Abstract
The Arabidopsis thaliana F-box gene HAWAIIAN SKIRT (HWS) affects organ growth and the timing of floral organ abscission. The loss-of-function hws-1 mutant exhibits fused sepals and increased organ size. To understand the molecular mechanisms of HWS during plant development, we mutagenized hws-1 seeds with ethylmethylsulphonate (EMS) and screened for mutations suppressing hws-1 associated phenotypes. We isolated the shs1/hws-1 (suppressor of hws-1) mutant in which hws-1 sepal fusion phenotype was suppressed. The shs1/hws-1 mutant carries a G→A nucleotide substitution in the MIR164 binding site of CUP-SHAPED COTYLEDON 1 (CUC1) mRNA. CUC1 and CUP-SHAPED COTYLEDON 2 (CUC2) transcript levels were altered in shs1, renamed cuc1-1D, and in hws-1 mutant. Genetic interaction analyses using single, double and triple mutants of cuc1-1D, cuc2-1D (a CUC2 mutant similar to cuc1-1D), and hws-1, demonstrate that HWS, CUC1 and CUC2 act together to control floral organ number. Loss of function of HWS is associated with larger petal size due to alterations in cell proliferation and mitotic growth, a role shared with the CUC1 gene.
Introduction
Understanding how organ growth is controlled is a pivotal step to manipulating crop yield through the production of bigger and more robust plants. The formation of mature organs requires coordinated regulation of cell proliferation and expansion [1, 2]; the regulatory pathways of lateral organ growth modulated by cell number and size has been reviewed [3].
A recent meta-analysis of growth parameters of leaves and roots of Gramineae and Eudicotyledonous, demonstrated that cell proliferation rather than cell expansion determines the final size of plant organs; the number of dividing cells rather than the rate of division determines cell production; smaller cells leaving the meristem elongate more than bigger cells; and the differences in mature cell size are determined by cell expansion duration [4].
Flowers are the reproductive units of a plant, understanding the regulatory mechanisms that form and shape them is paramount. Flowers arise from the undifferentiated stem cells of the floral apical meristem [5]; a mature Arabidopsis thaliana flower has four sepals, four petals, six stamens and two congenital fused carpels that form the gynoecium; these organs emerge from four concentric whorls and their number is highly conserved [6]. Complex regulatory pathways of homeotic and cadastral genes determine floral organ identity, morphology, position, size and number in an Arabidopsis flower [7]. Cadastral genes prevent organ fusion by defining expression boundaries of floral identity genes [8, 9]. The boundaries are shaped by a combination of a reduction in the frequency of cell division, a cessation of DNA synthesis, and an interruption in the expression of cell cycle related genes in a group of specialized cells forming the boundaries [10, 11].
Many genes defining floral whorls, affecting boundary formation and floral organ number have been described (for reviews see [12–14]). Among these genes, CUP SHAPED COTYLEDON1 and 2 (CUC1 and CUC2) NAC transcription factors, are essential for boundary formation and promotion of SHOOT MERISTEMLESS (STM) expression to initiate the shoot meristem [15–19].
MIR164 regulate boundary formation and floral organ number by establishing and maintaining the boundary domain by controlling post-transcriptional degradation of the CUC1 and CUC2 mRNAs [20–22]. The early extra petals1 (eep1), a mutant of MIR164C, has increased petal number in the youngest flowers [20]. Constitutively expressing the MIR164B gene results in plants showing partial fusion of cotyledons and floral organs comparable to double mutant cuc1/cuc2 plants. Furthermore, such plants show fused rosette leaves, and leaf-stem and stem-pedicel fusions [21, 22]. Plants containing mRNAs resistant versions of CUC1 or CUC2 to MIR164 display alterations in development including increased petal numbers and reduced sepal numbers [22], increased formation of carpel margin meristems [23], alterations in location and size of boundaries [21] and enlarged vegetative and reproductive lateral organs [24].
The characteristic organ fusions of the hws-1 mutant are similar to these observed in the cuc1/cuc2 double mutant [25] and the Pro35S:164B lines [21, 22], while the Pro35:HWS line displays sepal separation [26]. We hypothesize that HWS contributes to boundary formation and regulation of organ number by indirectly altering transcripts of MIR164, CUC1 and CUC2.
HWS forms part of a SCF (Skp, Cullin, F-box containing complex) E3 ligase that specifically binds to a target substrate destined for degradation via the 26S proteasome. Although the F-box HAWAIIAN SKIRT (HWS) gene is important to control organ growth and floral organ abscission timing in Arabidopsis thaliana [26], its mechanism of action remains unknown. To identify the protein that HWS is targeting for degradation, we mutagenized hws-1 seeds with ethylmethylsulphonate (EMS) to screen for mutants suppressing hws-1 associated phenotypes.
In this study, we describe the mapping and characterisation of cuc1-1D, in which the sepal fusion phenotype of hws-1 is suppressed. cuc1-1D is mutated in the MIR164 target site of CUC1 mRNA. Our data reveal that HWS controls floral organ number by modulating transcript accumulation levels of MIR164, CUC1 and CUC2 genes. We have also shown that HWS regulates cell proliferation and mitotic growth in Arabidopsis petals.
Materials and methods
Plant material
We obtained Arabidopsis Columbia-0 seeds from the Nottingham Arabidopsis Stock Center. Single, double and triple mutants between hws-1, cuc1-1D and cuc2-1D were generated by crossing the genotypes as described in [27]. The ffo1 mutant (Landsberg background) [28] was sourced from Elliot Meyerowitz, and the cuc2-1D mutant (Columbia-0 background) [24] from John Walker. The F1 plants were self-pollinated and homozygous F2 lines were identified using PCR. All lines were grown in a growth room with temperature of 22±2°C, and photoperiod of 22h light/2h darkness supplemented with fluorescent lights at a light intensity of 200 μmol m-2s-1 (Polylox XK 58W G-E 93331). The hws-1 EMS populations were maintained in a greenhouse, temperature: 23±2°C and photoperiod: 16h light/8h darkness.
To identify the nature of the mutation in allele hws-5 (ffo1), genomic DNA from seedlings from the ffo1 mutant was extracted (Qiagen, DNAeasy Plant Mini kit) and used to amplify the genomic region from the HWS gene using the primers At3g61590ForcDNA and At3g61590rev. PCR reactions were performed using Platinum® pfx DNA polymerase (Invitrogen). The amplified band was gel purified using Genelute™ Gel extraction kit (Sigma), and sequenced using primers At3g61590ForcDNA, At3g61590Rev, SSLPHSFor, SSLPHSRev, HS 5’endutrfor and HSmap3rev.
EMS mutagenesis of hws-1 and suppressor screening
Approximately 5000 seeds from the hws-1 mutant (Columbia-0 background) were treated with EMS as described by [29] and [30]. 269 pots of 13 cm with 10–20 M1 seeds each were sown and their seeds were collected in bulked M2 families. 308 plants per population were grown individually in 1cmX1cm pots and once flowering was initiated, potential suppressors of the sepal fusion phenotype from hws-1 were identified and isolated. Mutant lines were backcrossed four times with wild-type Columbia-0. The cuc1-1D homozygous line was identified by segregating away the hws-1 mutation, by growing independent lines and by ensuring that all plants displayed extra floral organs and none were hws-1 mutants (5 generations of plants). All putative suppressors were tested by PCR to confirm that they were in the hws-1 background using the primers SSLPHSFor /SSLPHSRev; an Extract-N-Amp PCR kit (Sigma) was used to extract genomic DNA.
The F1 progeny from a cross between the shs1 (shs1/hws-1) mutant and the hws-5 (ffo1) allele was allowed to self to generate the F2 mapping population. About 500 F2 plants were grown and DNA was extracted (Sigma-Aldrich, GenElute™ Plant Genomic DNA Miniprep Kit) from those that displayed a hws-1 like phenotype. Because the mutant allele of shs1 is dominant in a hws-1 mutant background, individuals with the wild type allele of shs1 had to be selected for the map-based cloning procedure. Hence, 94 out of 500 F2 individuals with the hws-1 mutant phenotype were selected.
Mapping analysis included a combination of InDels and SSLP markers [31]. When the region was narrowed to a 0.875MB segment, candidate genes in this region were identified and a 1.565kb genomic region of the At3g15170 (CUC1) gene was sequenced.
Plasmid constructions and plant transformation
For the complementation construct, a genomic region of the CUC1 gene of 2.498kb containing 1.386kb promoter upstream of the ATG plus 5’ and 3’ untranslated regions, introns and exons and including the substitution mutation identified in the cuc1-1D allele, was amplified from shs1/hws-1 using the primers CUC1prFor and CUC1Rev. Restriction sites were added to clone the genomic region into the PBI101.2 vector.
For the silent version construct (referenced as CUC1-SV), the equivalent genomic region was amplified from wild type plants and mutagenized using the QUikChange II XL site-Directed mutagenesis kit (Agilent Technologies) to introduce a silent single point mutation C→T at 1.236kb from the ATG of CUC1. Escherichia coli XL10- Gold Ultra competent cells were transformed, positive colonies were selected by PCR, and successful mutagenesis was confirmed by sequencing. The plasmid was electroporated into Agrobacterium tumefaciens C58 strain and Arabidopsis plants were transformed using the floral dip method described by [32].
For the MIR164B modified version, genomic DNA from Columbia-0 seedlings was used to amplify a region of 1.340kb comprising the MIR164B gene, using the primers Comp164bFor and Comp164bRev. The PCR reaction was performed using Platinum® pfx DNA polymerase (Invitrogen). The obtained band was sequenced and a product containing the BamHI and SacI restriction sites was generated using the primers Comp164bForBamHI and Comp164bRevSacI. This segment was sub-cloned in a MOG402 engineered vector comprising two copies of the CaMV 35S promoter [26]. A single nucleotide change was introduced in nucleotide number 9 (from the 5’ end) of the binding site from C →T, so that the new version had a restored binding affinity to the mRNA of the cuc1-1D mutant. The plasmid was mutagenized as described previously for the CUC1-SV. Twenty-four independent transformants from each construct were analysed.
Floral organs numbers, petal cell size measurements and statistical analyses
Five flowers from six plants (n = 30) stage 15a [6] were dissected. Floral organs were analyzed and counted using a Zeiss Stemi-SV6 stereomicroscope. Photographs of flowers (10 days after anthesis) were taken using a Canon Power Shot-A620 camera and captured with Canon ZoomBrowserEX5.5.0.190. Petal size and cell number were determined as described in [33] using twenty flowers from four independent plants of each genotype grown under the same conditions.
For fluorescence-activated cell sorting (FACS) analysis, 2 young leaves, 50 flowers or 200 petals from 5 independent plants were used for each genotype. Nuclei isolation and FACS analysis were performed as described in [34] using MACSQuant VYB (Miltenyi Biotec) cytometer.
Statistical analyses were undertaken and graphics created for all measurements. Regression analyses and ANOVA using generalized linear models were performed using GenStat 15.1.0.8035. Graphs were created using Microsoft Excel 2010 and annotated in Adobe Photoshop 7.0.1.
DAPI staining
Arabidopsis flowers from Columbia-0, hws-1, cuc1-1D, hws-1/cuc1-1D were harvested in water containing 0.05% (v/v) Triton X-100 (Sigma). Nuclei were stained by adding 1μg/ml DAPI (Sigma) to the solution for 15 min at RT. Samples were washed twice with distilled water containing 0.05% (v/v) Triton X-100 (Sigma). Petals were dissected and DAPI staining was observed using Nikon Optiphot-2 microscope equipped with a Leica DFC320 camera.
RT-qPCR analyses of gene expression
Total RNA from a cluster of buds and young flowers (up to stage 12, [6]) from Columbia-0, hws-1, cuc1-1D, hws-1/cuc1-1D, cuc2-1D mutants and Pro35:HWS line was extracted using TRIzol reagent (Life Technologies). Three biological replicates and two technical replicates from each sample were used to perform the RT-qPCR analyses.
Expression analyses were determined using the SuperScript III Reverse transcriptase kit (Invitrogen). The First-strand cDNA was synthesised in a 20 μL reaction containing 2μg of total RNA, 1 μL mL-1 oligo dT (500 ng μl-1) and 2 μL 5mM dNTPs. Each sample was diluted by eight fold. Standards were prepared pooling 65 μL of each sample to dilutions 1, 1:4, 1:16, 1:64 and 1:256. Three biological and two technical replicates were used per sample (n = 3).
qPCR reactions were performed using Sensimix SYBR Hi-Rox Kit (Bioline) in a final volume of 10 μL, using a Roche Light Cycler 480 II. Transcripts were quantified using the Light Cyler 480 software version 1.5 using TUBULIN 4 (At5g44340) gene as the endogenous reference. Graphs were created in Excel using the data generated from the Light Cycler 480 software.
All primers described in material and methods are listed in S1 Table.
Accession numbers
Sequence data can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: HWS, At3g61590; CUC1, At3g15170; CUC2, AT5G53950; MIR164A, At2g47585; MIR164B, At5g01747; and MIR164C, At5g27807.
Results
The mutant shs1 in hws-1 suppresses the sepal fusion phenotype and carries a transition in the MIR164 binding domain of CUC1
To further understand the molecular mechanism of HWS in plant development, we mutagenized hws-1 seeds (Columbia background) with ethylmethylsulphonate (EMS) and screened for revertants of the hws-1 floral sepal fusion phenotype. About 5,000 M1 plants were produced and their seeds were pooled in 269 bulked M2 families; 308 individuals of each M2 population were screened for reversion of the sepal fusion phenotype. We isolated the mutant suppressor of hws-1 (shs1) in the hws-1 background that exhibited no sepal fusion, suggesting suppression of the hws-1 phenotype. The suppressor shs1/hws-1 (Columbia-0) was crossed to hws-5 (ffo1, Ler background; S1 Fig) and all the F1 progeny showed the shs1/hws-1 mutant phenotype, indicating that the shs1 mutation is dominant. Selfing this F1 revealed a 3:1 segregation of shs1/hws-1: hws-1 in F2 (data not shown) confirming that the mutation in the shs1/hws-1 line is a dominant allele in the hws-1 background (Fig 1A, 1B, 1C, 1D, 1E, 1I, 1K, 1L, 1M, 1Q, 1R, 1S, 1T and 1U). This F2 progeny was used as a mapping population.
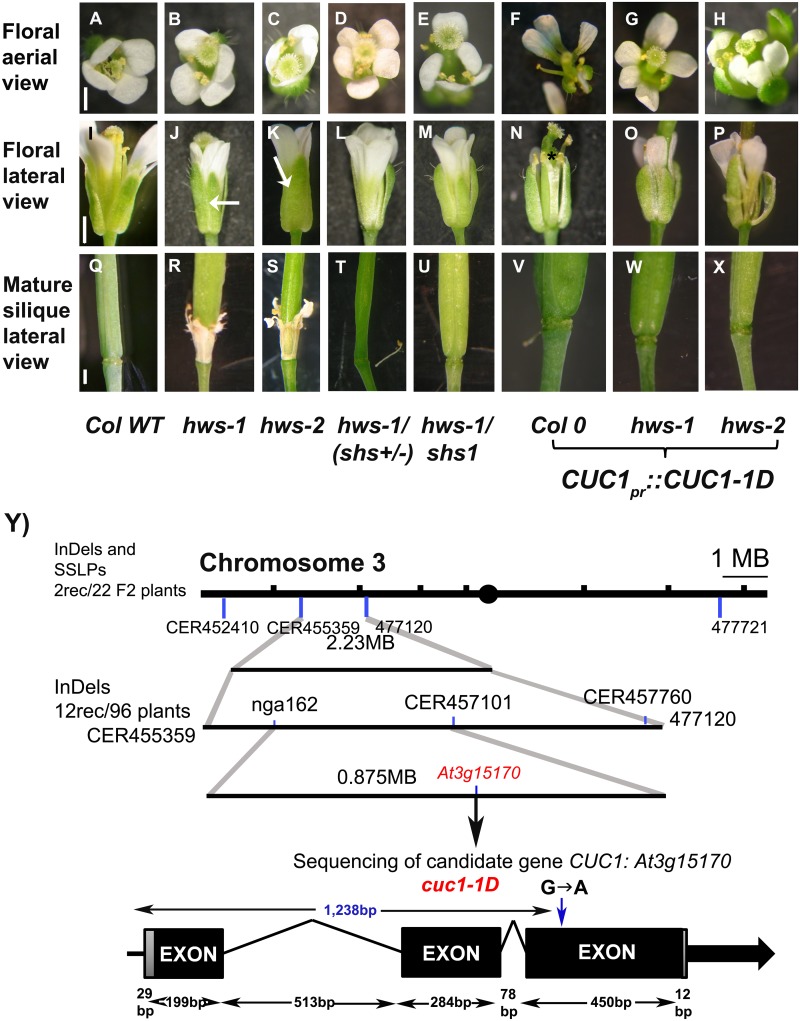
Fig 1. The shs1 mutant is an allele of CUC1.
(A-H), Aerial and (I-P), lateral views of flowers at stage 15a; and (Q-X), lateral view of mature green siliques. From: (A, I, Q), Columbia-0; (B, J, R), hws-1 (Columbia-0 background);(C, K, S), hws-2 (Ler background); (D, L, T), hws-1/shs+/-; (E, M. U), hws-1/shs1 (hws-1/cuc1-1D); and primary transformants of (F, N, V), Columbia-0; (G, O, W), hws-1; and (H, P, X), hws-2 complemented with a genomic region containing the CUC1pr::CUC1-1D gene. Scale bars: 1mm. Arrows show the sepal fusions. A petal in F and a sepal on P have been removed. * in N shows stamen fusion. (Y), Mapping strategy used to identify the cuc1-1D mutation. Structure of the gene and location of the transition substitution (G→A) 1,238bp from the ATG are included, intragenic regions are represented by thin lines and exons by black boxes.
A map-based cloning approach combining InDels and SSLP markers [31] located the shs1 mutation in a 0.875 MB region at the top of chromosome 3 (Fig 1Y). This region contains approximately 1,100 genes, including At3g15170 (CUC1). We analysed the genomic DNA region corresponding to CUC1 as this provided a strong putative candidate based on similarity of phenotypes between the double cuc1/cuc2 and the hws-1 mutants. Sequencing of this locus in the shs1/hws-1 mutant identified a transition mutation G→A, 1.238kb downstream of the ATG of CUC1. This mutation is located in the binding domain of the MIR164 target site of CUC1 [21, 25] and introduces the amino-acid substitution cysteine→ tyrosine in CUC1 (Fig 2A and 2B). Consequently, the shs1 mutant was renamed cuc1-1D. The double mutant hws-1/cuc1-1D was backcrossed with Columbia-0 to obtain a cuc1-1D single mutant for successive analyses which displayed sepal separation (Fig 3A and 3F).
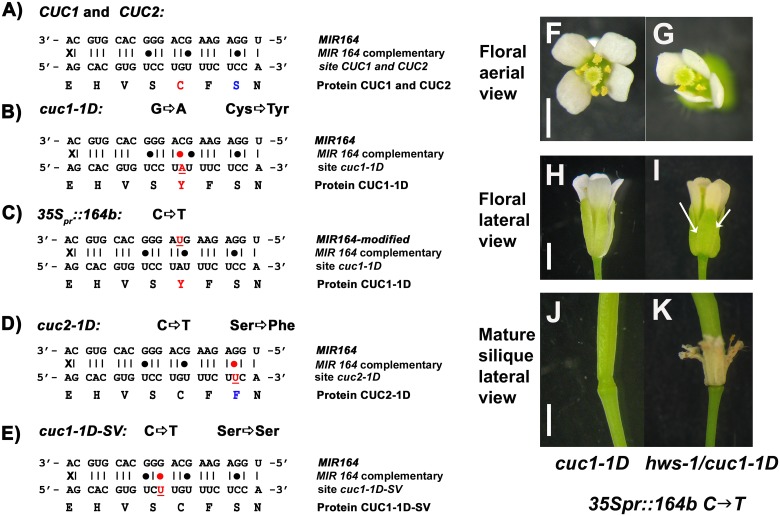
Fig 2. Mutations and constructs in CUC1, CUC2 and MIR164.
Schematic diagram of MIR164, MIR164 complementary binding sites in CUC1 and CUC2 mRNAs and CUC1, CUC2 proteins or their equivalent in generated constructs; (A), wild type (B), cuc1-1D mutation; (C), cuc1-1D mutation and MIR164 modified site introduced for complementation analyses; (D), cuc2-1D mutation (modified from [24]); (E), cuc1-1D silent version (cuc1-1D-SV). Mutations are underlined, the amino acid substitutions are identified in red/blue font, and changes in binding affinity from the MIR164 are indicated with a red dot. (F-K), Complementation analyses in primary transformants using a modified version of MIR164B; (F-G), aerial and (H-I), lateral view of flowers at stage 15a and (J-K), lateral view of mature siliques from complementation lines in cuc1-1D and hws-1/cuc1-1D backgrounds using the 35Spro::164B C→T construct, arrows show sepal fusion. Twenty-four primary independent transformants from each line were analysed. All transformants reverted or not the sepal fusion phenotype in the cuc1-1D and hws-1/cuc1-1D backgrounds respectively. Scale bars: 30 μm F-G and 1mm in H-K.
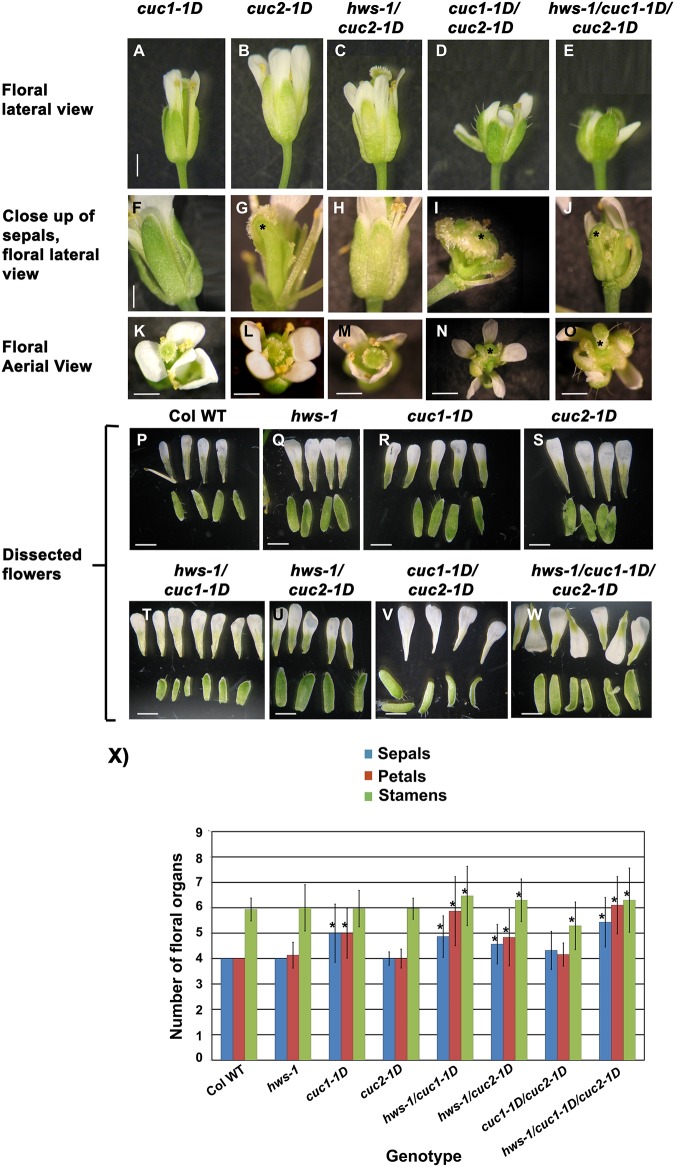
Fig 3. Floral organ number is affected in single, double and triple mutants of hws-1, cuc1-1D and cuc2-1D.
Comparative phenotypic analyses of flowers at developmental stage 15a. (A-E), lateral view of flowers; (F-J), close up of sepal separation; (K-O), aerial view at stage 15a from: (A, F, K) cuc1-1D; (B, G, L), cuc2-1D; (C, H, M), hws-1/cuc2-1D; (D, I, N), cuc1-1D/ cuc2-1D and (E, J, O), hws-1/cuc1-1D/cuc2-1D. (P-W), dissected flowers at stage 15a from: (P) Columbia-0, (Q) hws-1, (R) cuc1-1D, (S) cuc2-1D, (T) hws-1/cuc1-1D, (U) hws-1/cuc2-1D, (V) cuc1-1D/ cuc2-1D and (W) hws-1/cuc1-1D/cuc2-1D. Scale bars: 1 mm in (A-J) and 300 μm in (K-W), * show misshapen organs. (X), Five flowers from six plants of each genotype were dissected and their floral organs quantified and statistically analysed by regression analyses using generalized linear models. Stars indicate a significant difference in the mean at P≤0.05 n = 30. Bars indicate SD.
To confirm that the mutation identified in CUC1 was responsible for the shs1 phenotype, Columbia-0, hws-1 and hws-2 (T-DNA insertion; S1 Fig) plants were transformed with a 2.498kb genomic segment of the CUC1 gene containing a 1.386kb promoter region upstream of the ATG, 5’ and 3’ untranslated regions, introns and exons and included the mutation identified in the cuc1-1D allele. This segment was sufficient to suppress the sepal fusion phenotypes in hws-1 and hws-2 mutants (Fig 1O, 1P, 1W and 1X). These data support the assertion that the single point mutation in the binding domain of MIR164 is sufficient to suppress the floral fusion phenotype of hws mutants.
A mutated version of the MIR164B gene reverts the floral phenotypes in cuc1-1D and hws-1/cuc1-1D backgrounds
To investigate if the phenotypes observed in the cuc1-1D and hws-1/cuc1-1D mutants result from altering the binding affinity of MIR164 to the mRNA of CUC1, we generated a construct containing a 1.340kb genomic region of MIR164B gene expressed under the control of the CaMV 35S promoter. A point mutation was introduced to change the nucleotide C→T in the binding module, so that it matched the target in cuc1-1D exactly (Fig 2C). Transformation of either cuc1-1D or hws-1/ cuc1-1D with this modified MIR164 transgene confirmed that this point mutation was enough to restore the wild-type and hws-1 phenotypes of the cuc1-1D and hws-1/cuc1-1D mutants, respectively (Fig 2F, 2G, 2H, 2I, 2J and 2K); twenty-four independent transformants were analysed, all plants reverted to wild-type in cuc1-1D or hws-1 in hws-1/cuc1-1D mutants. These data suggest that the mutation in cuc1-1D is enough to disrupt the binding of the mature MIR164 molecule to the mRNA of CUC1 and it generates a version of CUC1 mRNA resistant to microRNA directed degradation.
Introgression of the cuc2-1D mutation into hws-1 suppresses the sepal fusion phenotype
The MIR164 binding sites are identical in CUC1 and CUC2 (Fig 2A). To investigate if mutation in the MIR164 binding site of CUC2 could also suppress the sepal fusion in hws-1, the hws-1 and cuc2-1D mutants were crossed (Figs 1B, 1J, 1R, 3B, 3G and 3L) [24]. cuc2-1D carries a transversion mutation (G→T) in the CUC2 mRNA regulatory binding domain of the MIR164 target site ([24]; Fig 2D). F2 population plants were genotyped by PCR and homozygous for hws-1 and either heterozygous (data not shown), or homozygous for the cuc2-1D mutation were identified.
Both heterozygous and homozygous cuc2-1D were able to suppress the sepal fusion phenotype in the hws-1 background; thus similar results to those observed with cuc1-1D (data shown for homozygous hws-1/cuc2-1D, Fig 3C, 3H and 3M).
These data show that a mutation in the regulatory binding domain of the MIR164 target site of the CUC2 mRNA is also able to suppress the hws fused sepal phenotype.
HWS and CUC1 regulate floral organ number in Arabidopsis
In addition to rescuing the sepal fusion phenotype of the hws-1 mutant, the new cuc1-1D allele in a hws-1 background (cuc1-1D/hws-1), displayed statistically significant increase of floral organs number when compared to Columbia-0. cuc1-1D/hws-1 exhibited 4.87±0.82 sepals, 5.87±1.36 petals and 6.47±1.17 stamens (Figs 1E, 3T and 3X). hws-1/cuc2-1D mutant plants also showed statistically significant increases in sepal, petal and stamens number of 4.57±0.77, 4.83±1.12 and 6.3±0.84, respectively, compared to the Columbia-0 (Fig 3U and 3X). Floral organ numbers were also investigated in hws-1, cuc1-1D, cuc2-1D, cuc1-1D/cuc2-1D and hws-1/cuc1-1D/cuc2-1D mutants. The hws-1 and cuc2-1D mutants showed wild-type flower organ number (Figs 1B, 3Q, 3L, 3S and 3X). The cuc1-1D mutant, in Columbia-0 background, had a statistically significant increase in sepal and petal numbers of 5±1.14 and 5±0.98, respectively, but not of stamens when compared to Columbia-0 (Fig 3K, 3R and 3X). A statistically significant decrease stamens number (5.29±0.94) was observed in cuc1-1D/cuc2-1D mutants when compared to Columbia-0 (Fig 3N, 3V and 3X). No statistically significant increase of petals and sepals was observed. In the hws-1/cuc1-1D/cuc2-1D triple mutant a statistically significant increase in the number of sepals, petals and stamens of 5.43±0.97, 6.1±1.25, and 6.3±1.26, respectively, was observed compared to the Columbia-0 (Fig 3O, 3W and 3X). We previously demonstrated that the Pro35S:HWS line displays 4 sepals, 4 petals and 6 stamens [26].
Interestingly, extra floral organs were also observed in the hws-1 and hws-2 plants complemented with the construct containing the cuc1-1D mutation, similar to shs1/hws-1 line identified in our EMS studies (Fig 1G and 1H). However, plants harboring cuc1-1D mutation in Columbia-0 background, exhibited no defects in floral organ number, but floral organ shape and size were altered (Fig 1F and 1N).
These data show that adding hws-1 to either cuc1-1D or cuc2-1D leads to increase in floral organ number. These results suggest that HWS together with CUC1 and CUC2 plays a role in regulating floral organ number in Arabidopsis.
The binding affinity of MIR164 to the CUC1 mRNA and the presence of HWS are crucial for sepal fusion rescue and for regulating floral organ number and identity
To investigate if the sepal fusion rescue and the extra floral organs phenotypes observed in the cuc1-1D/hws-1 mutant is due to altering the binding affinity of MIR164 to the mRNA of CUC1; or to the amino-acid substitution (Fig 2B) in the CUC1 protein; a silent mutation C→T was introduced 1.236kb downstream from the ATG of CUC1 where MIR164 binds. This modification does not change the amino-acid sequence, but likely weakens the binding affinity of the MIR164 (Fig 2E) to its target sequence of CUC1. If additional floral organs seen in the cuc1-1D mutant are the consequence of changing the amino-acid, it is expected that a silent mutation in the CUC1 gene would not rescue sepal fusion, nor produce extra floral organs in the hws-1 or Columbia-0 backgrounds. If the extra floral organs are the consequence of changing the binding affinity of MIR164 to the mRNA of CUC1, an increase in floral organ numbers in the hws-1 background and an equal or more extreme phenotype in the cuc1-1D and hws-1/cuc1-1D lines are expected. Plants from Columbia-0, hws-1, cuc1-1D and hws-1/cuc1-1D were transformed with this construct (CUC1-SV), twenty-four independent transformants were analysed. Results show that the silent version of CUC1 did not affect floral organ number in primary transformants in the Columbia-0 background (Fig 4A, 4B and 4C). We observed a similar effect in our complementation experiments of Columbia-0 with the cuc1-1D construct where organ number was not affected (Fig 1F, 1N and 1V), suggesting that the presence of an endogenous wild-type copy of CUC1 is enough to maintain the usual floral organ numbers. The CUC1-SV supressed the sepal fusion phenotype and increased floral organ number in the hws-1 background in a similar fashion as in the hws-1/cuc1-1D line (Fig 4D, 4E, 4F and 4M). Introducing CUC1-SV in the cuc1-1D and the hws-1/cuc1-1D backgrounds, resulted in increased floral organ number and more severe phenotypes were observed. In addition, in the cuc1-1D and hws-1/cuc1-1D lines, some petals appeared misshapen, some stamens appeared bifurcated and some organs showed sepal/petal chimera phenotype (Fig 4G, 4H, 4I, 4J, 4K and 4L). These results support the hypothesis that the rescued sepal fusion and increased number of floral organs in the hws-1/cuc1-1D line are due to the change in the binding affinity of MIR164 to the mRNA of CUC1 and not to the amino-acid substitution in the CUC1 protein.
Fig 4. A silent mutation in CUC1 does not change floral organ numbers in a Columbia-0 background but induces extra floral organs in hws-1, cuc1-1D and hws-1/cuc1-1D.
(A, D, G, J, M), Lateral view; (B, E, H, K), Aerial view; (C, F, I, L), dissected flowers of primary transformants in the following backgrounds: (A-C), Columbia-0; (D-F), hws-1; (G-I), cuc1-1D; and (J-L), hws-1/cuc1-1D, note bifurcated anther inidicated with a white star in panel L. (M), Mature siliques showing suppression of sepal fusion in hws-1: left silique originated from a hws-1 mutant, right silique originated from a primary transformant hws-1 plant transformed with CUC1-SV. Scale bars: 1mm. Black and white stars show altered floral organs.
HWS is involved in the control of cell proliferation in petals of Arabidopsis
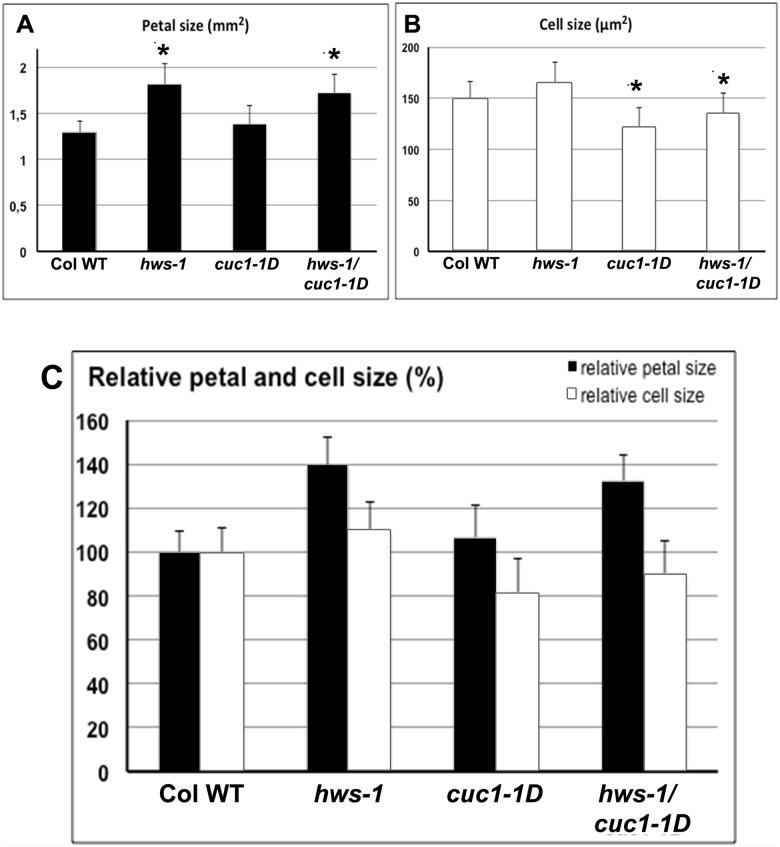
To investigate the difference in floral organ size observed between Columbia-0 and hws-1, cuc1-1D and hws-1/cuc1-1D, 80 petals from 20 flowers from each genotype were dissected to determine petal size and cell number. Compared to Columbia-0, petals of hws-1 were about 40% bigger in size (Figs 3Q and 5A). This difference was statistically significant (p<0.001). Petal cell size measurements showed no significant difference between wild-type and hws-1 (Fig 5B). Indicating that the increased size of hws-1 petals is likely to be associated with increased cell number. These data suggest a direct or an indirect negative role of HWS on cell proliferation (Fig 5C).
Fig 5. HWS affect cell proliferation in petals.
Analyses of (A), petals size (mm2) and (B), petal cell size (μm2) in Columbia-0, hws-1, cuc1-1D, and hws-1/cuc1-1D. Five flowers from four independent plants from each genotype were dissected and their size and the size of petal cells were determined. (C), Relative petal and cell sizes compared to Columbia-0 (100%). Stars indicate a significant difference in the mean at P≤0.001 n = 80.
Petals of cuc1-1D were similar to wild-type (Figs 3R, 5A and 5C). However, cell size measurement revealed that cuc1-1D cells were smaller than that of wild-type petals, suggesting that petals of cuc1-1D have more cells compared to the wild-type (Fig 5B). These data also support a role of CUC1 in cell proliferation, thus in agreement with previously reported data [35].
The double mutant hws-1/cuc1-1D exhibited a statistically significant increase in petal size (about 35%) similar to that observed in hws-1 (Fig 3T), showing that adding cuc1-1D mutation did not modify petal size. As mentioned above. Petal cell size in hws-1 was not different from wild-type, but the double mutant hws-1/cuc1-1D showed reduction in cell size similar to that observed in cuc1-1D mutant (Fig 5A, 5B and 5C). DNA content measurements showed a normal ploidy level in hws-1, cuc1-1D and hws-1/cuc1-1D leaves, flowers and petals with fractions of cells at 2N, 4N and 8N similar to Columbia-0 (S2 Fig). In situ DAPI staining in petals shows the presence of only one nucleus per cell in all cell types of hws-1, cuc1-1D and hws-1/cuc1-1D, and this was comparable to that observed in wild-type petals (S3 Fig). These data show that there is no endoreduplication in hws-1, cuc1-1D or hws-1/cuc1-1D mutants and suggest that both HWS and CUC1 likely negatively control cell proliferation and mitotic growth in Arabidopsis petals. However, more data are required to address more precisely how HWS and CUC1 cooperate to regulate cell proliferation and whether they act on the duration of cell proliferation and/or the rate of cell division.
A feedback loop mechanism between MIR164 and CUC is present in flowers
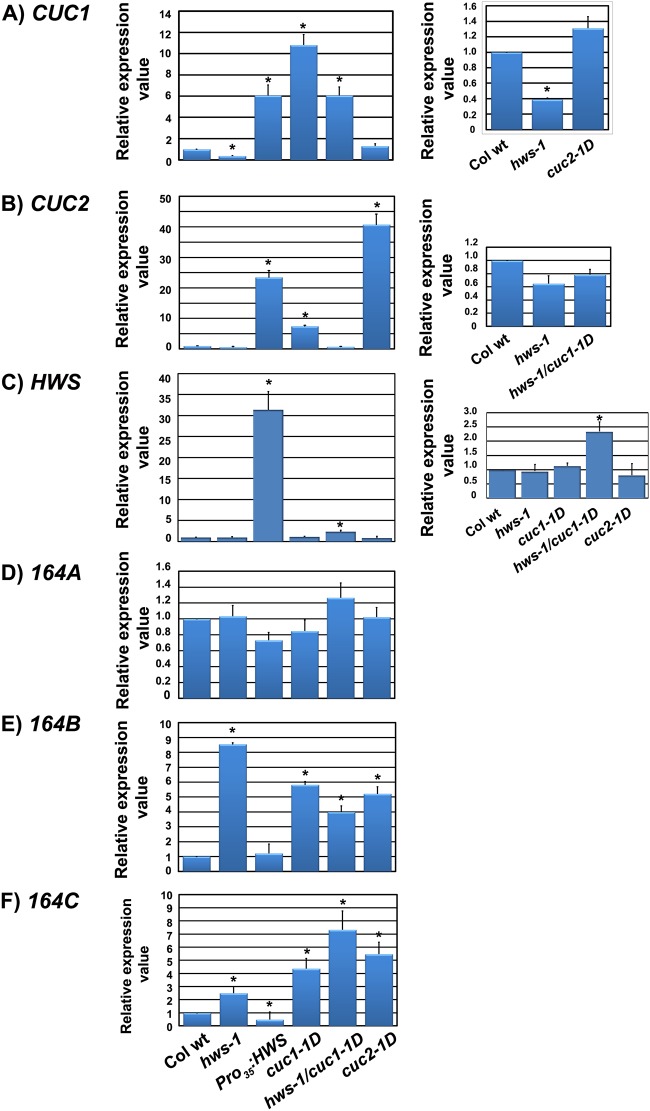
The cuc1-1D mutation introduces a substitution (G→A) in the CUC1 MIR164 target site (nucleotide 9/21 from the 5’ end; Fig 2B). It has been demonstrated that cleavage of CUC1 mRNA occurs between nucleotides pairing to residue10 of the MIR164 [36]. To investigate whether this change in the CUC1 mRNA modifies the regulatory effectiveness of MIR164, transcript levels of CUC1, CUC2, HWS1, MIR164A, MIR164B and MIR164C genes were analysed by RT-qPCR in buds and young flowers from wild-type, hws-1, Pro35S:HWS [26], cuc1-1D (in Columbia-0 background), hws-1/cuc1-1D and cuc2-1D lines.
Relative to the wild-type, a statistically significant over-accumulation of CUC1 mRNA was observed in Pro35S:HWS, hws-1/cuc1-1D (about six fold) and in cuc1-1D lines (about eleven fold). Conversely, in the hws-1 mutant line CUC1 mRNA accumulation was significantly reduced compared to the WT control (p<0.001). No statistically significant accumulation of CUC1 mRNA, compared to the wild-type, was observed in the cuc2-1D mutant (Fig 6A). CUC2 showed statistically significant higher transcript levels in Pro35S:HWS, cuc1-1D and cuc2-1D lines, with approximately twenty, seven and forty fold over-accumulation, respectively, compared to the wild-type (p<0.001). The difference in the levels of CUC2 mRNA in hws-1 and hws-1/cuc1-1D were not statistically significant compared to the control (Fig 6B). A statistically significant over-accumulation of HWS1 mRNA relative to wild-type of about 30 and 2.3 fold was observed in both, Pro35S:HWS, and hws-1/cuc1-1D lines, respectively (p<0.001). Slight differences in HWS transcript that were not statistically significant were observed in hws-1, cuc1-1D and cuc2-1D compared to the control (Fig 6C). The hws-1 mutant harbours a 28 nucleotides deletion mutation near the 3’end (S1 Fig) thus some transcript accumulation is expected in the hws-1 mutant line against a null mutant. These results suggest that the sepal fusion phenotype observed in the hws-1 mutant, which phenocopies the sepal fusion of the double mutant cuc1/cuc2 [25], is due to a reduction of CUC1 transcript. Overexpression of the HWS gene results in an increase of transcript levels of CUC1 and CUC2 genes which allows sepal separation in the Pro35:HWS line.
Fig 6. Transcript levels of CUC1 CUC2, MIR164A, MIR164B and MIR164C genes are affected in single and double mutants and in the Pro35:HWS lines.
RT-qPCR measurements of (A), CUC1; (B), CUC2; (C), HWS; (D), MIR164A; (E), MIR164B; (F), MIR164C RNA levels in Columbia-0, hws-1, 35Spro:HWS, cuc1-1D, hws-1/cuc1-1D and cuc2-1D. Stars indicate a significant difference in the mean at P≤0.001. Relative expression values represent the mean ± SD of three biological replicates and two technical replicates from each sample (n = 30).
Transcript levels of MIR164A were not significantly altered in any of the studied lines (Fig 6D). However, transcript levels of MIR164B and MIR164C were significantly increased by about 8 and 2, 6 and 4, 4 and 7 and 5 fold in the hws-1, cuc1-1D, hws-1/cuc1-1D and cuc2-1D mutants, respectively (p<0.001), while in the, Pro35S:HWS they were similar or lower than in the wild-type (Fig 6E and 6F). Our RT-PCR results in the Pro35S:HWS indicate that the target of HWS likely affects accumulation of MIR164.
In agreement with previously reported data [35], our results also show that in floral buds and flowers of our lines studied, CUC1 and CUC2 transcripts are regulated by MIR164B and MIR164C. It was reported that expression of MIR164A, MIR164B and MIR164C in inflorescences of wild type, single, double and triple MIR mutants show partial overlap [35]. In our material, we could not detect changes of expression of MIR164A, suggesting that regulation for each MIR164 gene may be different and MIR164B and MIR164C are regulated by HWS.
Mutation of the MIR164 target domain in CUC1 or CUC2 results in an over-accumulation of MIRNA164B and MIRNA164C transcripts, suggesting a feedback loop mechanism where higher levels of CUC1 or CUC2 mRNA trigger the accumulation of MIRNA164B and MIRNA164C to reduce the higher levels of CUC transcript. These results suggest that the sepal fusion phenotype observed in the hws-1 mutant, which phenocopies the sepal fusion of a MIR164B overexpressing line [21, 22], is due to an increase of MIR164B and MIR164C levels, while overexpressing the HWS gene results in a decrease of transcript levels of MIR164C. Moreover, the data indicate that the nucleotide changed in cuc1-1D is crucial for the regulation of transcript levels of the CUC1 mRNA by MIR164.
Discussion
Our work here has demonstrated that HWS contributes to the co-ordination of floral organ number and boundary formation by altering MIR164 and CUC1 transcript levels. We have shown that the regulatory domain of MIR164 is crucial for this event to take place. HWS and CUC1 also regulate cell number and size in petals, and influence cell proliferation.
We generated and characterized an EMS mutant suppressor line of the hws-1 mutant. To generate the mapping population and to identify our mutant gene, we used our discovery of a fifth mutant allele of the HWS gene in the previously identified ffo1 mutant. The ffo1 mutant was isolated from a genetic screen for modifiers of the F-box gene UNUSUAL FLORAL ORGANS (UFO) meristem activity. [28]. UFO controls meristem determination, organ primordia identity and cell propagation in the developing boundaries between floral organs [37].
cuc1-1D is a dominant mutant of CUC1 and was identified because it suppresses the hws-1 sepal fusion phenotype. We demonstrated that cuc2-1D [24] also suppresses the sepal fusion phenotype of hws-1, and both hws-1/cuc1-1D and hws-1/cuc2-1D have increased floral organ numbers. The cuc1-1D mutation has two major effects: (1) a single nucleotide change in the binding domain of MIR164 of the CUC1 mRNA transcript and (2) an amino-acid substitution in the CUC1 protein. Two hypotheses might account for how the cuc1-1D mutation leads to reversion of the sepal fusion phenotype. The amino-acid substitution in the CUC1 protein could change the conformation of the protein affecting its functionality, or alternatively, the nucleotide change could affect the binding affinity of the MIR164 to the CUC1 mRNA. Our results show that the sepal fusion rescue is a consequence of changing the binding affinity of MIR164. Moreover, it was previously reported that protein accumulation of CUC1 and CUC2 increase at boundary regions and in the center of meristems in plants containing miRNA cleavage-resistant versions of CUC1 and CUC2 translationally fused to GFP driven by their own promoters [35]. These published data taken together with our data, suggest that appropriate levels of CUC genes are necessary to maintain organ number and boundary formation in flowers. The observed phenotypes in hws-1, cuc1-1D and hws-1/cuc1-1D with either a mutated MIR164 or a silent mutation of CUC1 introgressed into them demonstrate that the efficacy in generating a MIR164 resistant allele of the CUC1 gene lies in the nucleotide change rather than the amino-acid substitution. Complementation analyses in hws-1, cuc1-1D and hws-1/cuc1-1D lines using a silent CUC1 construct not only rescued the sepal fusion phenotype of the hws-1 mutant, but phenocopied the increase of floral organ numbers observed in the original hws-1/cuc1-1D line. These findings support the hypothesis that correct binding of MIR164 to the CUC1 mRNA is important for sepal boundary formation and regulation of floral organ number. HWS is an F-box protein that targets for degradation a yet unidentified protein, so it is likely that its effect in floral organ number is indirect.
When a CUC1 silent mutant version was introduced into the cuc1-1D and hws-1/cuc1-1D backgrounds, floral organ number was increased and more severe phenotypes were observed: some misshapen petals, bifurcations in stamens, and some mosaic organs between sepals and petals in the same organ. These findings suggest that increased amounts of CUC1 protein affect the homeostasis for the correct number and development of floral organs, possibly by altering the spatial expression of B function homeotic genes or by interfering with primordia formation.
The size of cuc1-1D mutant petals is comparable to wild-type, but their cells are smaller than wild-type. It has been proposed that MIR164 limit the expansion of the boundary domain by degrading CUC1 and CUC2 mRNAs [21] and NAC genes repressing growth [38]. Though it has been demonstrated that CUC1 regulates STM and a feedback loop exists [38, 39], it is possible that CUC1 regulates the expression of other target genes yet to be identified. [40] reported that the role of CUC genes in the definition of inter-sepal boundaries is to suppress growth of sepal tissues. Our findings suggest that HWS and CUC1 regulate cell size and number in petals and perhaps within the floral organ boundaries and their interaction modulate organ size.
We have shown that transcript levels of CUC1 are down regulated in the hws-1 mutant and both CUC1 and CUC2 are up regulated in the Pro35:HWS line, supporting our hypothesis that HWS is indirectly involved in modulating organ size and boundary formation by regulating CUC1 and CUC2 genes. Interestingly, in a cuc1-1D mutant a seven-fold increase in expression of CUC2 can be observed, suggesting that in this particular mutant insufficient MIR164 is produced to maintain normal transcript levels of the CUC1 and CUC2 genes, or there can be a sequestration of MIR164 by the mutated CUC1 gene.
HWS indirectly contributes to boundary formation by regulating CUC1 and CUC2. In the hws-1 mutant, MIR164B and MIR164C levels are elevated, while levels MIR164A are unchanged. Remarkably, MIR164A is activated in leaf primordia under continuous expression of STM, a direct regulator of CUC1 and CUC2 during SAM formation and cotyledon separation [38, 39, 41], thus suggesting the existence of different mechanisms for boundary formation in leaves and flowers.
The sepal fusion phenotype observed in the hws-1 mutant, which phenocopies the sepal fusion of a MIR164B overexpression line [21, 22], is likely due to an increase of MIR164B and MIR164BC levels. Overexpressing HWS results in a decreased transcript level of MIR164, suggesting that the target of HWS play an important role in regulating MIR164, either in their production, function or degradation; indeed, our own work and that of [42] propose a role for HWS in the microRNA pathway.
Our results suggest a feedback loop mechanism where higher levels of CUC1 or CUC2 mRNA trigger the accumulation of MIR164B and MIR164C to reduce the higher levels of CUC1 transcript. We propose that HWS regulates a target that directly or indirectly modulates MIR164B and MIR164C which, in turn, negatively regulates CUC1 and CUC2 to control formation of sepal boundaries and floral organ number. Alternatively, the target for HWS might modulate CUC1 and CUC2 levels, which subsequently alter MIR164B and MIR164C levels. Our results suggest that CUC1 regulates MIR164 directly or indirectly via a feedback loop mechanism. There is a possibility that such mechanism also exists for CUC2. It is likely that these feedback loop mechanisms are disrupted in the Pro35:HWS line where the target for HWS is reduced or absent.
Supporting our results, [43] reported that the poplar HWS orthologue (PtaHWS) is part of a regulatory network involving PtaNAC1 and PtamiRNA164e, and proposed its importance for lateral root formation in response to low nitrogen conditions. [44] showed that HWS controls root meristem activity. The authors also suggested that HWS may regulate cell division in the transit amplifying cells adjacent to the quiescent centre of Arabidopsis roots.
RABBIT EARS (RBE) encodes a SUPERMAN-like zinc finger transcription factor that defines earlier sepal-petal whorl boundary and inter-sepal boundaries by repressing AGAMOUS, a keeper of spatial boundaries in Arabidopsis, and it acts in the same pathway and downstream of UFO [45]. [46] reported that RBE regulates the expression of all three MIR164 genes, and that RBE interacts with the promoter of MIR164C, a gene that has been reported to be involved in the regulation of petal number in Arabidopsis [20]. Moreover, PTL that acts upstream of RBE [47] represses growth in the boundaries between sepal primordia and the outermost whorl [48]. Loss of function of the PTL gene results in an increase in the size of the inter-sepal zone of floral buds by 35–40% due to cell proliferation but not due to changes in cell size. It has been hypothesized that the role of PTL in the boundary region is to keep its size in check [40].
Analyses of the expression of the PTL, RBE, AG and UFO genes in our mutant collection will identify whether the role of HWS in floral organogenesis and boundary formation occurs via this pathway. Protein-protein interaction studies are underway to identify if any of these genes interact with HWS.
It has been reported that CUC1 and CUC2 genes interact with SPATULA (SPT) to control carpel margin development [49] and promote the formation of carpel margin meristem during Arabidopsis gynoecium development [23]. Our studies suggest that HWS may also be a key regulator gene during fruit development as the triple mutant hws-1/cuc1-1D/cuc2-1D showed severe reduction in seed production (data not shown). These findings may be relevant to address food security issues.
In conclusion, our findings add a new layer of complexity to the gene regulatory mechanisms that are involved controlling cell growth, organ development and boundary formation in Arabidopsis flowers.
Supporting information
(A) hws-1 mutant. (B), floral fusion organs1 (ffo1) mutant in Landsberg erecta [28]. The ffo1 mutant, renamed here as hws-5, is an allele of HWS. (C), The double mutant ffo1/hws-1 exhibits fused sepal phenotype. (D), F1 progeny of the shs1/hws-1 suppressor line crossed to ffo1 (hws-5) shows a shs1/hws-1 phenotype. Side view of mature green siliques are shown. Scale bar: 1mm. (E), Structure of the HWS gene; the intragenic region in the 5’UTR is indicated as a fine line, positions of all HWS known alleles are indicated in this figure. We identified hws-2 in our previous study [26]. hws-3 and hws-4 were identified in a suppressor screen of the shortroot (shr) mutant [43]. Sequencing analyses confirmed that ffo1 (hws-5) carries a G to A mutation 630bp from the ATG resulting in a premature opal stop codon. This newly identified allele was used in the mapping analyses and positional cloning for the hws-1 suppressor mutant. Delta symbol indicates the deletion in the hws-1 allele. (F), Nucleotide and amino acid sequences of the HWS gene indicating the exact position of the five known HWS alleles; mutations are indicated in underlined capital fonts. Inverted delta indicates the site of the T-DNA insertion in the hws-2 allele.
(TIF)
Nuclei were isolated from young leaves, flowers and petals of Arabidopsis thaliana Columbia-0, hws-1, cuc1-1D, hws-1/cuc1-1D. After DAPI staining, samples were analysed by FACS. Peaks represent cells with 2N, 4N and 8N DNA content. The X axis represents DAPI intensity and the Y axis shows cell number.
(TIF)
(A), Petal of Arabidopsis thaliana. DAPI stained nuclei were observed in the top part conical cells, in the middle transition area and at the bottom part of the petal (white squares) Bar = 0.5mm. (B-E), DAPI stained nuclei in upper conical cells of Arabidopsis petal from Columbia-0, hws-1, cuc1-1D, hws-1/cuc1-1D, respectively. (F-I), DAPI stained nuclei in cells of the middle transition area of Arabidopsis petal from Columbia-0, hws-1, cuc1-1D, hws-1/cuc1-1D, respectively. (J-M), DAPI stained nuclei in cells of the bottom area of Arabidopsis petal from Columbia-0, hws-1, cuc1-1D, hws-1/cuc1-1D, respectively. Scale bar: 25μm.
(TIF)
Experiment or procedure, primer name and sequences of primers are included.
(DOC)
Acknowledgments
We thank E. Meyerowitz (California Institute of Technology) and J. Walker (University of Missouri) for providing seeds of the ffo and cuc2-1D mutants, respectively; Annemiek Smit-Tiekstra (Radboud University Nijmegen), for technical assistance with mapping; A. Lacroix & J. Berger (RDP-ENS-Lyon) for plant handling.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by funds from the University of Nottingham, Radboud University Nijmegen and the French “Agence Nationale de la Recherche” (grants ANR-09-BLAN-0006 and ANR- 13-BSV7-0014 to MB).
References
- 1.Bögre L, Magyar Z, Lopez-Juez E. New clues to organ size control in plants. Genome Biol. 2008; 9: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Powell AE, Lenhard M. Control of organ size in plants. Curr Biol. 2012; 22: R360–R467. doi: 10.1016/j.cub.2012.02.010 [DOI] [PubMed] [Google Scholar]
- 3.Hepworth J, Lenhard M. Regulation of plant lateral-organ growth by modulating cell number and size. Curr Op in Plant Biol. 2014; 17: 36–42. [DOI] [PubMed] [Google Scholar]
- 4.Gázquez A, Beemster G. What determines organ size differences between species? A meta-analysis of the cellular basis. New Phytol. 2017; 215: 299–308. doi: 10.1111/nph.14573 [DOI] [PubMed] [Google Scholar]
- 5.Ratcliffe O, Bradley D, Coen E. Separation of shoot and floral identity in Arabidopsis. Development. 1999; 126: 1109–1120. [DOI] [PubMed] [Google Scholar]
- 6.Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990; 2: 755–767. doi: 10.1105/tpc.2.8.755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowman JL, Smyth DR, Meyerowitz EM. Genes directing flower development in Arabidopsis. Plant Cell. 1989; 1: 37–52. doi: 10.1105/tpc.1.1.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyerowitz EM. Genetic control of cell division patterns in developing plants. Cell. 1997; 88: 299–308. [DOI] [PubMed] [Google Scholar]
- 9.Weigel D. The genetics of flower development: From floral induction to ovule morphogenesis. Ann Rev Genet. 1995; 29: 19–39. doi: 10.1146/annurev.ge.29.120195.000315 [DOI] [PubMed] [Google Scholar]
- 10.Breuli-Broyer S, Morle P, de Almeida-Engler J, Coustham V, Negutiu I, Trehin C. High-resolution boundary analysis during Arabidopsis thaliana flower development. Plant J. 2004; 38: 182–192. doi: 10.1111/j.1365-313X.2004.02026.x [DOI] [PubMed] [Google Scholar]
- 11.Rast MI, Simon R. The meristem-to-organ boundary: More than an extremity of anything. Curr Opin Genet Dev. 2008; 18: 287–294. doi: 10.1016/j.gde.2008.05.005 [DOI] [PubMed] [Google Scholar]
- 12.Gaillochet CH, Daum G, Lohmann JU. O Cell, Where Art Thou? The mechanisms of shoot meristem patterning. Curr Op Plant Biol. 2015; 23: 91–97. [DOI] [PubMed] [Google Scholar]
- 13.Yu H, Huang T. Molecular Mechanisms of Floral Boundary Formation in Arabidopsis. Int J Mol Sci. 2016; 17: 317 doi: 10.3390/ijms17030317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Žádníková P, Simon R. How boundaries control plant development. Curr Op Plant Biol. 2014; 17: 116–125. [DOI] [PubMed] [Google Scholar]
- 15.Ishida T, Aida M, Takada S, Tasaka M. Involvement of CUP-SHAPED COTYLEDON genes in gynoecium and ovule development in Arabidopsis thaliana. Plant Cell Physiol. 2000; 41: 60–67. [DOI] [PubMed] [Google Scholar]
- 16.Katayama N, Koi S, Kato M. Expression of SHOOT MERISTEMLESS, WUSCHEL, and ASYMMETRIC LEAVES1 homologs in the shoots of podostemaceae: Implications for the evolution of novel shoot organogenesis. Plant Cell. 2010; 22: 2131–2140. doi: 10.1105/tpc.109.073189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Souer E, Van Houwelingen A, Kloos D, Mol J, Koes R. The No Apical Meristem gene in petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell. 1996; 85: 159–170. [DOI] [PubMed] [Google Scholar]
- 18.Takada S, Hibara K, Ishida T, Tasaka M. The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development. 2001; 128: 1127–1135. [DOI] [PubMed] [Google Scholar]
- 19.Takada S, Hanano K, Kariya A, Shimizu S, Zhao Li, Matsui M, et al. CUP-SHAPED COTYLEDON1 transcription factor activates the expression of LHS4 and LSH3, two members of the ALOG gene family, in shoot organ boundary cells. Plant J. 2011; 66: 1066–1077. doi: 10.1111/j.1365-313X.2011.04571.x [DOI] [PubMed] [Google Scholar]
- 20.Baker CC, Sieber P, Wellmer F, Meyerowitz EM. The early extra petals1 mutant uncovers a role for MicroRNA miR164c in regulating petal number in Arabidopsis. Curr Biol. 2005;15: 303–315. doi: 10.1016/j.cub.2005.02.017 [DOI] [PubMed] [Google Scholar]
- 21.Laufs P, Peaucelle A, Morin H, Traas J. MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development. 2004; 131: 4311–4322. doi: 10.1242/dev.01320 [DOI] [PubMed] [Google Scholar]
- 22.Mallory AC, Dugas DV, Bartel DP, Bartel B. MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr Biol. 2004; 14: 1035–1046. doi: 10.1016/j.cub.2004.06.022 [DOI] [PubMed] [Google Scholar]
- 23.Kamiuchi Y, Yamamto K, Furutani M, Tasaka M, Aida M. The CUC1 and CUC2 genes promote carpel margin meristem formation during Arabidopsis gynoecium development. Frontiers in Plant Sci. 2014; 5: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larue CT, Wen J, Walker JC. A microRNA-transcription factor module regulates lateral organ size and patterning in Arabidopsis. Plant J. 2009; 58: 450–463. doi: 10.1111/j.1365-313X.2009.03796.x [DOI] [PubMed] [Google Scholar]
- 25.Aida M, Ishida T, Fukaki H, Fujishawa H, Tasaka M. Genes involved in organ separation in Arabidopsis: An Analysis of the cuc-shaped cotyledon Mutant. Plant Cell. 1997; 9: 841–857. doi: 10.1105/tpc.9.6.841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.González-Carranza ZH, Rompa U, Peters JL, Bhatt A, Wagstaff C, Stead AD, et al. HAWAIIAN SKIRT–an F-box gene that regulates organ fusion and growth in Arabidopsis. Plant Physiol. 2007; 144: 1370–1382. doi: 10.1104/pp.106.092288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weigel D, Glazebrook J. Genetic Analysis of mutants In Weigel D. and Glazebrook J., eds, Arabidopsis: A Laboratory Manual, Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2002. [Google Scholar]
- 28.Levin J, Fletcher J, Chen X, Meyerowitz E. A genetic screen for modifiers of UFO meristem activity identifies three novel FUSED FLORAL ORGANS genes required for early flower development in Arabidopsis. Genetics. 1998; 149: 579–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leyser HMO, Furner IJ. EMS mutagenesis of Arabidopsis In Arabidopsis: The Complete Guide, Version 1.4, Flanders D. and Dean C., eds. Norwich: UK AFRC Arabidopsis Programme, 1992. pp. 9–10. [Google Scholar]
- 30.Dinh TT, Luscher E, Li S, Liu X, Won SY, Chen X. Genetic Screens for Floral Mutants in Arabidopsis thaliana: Enhancers and Suppressors. Method in Mol Biol. 2014; 1110: 127–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peters JL, Cnudde F, Gerats T. Forward genetics and map-based cloning approaches. Trends Plant Sci. 2003; 8: 484–491. doi: 10.1016/j.tplants.2003.09.002 [DOI] [PubMed] [Google Scholar]
- 32.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium mediated transformation of Arabidopsis thaliana. Plant J. 1998; 16: 735–743. [DOI] [PubMed] [Google Scholar]
- 33.Szecsi J, Wippermann B, Bendahmane M. Genetic and phenotypic analyses of petal development in Arabidopsis. Method Mol Biol. 2014; 1110:191–202. [DOI] [PubMed] [Google Scholar]
- 34.Brioudes F, Thierry AM, Chambrier P, Mollereau B, Bendahmane M. Translationally controlled tumor protein is a conserved mitotic growth integrator in animals and plants. Proc Nat Acad Sci USA. 2010; 107: 16384–16389. doi: 10.1073/pnas.1007926107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sieber P, Wellmer F, Gheyselinck J, Riechmann JL, Meyerowitz EM. Redundancy and specialization among plant microRNAs: role of the MIR164 family in developmental robustness. Development. 2007; 134: 1051–1060. doi: 10.1242/dev.02817 [DOI] [PubMed] [Google Scholar]
- 36.Kasschau KD, Xie Z, Allen E, Llave C, Chapman EJ, Krizan KA, et al. P1/HC-Pro, a viral suppressor of RNA silencing, interfereswith Arabidopsis development and miRNA function. Dev Cell. 2003; 4: 205–217. [DOI] [PubMed] [Google Scholar]
- 37.Laufs P, Coen E, Kronenberger J, Traas J, Doonan J. Separable roles of UFO during floral development revealed by conditional restoration of gene function. Development. 2003; 130: 785–796. [DOI] [PubMed] [Google Scholar]
- 38.Aida M, Ishida T, Tasaka M. Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development.1999; 126: 1563–1570. [DOI] [PubMed] [Google Scholar]
- 39.Spinelli SV, Martin AP, Viola IL, González DH, Palatnik JF. A mechanistic link between STM and CUC1 during Arabidopsis development. Plant Phys. 2011; 156: 1894–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lampugnani ER, Kilinc A, Smyth DR. PETAL LOSS is a boundary gene that inhibits growth between developing sepals in Arabidopsis thaliana. Plant J. 2012; 71: 724–735. doi: 10.1111/j.1365-313X.2012.05023.x [DOI] [PubMed] [Google Scholar]
- 41.Landrein B, Kiss A, Sassi M, Chauvet A, Das P, Cortizo M, et al. Mechanical stress contributes to the expression of the STM homeobox gene in Arabidopsis shoot meristems. Elife 2015; 4:e07811 ; http://doi.org/10.7554/eLife.07811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lang PLM, Christie MD, Dogan E, Scwab R, Hagmann J, Van de Weyer A-L, et al. A role for the F-box protein HAWAIIAN SKIRT in plant miRNA function. BioRxiv. 2017; doi: 10.1101/123703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dash M, Yordanov YS, Georgieva T, Kumari S, Wei H, Busov V. A systems biology approach identifies new regulators of poplar root development under low nitrogen. Plant J. 2015; 84: 335–346. doi: 10.1111/tpj.13002 [DOI] [PubMed] [Google Scholar]
- 44.Kim E-S, Choe G, Sebastian J, Ryu KH, Mao L, Fei Z. et al. HAWAIIAN SKIRT regulates the quiescent center-independent meristem activity in Arabidopsis roots. Physiol Plantarum. 2016; 157: 221–233. [DOI] [PubMed] [Google Scholar]
- 45.Krizek BA, Lewis MW, Fletcher JC. RABBIT EARS is a second-whorl repressor of AGAMUS that maintains spatial boundaries in Arabidopsis flowers. Plant J. 2006; 45: 369–383. doi: 10.1111/j.1365-313X.2005.02633.x [DOI] [PubMed] [Google Scholar]
- 46.Huang T, Lopez-Giraldez F, Townsend JP, Iris VF. RBE controls microRNA 164 expression to effect floral organogenesis. Development. 2012; 139: 2161–2169. doi: 10.1242/dev.075069 [DOI] [PubMed] [Google Scholar]
- 47.Takada S, Matsumoto N, Okada K. RABBIT EARS, encoding a SUPERMAN-like zinc finger protein, regulates petal development in Arabidopsis thaliana. Development. 2004; 132: 1555–1565. [DOI] [PubMed] [Google Scholar]
- 48.Lampugnani ER, Kilinc A, Smyth DR. Auxin controls petal initiation in Arabidopsis. Development. 2013; 140:185–194 doi: 10.1242/dev.084582 [DOI] [PubMed] [Google Scholar]
- 49.Nahar M A-U, Ishida T, Smyth DR, Tasaka M, Aida M. Interactions of CUP-SHAPED COTYLEDON and SAPTULA genes control carpel margin development in Arabidopsis thaliana. Plant Cell Physiol. 2012; 53: 1134–1143. doi: 10.1093/pcp/pcs057 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) hws-1 mutant. (B), floral fusion organs1 (ffo1) mutant in Landsberg erecta [28]. The ffo1 mutant, renamed here as hws-5, is an allele of HWS. (C), The double mutant ffo1/hws-1 exhibits fused sepal phenotype. (D), F1 progeny of the shs1/hws-1 suppressor line crossed to ffo1 (hws-5) shows a shs1/hws-1 phenotype. Side view of mature green siliques are shown. Scale bar: 1mm. (E), Structure of the HWS gene; the intragenic region in the 5’UTR is indicated as a fine line, positions of all HWS known alleles are indicated in this figure. We identified hws-2 in our previous study [26]. hws-3 and hws-4 were identified in a suppressor screen of the shortroot (shr) mutant [43]. Sequencing analyses confirmed that ffo1 (hws-5) carries a G to A mutation 630bp from the ATG resulting in a premature opal stop codon. This newly identified allele was used in the mapping analyses and positional cloning for the hws-1 suppressor mutant. Delta symbol indicates the deletion in the hws-1 allele. (F), Nucleotide and amino acid sequences of the HWS gene indicating the exact position of the five known HWS alleles; mutations are indicated in underlined capital fonts. Inverted delta indicates the site of the T-DNA insertion in the hws-2 allele.
(TIF)
Nuclei were isolated from young leaves, flowers and petals of Arabidopsis thaliana Columbia-0, hws-1, cuc1-1D, hws-1/cuc1-1D. After DAPI staining, samples were analysed by FACS. Peaks represent cells with 2N, 4N and 8N DNA content. The X axis represents DAPI intensity and the Y axis shows cell number.
(TIF)
(A), Petal of Arabidopsis thaliana. DAPI stained nuclei were observed in the top part conical cells, in the middle transition area and at the bottom part of the petal (white squares) Bar = 0.5mm. (B-E), DAPI stained nuclei in upper conical cells of Arabidopsis petal from Columbia-0, hws-1, cuc1-1D, hws-1/cuc1-1D, respectively. (F-I), DAPI stained nuclei in cells of the middle transition area of Arabidopsis petal from Columbia-0, hws-1, cuc1-1D, hws-1/cuc1-1D, respectively. (J-M), DAPI stained nuclei in cells of the bottom area of Arabidopsis petal from Columbia-0, hws-1, cuc1-1D, hws-1/cuc1-1D, respectively. Scale bar: 25μm.
(TIF)
Experiment or procedure, primer name and sequences of primers are included.
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.