Abstract
Little is known about physiological factors that affect the sense of olfaction in dogs. The objectives of this study were to describe the canine nasal and oral microbiota in detection dogs. We sought to determine the bacterial composition of the nasal and oral microbiota of a diverse population of detection canines. Nasal and oral swabs were collected from healthy dogs (n = 81) from four locations—Alabama, Georgia, California, and Texas. Nasal and oral swabs were also collected from a second cohort of detection canines belonging to three different detection job categories: explosive detection dogs (SP-E; n = 22), patrol and narcotics detection dogs (P-NDD; n = 15), and vapor wake dogs (VWD-E; n = 9). To understand if the nasal and oral microbiota of detection canines were variable, sample collection was repeated after 7 weeks in a subset of dogs. DNA was extracted from the swabs and used for 454-pyrosequencing of the16S rRNA genes. Nasal samples had a significantly lower diversity than oral samples (P<0.01). Actinobacteria and Proteobacteria were higher in nasal samples, while Bacteroidetes, Firmicutes, Fusobacteria, and Tenericutes were higher in oral samples. Bacterial diversity was not significantly different based on the detection job. No significant difference in beta diversity was observed in the nasal samples based on the detection job. In oral samples, however, ANOSIM suggested a significant difference in bacterial communities based on job category albeit with a small effect size (R = 0.1079, P = 0.02). Analysis of the composition of bacterial communities using LEfSe showed that within the nasal samples, Cardiobacterium and Riemerella were higher in VWD-E dogs, and Sphingobacterium was higher in the P-NDD group. In the oral samples Enterococcus and Capnocytophaga were higher in the P-NDD group. Gemella and Aggregatibacter were higher in S-PE, and Pigmentiphaga, Chryseobacterium, Parabacteroides amongst others were higher within the VWD-E group. Our initial data also shows that there is a temporal variation in alpha diversity in nasal samples in detection canines.
Introduction
It is estimated that the olfactory perception of dogs is about 10,000 times more sensitive than that of man [1]. This remarkable ability of dogs is beneficial to humans in a variety of ways, as dogs can be trained and utilized for various detection jobs like detection of explosives and narcotics, search and rescue, as well as other duties that require an extremely developed olfactory system [2]. Over the past years, there has been a significant interest in research to improve the olfactory performance of working canines.
Studies have shown that the microbiota can have an impact on health, immune homeostasis, nutrient status [3] and the cognitive function [4] of the host. The majority of those studies have focused on the microbiota of the gastrointestinal tract; however, there is sufficient evidence suggesting that the microbiota associated with skin [5] and the oral cavity [6] also has crucial health implications on the host [5, 7, 8].
Microbes can play a role in influencing group-specific social orders mediated via olfaction. For example, in hyena populations, members of the same family clans harbor more similar microbial communities in their anal glands than members of other family clans [9]. The scent secretions from these microbial communities in the anal glands are factors that allow family members to recognize other members of their clan [9], indicating the importance of the microbiota in olfactory signaling. Studies also show that signals derived from gut microbiota can influence the olfactory receptors in tissues outside the olfactory system [10]. Recent literature shows that the manipulation of the gut microbiota in mice by changing the diet [11] or by introducing antimicrobials [12] could lead to changes in cognition, behavior, and gene expression levels in the brain. Studies in germ-free mice have shown that the microbiota can modulate the physiology of the olfactory epithelium [13]. Jenkins et al. reported that the oral administration of metronidazole altered the detection ability of explosive detection dogs [14]. These findings suggest that microbial communities could influence host physiology and behavior. Studies show that when compared to the gut microbiota, host skin microbiota is more dependent on geographic location [14]. A potentially unfavorable outcome, especially in the case of military working detection dogs, would be if transportation of detection dogs to different locations could alter their nasal and oral microbiota, and thereby affect their olfactory detection skills.
Previous studies have described the canine nasal [15, 16] and oral [16–19] microbiota, using culture dependent and culture independent methods. However, most of these studies were focused on pet dogs. The goals of the study were, 1) to further describe the resident nasal and oral microbiota in a larger and more diverse population of dogs from different geographic locations, and 2) to compare the nasal and oral microbiota of dogs with different detection jobs and to assess the temporal stability of nasal and oral microbiota in these dogs after 7 weeks.
Methods
Animal enrollment and sample collection
To describe the nasal and oral microbiota of healthy dogs from different settings, a total of 149 swabs were collected from 81 healthy dogs (cohort-1) from four locations: Alabama (military/law enforcement working dogs), Georgia (upland sporting dogs), California (service dogs) and Texas (pet dogs). From these, we collected 69 nasal and 80 oral swabs. Sterile swabs were inserted approximately half an inch in the nasal cavity and swabbed. For oral samples, buccal swabs were collected. From each site, two swabs were collected and immediately transferred to the lysis buffer from Mobio Power Soil DNA Extraction kit (MoBio Laboratories, Inc., CA). Paired samples were not collected from some of the enrolled dogs since they did not co-operate with the sample collection.
Nasal (n = 34) and oral (n = 46) samples were collected from a second cohort (cohort-2) of dogs to describe the microbiota of dogs with different detection jobs. These dogs were all from a single training facility and were on the same diet. These dogs were trained for specific detection tasks like explosives detection (SP-E), patrol and narcotics detection (P-NDD) and vapor wake detection (VWD-E). The SP-E dogs are more proficient in locating explosive devices, while the VWD-E dogs can work off the leash and can detect very dilute odors. Dogs from the P-NDD group are placed in law enforcement and are skilled in detecting narcotic substances.
To understand the temporal stability of the nasal and oral microbiota in detection dogs, nasal and oral samples were collected at a second-time point from cohort-2, approximately 7 weeks after the first collection. The metadata for all dogs enrolled in the study are listed in S1 and S2 Tables.
To control for contamination, an unused swab was also processed along with the nasal and oral swabs. The IACUC review committee at Texas A&M University (AUP 2014–0065 CA) and the Department of the Defense, Office of Naval Research (#NRD-903), approved the collection of samples.
DNA isolation
DNA was extracted from the swabs with a MoBio Power soil DNA isolation kit (MoBio Laboratories, USA) following the manufacturer’s instructions.
Sequencing of 16S rRNA genes
Bacterial tag-encoded FLX-Titanium amplicon pyrosequencing targeting the V4–V6 region of the 16S rRNA gene was performed at MR DNA Laboratory, Shallowater, TX, USA using the forward and reverse primers: 530F (5’-GTGCCAGCMGCNGCGG-3’) and 1100R (5’-GGGTTNCGNTCGTTG-3’). Raw sequence data was screened, trimmed, filtered, denoised and barcodes and chimera sequences were removed from the dataset using QIIME v1.8 [20] pipeline and UCHIME [21]. Operational Taxonomic Units (OTUs) were assigned based on at least 97% sequence similarity against the Greengenes 13_8 reference database [22]. For downstream analysis, sequences assigned as chloroplast, mitochondria and Unassigned were removed. Additionally, OTUs that were assigned to the phylum cyanobacteria were considered to be potential plant chloroplast contaminants and excluded from the analysis. Sequences were rarefied to an even sequencing depth of 4,600 sequences/sample in cohort-1 and 4,960 sequences/sample in cohort-2 to account for unequal sequencing depth across samples. The sequences were deposited in SRA under the accession numbers: SRP060357 and SRP072443.
Statistical analysis
Differences in bacterial communities between nasal and oral swabs were analyzed using the phylogeny-based UniFrac distance metric and visualized with PCoA plots; rarefaction curves showing alpha diversity indices (Chao1, Shannon and Observed OTUs) were generated within QIIME [20]. ANOSIM (Analysis of Similarity) test within PRIMER 6 software package (PRIMER-E Ltd., Luton, UK) was used to analyze significant differences in microbial communities between the nasal and oral samples. ANOSIM test compares microbial community composition between samples, wherein microbial communities that are different will have an R statistic near 1 and similar microbial communities will have an R statistic nearing 0. The data were tested for normality using the Shapiro-Wilk test (JMP Pro 11, SAS software Inc.). Most of the datasets did not meet the assumptions of normality hence Mann-Whitney U test or Kruskal-Wallis test was used. Subsequently, statistical analysis was performed using Calypso [23]. The resulting P-values were adjusted for multiple comparisons by Benjamini & Hochberg’s False Discovery Rate (FDR), and an adjusted P<0.05 was considered statistically significant [24].
Linear discriminant analysis effect size (LEfSe) was used to elucidate bacterial taxa (16S rRNA genes) associated with nasal or oral samples [25]. LEfSe was used online in the Galaxy workflow framework.
Results
Sequence analysis
The sequences obtained from cohort-1 were rarified to an even sequencing depth of 4,600 sequences per sample to adjust for uneven sequencing depth across the samples. From dogs in cohort-1 (n = 81), 69 nasal samples and 80 oral samples were collected. Three oral samples had to be excluded from the analysis as these samples had low sequence yield and did not reach the rarefaction depth. From dogs in cohort-2 (n = 46), 81 samples were collected nasal = 34; oral = 46) and were retained for downstream analysis. An additional 82 samples from time point 2 were collected (nasal = 29; oral = 41). From these only paired samples with a matching time point 1 sample were retained. The samples were rarefied to 4,960 sequences per sample. The summary of the sequence analysis is given in Table 1.
Table 1. Summary of sequencing runs.
Tabular data given for the number of samples sequenced, OTUs identified and high-quality sequencing reads retained after filtering.
| Summary | Cohort-1 | Cohort-2 | ||||
|---|---|---|---|---|---|---|
| Nasal | Oral | Time point 1 | Time point 2 | |||
| Nasal | Oral | Nasal | Oral | |||
| Number of samples | 69 | 80 | 34 | 46 | 29 | 41 |
| Total number of high-quality sequences after filtering | 1,493,128 | 1,363,045 | 754,974 | 1,107,926 | 990,879 | 1,067,354 |
| Sequencing reads per sample | ||||||
| Minimum | 4,127 | 1,534 | 5,116 | 7,704 | 5,784 | 2,216 |
| Maximum | 57,824 | 40,111 | 71,037 | 115,883 | 172,947 | 115,883 |
| Median | 18,130 | 15,149 | 18,148 | 15,514 | 17,831 | 15,103 |
| Mean | 21,330 | 16,827 | 22,205 | 24,085 | 28,310 | 23,203 |
| Standard deviation | 13,478 | 8,852 | 15,254 | 26,275 | 30,870 | 26,938 |
Microbiota of the nasal and oral cavities
Alpha diversity indices for all samples are summarized in Table 2. The rarefaction curve for observed OTUs is shown in Fig 1. Alpha diversity as described by Chao 1, Observed OTUs (species richness), and Shannon diversity index was significantly higher (P<0.001) in the oral samples when compared to the nasal samples.
Table 2. Summary of alpha diversity indices at a depth of 4,600 sequences per sample based on the site of sampling.
| OTU picking method | Nasal | Oral | P value* |
|---|---|---|---|
| Open reference | |||
| Chao1 | 240(82–1118) | 896(359–1656) | <0.001 |
| Observed species | 102(39–603) | 374(164–644) | <0.001 |
| Shannon | 2.6(0.2–6.5) | 5.5(3.3–6.7) | <0.001 |
| Closed reference | |||
| Chao1 | 71(2–708) | 193(78–330) | <0.001 |
| Observed species | 55(2–475) | 148(57–252) | <0.001 |
| Shannon | 2(0.06–6.2) | 4.88(2.6–6.2) | <0.001 |
*P values determined by Mann-Whitney U test significance level <0.05)
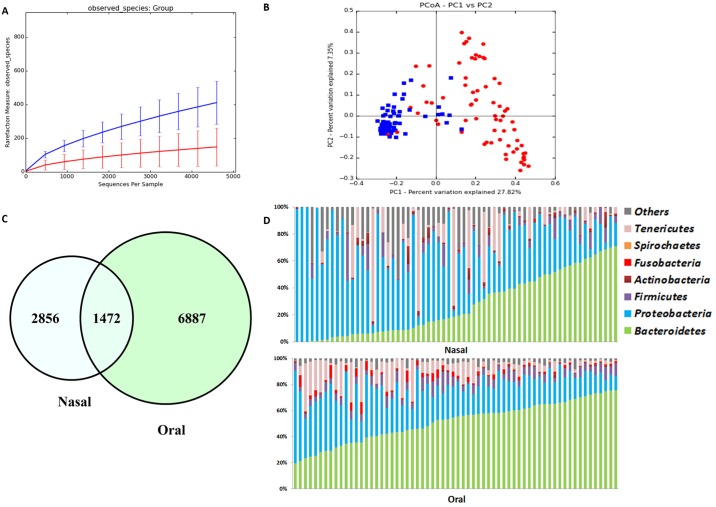
Fig 1. Bacterial diversity measures for all samples from cohort-1.
(A) Alpha diversity: rarefaction analysis (number of observed species) of 16S rRNA gene sequences. Lines represent the mean of each group, while the error bars represent the standard deviations. (B) Beta diversity: Principal coordinate analysis (PCoA) of unweighted UniFrac distances of 16S rRNA genes. Analysis of similarity (ANOSIM) revealed clustering between oral and nasal samples (R = 0.6980; P = 0.01). (C) Comparison of OTUs between nasal and oral samples. A Venn diagram showing overlapping OTU’s between nasal and oral samples. (D) Bacterial phyla in nasal versus oral samples. Most common bacterial phyla identified in nasal and oral samples from healthy dogs sorted by the phylum Bacteroidetes.
PCoA analysis (Fig 1, Panel B) based on unweighted UniFrac distance metric showed a distinct separation between nasal and oral samples. Bacterial community composition was significantly different as revealed by ANOSIM (R = 0.5736, P = 0.01). Individual bacterial groups were analyzed using a Kruskal-Wallis test (S3 Table), and several bacterial taxa were significantly different between nasal and oral samples. The similarities and differences in bacterial composition between the nasal and oral samples were visualized using a Venn diagram (Fig 1, Panel C) and showed that there were 1,472 OTUs shared between the two groups of samples. OTUs unique to the nasal and oral samples were 2,856 and 6,887 respectively. Community composition at the phylum level showed a dog-to-dog variation (Fig 1, Panel D). The predominant bacterial phyla in nasal samples were Proteobacteria followed by Bacteroidetes, whereas oral samples were mostly composed of Bacteroidetes followed by Proteobacteria. Alphaproteobacteria, Actinobacteria, and Gammaproteobacteria were significantly higher in nasal samples while Bacteroidia, Mollicutes, Betaproteobacteria, and Flavobacteriia were higher in oral samples (P<0.001). At the genus level, Moraxella, Leucobacter, Helcococcus, and Cardiobacterium were significantly higher in nasal samples while the genera Porphyromonas, Capnocytophaga, Mycoplasma, and unclassified genera in the families Neisseriaceae and Pasteurellaceae were significantly higher in oral samples (P<0.001).
Effect of geographical location on the canine nasal microbiota
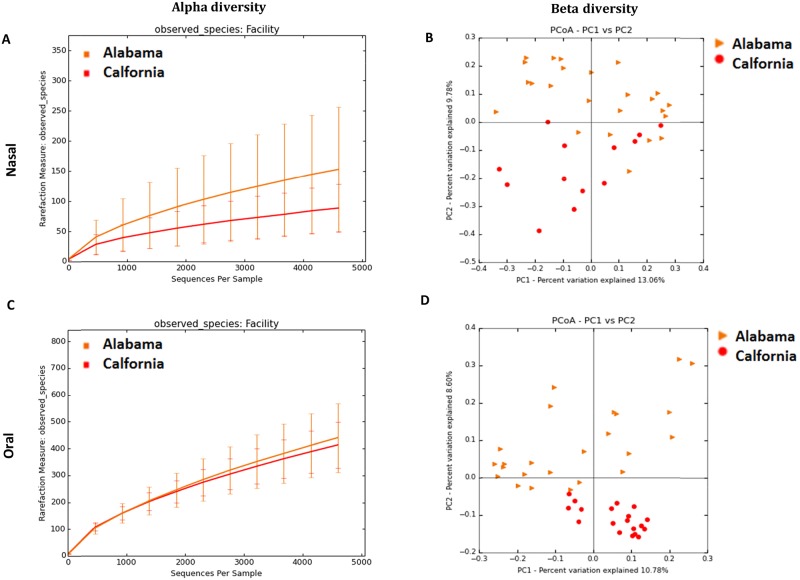
When all the dogs in cohort-1 were analyzed, there was a significant difference in alpha (P<0.05) and beta diversity (ANOSIM; R = 0.2598; P = 0.01) based on location. However, to disentangle the effect of breeds vs. location, we analyzed only Labradors from two locations, Alabama and California, as these were the only two locations with a sufficient number of dogs of the same breed. The diversity indices, Observed OTUs and Chao1 (richness) were significantly lower (P<0.05) for the nasal samples from California when compared to Alabama. Alpha diversity measures are summarized in Table 3 and rarefaction plot for observed OTUs is shown in Fig 2 (Panel A). PCoA plot (Fig 2, Panel B) visually show that the bacterial communities cluster significantly different based on location (ANOSIM; R = 0.2275; P = 0.01).
Table 3. Summary of alpha diversity indices of samples from Labradors at a depth of 4,600 sequences per sample based on location.
| Median (Min-Max) | |||
|---|---|---|---|
| Nasal | Alabama (n = 24) | California (n = 13) | P value* |
| Observed species | 107(77–603) | 74(39–159) | 0.003 |
| Shannon | 2.69(1.7–6.52) | 2.4(0.3–5.1) | 0.309 |
| Chao1 | 260(145–1089) | 205(93–317) | <0.004 |
| Oral | Alabama (n = 24) | California (n = 19) | P value |
| Observed species | 416(195–545) | 350(290–531) | 0.385 |
| Shannon | 5.6(3.3–6.6) | 5.7(5.3–6.7) | 0.221 |
| Chao1 | 977(457–1379) | 863(524–1360) | 0.163 |
*P values determined by Kruskal Wallis test (significance level <0.05)
Fig 2. Bacterial diversity measures of samples from Labradors based on location.
Alpha diversity measures: rarefaction analysis (number of observed species) of 16S rRNA gene sequences in nasal and oral samples. Lines represent the mean of each group, while the error bars represent the standard deviations. Beta diversity: Principal coordinate analysis (PCoA) of unweighted UniFrac distances of 16S rRNA genes. Analysis of similarity (ANOSIM) revealed clustering based on location for B) Nasal (R = 0.2275; P = 0.01) and D) Oral samples (R = 0.2008; P = 0.01).
Effect of geographical location on the canine oral microbiota
When the oral samples from dogs in cohort-1 from four different locations were analyzed, Shannon diversity index, a measure of alpha diversity and beta diversity were significantly different based on location (R = 0.146; P = 0.01). To understand if this was a true location vs. breed effect, as described above for the nasal samples from these dogs, chose only the Labrador dogs from these two locations (Alabama and California). There was no significant difference in alpha diversity indices (Table 3) in oral samples; alpha rarefaction plot for observed OTUs is shown in Fig 2 (Panel C). PCoA plots (Fig 2, Panel D) show that the bacterial communities cluster significantly different based on location (ANOSIM; R = 0.2008; P = 0.01) for oral samples.
There were no significant differences based on gender in the bacterial diversity or community composition (P>0.05) in nasal and oral samples in the same set of Labradors within cohort-1.
Nasal and oral microbiota of dogs with different detection jobs
To understand the nasal and oral microbiota of dogs with specific detection jobs, the following analyses were performed on samples from a second cohort (cohort-2) of dogs that lived in a single training facility and were on the same diet. As in cohort-1, the bacterial diversity in nasal samples was significantly lower (P<0.05) than that of oral samples (S1 Fig, Panel A). The samples also clustered based on the sampling site (nasal or oral) in cohort-2 as well (S1 Fig, Panel B). There was also an inter-dog variability in nasal and oral microbiota (S2 Fig) in the detection dogs similar as observed in dogs from cohort-1.
Nasal samples
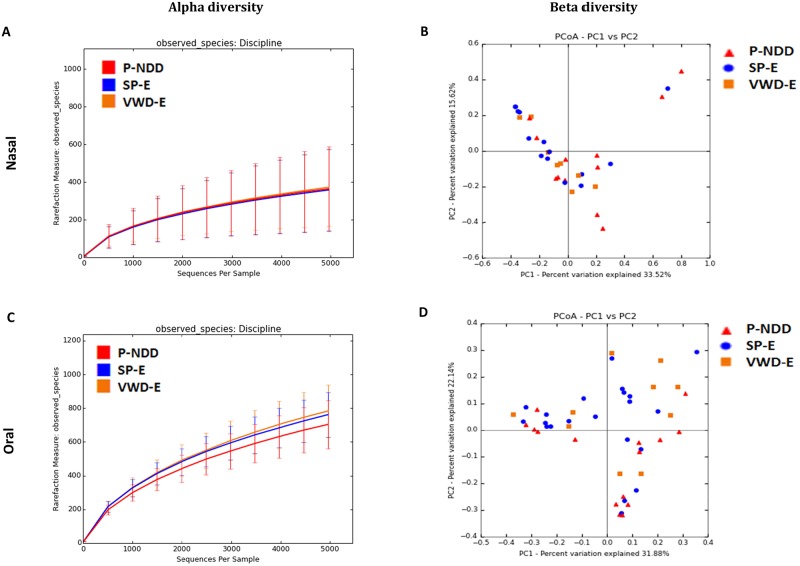
In cohort 2, there was no significant difference in alpha diversity based on detection jobs (Fig 3, Panel A). Based on PCoA plots (Fig 3, Panel B) there was no significant difference in microbial communities between the dogs belonging to the three different detection job categories based on ANOSIM test (R = 0.0469; P = 0.19). Based on LEfSe (Table 4), the family Cardiobacteriaceae was enriched in the VWD-E group of dogs while the family Enterococcaeceae was enriched in P-NDD group of dogs. At the genus level, the genera Cardiobacterium, Riemerella and an unclassified genus within the order Cardiobacteriales was more abundant in the P-NDD dogs while Sphingobacterium and an unclassified genus within Enterobacteriaceae were associated with the VWD-E group of dogs. However, univariate statistics did not show a significant difference in bacterial taxa (S4 Table).
Fig 3. Bacterial diversity measures of samples based on job category of detection dogs in cohort 2 from nasal and oral samples.
Lines represent the mean of each group, while the error bars represent the standard deviations. Beta diversity: Principal coordinate analysis (PCoA) of unweighted UniFrac distances of 16S rRNA genes. Analysis of similarity (ANOSIM) for B) nasal (R = 0.05; P = 0.19) and D) oral samples (R = 0.11, P = 0.02).
Table 4. Linear discriminant analysis of bacterial genera and their associations with detection jobs.
Only LDA scores of >3.0 are shown.
| LDA | Category | |
|---|---|---|
| Taxa | ||
| Nasal | ||
| Unclassified_Cardiobacteriales | 3.69 | VWD-E |
| Sphingobacterium | 3.72 | P-NDD |
| Unclassified_Enterobacteriaceae | 3.93 | P-NDD |
| Riemerella | 4.18 | VWD-E |
| Cardiobacterium | 4.67 | VWD-E |
| Oral | ||
| Unclassified_Leptotrichiaceae | 2.94 | VWD-E |
| Unclassified_Aerococcaceae | 3.35 | SP-E |
| Aggregatibacter | 3.12 | SP-E |
| Enterococcus | 3.26 | P-NDD |
| Gemella | 3.27 | S-PE |
| Parabacteroides | 3.3 | VWD-E |
| Unclassified_Clostridiales | 3.42 | VWD-E |
| Chryseobacterium | 3.61 | VWD-E |
| Pigmentiphaga | 3.63 | VWD-E |
| Unclassified_BD15 | 3.77 | VWD-E |
| Unclassified_Neisseriaceae | 4.18 | VWD-E |
| Mycoplasma | 4.21 | VWD-E |
| Unclassified_Lachnospiraceae | 4.4 | P-NDD |
| Conchiformibius | 4.41 | VWD-E |
| Capnocytophaga | 4.77 | P-NDD |
Oral samples
There was no significant difference in alpha diversity indices in oral samples based on detection jobs (Fig 3, Panel C). PCoA plots did not show a clear clustering based on the detection jobs (Fig 3, Panel D). However, based on ANOSIM test there was a significant difference in microbial communities between the dogs belonging to the three different detection job categories (R = 0.1079, P = 0.02). Based on LEfSe (Table 4), the genera Capnocytophaga, Enterococcus, and an unclassified genus within the family Lachnospiracea were associated with the P-NDD group of dogs while Gemella, Aggregatibacter, and an unclassified genus within Aerocoocaceae were associated with the SP-E group of dogs. Conchiformibius, Pigmentiphaga, Chryseobacterium, Mycoplasma, and Parabacteroides were enriched in the VWD-E dogs. However, univariate statistics did not show a significant difference in bacterial taxa (S5 Table).
The effect of age, breed, gender on nasal and oral microbiota in detection dogs
The effect of confounding factors such as the effect of age, breed, and gender was assessed in cohort-2, as all dogs lived in the same environment and were on the same diet. A total of 47 dogs were sampled and ranged in age from 0.5 to 6 years. The majority of the dogs belonged to the breeds Belgian Malinois (n = 14), German Shepherd (n = 10), and Labrador Retriever (n = 17). No significant differences were observed in bacterial communities in nasal samples based on age group, breed, and sex of the detection dogs. A significant difference was seen in bacterial communities in oral samples based on age group and breed (P<0.05) while there was no significant difference based on the sex of the detection dogs (S6 Table).
Temporal variability of nasal and oral microbiota of detection canines
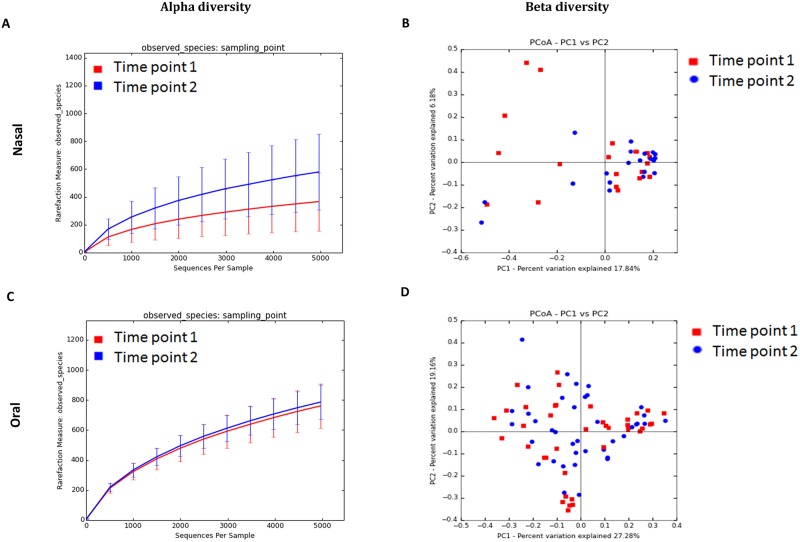
In order to understand the temporal stability of nasal and oral microbiota of detection canines, a second sample was collected from dogs in cohort-2, seven weeks after the first collection. Paired samples from both time points from 20 dogs were analyzed for nasal samples and 40 dogs for oral samples. In nasal samples, there was a significant difference in diversity based on the sampling time point (Fig 4, Panel A). There was a higher diversity in the samples when sampled at the second time point (P<0.01). PCoA plots do not show a clustering pattern based on sampling time point (Fig 4, Panel B), which was further confirmed with ANOSIM (R = 0.0272; P = 0.160). Based on LEfSe, a few bacterial genera were higher at time point 2 (Table 5). Chryseobacterium, Fimbriimonas, Halomonas, Paludibacter, and Porphorymonas were amongst the bacteria that were higher in the nasal samples at the second time point. However, analysis of bacterial groups using univariate testing (Wilcoxon signed rank test) did not show any significant differences in the abundances of bacterial groups between time points 1 and 2 at different taxonomic levels (data not shown).
Fig 4. Bacterial diversity measures of samples based on sampling time point in cohort-2 from nasal and oral samples.
Lines represent the mean of each group, while the error bars represent the standard deviations. Beta diversity: Principal coordinate analysis (PCoA) of unweighted UniFrac distances of 16S rRNA genes. Analysis of similarity (ANOSIM) for B) (R = 0.0272; P = 0.160) and D) oral samples (R = -0.0071, P = 0.582).
Table 5. Linear discriminant analysis of bacterial genera and their associations with the sampling time.
Only LDA scores of >3.0 are shown.
| Taxa | LDA | Category |
|---|---|---|
| Nasal | ||
| Unclassified_Sphingomonadaceae | 3.02 | Time point 2 |
| Unclassified_F16 | 3.13 | Time point 2 |
| Unclassified_Peptostreptococcaceae | 3.15 | Time point 2 |
| Agrobacterium | 3.19 | Time point 2 |
| Caulobacter | 3.22 | Time point 2 |
| Unclassified_Oxalobacteraceae | 3.25 | Time point 2 |
| Unclassified_CW040 | 3.28 | Time point 2 |
| Fimbriimonas | 3.29 | Time point 2 |
| Unclassified_TM71 | 3.31 | Time point 2 |
| Unclassified_Sphingobacteriaceae | 3.33 | Time point 2 |
| Unclassified_Comamonadaceae | 3.35 | Time point 2 |
| Erwinia | 3.39 | Time point 2 |
| Pseudomonas | 3.47 | Time point 2 |
| Pseudoxanthomonas | 3.47 | Time point 2 |
| Unclassified_Caulobacteraceae | 3.53 | Time point 2 |
| Halomonas | 3.58 | Time point 2 |
| Sphingomonas | 3.58 | Time point 2 |
| Unclassified_Xanthomonadaceae | 3.58 | Time point 2 |
| Chryseobacterium | 3.59 | Time point 2 |
| Unclassified_Moraxellaceae | 4.16 | Time point 2 |
| Porphyromonas | 4.56 | Time point 2 |
| Oral | ||
| Unclassified_Erysipelotrichaceae | 2.79 | Time point 1 |
| Fusibacter | 2.85 | Time point 2 |
| Unclassified_CW040 | 3.14 | Time point 2 |
| Tannerella | 3.15 | Time point 2 |
| Pasteurella | 3.31 | Time point 1 |
In oral samples (n = 80), there was no significant difference in alpha diversity based on the sampling time point (Fig 4, Panel C). As evident by the lack of clustering in the PCoA plots (Fig 4, Panel D), no significant difference in microbial communities was observed with ANOSIM (R = -0.0071, P = 0.582). LEfSe identified the genus Pasteurella to be enriched in the first sampling time point and Fusibacter and Tannerella at the second time point (Table 5). Univariate statistics (Wilcoxon signed rank test) did not show any significant differences in the abundances of bacterial groups between time points 1 and 2 (data not shown).
Discussion
The focus of this study was to describe the canine nasal and oral microbiota in a larger population of dogs from different geographic locations. Furthermore, we analyzed samples from dogs living in the same environment (cohort-2) to compare the nasal and oral microbiota of dogs with different detection jobs and to assess the temporal stability of nasal and oral cavity microbiota.
The oral cavity had a higher bacterial diversity than the nasal cavity. Principal coordinate analysis (PCoA) plots revealed significant differences in microbial communities between the nasal and oral microbiota. In addition, this study was able to show the inter-individual variability in the nasal and oral microbiota of dogs, which can be seen in cohort-1. Though it can be argued that the location of the dogs could attribute to this variability, a similar pattern was also observed in the second cohort of dogs who were all from the same location (cohort-2). As observed in a previous study looking at canine skin microbiota, even the distribution of sequences from chloroplasts though removed from the final analysis, were not uniform between the dogs [26]. The high degree of inter-individual variability in the canine nasal and oral microbiota seen in this study relates to the “personal microbiome” concept suggested in previous studies [26–28].
We also examined the potential role of geographical location on the canine nasal and oral microbiota. Our results suggest that the nasal and oral microbiota can differ based on location. To remove the effect of breed as a potential confounding factor, we analyzed only Labradors from two locations, Alabama and California. Alpha diversity was significantly different based on location for the nasal samples but was not different for the oral samples. Beta diversity was significantly different based on the location in nasal and oral samples from Labradors. Previous studies show that when compared to the gut microbiota, host skin microbiota is more dependent on geographic location [15, 29]. Hence, more detailed and controlled studies are needed to understand if an altered microbiota due to a change in geographical location could bring about changes in physiological functions of the host.
Previous studies had detected Proteobacteria and Firmicutes as the predominant phyla inhabiting the canine nasal cavity [15, 30]. The main phyla in canine nasal cavity in this study were Proteobacteria and Bacteroidetes. Interestingly, there was a large variation based on the individual, with some dogs having Proteobacteria as the abundant phylum and some with Bacteroidetes. Other phyla detected in the canine nasal cavity were Firmicutes, Tenericutes, and GN02 at a lower abundance. The main bacterial classes associated with nasal samples was Gammaproteobacteria, Flavobacteria, and Bacteroidia. A culture-independent study [15] reported the same observation in canine nostrils. Tress et al., reported that Moraxellaceae is abundant in the nasal microbiota of healthy dogs, dogs with nasal neoplasia, and dogs with chronic lymphoplasmacytic or neutrophilic rhinitis [30]. The most frequently observed genera in the nasal cavity of healthy dogs in this study were Moraxella, Mycoplasma, Prevotella, Helcoccus, Cardiobacterium and unclassified genera within the phylum BD1-5 and the family [Weeksellaceae] respectively. BD1-5 is a candidate phylum which has been reported to have a small genome and is dependent on other members of the community for nutrients. It is a phylum that has not been isolated by culture successfully [31] and it has been previously reported in the canine nasal cavity [30].
In the current study, the predominant phyla found in the canine oral microbiota were Bacteroidetes, Proteobacteria, Firmicutes, and Fusobacteria. This is in concurrence with a previous study by Sturgeon et al., which describes the canine oral microbiota of 6 healthy pet dogs of various breeds based on culture-independent methods [17]. Porphyromonas was the predominant genus in oral samples in our study, which is in agreement with previous literature using culture-independent methods [16–18]. In most culture-based studies, Porphyromonas has been reported to be of low abundance [19, 32], as noted in a previous study [17], this could be due to the fastidious growth requirements of this organism. Some of the taxa found in the nasal and oral cavity of dogs in this study have been associated with mostly soil or plant samples previously [33, 34]. It is most likely that presence of these taxa is due to the interactions of the host with the outdoor environment.
There are no previous studies detailing the nasal and oral microbiota of working dogs or dogs trained for tasks that require special olfactory skills. In addition, there are no studies examining the variation of canine nasal and oral microbiota over time. The dogs in the cohort-2 were all from a single training facility and were on the same diet. This study did not reveal any significant differences in alpha diversity based on the detection category of dogs. However, there was a significant difference in beta diversity in oral samples based on the type of detection job. LEfSe was able to identify a few bacterial groups that were different between dogs belonging to the three different detection categories. Cardiobacterium and Riemerella were amongst the taxa that were enriched in the nasal samples from P-NDD and VWD-E dogs. These taxa have been reported as members of the healthy canine nasal microbiota [30, 35]. The genus Capnocytophaga was found to be enriched in the oral cavity of P-NDD group of dogs, a member of this genus has been associated with bite wound infections [36]. Similarly, Aggregatibacter and Gemella were found to be abundant in the oral cavity of SP-E dogs and were detected in healthy canine oral microbiota [37].
Even though we were able to look at only one additional time point approximately 7 weeks from the first sampling, the results suggest that in detection canines, the nasal microbiota may be variable while the oral microbiota is less so. The dogs have been in this facility for at least 6 months; however, most of these dogs were at different locations (Afghanistan, Europe, Iraq, Mexico and various locations in the United States) before being acquired by the facility. The authors could only propose at this point that this could be a driver for the variation, but further studies are needed to validate this.
This study had limitations. A variety of factors including diet, sex, breed, age, and the geographical location could affect the nasal and oral microbiota of dogs. We tried to examine the effect of location in cohort-1 by excluding the effects of confounding factors by focusing on dogs, which were of the same breed. Unfortunately, breed comparisons were not possible in the cohort-1, as the number of animals/breed in each location were very low to make meaningful statistical comparisons. We could not explore the effect of diet, since diet was linked to the location. Most individuals from the same location were on the same diet. Similarly, the effect of age was also difficult to conclude since it was also linked with location. In the second cohort of detection dogs, which were all from the same location and on the same diet, there was a difference in the oral microbiota based on the age and breed of the dogs. Since samples from cohort-1 and cohort-2 were sequenced on different runs; we did not merge the datasets together and introduce a sequencing run bias. Another limitation of the study was that there were fewer animals in each group to perform statistical comparisons and make stronger conclusions from the results. An interesting aspect would be to assess the differences if any in the nasal and oral microbiota of detection dogs to pet dogs. Further studies are needed to know if there are specific groups that are influencing olfactory detection skills in working dogs.
Conclusion
In this study, we report further characterization of the nasal and oral microbiota of dogs from different geographic locations using next generation sequencing. The results show that the nasal and oral microbiota is distinct and unique in bacterial diversity and community structure. Species richness, bacterial communities, and specific taxa were significantly different between nasal and oral samples. Our results suggest that geographical location may influence the nasal and oral cavity microbiota. This study is also the first study to analyze the nasal and oral microbiota of dogs with specific detection jobs. Initial data shows that there is no significant difference in diversity based on olfactory skills in canine nasal and oral cavity. However, a few distinct bacterial groups were identified using the LEfSe algorithm to be associated with the detection job categories. Our initial data suggest that nasal microbiota may be temporally more variable than oral microbiota in detection canines. Further studies are required to understand the role of the nasal and oral microbiota in canine olfaction.
Supporting information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(A) Alpha diversity: rarefaction analysis (number of observed species) of 16S rRNA gene sequences. Lines represent the mean of each group, while the error bars represent the standard deviations. (B) Beta diversity: Principal coordinate analysis (PCoA) of unweighted UniFrac distances of 16S rRNA genes. Analysis of similarity (ANOSIM) revealed clustering between nasal and oral samples (R = 0.58; P = 0.01).
(TIF)
Most common bacterial phyla identified in nasal and oral samples from healthy dogs sorted by the phylum Bacteroidetes.
(TIF)
Abbreviations
- ANOSIM
analysis of similarity
- FDR
false discovery rate
- LDA
linear discriminant analysis
- LEfSe
linear discriminant analysis effect size
- OTU
operational taxonomic units
- PCoA
principal coordinates analysis
- P-NDD
patrol and narcotics detection dogs
- SP-E
explosive detection dogs
- VWD-E
vapor wake dogs
Data Availability
The sequences analyzed in this study were deposited in SRA under accession number SRP060357 and SRP072443.
Funding Statement
This study was supported by a grant from the Office of Naval Research, grant # N000141410256, http://www.onr.navy.mil/.
References
- 1.Craven BA, Paterson EG, Settles GS. The fluid dynamics of canine olfaction: unique nasal airflow patterns as an explanation of macrosmia. J R Soc Interface. 2010;7(47):933–43. doi: 10.1098/rsif.2009.0490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hackner K, Pleil J. Canine olfaction as an alternative to analytical instruments for disease diagnosis: understanding 'dog personality' to achieve reproducible results. J Breath Res. 2017;11(1):012001 doi: 10.1088/1752-7163/aa5524 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cenit MC, Matzaraki V, Tigchelaar EF, Zhernakova A. Rapidly expanding knowledge on the role of the gut microbiome in health and disease. Biochim Biophys Acta. 2014;1842(10):1981–92. doi: 10.1016/j.bbadis.2014.05.023 . [DOI] [PubMed] [Google Scholar]
- 4.Magnusson KR, Hauck L, Jeffrey BM, Elias V, Humphrey A, Nath R, et al. Relationships between Diet-Related Changes in the Gut Microbiome and Cognitive Flexibility. Neuroscience. 2015;300:128–40. doi: 10.1016/j.neuroscience.2015.05.016 [DOI] [PubMed] [Google Scholar]
- 5.Sanford JA, Gallo RL. Functions of the skin microbiota in health and disease. Semin Immunol. 2013;25(5):370–7. doi: 10.1016/j.smim.2013.09.005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zarco MF, Vess TJ, Ginsburg GS. The oral microbiome in health and disease and the potential impact on personalized dental medicine. Oral Dis. 2012;18(2):109–20. doi: 10.1111/j.1601-0825.2011.01851.x . [DOI] [PubMed] [Google Scholar]
- 7.Weese JS. The canine and feline skin microbiome in health and disease. Vet Dermatol. 2013;24(1):137–45 e31. doi: 10.1111/j.1365-3164.2012.01076.x . [DOI] [PubMed] [Google Scholar]
- 8.Wade WG. The oral microbiome in health and disease. Pharmacol Res. 2013;69(1):137–43. doi: 10.1016/j.phrs.2012.11.006 . [DOI] [PubMed] [Google Scholar]
- 9.Theis KR, Schmidt TM, Holekamp KE. Evidence for a bacterial mechanism for group-specific social odors among hyenas. Sci Rep. 2012;2:615 doi: 10.1038/srep00615 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pluznick JL, Protzko RJ, Gevorgyan H, Peterlin Z, Sipos A, Han J, et al. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc Natl Acad Sci U S A. 2013;110(11):4410–5. doi: 10.1073/pnas.1215927110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruce-Keller AJ, Salbaum JM, Luo M, Blanchard Et, Taylor CM, Welsh DA, et al. Obese-type gut microbiota induce neurobehavioral changes in the absence of obesity. Biol Psychiatry. 2015;77(7):607–15. doi: 10.1016/j.biopsych.2014.07.012 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bercik P, Denou E, Collins J, Jackson W, Lu J, Jury J, et al. The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology. 2011;141(2):599–609, e1–3. doi: 10.1053/j.gastro.2011.04.052 . [DOI] [PubMed] [Google Scholar]
- 13.Francois A, Grebert D, Rhimi M, Mariadassou M, Naudon L, Rabot S, et al. Olfactory epithelium changes in germfree mice. Sci Rep. 2016;6:24687 doi: 10.1038/srep24687 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenkins EK, Lee-Fowler TM, Angle TC, Behrend EN, Moore GE. Effects of oral administration of metronidazole and doxycycline on olfactory capabilities of explosives detection dogs. Am J Vet Res. 2016;77(8):906–12. doi: 10.2460/ajvr.77.8.906 . [DOI] [PubMed] [Google Scholar]
- 15.Hoffmann AR, Patterson AP, Diesel A, Lawhon SD, Ly HJ, Stephenson CE, et al. The Skin Microbiome in Healthy and Allergic Dogs. Plos One. 2014;9(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Misic AM, Davis MF, Tyldsley AS, Hodkinson BP, Tolomeo P, Hu B, et al. The shared microbiota of humans and companion animals as evaluated from Staphylococcus carriage sites. Microbiome. 2015;3:2 doi: 10.1186/s40168-014-0052-7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sturgeon A, Stull JW, Costa MC, Weese JS. Metagenomic analysis of the canine oral cavity as revealed by high-throughput pyrosequencing of the 16S rRNA gene. Vet Microbiol. 2013;162(2–4):891–8. doi: 10.1016/j.vetmic.2012.11.018 [DOI] [PubMed] [Google Scholar]
- 18.Oh C, Lee K, Cheong Y, Lee SW, Park SY, Song CS, et al. Comparison of the Oral Microbiomes of Canines and Their Owners Using Next-Generation Sequencing. Plos One. 2015;10(7). doi: 10.1371/journal.pone.0131468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elliott DR, Wilson M, Buckley CM, Spratt DA. Cultivable oral microbiota of domestic dogs. J Clin Microbiol. 2005;43(11):5470–6. doi: 10.1128/JCM.43.11.5470-5476.2005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–6. doi: 10.1038/nmeth.f.303 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–200. doi: 10.1093/bioinformatics/btr381 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–72. doi: 10.1128/AEM.03006-05 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zakrzewski M, Proietti C, Ellis JJ, Hasan S, Brion MJ, Berger B, et al. Calypso: a user-friendly web-server for mining and visualizing microbiome-environment interactions. Bioinformatics. 2017;33(5):782–3. doi: 10.1093/bioinformatics/btw725 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate—a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57(1):289–300. [Google Scholar]
- 25.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6). doi: 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cusco A, Sanchez A, Altet L, Ferrer L, Francino O. Individual Signatures Define Canine Skin Microbiota Composition and Variability. Front Vet Sci. 2017;4:6 doi: 10.3389/fvets.2017.00006 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326(5960):1694–7. doi: 10.1126/science.1177486 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meason-Smith C, Diesel A, Patterson AP, Older CE, Mansell JM, Suchodolski JS, et al. What is living on your dog's skin? Characterization of the canine cutaneous mycobiota and fungal dysbiosis in canine allergic dermatitis. Fems Microbiology Ecology. 2015;91(12). doi: 10.1093/femsec/fiv139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng Y, Fox S, Pemberton D, Hogg C, Papenfuss AT, Belov K. The Tasmanian devil microbiome-implications for conservation and management. Microbiome. 2015;3(1):76 doi: 10.1186/s40168-015-0143-0 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tress B, Dorn ES, Suchodolski JS, Nisar T, Ravindran P, Weber K, et al. Bacterial microbiome of the nose of healthy dogs and dogs with nasal disease. Plos One. 2017;12(5):e0176736 doi: 10.1371/journal.pone.0176736 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garza DR, Dutilh BE. From cultured to uncultured genome sequences: metagenomics and modeling microbial ecosystems. Cell Mol Life Sci. 2015;72(22):4287–308. doi: 10.1007/s00018-015-2004-1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riggio MP, Lennon A, Taylor DJ, Bennett D. Molecular identification of bacteria associated with canine periodontal disease. Vet Microbiol. 2011;150(3–4):394–400. doi: 10.1016/j.vetmic.2011.03.001 . [DOI] [PubMed] [Google Scholar]
- 33.Zarraonaindia I, Owens SM, Weisenhorn P, West K, Hampton-Marcell J, Lax S, et al. The soil microbiome influences grapevine-associated microbiota. MBio. 2015;6(2). doi: 10.1128/mBio.02527-14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newman MM, Hoilett N, Lorenz N, Dick RP, Liles MR, Ramsier C, et al. Glyphosate effects on soil rhizosphere-associated bacterial communities. Sci Total Environ. 2016;543(Pt A):155–60. doi: 10.1016/j.scitotenv.2015.11.008 . [DOI] [PubMed] [Google Scholar]
- 35.Ericsson AC, Personett AR, Grobman ME, Rindt H, Reinero CR. Composition and Predicted Metabolic Capacity of Upper and Lower Airway Microbiota of Healthy Dogs in Relation to the Fecal Microbiota. Plos One. 2016;11(5):e0154646 doi: 10.1371/journal.pone.0154646 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Butler T. Capnocytophaga canimorsus: an emerging cause of sepsis, meningitis, and post-splenectomy infection after dog bites. Eur J Clin Microbiol Infect Dis. 2015;34(7):1271–80. doi: 10.1007/s10096-015-2360-7 . [DOI] [PubMed] [Google Scholar]
- 37.Dewhirst FE, Klein EA, Thompson EC, Blanton JM, Chen T, Milella L, et al. The Canine Oral Microbiome. Plos One. 2012;7(4). doi: 10.1371/journal.pone.0036067 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(A) Alpha diversity: rarefaction analysis (number of observed species) of 16S rRNA gene sequences. Lines represent the mean of each group, while the error bars represent the standard deviations. (B) Beta diversity: Principal coordinate analysis (PCoA) of unweighted UniFrac distances of 16S rRNA genes. Analysis of similarity (ANOSIM) revealed clustering between nasal and oral samples (R = 0.58; P = 0.01).
(TIF)
Most common bacterial phyla identified in nasal and oral samples from healthy dogs sorted by the phylum Bacteroidetes.
(TIF)
Data Availability Statement
The sequences analyzed in this study were deposited in SRA under accession number SRP060357 and SRP072443.