Abstract
Objective
One of the critical mechanisms of gastrointestinal cancer pathogenesis is the silencing of death associated protein kinase 1 (DAPK1), which could be caused by aberrant methylation of the promoter. However, the relationship between DAPK1 methylation and the risk of gastrointestinal cancer is still controversial. Hence, we conducted this study to determine the potential correlation.
Methods
Eligible publications were searched in the Pubmed, Embase, and Cochrane Library through November 2016 according to the inclusion criteria and exclusion criteria. Revman 5.3 and Stata 12.0 software were used to analyze the relevant data regarding the association between the frequency of DAPK1 methylation and gastrointestinal cancer.
Results
A total of 22 studies with 2406 patients were included in this meta analysis. Methylation of DAPK1 was positively related with the risk of gastrointestinal cancer (odds ratio [OR] = 5.35, 95% confidence interval [CI]: 2.76–10.38, P<0.00001, random effects model). The source of heterogeneity was analyzed by sensitivity analysis and subgroup analysis. After omitting one heterogeneous study, the I2 decreased and the OR increased in pooled analysis. Also, the heterogeneity decreased most significantly in the subgroup of studies that had a sample size of less than 60 cases. Then, the correlations between DAPK1 methylation and clinicopathological features of gastrointestinal cancer were assessed. DAPK1 methylation was positively correlated with the lymph node (N) stage (positive vs. negative, OR = 1.45, 95%CI: 1.01–2.06, P = 0.04, fixed effects model) and poor differentiation (OR = 1.55, 95%CI: 1.02–2.35, P = 0.04, fixed effects model) in gastric cancer, and the association was significant among Asian patients. However, among cases of gastrointestinal cancer, the association between DAPK1 methylation and tumor (T) stage, N stage, distant metastasis (M) stage, and cancer differentiation were not statistically significant.
Conclusions
DAPK1 methylation is a potential biomarker for the early diagnosis of gastrointestinal cancer. Further analysis of the clinicopathological features indicated that aberrant methylation of DAPK1 is positively associated with the tumorigenesis of gastrointestinal cancer, and metastasis of gastric cancer.
Introduction
Despite advances in the treatment of gastrointestinal cancer, it is still the leading cause of cancer-related mortality. For instance, gastric cancer (GC) ranks second, colorectal cancer (CRC) ranks fourth, and esophageal cancer (EC) ranks sixth as the most deadly cancers globally[1]. Increasing numbers of studies have been performed to demonstrate the mechanism of carcinogenesis, and to identify biomarkers for early diagnosis of gastrointestinal cancer[2].
Methylation of DNA is dramatically altered in cancers. Promoter CpG islands methylation is one type of DNA methylation that could result in the inactivation of tumor suppressor genes[3], such as death-associated protein kinase 1 (DAPK1). DAPK1 is a member of the Ser/Thr kinase family, and was found originally in interferon gamma (INF-γ)–induced death in HeLa cells [4]. Its critical role in regulating cell death and autophagy has been demonstrated[5]. In addition, DAPK1 could be involved in multiple cell death processes induced by a variety of internal and external apoptotic stimulants, such as tumor necrosis factor-alpha and Fas ligand, and could mediate the pro-apoptotic pathway[6].
As a well-known tumor suppressor gene, DAPK1 expression can suppress tumor growth and metastasis[7]. It has been confirmed that DAPK1 is epigenetically silenced through methylation of its promoter in various human cancers including gastrointestinal cancer[8–10]. However, it remains controversial whether DAPK1 promoter methylation is related to the risk of gastrointestinal cancer. Previous studies have reported that the DAPK1 promoter methylation is much more frequent in EC, GC, CRC cancer tissues than that in control tissue[8, 10–12]. However, in some other studies, the frequency of DAPK1 methylation showed no obvious increase[13] or even a reverse trend[14] in cancer samples. Therefore, we conducted this meta analysis to investigate the correlativity between DAPK1 promoter methylation and gastrointestinal cancer.
Methods
Search strategy
We searched the Pubmed, Embase, and Cochrane Library electronic databases to, find the eligible articles using the search terms “DAPK1”, “death-associated protein kinase 1”, “DAPK”, “DAP kinase”, or “DAPK protein” with “neoplasms”, “cancer”, “tumor”, or “neoplasia” through November04, 2016. We also searched the reference lists of relevant articles to find additional qualified articles. Only publications written in English were selected. Among all the articles that we had searched, unrelated studies were excluded by reading the title and abstract. Then, full texts of the candidate studies were inspected thoroughly to determine whether they met the inclusion and exclusion criteria.
The inclusion criteria were as follows: 1. studies that evaluated the association between DAPK1 methylation and gastrointestinal cancer, including EC, GC and CRC; 2. diagnosis of gastrointestinal cancer was histologically confirmed; 3. methylation status was examined by methylation-specific polymerase chain reaction (MSP); and 4. definitive data for the frequency of DAPK1 methylation were provided.
We excluded unsuitable studies according to the following criteria: 1. the studies were performed without a control group; 2. the cancer group included cases of diverse precancerous lesions; 3. peripheral blood or other non-epithelium tissue was used as the object of detection; and 4. data regarding the frequency of DAPK1 methylation could not be extracted from the raw data.
The quality of the included studies was assessed on the basis of the Newcastle-Ottawa Quality Assessment Scale (NOS). Four stars were used to evaluate the selection of study groups. Two stars were used to estimate the comparability of cases and controls. and three stars were used to value the exposure. Publications that scored less than 6 stars were excluded[15].
Data extraction
Data in the text, figures, and tables of included studies were extracted by two authors using a data collection form that included author names, publication year, country, geographic area, method for detecting DNA methylation, source of the control group, number of patients, age distribution, gender distribution, and clinicopathological features (tumor stage, lymph node stage, distant metastasis and differentiation), follow-up time, and 5-year overall survival (OS) and disease-free survival (DFS) rates. The GetData Graph Digitizer v2.24 was used to extract the data from figures[16]. Discussions were held by three authors when uncertainty was encountered in data extraction.
Statistical analysis
Review Manager 5.3 and Stata 12.0 software were used to analyze the data. Forest plots were generated to analyze the ORs and 95%CIs. Heterogeneity among studies was assessed by Q and I2 tests. An I2 value of 0% indicates no observed heterogeneity, whereas, 25% indicates low, 50% indicates moderate and 75% indicates high heterogeneity[17]. A random effects model was utilized when the heterogeneity is high, otherwise, the fixed effects model was applied. Sensitivity analysis and subgroup analysis were conducted to find the potential source of heterogeneity. Publication bias was qualitatively assessed by funnel plot generation which was conducted using Revman 5.3, and quantitatively evaluated by Egger weighted regression test and Begg rank correlation test, which were calculated using Stata 12.0 software. A P value ≤0.05 was regarded as statistically significant.
Results
Inclusion of studies in meta-analysis
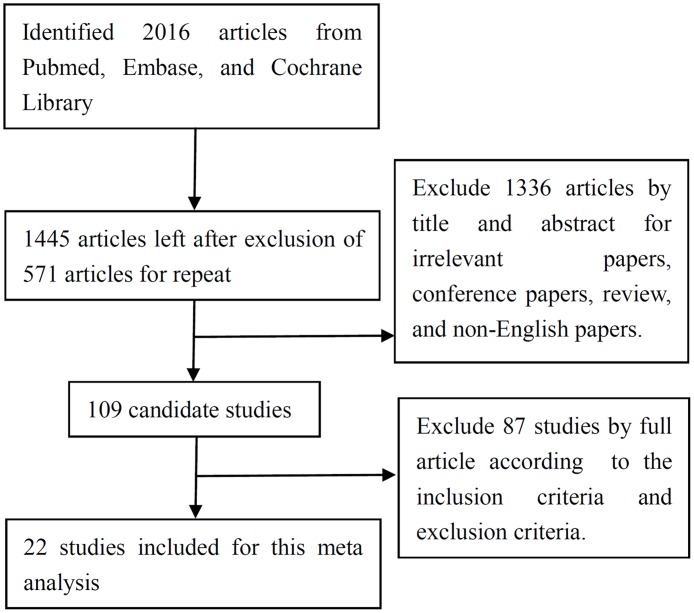
A total of 2016 articles were identified initially from the searched databases. Among these, 571 articles were excluded as repeated publications. Then we excluded 1336 articles as being irrelevant, conference papers, review articles, and manuscripts not published in English paper based on reading the title and abstract. Afterward, 109 candidate studies were further reviewed by reading of the full articles. In the end, 87 studies were excluded according to the inclusion and exclusion criteria, and 22 studies with 2406 patients were included for this review (Fig 1).
Fig 1. Flow chart of study selection for this meta analysis.
Among all the included studies, 2 studies assessed the frequency of DAPK1 methylation in EC, 10 in GC, 8 in CRC, 1 in both GC and CRC, and 1 in both EC and GC. Fourteen studies were performed in Asia, five in Europe, two in South America, and one in Africa. The control group was from normal tissue in 11 studies, whereas others were from normal tissue adjacent to the tumor. All the studies were retrospective studies, and the MSPCR was used to assess the methylation of DAPK1 in the tissue sample. The associations between DAPK1 methylation and T stage, N stage, M stage and differentiation were presented in 9, 13, 6, and 10 studies, respectively. The characteristics of the included studies are shown in Table 1.
Table 1. Characteristics of the included studies.
| No. | Author | Year | Country | Cancer Type | Case(cancer/ control) | Source of Control | Methylation in tumor | Methylation in Control |
|---|---|---|---|---|---|---|---|---|
| 1 | Bagci[18] | 2016 | Turkey(Asia) | CRC | 93/14 | AT | 42/93 | 4/14 |
| 2 | Laskar[19] | 2015 | India(Asia) | CRC | 80/20 | AT | 27/80 | 6/20 |
| 3 | Almeida[20] | 2015 | Brazil(South America) | CRC | 5 | AT | 4/5 | 5/5 |
| 4 | Kupčinskaitė-Noreikienė[21] | 2013 | Lithuanian (Europe) | GC | 69 | AT | 33/69 | 32/69 |
| 5 | Nomura[11] | 2013 | Japan(Asia) | GC | 115/412 | NT | 95/115 | 201/412 |
| 6 | Ye[8] | 2012 | China(Asia) | GC | 62 | AT | 34/62 | 11/62 |
| 7 | Li [22] | 2011 | China(Asia) | EC | 47 | AT | 22/47 | 6/47 |
| 8 | Hu[23] | 2010 | China(Asia) | GC | 70/30 | NT | 42/70 | 0/30 |
| AT | 42/70 | 10/70 | ||||||
| 9 | Lee[12] | 2009 | Korea(Asia) | CRC | 243/148 | NT | 81/243 | 0/148 |
| 10 | Zou[24] | 2009 | China(Asia) | GC | 16/20 | NT | 7/16 | 0/20 |
| 11 | Ksiaa[25] | 2009 | Tunisia (Africa) | GC | 68/53 | AT | 21/68 | 13/53 |
| 12 | Kato[26] | 2008 | Japan(Asia) | GC | 81/43 | AT | 18/81 | 4/43 |
| 13 | Kuester[10] | 2007 | Germany (Europe) | EC | 35/20 | NT | 21/35 | 4/20 |
| 14 | Mittag [9] | 2006 | Germany (Europe) | CRC | 22/8 | AT | 18/22 | 2/8 |
| 15 | Anacleto[27] | 2005 | Brazil(South America) | CRC | 106/30 | AT | 21/106 | 0/30 |
| 16 | Chan[28] | 2005 | China(Asia) | GC | 107/23 | NT | 74/107 | 0/23 |
| 17 | Schildhaus[29] | 2005 | Germany (Europe) | GC | 7 | AT | 6/7 | 2/7 |
| EC | 10 | AT | 7/10 | 4/10 | ||||
| 18 | Lee [30]. | 2004 | Korea(Asia) | CRC | 149/24 | NT | 71/149 | 0/24 |
| 19 | Sabbioni[31] | 2003 | Italy(Europe) | GC | 21/6 | NT | 19/21 | 2/6 |
| CRC | 47/4 | NT | 35/47 | 0/4 | ||||
| 20 | Waki[14] | 2003 | Japan(Asia) | GC | 93 | AT | 40/93 | 68/93 |
| 21 | Yamaguchi[32] | 2003 | Japan(Asia) | CRC | 122/10 | NT | 67/122 | 0/10 |
| 22 | To[33] | 2002 | China(Asia) | GC | 31/10 | NT | 22/31 | 0/10 |
NT: normal tissue
AT: normal tissue adjacent to the tumor
Association between DAPK1 methylation and gastrointestinal cancer
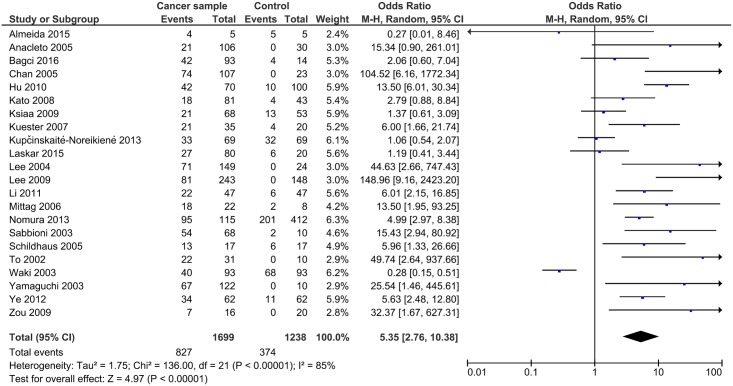
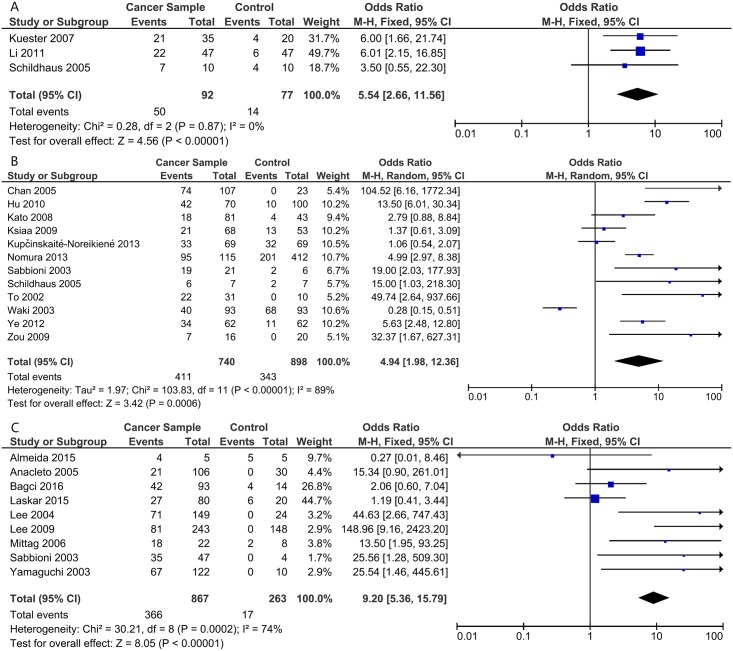
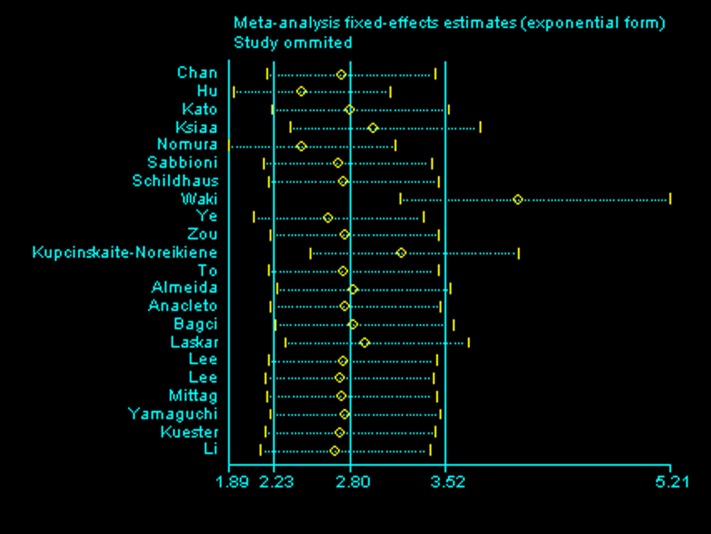
Generally, the methylation of DAPK1 was positively related to the risk of gastrointestinal cancer, with a pooled OR of 5.35 (95%CI: 2.76–10.38, P<0.00001) using the random effects model due to high heterogeneity (I2 = 85%, P<0.00001; Fig 2). The association was more obvious in CRC (OR = 9.20, 95%CI: 5.36–15.79, P<0.00001, fixed effects model; Fig 3). Meanwhile, the ORs were 5.54 in EC (95%CI:2.66–11.56, P<0.00001, fixed effects model), and 4.94 in GC (95%CI: 1.98–12.36, P = 0.006, random effects model; Fig 3). To find the source of heterogeneity, a sensitivity analysis was applied. As shown in Fig 4, the study conducted by Waki et al.[14] could affect the result remarkably (Fig 4). After omitting this study, the I2 decreased and the OR increased in both the pooled analysis (I2 = 72%, OR = 5.40, 95%CI: 4.30–6.78, P<0.00001, fixed effects model; S1 Fig) and GC analysis (I2 = 78%, OR = 5.93, 95%CI: 2.84–12.38, P<0.00001, random effects model; S2 Fig). Then subgroup analysis according to the source of the control group, geographic area, and sample size of cases were applied to further analyze the source of heterogeneity. The heterogeneity decreased most significantly in the subgroup of studies with a sample size of cases was less than 60 (I2 = 12% in pooled analysis). Also, in the subgroup of studies that took normal tissue as a control group, the I2 was lower (I2 = 61%) and the OR greater (OR = 12.94, 95%CI: 8.65–19.36, P<0.00001, fixed effects model). In addition, analysis in Asian patients produced a significantly increased OR (OR = 7.64, 95%CI: 2.89–20.20, P<0.0001; Table 2)
Fig 2. DAPK1 methylation and the risk of gastrointestinal cancer.
Fig 3. DAPK1 methylation and the risk of different type of gastrointestinal cancer: A. esophageal cancer (EC); B. gastric cancer (GC); and C. colorectal cancer (CRC).
Fig 4. Sensitivity analysis.
Table 2. Subgroup analysis of studies reporting on the association of DAPK1 methylation and gastrointestinal cancer.
| Source of the control group | Geographic area | Sample size of case group | |||||
|---|---|---|---|---|---|---|---|
| Normal tissue subgroup | Normal tissue adjacent to the tumor | Asian subgroup | Non-Asian subgroup | >60 | ≤60 | ||
| Overall | Study(n) | 10 | 12 | 13 | 8 | 14 | 7 |
| OR (95%CI) |
12.94 (8.65, 19.36) |
2.92 (2.19, 3.90) |
7.74 (5.78, 10.36) |
2.40 (1.62, 3.55) |
5.50 (2.77, 10.92) |
7.37 (4.08, 13.33) |
|
| Model | Fixed | Fixed | Fixed | Fixed | Random | Fixed | |
| I2 | 61% | 67% | 67% | 68% | 79% | 12% | |
| P | <0.00001 | <0.00001 | <0.00001 | <0.0001 | <0.00001 | <0.00001 | |
| EC | Study(n) | 1 | 2 | 1 | 2 | - | 3 |
| OR (95%CI) |
6.00 (1.66, 21.74) |
5.33 (2.17, 13.05) |
6.01 (2.15, 16.85) |
5.07 (1.77, 14.52) |
- | 5.54 (2.66, 11.56) |
|
| Model | Fixed | Fixed | - | Fixed | - | Fixed | |
| I2 | - | 0% | - | 0% | - | 0% | |
| P | 0.006 | 0.0003 | 0.0006 | 0.002 | - | <0.00001 | |
| GC | Study(n) | 6 | 6 | 7 | 4 | 7 | 4 |
| OR (95%CI) |
9.04 (5.75, 14.21) |
3.21 (1.39, 7.40) |
7.26 (5.12, 10.29) |
1.60 (1.01, 2.54) |
4.12 (1.85, 9.21) |
27.54 (7.23, 104.86) |
|
| Model | Fixed | Random | Fixed | Fixed | Random | Fixed | |
| I2 | 59% | 78% | 55% | 68% | 84% | 0% | |
| P | <0.00001 | 0.006 | <0.00001 | 0.05 | 0.0006 | <0.00001 | |
| CRC | Study(n) | 4 | 5 | 5 | 4 | 6 | 3 |
| OR (95%CI) |
64.96 (15.20, 277.62) |
2.64 (0.84, 8.25) |
10.18 (1.25, 83.19) |
8.36 (2.55, 27.43) |
10.53 (1.67, 66.37) |
6.34 (1.80, 22.42) |
|
| Model | Fixed | Fixed | Random | Fixed | Random | Fixed | |
| I2 | 0% | 51% | 85% | 37% | 81% | 57% | |
| P | <0.00001 | 0.09 | 0.03 | 0.0005 | 0.01 | 0.004 | |
Relationship between DAPK1 methylation and clinicopathological features of gastrointestinal cancer
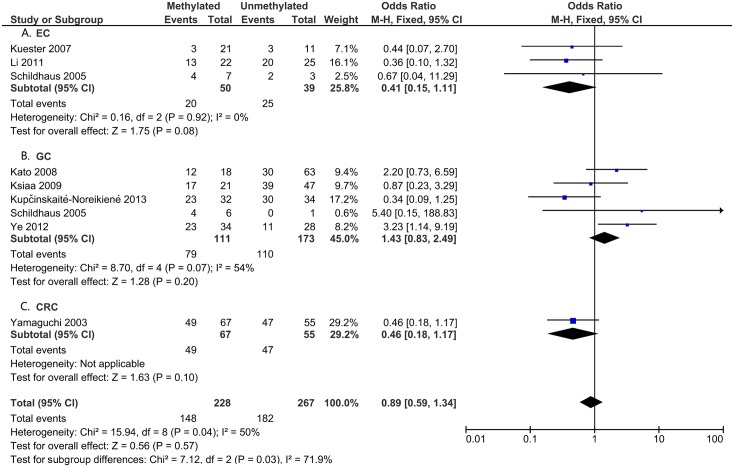
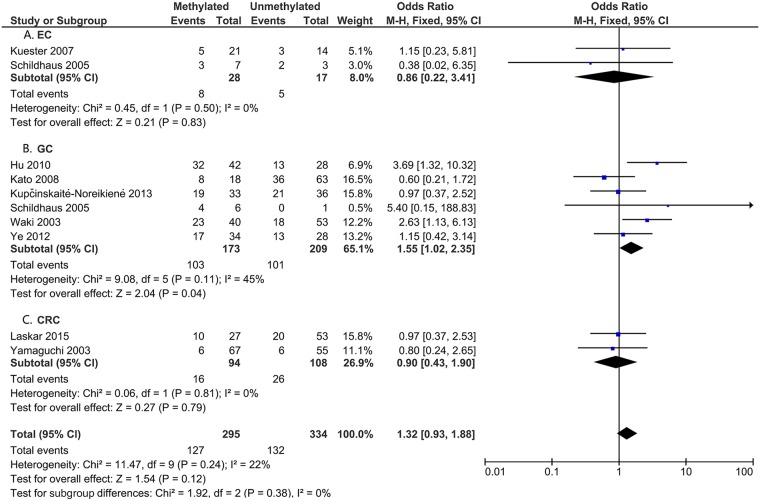
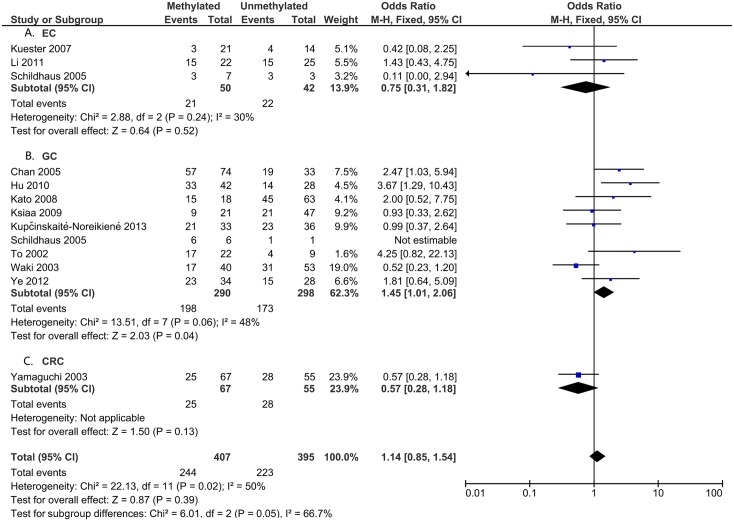
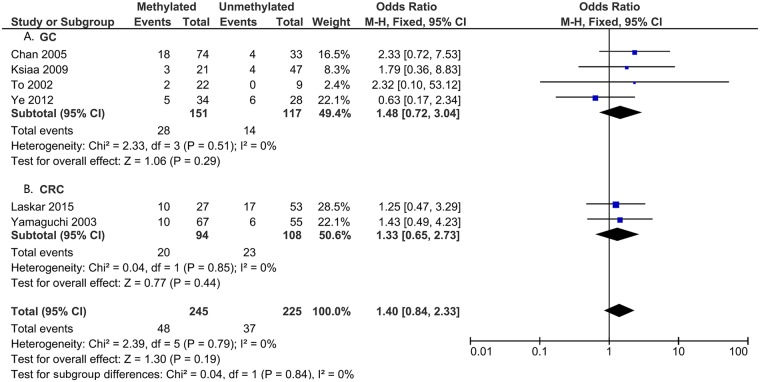
To analyze the role of DAPK1 in the pathogenesis of gastrointestinal cancer, the correlations between DAPK1 methylation and clinicopathological features were assessed (Figs 5–8). As is shown in Fig 5, DAPK1 methylation was not correlated with the T stage of gastrointestinal cancer (T3+T4 vs. T1+T2, OR = 0.89, 95%CI:0.59–1.34, P = 0.57, fixed effects model), nor with that of EC, GC, or CRC (Fig 5). As for N stage, DAPK1 methylation was positively related to the N stage of GC (positive vs. negative, OR = 1.45, 95%CI: 1.01–2.06, P = 0.04, fixed effects model), but not that of gastrointestinal cancer, nor EC or CRC (Fig 6). In addition, no obvious association has been found between the methylation of DAPK1 and the M stage of gastrointestinal cancer (Fig 7). Moreover, DAPK1 methylation was associated with the poor differentiation of GC (G3 vs. G1+G2, OR = 1.55, 95%CI: 1.02–2.35, P = 0.04, fixed effects model; Fig 8). However, DAPK1 methylation was not related to the age (>60 vs.<60, OR = 0.83, 95%CI: 0.54–1.27, P = 0.40, fixed effects model) or gender (male vs. female, OR = 0.48, 95%CI: 0.16–1.44, P = 0.19, fixed effects model) of gastrointestinal cancer patients (Table 3). Also, it was not correlated with the Lauren Classification of GC (intestinal vs. diffuse, OR = 1.12, 95%CI: 0.71–1.77, P = 0.63; Table 3).
Fig 5. DAPK1 methylation and T stage (T3+T4 vs.T1+T2) in: A. EC; B. GC; and C. CRC.
Fig 8. DAPK1 methylation and cancer differentiation (G3 vs. G1+G2) in: A. EC; B. GC; and C. CRC.
Fig 6. DAPK1 methylation and N stage (positive vs. negative) in: A. EC; B. GC; and C. CRC.
Fig 7. DAPK1 methylation and M stage (M1 vs. M0): A. GC; and B. CRC.
Table 3. Associations between DAPK1 methylation and the clinicopathological features of gastrointestinal cancer.
| Age (>60 vs. <60) |
Gender (Male vs. Female) |
Lauren Classification (intestinal vs. diffuse) |
Asian T stage (T3+T4 vs.T1+T2) |
Asian N stage (positive vs. negative) |
Asia M stage (M1 vs. M0) |
Asia Differentiation (G3 vs. G1+G2) |
||
|---|---|---|---|---|---|---|---|---|
| Overall | Study(n) | 9 | 3 | - | 4 | 8 | 5 | 6 |
|
OR (95%CI) |
0.83 (0.54, 1.27) |
0.48 (0.16, 1.44) |
- | 1.06 (0.64, 1.74) |
1.29 (0.91, 1.81) |
1.37 (0.80, 2.34) |
1.41 (0.95, 2.10) |
|
| Model | Fixed | Fixed | - | Fixed | Fixed | Fixed | Fixed | |
| I2 | 0% | 0% | - | 75% | 61% | 0% | 48% | |
| P | 0.40 | 0.19 | - | 0.83 | 0.15 | 0.25 | 0.09 | |
| EC | Study(n) | 3 | 9 | - | 1 | 1 | - | - |
|
OR (95%CI) |
0.69 (0.29, 1.67) |
1.16 (0.81, 1.68) |
- | 0.36 (0.10, 1.32) |
1.43 (0.43, 4.75) |
- | - | |
| Model | Fixed | Fixed | - | Fixed | Fixed | - | - | |
| I2 | 0% | 0% | - | - | - | - | - | |
| P | 0.41 | 0.42 | - | 0.12 | 0.56 | - | - | |
| GC | Study(n) | 5 | 3 | 5 | 2 | 6 | 3 | 4 |
|
OR (95%CI) |
0.88 (0.54, 1.45) |
1.01 (0.57, 1.79) |
1.12 (0.71, 1.77) |
2.68 (1.26, 5.72) |
1.66 (1.10, 2.51) |
1.41 (0.63, 3.16) |
1.69 (1.06, 2.72) |
|
| Model | Fixed | Fixed | Fixed | Fixed | Fixed | Fixed | Fixed | |
| I2 | 15% | 0% | 24% | 0% | 57% | 11% | 60% | |
| P | 0.62 | 0.98 | 0.63 | 0.01 | 0.02 | 0.40 | 0.03 | |
| CRC | Study(n) | 1 | 3 | - | 1 | 1 | 2 | 2 |
|
OR (95%CI) |
0.87 (0.07,10.42) |
1.01 (0.57, 1.79) |
- | 0.46 (0.18, 1.17) |
0.57 (0.28, 1.18) |
1.33 (0.65, 2.73) |
0.90 (0.43, 1.90) |
|
| Model | Fixed | Fixed | - | Fixed | Fixed | Fixed | Fixed | |
| I2 | - | 0% | - | - | 0% | 0% | ||
| P | 0.91 | 0.98 | 0.10 | 0.13 | 0.44 | 0.79 |
Since the relationship between DAPK1 methylation and gastrointestinal cancer was stronger in Asian patients, further analysis was performed in the subgroup of Asian patients to reveal the association between DAPK1 methylation and the clinicopathological features of gastrointestinal cancer. A closer association was revealed between DAPK1 methylation and the T stage, N stage, and differentiation of GC, for which the ORs were 2.68 (T3+T4 vs.T1+T2, 95%CI: 1.26–5.72, fixed effects model), 1.66 (positive vs. negative 95%CI: 1.10–2.51, fixed effects model), and 1.69 (G3 vs. G1+G2, 95%CI: 1.06–2.72), respectively (Table 3). However, the associations between DAPK1 methylation and clinicopathological features were not significant in the overall analysis (Table 3).
The data for 5-year OS/DFS rates were insufficient to conduct a survival analysis.
Publication bias
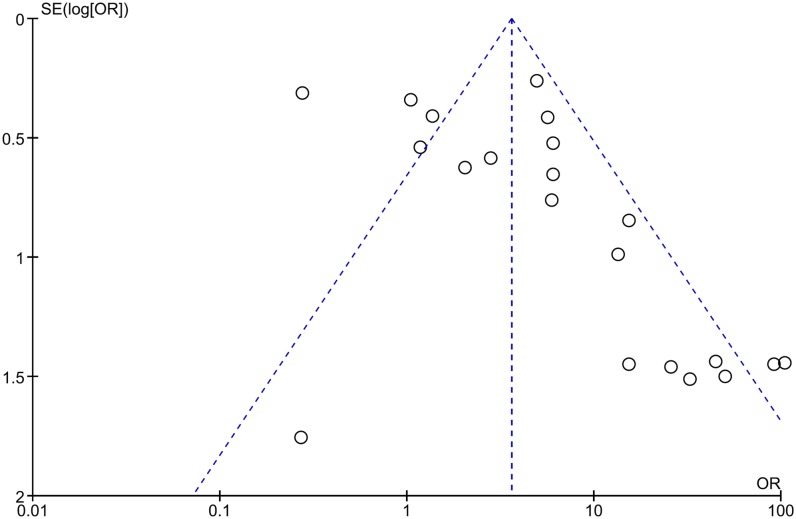
The shape of the generated funnel plot seemed asymmetrical in the pooled analysis (Fig 9). In addition, P values<0.05 were calculated for Egger’s tests and Begg’s tests in the overall analysis, which suggests the existence of publication bias (Table 4). However, in the analysis of the association between DAPK1 methylation and the clinicopathological features of gastrointestinal cancer, the P values on Egger’s tests and Begg’s tests were greater than 0.05, except for the Egger’s test result for the T stage of EC (P = 0.007; Table 4 and S2 Table).
Fig 9. Funnel plot of the result of pooled analysis.
Table 4. Analysis of publication bias among included studies.
| Study(n) | P value of Egger’s test | P value of Begg’s test | ||
|---|---|---|---|---|
| Pooled analysis | Overall | 22 | 0.024 | 0.032 |
| EC | 3 | 0.259 | 1.000 | |
| GC | 12 | 0.141 | 0.244 | |
| CRC | 9 | 0.040 | 0.005 | |
|
T stage (T3+T4 vs. T1+T2) |
Overall | 9 | 0.992 | 0.251 |
| EC | 3 | 0.007 | 1.000 | |
| GC | 5 | 0.968 | 1.000 | |
|
N stage (positive vs. negative) |
Overall | 13 | 0.806 | 0.373 |
| EC | 3 | 0.233 | 1.000 | |
| GC | 9 | 0.191 | 1.000 | |
|
Metastasis (M1 vs. M0) |
Overall | 6 | 0.730 | 0.707 |
| GC | 4 | 0.863 | 0.734 | |
| CRC | 2 | - | 1.000 | |
|
Differentiation (G3 vs. G1+G2) |
Overall | 9 | 0.723 | 1.000 |
| EC | 2 | - | 1.000 | |
| GC | 6 | 0.823 | 1.000 | |
| CRC | 2 | - | 1.000 | |
|
Age (>60 vs. <60) |
Overall | 8 | 0.491 | 0.446 |
|
Gender (Male vs. Female) |
Overall | 14 | 0.993 | 0.661 |
|
Lauren Classification (intestinal vs. diffuse) |
GC | 5 | 0.618 | 0.462 |
Discussion
Consistent with the goal of precision medicine, molecular pathological epidemiology (MPE) based on molecular classification of disease is becoming increasingly attractive[34]. This approach can discover molecular biomarkers, identify relevant subtypes, and establish the relationship between the risk factors with specific subtype[35]. Various environmental and lifestyle factors such as one-carbon metabolism, cigarette smoking, and diet could be associated with aberrant DNA methylation, which was found to be an important biomarker and novel target for treatment in various cancers[36]. Abnormal methylation of the promoter is a critical mechanism for the down-regulation of genes including DAPK1[26].
DAPK1, as a classical anti-oncogene, has been demonstrated to play an important role in the development, progression and metastasis of tumors[7]. Down-regulation of DAPK1 expression has been correlated with the severity of malignancy and lymph node metastasis in various cancers including lung cancer[37], urinary tract carcinoma[38], and esophageal squamous cell carcinoma[39]. It has been shown that DAPK1 can influence cell survival and apoptosis by activating the mammalian target of rapamycin complex1 (mToRC1)[40]. Up-regulation of DAPK1 alleviates the malignant behavior of pancreatic carcinoma through the PI3K/Akt and ERK pathway[41]. In addition, DAPK1 is involved in activating the mTOR pathway by breaking the TSC1/TSC2 complex in the p53-mutant triple receptor–negative breast cancer[42].
Hypermethylation of DAPK1 has been found to be involved in head and neck cancers[43], papillary thyroid cancer[44], and even brain metastases of various solid tumors[45]. Recently, several studies have investigated the roles of DAPK1 methylation in cervical cancer[46], lung cancer[47] and GC[48]. However, a systematical analysis of its role in gastrointestinal cancer has not been reported. Therefore, the present study was needed to uncover the potential value of DAPK1 methylation in the diagnosis and pathogenesis of gastrointestinal cancer.
The pooled OR indicated that DAPK1 methylation was positively correlated with the risk of gastrointestinal cancer, which suggests the potential value of DAPK1 methylation in the diagnosis of gastrointestinal cancer, especially in CRC. In addition, in the subgroup analysis of studies that used normal tissue as the control group, a tighter relationship was demonstrated. The dissimilar results for the different sources for the control group suggested that the degree of DAPK1 methylation in the normal tissue adjacent to tumor tissue was higher than that in normal tissues. These findings were consist with previous results showing that DAPK1 methylation is significantly related to the risk of precancerous lesions such as intestinal metaplasia (IM) [49] and Barrett’s metaplasia[10]. Moreover, the frequency of DAPK1 methylation was shown to gradually increased from precancerous lesions to cancer[10, 24]. Therefore, detection of DAPK1 methylation could be used for the early diagnosis of gastrointestinal cancer. In addition, the association of DAPK1 methylation with the risk of gastrointestinal cancer was most notable in Asian patients and in CRC patients, which suggests that the pathogenic role of DAPK1 methylation in different geographical regions and tumor locations of gastrointestinal cancer vary.
Furthermore, we investigated the associations between the frequency of DAPK1 methylation and the clinicopathological features of gastrointestinal cancer. Our results showed that DAPK1 methylation was unrelated to cancer differentiation, T stage, N stage, or M stage in gastrointestinal cancer. Such results indicated that DAPK1 methylation could promote the carcinogenesis process but not the processes of invasion and metastasis[32]. When stratified by location, DAPK1 methylation was positively correlated with lymph node metastasis and poor differentiation in GC, moreover the correlation was more significant among Asian patients, which suggests that DAPK1 methylation was involved in the metastasis of GC in Asian patients. In addition, it is more accurate to assess the prognosis of gastrointestinal cancer by combining analysis of DAPK1 and other genes, because the number of methylated gene gradually increases from 0.12 and 0.8 in adjacent normal tissues to 3.3 and 2.5 in GC[25] and EC tissues[22], respectively. Although the frequency of DAPK1 methylation was found to increase with ages[50], we found that methylation of DAPK1 was not correlated with age in gastrointestinal cancer patients.
The survival analysis showed that DAPK1 methylation was correlated with the susceptibility of recurrence, metastasis and disease-related death (67.6% in methylated group vs. 41.9% in unmethylated group) in GC[28]. However, in other studies, DAPK1 methylation was not associated with OS in GC [14] or EC[51]. Such disagreement suggests that more studies are needed for more conclusive survival analysis.
Inevitably, there are some limitations in this meta analysis. First, heterogeneity existed in some analyses, though it could be alleviated by the sensitivity analysis and subgroup analysis according to the potential heterogeneous factors, such as the source of the control group, geographic area, and tumor location. To better analyze the association between DAPK1 methylation and gastrointestinal cancer, a more precise method like the qMSP should be used in future studies to distinguish the degree of the methylation [52]. In addition, potential publication bias is inevitable, and the existence of publication bias in the overall analysis may reduce the power and accuracy of the relationship between DAPK1 methylation and gastrointestinal cancer. Last but not least, the association between DAPK1 methylation and the survival of patients could not be estimated due to an insufficient amount of related data. The above limitations may partially influence the significance of DAPK1 methylation and the clinicopathological analyses. Therefore, larger prospective studies are needed to validate our results.
In summary, the findings of this meta-analysis indicate that the methylation of DAPK1 may be valuable biomarker in the diagnosis and the tumorgenesis of gastrointestinal cancer. However, DAPK1 methylation was not correlated with the clinicopathological features of gastrointestinal cancer, but was associated with the N stage and cancer differentiation of GC. Thus, further studies of DAPK1 and its potential role in the progression of gastrointestinal cancer are needed.
Supporting information
(TIF)
(TIF)
(DOC)
(DOCX)
Acknowledgments
This study was supported by the National Nature Science Foundation of China (No. 81470789 and 81271199)(http://www.nsfc.gov.cn/), and Research Fund of Public welfare in Health Industry (No.201402015), Health and Family Plan Committee of China, 2014 (http://www.nhfpc.gov.cn/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank Medjaden Bioscience Limited (Hong Kong, China) for proofreading this manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by the National Nature Science Foundation of China (No. 81470789 and 81271199) (http://www.nsfc.gov.cn/), and Research Fund of Public welfare in Health Industry (No. 201402015), Health and Family Plan Committee of China, 2014 (http://www.nhfpc.gov.cn/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Global Burden of Disease Cancer C, Fitzmaurice C, Dicker D, a A, Hamavid H, Moradi-Lakeh M, et al. The Global Burden of Cancer 2013. JAMA oncology. 2015. July;1(4):505–27. doi: 10.1001/jamaoncol.2015.0735 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang B, Chen JH, Yuan WZ, Ji JT, Liu ZY, Wu L, et al. Prognostic and clinical value of Sirt1 expression in gastric cancer: A systematic meta-analysis. Journal of Huazhong University of Science and Technology Medical sciences = Hua zhong ke ji da xue xue bao Yi xue Ying De wen ban = Huazhong keji daxue xuebao Yixue Yingdewen ban. 2016. April;36(2):278–84. doi: 10.1007/s11596-016-1580-0 . [DOI] [PubMed] [Google Scholar]
- 3.Tahara T, Arisawa T. DNA methylation as a molecular biomarker in gastric cancer. Epigenomics. 2015. 1 June;7(3):475–86. doi: 10.2217/epi.15.4 . [DOI] [PubMed] [Google Scholar]
- 4.Deiss LP, Feinstein E, Berissi H, Cohen O, Kimchi A. Identification of a novel serine/threonine kinase and a novel 15-kD protein as potential mediators of the gamma interferon-induced cell deathc. Genes Dev. 1995;1(9):15–30. [DOI] [PubMed] [Google Scholar]
- 5.Singh P, Ravanan P, Talwar P. Death Associated Protein Kinase 1 (DAPK1): A Regulator of Apoptosis and Autophagy. Frontiers in molecular neuroscience. 2016;9:46 doi: 10.3389/fnmol.2016.00046 . Epub 2016/07/23. Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Celik S, Akcora D, Ozkan T, Varol N, Aydos S, Sunguroglu A. Methylation analysis of the DAPK1 gene in imatinib-resistant chronic myeloid leukemia patients. Oncology letters. 2015. January;9(1):399–404. doi: 10.3892/ol.2014.2677 . Epub 2014/12/02. Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schneider-Stock R. Death-associated kinase (DAPK): a cancer "gene chameleon". Apoptosis: an international journal on programmed cell death. 2014. February;19(2):285 doi: 10.1007/s10495-013-0932-5 . Epub 2013/11/08. Eng. [DOI] [PubMed] [Google Scholar]
- 8.Ye M, Li D, Zhou F, Guo Q, Xia B. Epigenetic regulation of death-associated protein kinase expression in primary gastric cancers from Chinese patients. European journal of cancer prevention: the official journal of the European Cancer Prevention Organisation (ECP). 2012. May;21(3):241–6. doi: 10.1097/CEJ.0b013e32834c9caa . Epub 2011/10/08. Eng. [DOI] [PubMed] [Google Scholar]
- 9.Mittag F, Kuester D, Vieth M, Peters B, Stolte B, Roessner A, et al. DAPK promotor methylation is an early event in colorectal carcinogenesis. Cancer letters. 2006. August 18;240(1):69–75. doi: 10.1016/j.canlet.2005.08.034 . Epub 2005/10/26. Eng. [DOI] [PubMed] [Google Scholar]
- 10.Kuester D, Dar AA, Moskaluk CC, Krueger S, Meyer F, Hartig R, et al. Early involvement of death-associated protein kinase promoter hypermethylation in the carcinogenesis of Barrett's esophageal adenocarcinoma and its association with clinical progression. 2007. March;9(3):236–45. . Epub 2007/04/03. Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nomura T, Tahara T, Shiroeda H, Minato T, Matsue Y, Hayashi R, et al. Influence of HRH2 promoter polymorphism on aberrant DNA methylation of DAPK and CDH1 in the gastric epithelium. BMC gastroenterology. 2013. January 2;13:1 doi: 10.1186/1471-230X-13-1 . Epub 2013/01/03. Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee BB, Lee EJ, Jung EH, Chun HK, Chang DK, Song SY, et al. Aberrant methylation of APC, MGMT, RASSF2A, and Wif-1 genes in plasma as a biomarker for early detection of colorectal cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009. October 1;15(19):6185–91. doi: 10.1158/1078-0432.CCR-09-0111 . Epub 2009/09/24. Eng. [DOI] [PubMed] [Google Scholar]
- 13.Xu XL, Yu J, Zhang HY, Sun MH, Gu J, Du X, et al. Methylation profile of the promoter CpG islands of 31 genes that may contribute to colorectal carcinogenesis. World journal of gastroenterology. 2004. December 1;10(23):3441–54. doi: 10.3748/wjg.v10.i23.3441 . Epub 2004/11/05. Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waki T, Tamura G, Sato M, Terashima M, Nishizuka S, Motoyama T. Promoter methylation status of DAP-kinase and RUNX3 genes in neoplastic and non-neoplastic gastric epithelia. Cancer science. 2003. April;94(4):360–4. . Epub 2003/06/26. Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiong J, Li Y, Huang K, Lu M, Shi H, Ma L, et al. Association between DAPK1 promoter methylation and cervical cancer: a meta-analysis. PloS one. 2014;9(9):e107272 doi: 10.1371/journal.pone.0107272 . Epub 2014/10/01. Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, Xia Q, Jiang B, Chang W, Yuan W, Ma Z, et al. Prognostic Value of Cancer Stem Cell Marker ALDH1 Expression in Colorectal Cancer: A Systematic Review and Meta-Analysis. PloS one. 2015;10(12):e0145164 doi: 10.1371/journal.pone.0145164 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Statistics in medicine. 2002. June 15;21(11):1539–58. doi: 10.1002/sim.1186 . [DOI] [PubMed] [Google Scholar]
- 18.Bagci B, Sari M, Karadayi K, Turan M, Ozdemir O, Bagci G. KRAS, BRAF oncogene mutations and tissue specific promoter hypermethylation of tumor suppressor SFRP2, DAPK1, MGMT, HIC1 and p16 genes in colorectal cancer patients. Cancer biomarkers: section A of Disease markers. 2016. June 24;17(2):133–43. doi: 10.3233/CBM-160624 . Epub 2016/08/20. Eng. [DOI] [PubMed] [Google Scholar]
- 19.Laskar RS, Ghosh SK, Talukdar FR. Rectal cancer profiling identifies distinct subtypes in India based on age at onset, genetic, epigenetic and clinicopathological characteristics. Molecular carcinogenesis. 2015. December;54(12):1786–95. doi: 10.1002/mc.22250 . Epub 2014/11/25. Eng. [DOI] [PubMed] [Google Scholar]
- 20.Almeida FG, de Aquino PF, de Souza AD, de Souza AQ, do Carmo Vinhote S, Mac-Cormick TM, et al. Colorectal cancer DNA methylation patterns from patients in Manaus, Brazil. Biological research. 2015. September 12;48:50 doi: 10.1186/s40659-015-0042-7 . Epub 2015/09/14. Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kupcinskaite-Noreikiene R, Skieceviciene J, Jonaitis L, Ugenskiene R, Kupcinskas J, Markelis R, et al. CpG island methylation of the MLH1, MGMT, DAPK, and CASP8 genes in cancerous and adjacent noncancerous stomach tissues. Medicina (Kaunas, Lithuania). 2013;49(8):361–6. . Epub 2014/02/11. Eng. [PubMed] [Google Scholar]
- 22.Li B, Wang B, Niu LJ, Jiang L, Qiu CC. Hypermethylation of multiple tumor-related genes associated with DNMT3b up-regulation served as a biomarker for early diagnosis of esophageal squamous cell carcinoma. Epigenetics. 2011. March;6(3):307–16. doi: 10.4161/epi.6.3.14182 . Epub 2010/12/15. Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu SL, Kong XY, Cheng ZD, Sun YB, Shen G, Xu WP, et al. Promoter methylation of p16, Runx3, DAPK and CHFR genes is frequent in gastric carcinoma. Tumori. 2010. Sep-Oct;96(5):726–33. . Epub 2011/02/10. Eng. [DOI] [PubMed] [Google Scholar]
- 24.Zou XP, Zhang B, Zhang XQ, Chen M, Cao J, Liu WJ. Promoter hypermethylation of multiple genes in early gastric adenocarcinoma and precancerous lesions. Human pathology. 2009. November;40(11):1534–42. doi: 10.1016/j.humpath.2009.01.029 . Epub 2009/08/22. Eng. [DOI] [PubMed] [Google Scholar]
- 25.Ksiaa F, Ziadi S, Amara K, Korbi S, Trimeche M. Biological significance of promoter hypermethylation of tumor-related genes in patients with gastric carcinoma. Clinica chimica acta; international journal of clinical chemistry. 2009. June 27;404(2):128–33. doi: 10.1016/j.cca.2009.03.044 . Epub 2009/04/02. Eng. [DOI] [PubMed] [Google Scholar]
- 26.Kato K, Iida S, Uetake H, Takagi Y, Yamashita T, Inokuchi M, et al. Methylated TMS1 and DAPK genes predict prognosis and response to chemotherapy in gastric cancer. International journal of cancer. 2008. February 1;122(3):603–8. doi: 10.1002/ijc.23143 . Epub 2007/10/19. Eng. [DOI] [PubMed] [Google Scholar]
- 27.Anacleto C, Leopoldino AM, Rossi B, Soares FA, Lopes A, Rocha JC, et al. Colorectal cancer "methylator phenotype": fact or artifact? Neoplasia (New York, NY). 2005. April;7(4):331–5. . Epub 2005/06/22. Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan AW, Chan MW, Lee TL, Ng EK, Leung WK, Lau JY, et al. Promoter hypermethylation of Death-associated protein-kinase gene associated with advance stage gastric cancer. Oncology reports. 2005. May;13(5):937–41. . Epub 2005/04/06. Eng. [PubMed] [Google Scholar]
- 29.Schildhaus HU, Krockel I, Lippert H, Malfertheiner P, Roessner A, Schneider-Stock R. Promoter hypermethylation of p16INK4a, E-cadherin, O6-MGMT, DAPK and FHIT in adenocarcinomas of the esophagus, esophagogastric junction and proximal stomach. International journal of oncology. 2005. June;26(6):1493–500. . Epub 2005/05/05. Eng. [PubMed] [Google Scholar]
- 30.Lee S, Hwang KS, Lee HJ, Kim JS, Kang GH. Aberrant CpG island hypermethylation of multiple genes in colorectal neoplasia. Laboratory investigation; a journal of technical methods and pathology. 2004. July;84(7):884–93. doi: 10.1038/labinvest.3700108 . Epub 2004/05/04. Eng. [DOI] [PubMed] [Google Scholar]
- 31.Sabbioni S, Miotto E, Veronese A, Sattin E, Gramantieri L, Bolondi L, et al. Multigene methylation analysis of gastrointestinal tumors: TPEF emerges as a frequent tumor-specific aberrantly methylated marker that can be detected in peripheral blood. Molecular diagnosis: a journal devoted to the understanding of human disease through the clinical application of molecular biology. 2003;7(3–4):201–7. . Epub 2004/04/08. Eng. [DOI] [PubMed] [Google Scholar]
- 32.Yamaguchi S, Asao T, Nakamura J, Ide M, Kuwano H. High frequency of DAP-kinase gene promoter methylation in colorectal cancer specimens and its identification in serum. Cancer letters. 2003. May 8;194(1):99–105. . Epub 2003/04/23. Eng. [DOI] [PubMed] [Google Scholar]
- 33.To KF, Leung WK, Lee TL, Yu J, Tong JH, Chan MW, et al. Promoter hypermethylation of tumor-related genes in gastric intestinal metaplasia of patients with and without gastric cancer. International journal of cancer. 2002. December 20;102(6):623–8. doi: 10.1002/ijc.10783 . Epub 2002/11/26. Eng. [DOI] [PubMed] [Google Scholar]
- 34.Ogino S, Nishihara R, VanderWeele TJ, Wang M, Nishi A, Lochhead P, et al. The Role of Molecular Pathological Epidemiology in the Study of Neoplastic and Non-Neoplastic Diseases in the Era of Precision Medicine. Epidemiology. 2016;27(4):602–11. doi: 10.1097/EDE.0000000000000471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogino S, Chan AT, Fuchs CS, Giovannucci E. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut. 2010;60(3):397–411. doi: 10.1136/gut.2010.217182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hughes LAE, Melotte V, de Schrijver J, de Maat M, Smit VTHBM, Bovee JVMG, et al. The CpG Island Methylator Phenotype: What's in a Name? Cancer research. 2013;73(19):5858–68. doi: 10.1158/0008-5472.CAN-12-4306 [DOI] [PubMed] [Google Scholar]
- 37.Harden SV, Tokumaru Y, Westra WH, Goodman S, Ahrendt SA, Yang SC, et al. Gene promoter hypermethylation in tumors and lymph nodes of stage I lung cancer patients. Clinical cancer research: an official journal of the American Association for Cancer Research. 2003. April;9(4):1370–5. . Epub 2003/04/10. Eng. [PubMed] [Google Scholar]
- 38.Catto JW, Azzouzi AR, Rehman I, Feeley KM, Cross SS, Amira N, et al. Promoter hypermethylation is associated with tumor location, stage, and subsequent progression in transitional cell carcinoma. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2005. May 1;23(13):2903–10. doi: 10.1200/JCO.2005.03.163 . Epub 2005/03/09. Eng. [DOI] [PubMed] [Google Scholar]
- 39.Zhai J, Yang X, Zhang Y, Qi Q, Hu J, Wang Q. Reduced expression levels of the death-associated protein kinase and E-cadherin are correlated with the development of esophageal squamous cell carcinoma. Experimental and therapeutic medicine. 2013. March;5(3):972–6. . Epub 2013/02/15. Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stevens C, Lin Y, Harrison B, Burch L, Ridgway RA, Sansom O, et al. Peptide combinatorial libraries identify TSC2 as a death-associated protein kinase (DAPK) death domain-binding protein and reveal a stimulatory role for DAPK in mTORC1 signaling. The Journal of biological chemistry. 2009. January 2;284(1):334–44. doi: 10.1074/jbc.M805165200 . Epub 2008/11/01. Eng. [DOI] [PubMed] [Google Scholar]
- 41.Qin Y, Ye GX, Wu CJ, Wang S, Pan DB, Jiang JY, et al. Effect of DAPK1 gene on proliferation, migration, and invasion of carcinoma of pancreas BxPC-3 cell line. International journal of clinical and experimental pathology. 2014;7(11):7536–44. . Epub 2015/01/01. Eng. [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao J, Zhao D, Poage GM, Mazumdar A, Zhang Y, Hill JL, et al. Death-associated protein kinase 1 promotes growth of p53-mutant cancers. The Journal of clinical investigation. 2015. July 1;125(7):2707–20. doi: 10.1172/JCI70805 . Epub 2015/06/16. Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanchez-Cespedes M, Esteller M, Wu L, Nawroz-Danish H, Yoo GH, Koch WM, et al. Gene promoter hypermethylation in tumors and serum of head and neck cancer patients. Cancer research. 2000. February 15;60(4):892–5. . Epub 2000/03/08. Eng. [PubMed] [Google Scholar]
- 44.Hu S, Liu D, Tufano RP, Carson KA, Rosenbaum E, Cohen Y, et al. Association of aberrant methylation of tumor suppressor genes with tumor aggressiveness and BRAF mutation in papillary thyroid cancer. International journal of cancer. 2006. November 15;119(10):2322–9. doi: 10.1002/ijc.22110 . Epub 2006/07/22. Eng. [DOI] [PubMed] [Google Scholar]
- 45.Gonzalez-Gomez P, Bello MJ, Alonso ME, Lomas J, Arjona D, Aminoso C, et al. Frequent death-associated protein-kinase promoter hypermethylation in brain metastases of solid tumors. Oncology reports. 2003. Jul-Aug;10(4):1031–3. . Epub 2003/06/07. Eng. [PubMed] [Google Scholar]
- 46.Agodi A, Barchitta M, Quattrocchi A, Maugeri A, Vinciguerra M. DAPK1 Promoter Methylation and Cervical Cancer Risk: A Systematic Review and a Meta-Analysis. PloS one. 2015;10(8):e0135078 doi: 10.1371/journal.pone.0135078 . Epub 2015/08/13. Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y, Zhu M, Zhang X, Cheng D, Ma X. Clinical significance of DAPK promoter hypermethylation in lung cancer: a meta-analysis. Drug design, development and therapy. 2015;9:1785–96. doi: 10.2147/DDDT.S78012 . Epub 2015/04/08. Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jia W, Yu T, Cao X, An Q, Yang H. Clinical effect of DAPK promoter methylation in gastric cancer. Medicine. 2016;95(43):e5040 doi: 10.1097/MD.0000000000005040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tahara T, Shibata T, Nakamura M, Yamashita H, Yoshioka D, Okubo M, et al. Increased number of CpG island hypermethylation in tumor suppressor genes of non-neoplastic gastric mucosa correlates with higher risk of gastric cancer. Digestion. 2010;82(1):27–36. doi: 10.1159/000252766 . Epub 2010/02/13. Eng. [DOI] [PubMed] [Google Scholar]
- 50.Qu Y, Dang S, Hou P. Gene methylation in gastric cancer. Clinica chimica acta; international journal of clinical chemistry. 2013. September 23;424:53–65. doi: 10.1016/j.cca.2013.05.002 . Epub 2013/05/15. Eng. [DOI] [PubMed] [Google Scholar]
- 51.Brock MV, Gou M, Akiyama Y, Muller A, Wu TT, Montgomery E, et al. Prognostic importance of promoter hypermethylation of multiple genes in esophageal adenocarcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2003. August 1;9(8):2912–9. . Epub 2003/08/13. Eng. [PubMed] [Google Scholar]
- 52.Fackler MJ, McVeigh M, Mehrotra J, Blum MA, Lange J, Lapides A, et al. Quantitative multiplex methylation-specific PCR assay for the detection of promoter hypermethylation in multiple genes in breast cancer. Cancer research. 2004. July 1;64(13):4442–52. doi: 10.1158/0008-5472.CAN-03-3341 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(DOC)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.