tab in the FIJI program and finally select the Select Scale function. A window will appear that will require the setting of the known distance based on the scale bar. Change the unit of length as well and click OK.
Now that the program is calibrated for distance, use the line function to measure cell length. To obtain the length, use the line function to trace the cell in its entirety. Select the Analyze tab again and then click on Measure. The length should appear in the units denoted previously.
Representative Results
Clostridium difficile is a spore-forming bacterium and thus it is essential to determine the morphology differences between vegetative and spore cells prior to any functional analysis. Figure 1 demonstrates representative images of vegetative cells that were captured during the exponential phase of the growth curve and spore cells. As depicted, vegetative cells are long, smooth, rod-shaped structures whereas spores are small, oval structures that have a rough exterior. Functionally, vegetative cells grow and divide rapidly and are responsible for the virulence of C. difficile infections by secreting toxins, whereas spores are normally dormant with little activity. Hence, antibiotics are mostly active against vegetative cells, while spores are usually resistant to drug treatment, due to several layers protecting the core with DNA, and lack of physiologic activity. Due to these facts, the majority of the morphologic analysis is focused on the vegetative cells.11,12
An antibiotic killing curve is the standard methodology to demonstrate antibiotic efficacy against growing bacteria. Concentrations used with C. difficile strain R20291 were based on the MICs values, 1 µg/mL for vancomycin and 0.51 µg/mL for metronidazole.10 As shown in Figure 2, the control cells grow and reach a plateau, whereas the treated cells decrease in total colony forming units (CFU) to the limit of detection (LOD) demonstrating bactericidal effect. As demonstrated, vancomycin (Figure 2A) and metronidazole (Figure 2B) are effective at killing C. difficile at supraMIC concentrations (4xMIC). One may notice that the antibiotic killing curves look similar, yet the drugs have different mechanisms of action (vancomycin prevents cell wall synthesis while metronidazole affects DNA replication). Therefore, we propose that antibiotic killing curves may not have enough of a discriminatory power to provide detailed differences between these antibiotics. We used microscopy to provide more discrimination.
Because of its microscopic size, C. difficile is not possible to image without high magnification. This has provided an opportunity for the use of scanning electron microscopy (SEM), an imaging method that can obtain resolution at the nanometer scale, to assess the detailed effects of antibiotic treatment directly on the cells. To demonstrate the utility of this approach, cells were imaged before and after drug treatment to determine how the morphology changed (Figure 3A). As demonstrated, some of the cells' walls were affected, as in the case of vancomycin, and some of the cells were smaller in size, as was the case for metronidazole. To test whether cell size was affected, we analyzed cell length using the program FIJI. Vegetative cell size can vary some in the control case, but most are roughly 6 µm in length (Figure 3B). When considering many cells from the control and treated settings, one can quickly notice that the cell length is preferentially affected in metronidazole but is not affected by vancomycin treatment (Figure 3C).
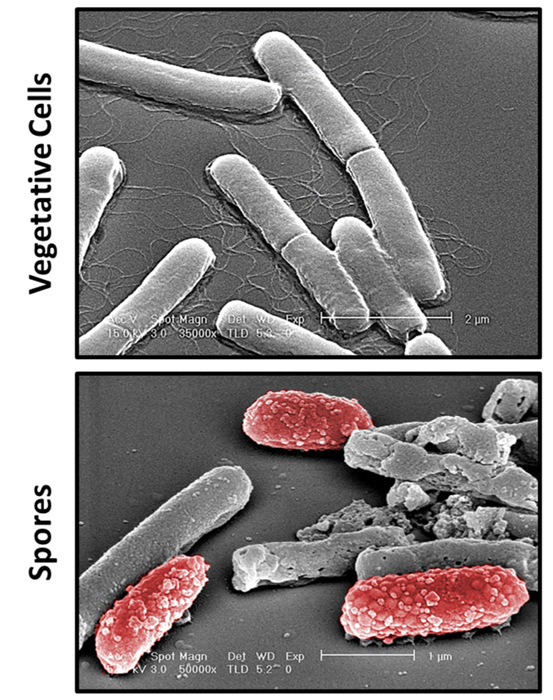 Figure 1:Differentiating Between Cell Types by Scanning Electron Microscopy. Shown are representative images of Clostridium difficile vegetative and spore cells. Vegetative cells are rod structures that are long and flagellated. In contrast, spore cells are small and have a rough and bumpy coat. Spores are pseudo-colored red for image purposes only. Scale bars are shown on the images. Please click here to view a larger version of this figure.
Figure 1:Differentiating Between Cell Types by Scanning Electron Microscopy. Shown are representative images of Clostridium difficile vegetative and spore cells. Vegetative cells are rod structures that are long and flagellated. In contrast, spore cells are small and have a rough and bumpy coat. Spores are pseudo-colored red for image purposes only. Scale bars are shown on the images. Please click here to view a larger version of this figure.
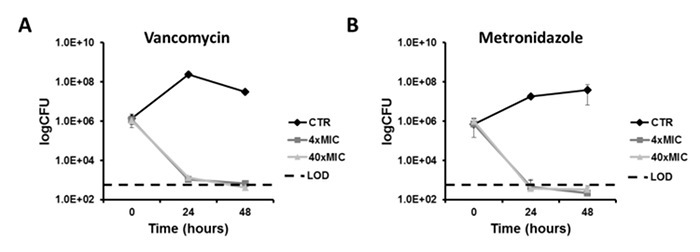 Figure 2:Antibiotic Time-Kill Curves. SupraMIC killing curves were carried for four different antibiotics: vancomycin (A), metronidazole (B). These antibiotics had significant killing action against C. difficile strain R20291 and approached the limit of detection (LOD) for counting colony forming units (CFU). Please click here to view a larger version of this figure.
Figure 2:Antibiotic Time-Kill Curves. SupraMIC killing curves were carried for four different antibiotics: vancomycin (A), metronidazole (B). These antibiotics had significant killing action against C. difficile strain R20291 and approached the limit of detection (LOD) for counting colony forming units (CFU). Please click here to view a larger version of this figure.
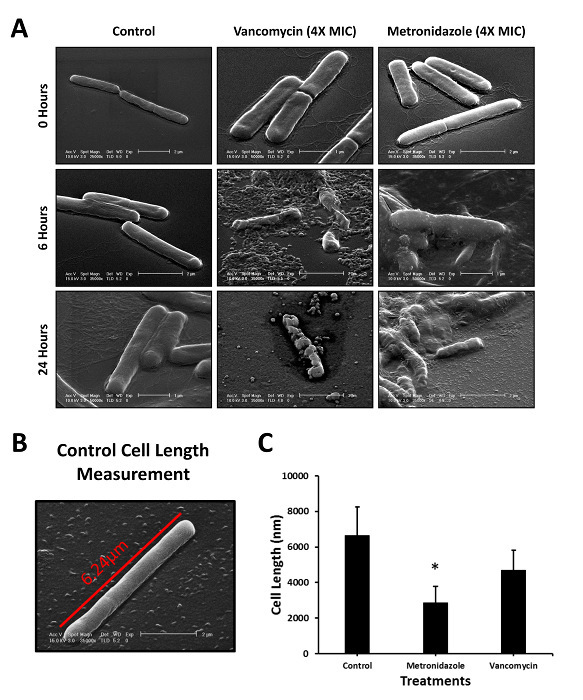 Figure 3: Examining the Pharmacological Effects of Antibiotic Treatment. Images of treated and non-treated cells were taken at multiple time points throughout the killing kinetic studies (A). Shown is a representative example of a control vegetative cell that was measured for length (B). Treated and non-treated cells were measured and compared (C). Experiments were performed in at least duplicate and > 17 cells were measured per group. *P< 0.05 compared to control and vancomycin treatment groups. Please click here to view a larger version of this figure.
Figure 3: Examining the Pharmacological Effects of Antibiotic Treatment. Images of treated and non-treated cells were taken at multiple time points throughout the killing kinetic studies (A). Shown is a representative example of a control vegetative cell that was measured for length (B). Treated and non-treated cells were measured and compared (C). Experiments were performed in at least duplicate and > 17 cells were measured per group. *P< 0.05 compared to control and vancomycin treatment groups. Please click here to view a larger version of this figure.
Discussion
The goal of the current study was to create a high-throughput method for isolating C. difficile and testing antibiotic susceptibility using scanning electron microscopy (SEM) as a means for a more thorough characterization of the antibiotic's pharmacological action. Using the protocols outlined here, we have demonstrated that imaging the cell's phenotypic response to antibiotic treatment can reveal insight into the pharmacological action of the drug. In total, the imaging portion of this protocol takes roughly 2 h in duration after collecting the cells, but can be much more discriminatory than typical killing kinetic studies alone. While learning to use an SEM can be technically demanding, preparation of samples is relatively simple and fast. We believe these protocols outlined here will provide an objective approach to evaluating the pharmacological action of antibiotic treatment.
Considering the similarities among the antibiotic killing curves (Figure 2), we sought to determine whether there were differences among the cell's response to pharmacological treatment. Using SEM, we determined that metronidazole had effects on cell length at supraMIC concentrations of the drug. While these phenotypes have not been reported previously, they are consistent with the antibiotics' mechanisms of actions and suggest an effect on cell metabolism and growth. In contrast, vancomycin had significant effects on the cell wall, which is also consistent with its mechanism of action.13 While there are similarities among the killing kinetics between these antibiotics, it is apparent from the SEM images that there are significant phenotypic differences. Creating an antibiotic phenotype library will allow for a more thorough characterization of drugs that may not have a clear mechanism of action identified, as is the case of ridinilazole.
Because this method is technically challenging, modifications may need to be made in order to address the scientific question including consistent cell preparation and alterations in sputtering of gold coating. Too much coating may blur any effects to the exterior of the cells, but not enough coating can result in charging of the beam and distort the images. To avoid this, prepare samples with different amounts of gold coating to optimize sputtering time. Lastly, consistent methodology between studies will allow inter-experimental comparisons to be made.
A limitation of using SEM is that it is restricted to observing cell morphology changes; therefore, functional studies are necessary to confirm any suspected response. Because of this, we believe that this imaging protocol can be used as a means of directing functional studies. Despite this limitation, immunogold labeling can be done using SEM to confirm any suspected changes in protein trafficking or aggregation; however, we have not yet conducted these experiments.
Due to the active pipeline of new antibiotics for C. difficile treatment, a more thorough evaluation of drug action is necessary. As presented here, SEM offers a unique, high-throughput, and reliable opportunity for characterizing pharmacological action. By imaging many different antibiotic effects among different strains of C. difficile, we will be able to understand why some antibiotics are more efficacious than others against specific pathogenic strains like the 027/NAP1/BI epidemic strain.14 Moreover, SEM analysis may be helpful to discriminate antibiotic effects between ribotypes or strains and could be used to study other bacterial species demonstrating its broad applicability.
Disclosures
KWG has received past and current research support from Merck & Co. and Summit, PLC.
Acknowledgments
These experiments have been supported by research grants from Merck and Co. and Summit, PLC.
References
- Lessa FC, et al. Burden of Clostridium difficile infection in the United States. N Engl J Med. 2015;372(9):825–834. doi: 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers RJ, et al. Ridinilazole: a novel therapy for Clostridium difficile infection. Int J Antimicrob Agents. 2016;48(2):137–143. doi: 10.1016/j.ijantimicag.2016.04.026. [DOI] [PubMed] [Google Scholar]
- Shah D, et al. Clostridium difficile infection: update on emerging antibiotic treatment options and antibiotic resistance. Expert Rev Anti Infect Ther. 2010;8(5):555–564. doi: 10.1586/eri.10.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose PG, et al. New EMA guideline for antimicrobial development. Lancet Infect Dis. 2012;12(4):265–266. doi: 10.1016/S1473-3099(12)70046-4. [DOI] [PubMed] [Google Scholar]
- Bassères E, et al. Impact on toxin production and cell morphology in Clostridium difficile by ridinilazole (SMT19969), a novel treatment for C. difficile infection. J Antimicrob Chemother. 2016;71(5):1245–1251. doi: 10.1093/jac/dkv498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endres BT, et al. A novel method for imaging the pharmacological effects of antibiotic treatment on Clostridium difficile. Anaerobe. 2016;40:10–14. doi: 10.1016/j.anaerobe.2016.04.013. [DOI] [PubMed] [Google Scholar]
- Endres BT, et al. Evaluating the Effects of Surotomycin Treatment on Clostridium difficile Toxin A and B Production, Immune Response, and Morphological Changes. Antimicrob Agents Chemother. 2016;60(6):3519–3523. doi: 10.1128/AAC.00211-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam MJ, Anu A, Walk ST, Garey KW. Investigation of potentially pathogenic Clostridium difficile contamination in household environs. Anaerobe. 2014;27:31–33. doi: 10.1016/j.anaerobe.2014.03.002. [DOI] [PubMed] [Google Scholar]
- Aitken SL, et al. In the Endemic Setting, Clostridium difficile Ribotype 027 Is Virulent But Not Hypervirulent. Infect Control Hosp Epidemiol. 2015. pp. 1–6. [DOI] [PMC free article] [PubMed]
- Basseres E, et al. Impact on toxin production and cell morphology in Clostridium difficile by ridinilazole (SMT19969), a novel treatment for C. difficile infection. J Antimicrob Chemother. 2016;71(5):1245–1251. doi: 10.1093/jac/dkv498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters BA, Roberts R, Stafford R, Seneviratne E. Relapse of antibiotic associated colitis: endogenous persistence of Clostridium difficile during vancomycin therapy. Gut. 1983;24(3):206–212. doi: 10.1136/gut.24.3.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilton CH, et al. Evaluation of the effect of oritavancin on Clostridium difficile spore germination, outgrowth and recovery. J Antimicrob Chemother. 2013;68(9):2078–2082. doi: 10.1093/jac/dkt160. [DOI] [PubMed] [Google Scholar]
- Ofosu A. Clostridium difficile infection: a review of current and emerging therapies. Ann Gastroenterol. 2016;29(2):147–154. doi: 10.20524/aog.2016.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald LC, et al. An epidemic, toxin gene-variant strain of Clostridium difficile. New Eng J Med. 2005;353(23):2433–2441. doi: 10.1056/NEJMoa051590. [DOI] [PubMed] [Google Scholar]


