Abstract
Slab gel electrophoresis (SGE) is the most common method for the separation of DNA fragments; thus, it is broadly applied to the field of biology and others. However, the traditional SGE protocol is quite tedious, and the experiment takes a long time. Moreover, the chemical consumption in SGE experiments is very high. This work proposes a simple method for the separation of DNA fragments based on an SGE chip. The chip is made by an engraving machine. Two plastic sheets are used for the excitation and emission wavelengths of the optical signal. The fluorescence signal of the DNA bands is collected by smartphone. To validate this method, 50, 100, and 1,000 bp DNA ladders were separated. The results demonstrate that a DNA ladder smaller than 5,000 bp can be resolved within 12 min and with high resolution when using this method, indicating that it is an ideal substitute for the traditional SGE method.
Keywords: Biochemistry, Issue 124, Genetics, Gel electrophoresis, Agarose, DNA separation, SYBR Green I, Fluorescence
Introduction
Slab gel electrophoresis (SGE) is the most effective method for DNA fragment separation1,2,3,4,5 and thus it is deemed a versatile tool in biochemical and biological analyses6,7,8. However, many experiments indicate that SGE is restricted by the following four problems: (1) the separations take many hours, and even days; (2) the chemical consumption is very high; (3) it requires a complicated apparatus (e.g., 2D electrophoresis cell, electrophoresis power supply, and gel imaging system); (4) the gel imaging system can only observe the separated DNA fragments when the experiment is finished. Furthermore, ethidium bromide (EtBr), which is commonly used in SGE9,10, is mutagenic and cancerogenic11,12. Thus, gloves should always be worn when handing gels containing EtBr.
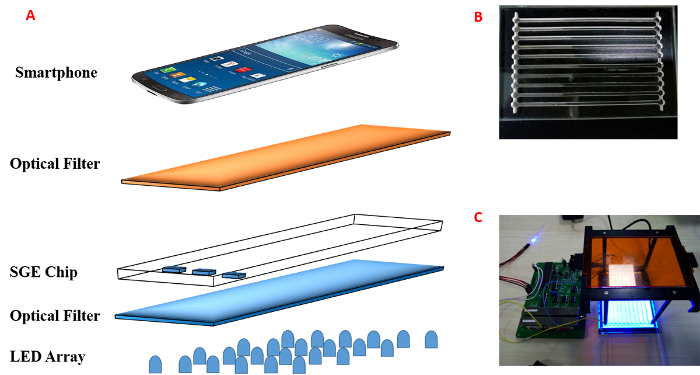
Capillary electrophoresis (CE) has numerous advantages13,14,15,16,17 compared to SGE, such as automatic operation, short separation time, and lower consumption. However, the CE instrument is quite expensive. Therefore, to overcome those limitations, a system has been developed (Figure 1) for the separation of DNA. Such a system can not only greatly reduce the chemical consumption and save on SGE experiment time (<8 min), but it can also perform real-time tracking of the DNA separation process in the agarose gel by smartphone. By following the procedures described in this protocol, students should be able to design and fabricate the SGE chip, prepare the agarose gel in the chip, set up a simple SGE system with a smartphone, and record the DNA migration process in the agarose gel.
Protocol
1. Basic Design of the SGE Chip
Use any transparent plastic, such as polymethylmethacrylate (PMMA) or polycarbonate. Note: The SGE chip is demonstrated in Figure 1B. The SGE chip consists of cylindrical holes for the TBE buffer, channels for DNA separation, and two lanes embedded along the holes for the electrode.
Fabricate arrays of SGE channels in the PMMA block using a laser engraving machine. Note: The geometric parameters of the chip are dependent upon the size of the DNA to be separated. For example, the optimal channel parameters for the SGE chip are 2.5 mm x 4.0 mm x 90 mm (width x depth x length) if the DNA fragment is smaller than 1,000 bp.
Based on the SGE design, fabricate combs to create the wells in the chip for loading the DNA sample.
2. Preparation of the Agarose Gel
Prepare 0.5× TBE by mixing 10× TBE (1× TBE = 89 mM Tris, 89 mM boric acid, and 2 mM EDTA; pH 8.4) and distilled water in a 1:19 ratio.
Prepare 1.0% agarose gel for the separation of 100-bp DNA ladders. Note: The concentration of agarose in 0.5x TBE buffer is dependent upon the sizes of the DNA fragments to be separated.
Place 0.1 g of agarose into a flask and add 10 mL of 0.5x TBE buffer. Note: The volume of the prepared solution should be less than 1/3 of the capacity of the flask.
Seal the flask with preservative film and then heat the agarose/buffer mixture in a microwave (medium, 1.0 min). Prior to putting the flask into the microwave, make some holes in the preservative film in case of an explosion.
Pour 2.7 mL of melted agarose solution into 4 channels of the SGE chip. Place the comb into the agarose solution to create the wells. Note: The melted agarose solution will cool down to the gel state at room temperature 3 min later.
Remove the comb from the gels and add 0.9 mL of 0.5x TBE buffer to cover the gel.
3. Running the Electrophoresis in the SGE Chip
Turn on the LED light source and place a 425 to 505 nm bandpass filter above the light source.
Mix 14.4 µL of a 100 bp DNA ladder and 1.6 µL of SYBR Green using a vortexer.
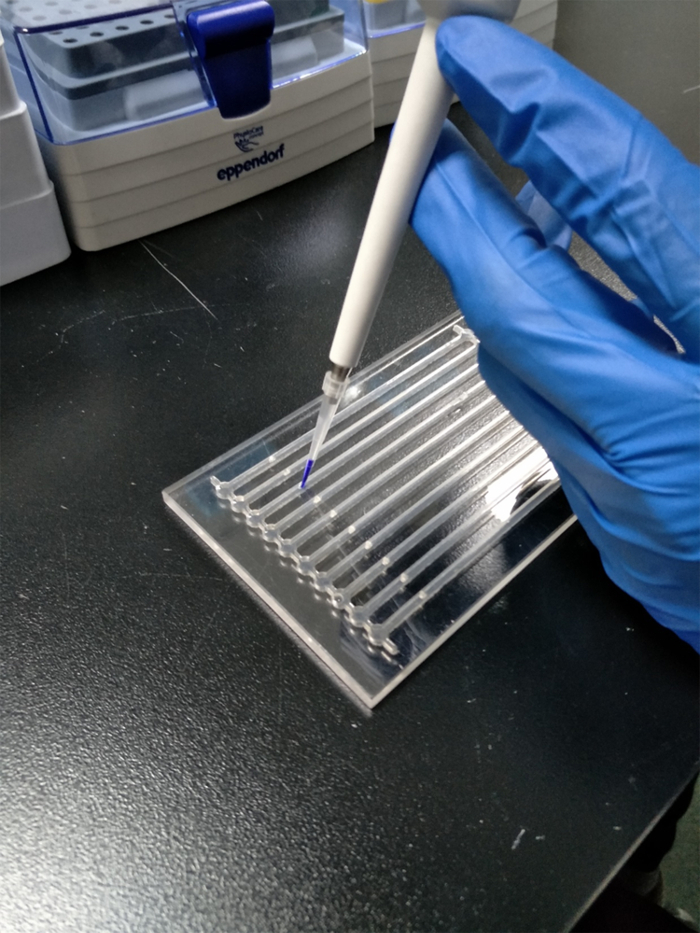
Load 4 µL of mixture into each well of the agarose gel in the SGE chip (Figure 2).
Put the electrode into the two lanes of the SGE chip.
Place a 550 to 700 nm bandpass filter above the SGE chip.
Switch on the power source. Set the electric voltage to 180 V (20 V/cm). Turn on the smartphone to record the DNA fragment separation process (Figure 3).
When the electrophoresis is finished 10 min later, turn off the power source and the LED light.
Clean the SGE chip.

Representative Results
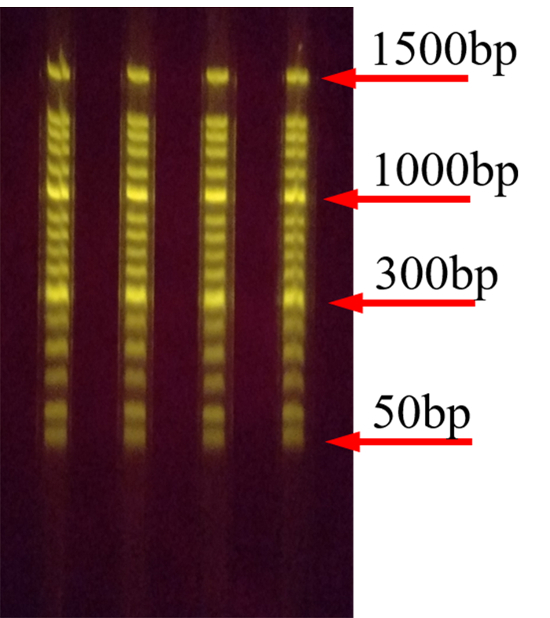
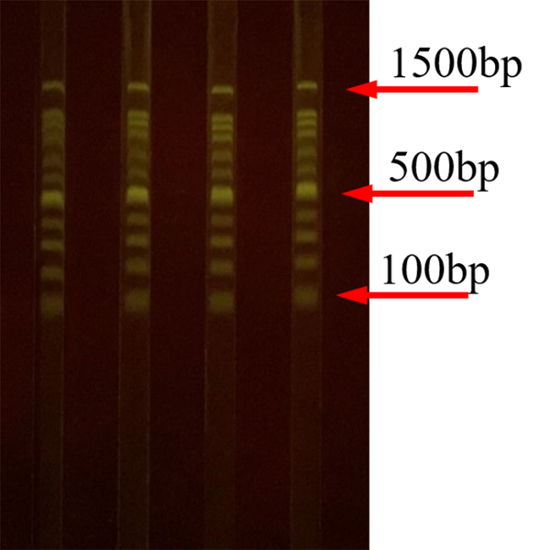
Figure 4, Figure 5, and Figure 6 represent a typical result after the gel electrophoresis of 50, 100, and 1,000 bp DNA ladders. After the experiment, the DNA fragments were well-separated. Furthermore, the same samples were separated in the 4 channels of the SGE chip, showing that DNA fragments of the same size move the same distance in each experiment.
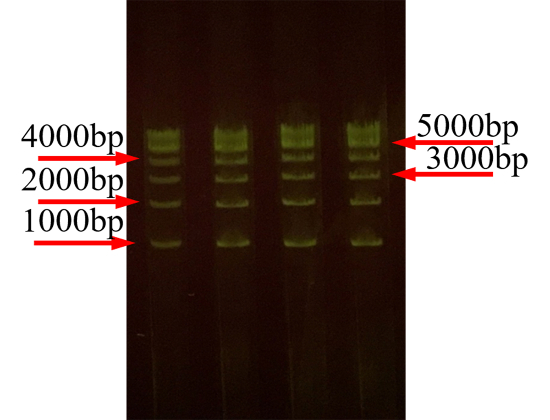
The separation performance of the DNA ladder can be evaluated by analyzing the relationship of the migration distance and the logarithm of DNA size; thus, SGE can be applied to calculating the size of unknown DNA fragments. In Figure 3, it was found that the migration distance is linearly related to the logarithm of DNA size when the DNA size is smaller than 700 bp.
Figure 1: Schematic and images. (A) A schematic diagram of the real-time tracking DNA separation system. It includes the LED array as the light source, the optical filter, and the CGE chip. The smartphone can record the DNA migration process. (B) The SGE chip. (C) The device assembled for electrophoresis. Please click here to view a larger version of this figure.
Figure 2 : A student loading the gel. Please click here to view a larger version of this figure.
Figure 3 : A student recording the migration on a smartphone. Please click here to view a larger version of this figure.
Figure 4: Separation of 50-bp DNA ladders on the SGE chip. The geometric parameters for the channel in the SGE chip are 2.5 mm x 4.0 mm x 90 mm (width x depth x length). The separation voltage is 120 V. Please click here to view a larger version of this figure.
Figure 5: Separation of 100-bp DNA ladders on the SGE chip. The geometric parameters for the channel in the SGE chip are 2.5 mm x 4.0 mm x 90 mm (width x depth x length). The separation voltage is 120 V. Please click here to view a larger version of this figure.
Figure 6: Separation of 1,000-bp DNA ladders on the SGE chip. The geometric parameters for the channel in the SGE chip are 3.6 mm x 4.0 mm x 90 mm (width x depth x length). The separation voltage is 180 V. Please click here to view a larger version of this figure.
Discussion
Agarose gel electrophoresis is widely employed for the separation of DNA, RNA, and protein. This work proposes a new method to replace the traditional gel electrophoresis protocol. Results demonstrate that 50, 100, and 1,000 bp DNA ladders can be separated well in such a small assembled device. The great advantage of this method is that not only can it separate the nucleic acids with little chemical consumption, but it can also record the separation process. Although the DNA fragments looks wide in Figure 2, the results can be improved by optimizing the parameters of the SGE chip and the concentration of the agarose gel. Furthermore, the total SGE process takes no more than 10 min if precast gels are used in the chip.
The separation performance of the DNA ladder is quite different if the channel width in the chip is changed. For example, when a 1,000-bp DNA ladder is separated, the DNA band looks wider if the width of the channel is small. This is possibly because the joule heating in the agarose gel is very high18, which affects the separation performance.
The SGE method proposed here is very simple. A portable mini-SGE device can be developed if the system is optimized. Such a device can be applied to point-of-care laboratory testing.
Disclosures
No conflicts of interest are declared.
Acknowledgments
We gratefully acknowledge support from National Natural Science Foundation of China (No. 21205078) and the Research Fund for the Doctoral Program of Higher Education of China (No.20123120110002). This work was partially supported by the National Key Research and Development Program of China (2016YFB1102303), the National Basic Research Program of China (973Program; 2015CB352001), and the National Natural Science Foundation of China (61378060).
References
- Carle GF, Olson MV. Separation of chromosomal DNA molecules from yeast by orthogonal-field-alternation gel electrophoresis. Nucleic. Acids. Res. 1984;12(14):5647–5664. doi: 10.1093/nar/12.14.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonell MW, Simon MN, Studier FW. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J. Mol. Biol. 1977;110(1):119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- Fangman WL. Separation of very large DNA molecules by gel electrophoresis. Nucleic. Acids. Res. 1978;5(3):653–665. doi: 10.1093/nar/5.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum H, Beier H, Gross HJ. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8(2):93–99. [Google Scholar]
- Gödde R, et al. Electrophoresis of DNA in human genetic diagnostics-state-of-the-art, alternatives and future prospects. Electrophoresis. 2006;27(5-6):939–946. doi: 10.1002/elps.200500675. [DOI] [PubMed] [Google Scholar]
- Studier FW. Analysis of bacteriophage T7 early RNAs and proteins on slab gels. J. Mol. Biol. 1973;79(2):237–248. doi: 10.1016/0022-2836(73)90003-x. [DOI] [PubMed] [Google Scholar]
- Sugden B, De Troy B, Roberts RJ, Sambrook J. Agarose slab-gel electrophoresis equipment. Anal. Biochem. 1975;68(1):36–46. doi: 10.1016/0003-2697(75)90676-4. [DOI] [PubMed] [Google Scholar]
- Rabilloud T, Chevallet M, Luche S, Lelong C. Two-dimensional gel electrophoresis in proteomics: Past, present and future. J. Proteomics. 2010;73(11):2064–2077. doi: 10.1016/j.jprot.2010.05.016. [DOI] [PubMed] [Google Scholar]
- Lee PY, Costumbrado J, Hsu CY, Kim YH. Agarose gel electrophoresis for the separation of DNA fragments. J. Vis. Exp. 2012. p. e3923. [DOI] [PMC free article] [PubMed]
- Gasser RB, et al. Single-strand conformation polymorphism (SSCP) for the analysis of genetic variation. Nat. Protoc. 2006;1(6):3121–3128. doi: 10.1038/nprot.2006.485. [DOI] [PubMed] [Google Scholar]
- Matselyukh BP, Yarmoluk SM, Matselyukh AB, Kovalska VB, Kocheshev IO, Kryvorotenko DV, Lukashov SS. Interaction of cyanine dyes with nucleic acids: XXXI. Using of polymethine cyanine dyes for the visualization of DNA in agarose gels. J. Biochem. Biophys. Methods. 2003;57(1):35–43. doi: 10.1016/s0165-022x(03)00036-8. [DOI] [PubMed] [Google Scholar]
- Lunn G, Sansone EB. Ethidium bromide: destruction and decontamination of solutions. Anal. Biochem. 1987;162(2):453–458. doi: 10.1016/0003-2697(87)90419-2. [DOI] [PubMed] [Google Scholar]
- Li Z, Liu C, Zhang D, Luo S, Yamaguchi Y. Capillary electrophoresis of RNA in hydroxyethylcellulose polymer with various molecular weights. J. Chormatogr. B. 2016;1011:114–120. doi: 10.1016/j.jchromb.2015.12.057. [DOI] [PubMed] [Google Scholar]
- Li Z, Liu C, Ma S, Zhang D, Yamaguchi Y. Analysis of the inhibition of nucleic acid dyes on polymerase chain reaction by capillary electrophoresis. Anal. Methods-UK. 2016;8(11):2330–2334. [Google Scholar]
- Liu F, et al. Developing a fluorescence-coupled capillary electrophoresis based method to probe interactions between QDs and colorectal cancer targeting peptides. Electrophoresis. 2016;37(15-16):2170–2174. doi: 10.1002/elps.201600165. [DOI] [PubMed] [Google Scholar]
- Wang J, et al. A capillary electrophoresis method to explore the self-assembly of a novel polypeptide ligand with quantum dots. Electrophoresis. 2016;37(15-16):2156–2162. doi: 10.1002/elps.201600164. [DOI] [PubMed] [Google Scholar]
- Miao P, et al. Nuclease assisted target recycling and spherical nucleic acids gold nanoparticles recruitment for ultrasensitive detection of microRNA. Electrochim. Acta. 2016;190:396–401. [Google Scholar]
- Heller C. Principles of DNA separation with capillary electrophoresis. Electrophoresis. 2001;22(4):629–643. doi: 10.1002/1522-2683(200102)22:4<629::AID-ELPS629>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]


