Abstract
The filamentous fungus Chromocrea spinulosa (Trichoderma spinulosum) exhibits both self-fertile (homothallic) and self-sterile (heterothallic) sexual reproductive behavior. Self-fertile strains produce progeny cohorts that are 50% homothallic, 50% heterothallic. Heterothallic progeny can mate only with homothallic strains, and progeny also segregate 50% homothallic, 50% heterothallic. Sequencing of the mating type (MAT) region of homothallic and heterothallic strains revealed that both carry an intact MAT1-1 locus with three MAT1-1 genes (MAT1-1-1, MAT1-1-2, MAT1-1-3), as previously described for the Sordariomycete group of filamentous fungi. Homothallic strains, however, have a second version of MAT with the MAT1-2 locus genetically linked to MAT1-1. In this version, the MAT1-1-1 open reading frame is split into a large and small fragment and the truncated ends are bordered by 115bp direct repeats (DR). The MAT1-2-1 gene and additional sequences are inserted between the repeats. To understand the mechanism whereby C. spinulosa can exhibit both homothallic and heterothallic behavior, we utilized molecular manipulation to delete one of the DRs from a homothallic strain and insert MAT1-2 into a heterothallic strain. Mating assays indicated that: i) the DRs are key to homothallic behavior, ii) looping out of MAT1-2-1 via intra-molecular homologous recombination between the DRs in self-fertile strains results in two nuclear types in an individual (one carrying both MAT1-1 and MAT1-2 and one carrying MAT1-1 only), iii) self-fertility is achieved by inter-nuclear recognition between these two nuclear types before meiosis, iv) the two types of nuclei are in unequal proportion, v) having both an intact MAT1-1-1 and MAT1-2-1 gene in a single nucleus is not sufficient for self-fertility, and vi) the large truncated MAT1-1-1 fragment is expressed. Comparisons with MAT regions of Trichoderma reesei and Trichoderma virens suggest that several crossovers between misaligned parental MAT chromosomes may have led to the MAT architecture of homothallic C. spinulosa.
Author summary
Fungi employ one of two mating tactics for sexual reproduction: self-sterile/heterothallic species can mate only with a genetically distinct partner while self-fertile/homothallic species do not require a partner. In ascomycetes, sexual reproduction is controlled by master regulators encoded by the mating-type (MAT) locus. The architecture of MAT differs in heterothallic versus homothallic species; heterothallics carry one of two forms (MAT1-1 or MAT1-2) per nucleus, whereas most homothallics carry both MAT forms in a single nucleus. There are intriguing exceptions. For example, the yeast models, Saccharomyces cerevisiae, and Schizosaccharomyces pombe undergo reversible MAT switching, not demonstrated in filamentous fungi. Here, we describe the mating mechanism in Chromocrea spinulosa (Trichoderma spinulosum), a filamentous ascomycete that exhibits both homothallic and heterothallic behavior. Self-fertile strains produce progeny cohorts that are 50% homothallic, 50% heterothallic. Self-sterile strains can mate only with homothallic strains, and when this occurs, homothallic and heterothallic progeny are also produced in a 1:1 ratio. By MAT sequencing and manipulation, we discovered unique MAT architecture and determined that self-fertility is achieved by deletion of MAT1-2 from most homothallic nuclei and subsequent inter-nuclear recognition between the resulting two, unevenly present, nuclear types in a common cytoplasm.
Introduction
Most fungi use one of two sexual reproductive strategies, i.e., heterothallism (self-sterility) or homothallism (self-fertility). A heterothallic fungus requires a genetically distinct partner to complete the sexual process, whereas a homothallic one does not require a partner [1]. In ascomycetes, sexual reproduction of both heterothallic and homothallic species is controlled by a single master regulatory locus called the mating-type (MAT) locus [1]. All heterothallic ascomycetes examined to date carry one of two MAT forms (idiomorphs [2]) per nucleus, that encode apparently unrelated, but, in fact, ancestrally related transcription factors [3]. Most homothallic species carry both MAT forms in a single nucleus [1,4,5]. There is a significant difference in the use of the term “homothallism” in filamentous fungi versus in yeasts such as Saccharomyces cerevisiae. Homothallism in the latter refers to a change in mating type/identity of some cells within a culture of a formerly uniform mating identity, followed by mating of “switched” with “unswitched” cells [6]. In S. cerevisiae, three MAT loci (one active and two silent loci containing opposite MAT genes) are linked on the same chromosome. Switching is achieved by homologous intramolecular recombination-mediated replacement of the active copy at MAT with a formerly silent copy of opposite mating type [6,7]. Methylotrophic yeasts, such as Hansenula polymorpha and Pichia pastoris have a simpler switching system, achieved by an inversion between two MAT loci (one active, one silent), mediated by inverted repeats located nearby [8,9]. This mechanism silences the formerly active MAT, thus altering mating type [10]. Although switching of mating type observed in the yeasts has not been demonstrated in filamentous ascomycetes, mating type instability has been reported for several, including Chromocrea (= Hypocrea) spinulosa (Trichoderma spinulosum) [11] (S1 Fig), Glomerella cingulata [12], various Ceratocystis species [13,14], Sclerotinia trifoliorum [15], Fusarium subglutinans [16], and Botrytinia fuckeliana [17]. Interestingly, only unidirectional switching of mating type has been observed in these cases [18]. For example, selfing of C. spinulosa and S. trifoliorum, both of which are assumed to be haploid, yields equal ratios of large and small ascospores (8:8 in C. spinulosa and 4:4 in S. trifoliorum) in a single ascus; mating ability segregates with size. Colonies derived from large spores (L) are self-fertile whereas those from small spores (S) are self-sterile; selfing of the former results in asci containing, again, large and small spores in equal ratio [11,19]. Unlike the well-described mechanism of mating-type switching characterized for e.g., S. cerevisiae, the molecular details of unidirectional switching in fungi such as C. spinulosa, Ceratocystis fimbriata, and S. trifoliorum are unclear, however deletion of MAT1-2-1 is involved in all three cases [20–23].
To explore this question in depth in C. spinulosa, we employed sequence and expression analyses of the MAT region, plus functional manipulation of genes. We present evidence supporting our hypothesis that, in order to self, homothallic strains must generate two versions of the MAT locus. One is the well-described MAT1-1 locus of Sordariomycetes encoding three genes, MAT1-1-1, MAT1-1-2 and MAT1-1-3, while the other is an altered MAT1-1 locus in which MAT1-2 is situated between truncated fragments of MAT1-1-1 (MAT1-1;MAT1-2). An 115 bp stretch of the MAT1-1-1 coding sequence is duplicated in each truncated MAT1-1-1 fragment. We argue that an intramolecular recombination between these repeats occurs pre-meiotically in chromosomes of nuclei carrying the MAT1-1;MAT1-2 locus, and results in a majority of nuclei with chromosomes carrying the typical MAT1-1 locus. We propose that recognition between these two types of nuclei occurs prior to karyogamy, after which a typical meiosis produces self-fertile (MAT1-1;MAT1-2):self-sterile (MAT1-1) progeny in a ratio of 1:1. These structural discoveries and molecular manipulation allowed us to propose a mechanism whereby homothallic strains can yield both homothallic and heterothallic progeny.
Results
Molecular architecture of MAT loci in self-fertile and self-sterile strains of C. spinulosa
TAIL and inverse PCR-based chromosome walking strategies generated 21.9-kb and 18.3-kb contiguous DNA fragments from Cs23 and Cs27 strains respectively (Fig 1, S1 and S2 Figs, S1 and S2 Tables). Both Cs23 and Cs27 carry MAT1-1 encoding the three canonical MAT1-1 genes (MAT1-1-1, MAT1-1-2, and MAT1-1-3). Cs23 also carries a second version of MAT (MAT1-1;MAT1-2) with MAT1-2 tightly linked to, and interrupting MAT1-1-1 (described in detail below).
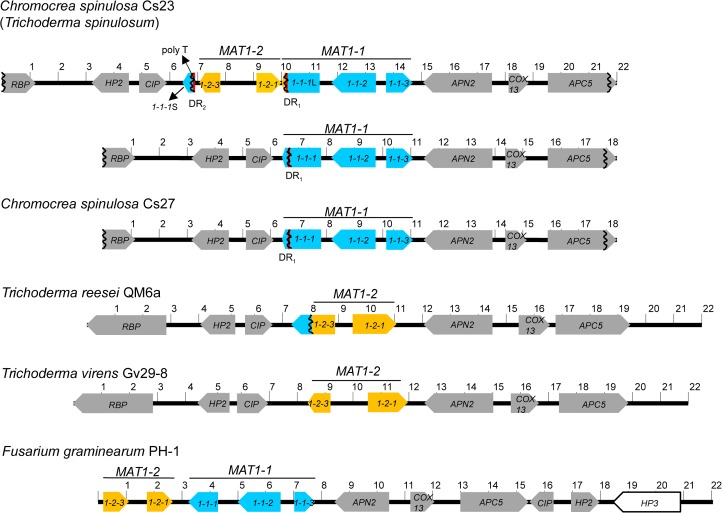
Fig 1. Structural organization of the MAT regions in homothallic and heterothallic C. spinulosa strains and comparisons with MAT in closely related heterothallic Trichoderma spp., and homothallic F. graminearum.
Homothallic Cs23 has two MAT structures, while heterothallic Cs27 has one. Open reading frames (ORF) are depicted as large arrows indicating the direction of transcription. Homologs are indicated by the same gene name and color. Direct repeats (DRs) are shown as vertical red bars. 1-1-1, 1-1-2, 1-1-3, 1-2-1, 1-2-3: MAT1-1-1, MAT1-1-2, MAT1-1-3, MAT1-2-1, MAT1-2-3, respectively. MAT1-1-1 partial ORFs are indicated by a vertical wavy line. The MAT1-1 genes (MAT1-1-1, MAT1-1-2, MAT1-1-3) and MAT1-2 genes (MAT1-2-1, MAT1-2-3) are indicated by a horizontal bar. HP: encodes a hypothetical protein; APN2: similar to DNA lyase 2; COX13: similar to cytochrome c oxidase polypeptide VIa; APC5: similar to anaphase-promoting complex subunit 5; CIP: similar to complex I intermediate-associated protein 30; RBP: similar to a protein carrying RNA-binding motif. Note MAT1-2-3 is fused to a fragment of MAT1-1-1 in T. reesii QM6a. Numbers represent kilobases.
Six additional ORFs were identified in the MAT flanks and noted to be conserved and syntenic when compared to MAT flanks of closely related heterothallic Trichoderma reesei and Trichoderma virens (Fig 1). These genes are also conserved in homothallic F. graminearum, although with some rearrangement (Fig 1, bottom line). Interestingly, the T. reesei QM6a MAT1-2 strain, which carries an intact MAT1-2-1 ORF, contains a partial MAT1-1-1 sequence (corresponding to 131 amino acids of the 3’ end) translationally fused to a hypothetical protein with similarity to F. graminearum MAT1-2-3 (28% over 55 amino acids out of 264 aa) [24] whose function is dispensable for self-fertility [25] (Fig 1). This MAT1-1 locus structure is conserved among MAT1-1 strains of Hypocrea jecorina, the teleomorphic stage of T. reesei [26]. In contrast, MAT1-2-3 is present as a standalone ORF near MAT in T. virens strain Gv29-8 (protein ID 221797 JGI MycoCosm database (http://genome.jgi.doe.gov/TriviGv29_8_2/TriviGv29_8_2.home.html). Identification of two different architectures of MAT1-2-3 in Trichoderma allowed us to speculate about MAT evolution in C. spinulosa self-fertile strain Cs23 (Discussion).
As noted above, two versions of Cs23 MAT are found; one version (MAT1-1) includes the three canonical MAT1-1 genes while the second (MAT1-1;MAT1-2) includes four MAT genes, three of which, MAT1-2-1, MAT1-1-2, and MAT1-1-3 are intact. The fourth, MAT1-1-1, is in two fragments, separated by ~3.5 kb of DNA that includes MAT1-2-1. The 5’ fragment of the MAT1-1-1 ORF (MAT1-1-1L) is the largest (1140bp/380 amino acids) and contains the entire alpha 1 domain box. The 3’ fragment (MAT1-1-1S) is much smaller (76 aa). Two identical 115-bp DNA stretches (designated DR1 and DR2) reading in the same direction were identified. DR1 is 126 bp from the 3’ end of MAT1-1-1L (Fig 2). DR2 is at the 5’ end of MAT1-1-1S. Additionally, a homopolymeric tract (poly T), 155 bp from DR2 and an ORF encoding MAT1-2-3 are between MAT1-1-1S and MAT1-2-1 (Figs 1 and 2). In contrast, in heterothallic strain Cs27, only one (DR1) of the two repeats is present at MAT1-1, MAT1-1-1 is not fragmented, and no poly T tract or MAT1-2-3 protein are present (Figs 1 and 2).
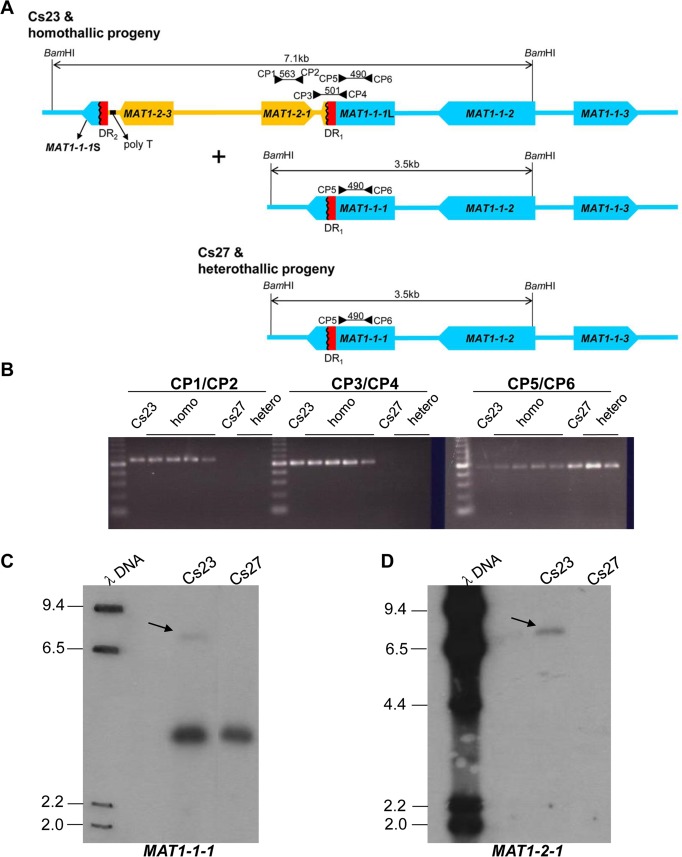
Fig 2. Structural organization of MAT in C. spinulosa homothallic and heterothallic strains.
(A). Homothallic strains carry two versions of the MAT region, as shown. In the version with four MAT genes, two identical 115-bp direct repeats (designated DR1 and DR2) are present. DR1 is at the 3’ end of the larger truncated fragment of the MAT1-1-1 ORF (MAT1-1-1L). DR2 is associated with the smaller truncated MAT1-1-1 ORF (MAT1-1-1S) that is adjacent to MAT1-2-3. A homopolymeric tract (poly T, indicated by a black bar) is found in the region between MAT1-1-1 and the MAT1-2-3 ORF. In contrast, heterothallic strain Cs27 has only one (DR1) of the two direct repeats and no poly T tract. A second version of MAT with a structure similar to Cs27 MAT is also found in Cs23. MAT1-1-1 is intact in this version. Primer positions, expected sizes of amplified fragments, and direction (arrowheads) are indicated. BamHI sites and expected fragment sizes are shown.(B). PCR amplification of MAT genes from Cs23, Cs27, and progeny of a self of Cs23 using MAT-specific primers (indicated in A). Primer pairs CP1/2 and CP3/4 amplified products from homothallic strains only. Agarose gels: leftmost lanes in each panel: 100 bp ladder, homo: homothallic progeny, hetero: heterothallic progeny. (C-D). DNA gel blot of genomic DNAs from Cs23 and Cs27 hybridized with either MAT1-1-1 (C) or MAT1-2-1 (D) probes. λ DNA: lambda DNA digested with HindIII, Cs23 and Cs27: BamHI-digested genomic DNA from C. spinulosa Cs23 and Cs27 respectively. The faint 7.1 kb band (C, arrows) was visible in DNA from Cs23 probed with either MAT1-1-1 or MAT1-2-1. Cs27 hybridized (3.5 kb) to the MAT1-1-1 probe but not the MAT1-2-1 probe. The 3.5 kb fragment is also evident in Cs23, supporting the notion that there are two MAT structures in homothallic strains. The lane Cs23 (D) was from the same gel as the other lanes, but with intervening lanes removed. Sizes (in kb) are indicated to the left of the gel.
The MAT1-1;MAT1-2 and MAT1-1 loci are found in unequal proportions
Primer sets matching MAT1-2-1 (CP1/CP2), MAT1-1-1L (CP5/CP6) and bridging DR1 (CP3/CP4) (S2 Table) amplified both MAT1-2-1 and MAT1-1-1 fragments and the DR1 region from DNA of C23 and self-fertile progeny, but only the MAT1-1-1 fragment from DNA of C27 and self-sterile progeny (Fig 2A and 2B). In addition, diagnostic MAT fragments hybridized to DNA gel blots of self-fertile Cs23 and self-sterile Cs27 strains in a manner consistent with the PCR amplification pattern (Fig 2C and 2D). In Cs23, two fragments,7. 1 and 3.5 kb, corresponding to the MAT1-1;MAT1-2 and MAT1-1 versions of MAT, respectively, were visible when probed with MAT1-1-1, while only the 3.5 kb fragment was visible in Cs27 (Fig 2A, 2C and 2D). Notably, in the Cs23 lane, the intensity of the 7.1 kb signal (arrow) was much lower than that of the 3.5 kb signal (Fig 2C). The same gel, when probed with MAT1-2-1, hybridized to the 7.1 kb band in Cs23, also with low intensity (Fig 2D, arrow). The MAT1-2-1 probe did not hybridize to Cs27. This is compelling evidence that the 7.1 kb band carries MAT1-1-1 (MAT1-1-1L) and MAT1-2-1 and that the copy number of MAT1-1;MAT1-2 is less than that of MAT1-1-1 on the 3.5 kb fragment. If the 3.5 kb MAT1-1-1 and 7.1 kb MAT1-1-1 and MAT 1-2-1 signals were from DNA on the same chromosome in the same nucleus, or if Cs23 were heterokaryotic and the two types of nuclei were in equal numbers, the signals would be expected to be approximately the same intensity. We hypothesize that these unequal signals arise from DNA at the MAT locus and that the MAT1-1;MAT1-2 locus and the MAT1-1 locus reside in different nuclei that are unequally distributed in the population. To achieve this configuration, we propose that an intramolecular recombination occurs between the direct repeats in homothallic Cs23, eliminating MAT1-2 and leaving an intact MAT1-1-1 ORF as shown in S3 Fig. Note that the 3’ end of MAT1-1-1L extends 126 nucleotides beyond DR1, and this extra sequence would be eliminated in the loop out (S3 Fig). Following the loop out, the combined MAT1-1-1 protein aligns well with other MAT1-1-1 proteins particularly those from Trichoderma species. Whether or not the complete MAT1-1-1 protein is required for homothallic function or the MAT1-1-1L fragment is sufficient is addressed below.
Does deletion of a direct repeat from the genome of Cs23 impact self-fertility?
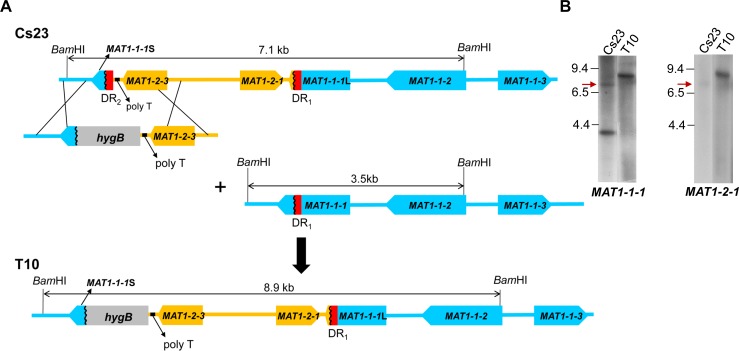
To determine if the direct repeats play a role in homothallic capability in Cs23, we deleted DR2 using the strategy shown in Fig 3A. DNA gel blot hybridization confirmed targeted replacement of the DR2 region with the hygB cassette via double crossover homologous recombination (Fig 3B). Specifically, BamHI-digested genomic DNA of candidate deletion strain T10 showed a single 8.9-kb band hybridizing to both MAT1-1-1 and MAT1-2-1 (Fig 3B, lane 2) instead of the 7.1-kb band in progenitor Cs23 (Fig 3B, lane 1, arrow), confirming that DR2 had been replaced by hygB. Remarkably, the signal in T10 DNA hybridizing to either MAT1-1-1 or MAT1-2-1 was equally strong in contrast to the faint 7.1 kb signals in DNA of Cs23 (Fig 3B, arrows). Note also that the intense MAT1-1-1 3.5 kb signal in progenitor Cs23 is missing in T10 (Fig 3B), which suggests that T10 lacks the version of MAT1-1 that is intact.
Fig 3. Targeted deletion of DR2 from Cs23.
(A). A construct carrying the selectable marker hygB plus regions of homology flanking DR2 was introduced into WT self-fertile strain Cs23. Double crossover by homology would replace DR2 with hygB, otherwise the genomic architecture of the region is unchanged. T10, a strain with this type of replacement, also lacks the second version of MAT carrying only MAT1-1.(B) Gel blot of BamHI-digested genomic DNA of Cs23 and candidate DR2-deletion transformant T10, hybridized with MAT1-1-1 or MAT1-2-1. A band at 8.9 kb is evident in T10 with either probe and is of equivalent intensity. The original BamHI-digested Cs23 MAT1-2-1 fragment (7.1 kb) shown in (A) is indicated by red arrows. The 3.5 kb band is present only in Cs23 and only when probed with MAT1-1-1. Sizes (in kb) are indicated to the left of the gel. All lanes are from the same gel; irrelevant lanes between the lanes Cs23 and T10 were cut out of the image.
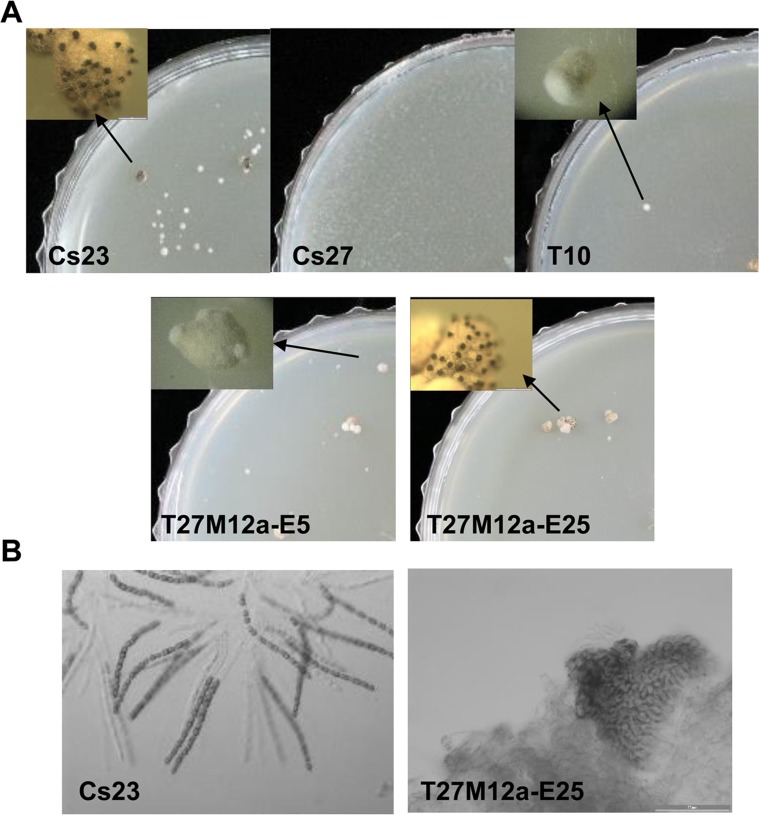
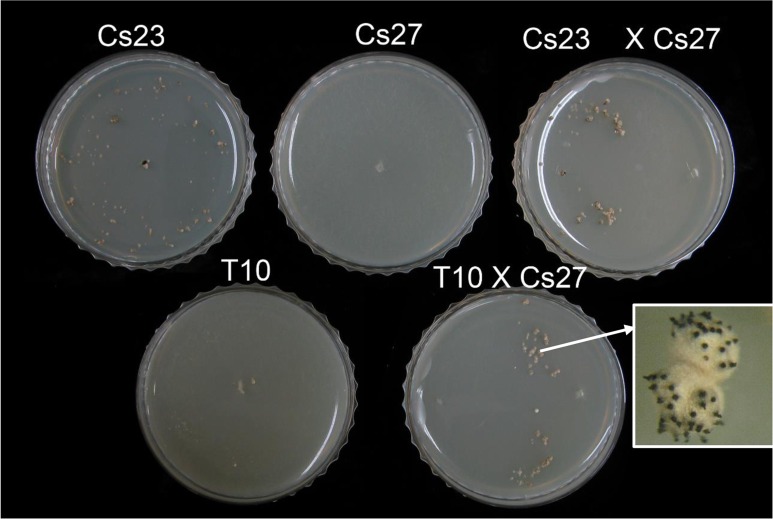
The T10 strain maintained resistance to hygromycin B through 10 successive transfers on drug-free medium, indicating that it was mitotically stable. Unexpectedly, however, and in contrast to self-fertile Cs23, T10 was self-sterile on both PDA and corn meal agar (CMA) media [11], although it did form white, rounded hyphal aggregates (Fig 4A, top, insert). The T10 aggregates did not differentiate into stroma bearing sexual fruiting bodies (perithecia) as did the control Cs23 strain (contrast Fig 4A left and right, top row).
Fig 4. Self-fertility of WT and transgenic strains.
(A). Self-fertile Cs23 forms stroma bearing perithecia (left panel inset, black dots at arrow) when grown on CMA, while self-sterile Cs27 does not. Strain T10 was able to form some stroma-like hyphal aggregates but no perithecia (top row inset, at arrow). Strains T27M12a-E5 and T27M12a-E25 formed stroma-like structures similar to those in Cs23, and, in the latter case, produced black perithecia immersed in the stroma, (but no asci/ascospores, B). (B). Self-fertile Cs23 forms stroma bearing perithecia with asci with 16 ascospores. Transgenic T27M12a-E25 forms stroma with perithecia but no ascospores.
These findings suggest that the DRs are a key feature of the molecular mechanism generating the two versions of MAT in Cs23. Our demonstration that the DNA hybridization signals are unequal supports the notion that the two versions of MAT are in different nuclei.
Can self-sterile Cs27 be made self-fertile by introduction of an intact copy of MAT1-2?
Two different Cs23 DNA fragments carrying MAT1-2 were introduced into the genome of Cs27 which carries only MAT1-1. In the first case, a plasmid (pMAT2) bearing the MAT1-2-1 ORF, truncated MAT1-1-1S/DR2/poly T/MAT1-2-3 and 1.0 and 0.5-kb of the 5’ and 3’ sequences flanking these regions, respectively, was introduced (Fig 5A). In the second case, a plasmid (pM2M1) carrying all three MAT1-1 genes (truncated MAT1-1-1L, MAT1-1-2, MAT1-1-3), as well as MAT1-2-1, was inserted (Fig 5C). Both homologous and ectopic integrants were obtained (Fig 5B and 5D). DNA gel blot analysis confirmed insertion of MAT1-2-1 into the Cs27 genome (Fig 5B and 5D). Homologous integrants generated using each strategy are represented by T27M12a-H1 (Fig 5A, Fig 5B, lane 7, asterisk, Table 1) and T27M12b-H3 (Fig 5C, Fig 5D lane 4, asterisk, Table 1), respectively.
Fig 5. Introduction of the MAT1-2 region from Cs23 into the genome of C. spinulosa Cs27.
(A and C). Schemes to introduce MAT1-2-1. Plasmids pMAT2 and pM2M1 are described in the text. T27M12a-H1 (B, lane H1, asterisk) and T27M12b-H3 (D, lane H3, asterisk), represent homologous integrants from transformation with each type of plasmid. (B). DNA gel blot of BamHI-digested DNA from control strains Cs23 and Cs27, six transgenic strains (lanes E1, E25, 1–4) carrying MAT1-2-1 at an ectopic position (ectopic), and five transgenic strains (lanes H1, 5–8) generated by homologous recombination as drawn in (A) (homologous), hybridized with a MAT1-2-1 probe. Lane E1: T27M12a-E1; lane E25: T27M12a-E25; lane 1: T27M12a-E7; lane 2: T27M12a-E5; lane 3: T27M12a-E9; lane 4: T27M12a-E18; lane H1: T27M12a-H1; lane 5: T27M12a-H3; lane 6: T27M12a-H4; lane 7: T27M12a-H12; lane 8: T27M12a-H15. All homologous recombinants showed a single hybridizing band of expected size (5.4 kb). Asterisks highlight strains specifically described in the text. (D). Gel blots of the SphI-digested genomic DNAs from the control strain Cs27 and four transgenic strains carrying MAT1-2-1, hybridized with a hygB probe. Lane 1: T27M12b-H1; lane 2: T27M12b-H4; lane H3: T27M12b-H3; lane 3: T27M12b-H7. Integration by homologous recombination as drawn in (C) should generate a single hybridizing band of 13.0 kb (e.g., lane H3). Note some lanes had more complicated patterns likely reflecting both homologous and ectopic integration events. Sizes (in kb) are indicated on the left.
Table 1. Number of outcross progeny with each phenotype.
| Outcross 1 | T10 (HR;SS) × Cs27 (SS) | |||
| Parental type | Recombinant type | |||
| HR;SS | SS | SF | HR;SF | |
| (T10 type) | (Cs27 type) | (Cs23 type) | (Cs23 type) | |
| 24 | 13 | 3 | 7 | |
| Outcross 2 | T10-P48* (HR;SS) × Cs27 (SS) | |||
| Parental type | Recombinant type | |||
| HR;SS | SS | HR;SF | ||
| (T10 type) | (Cs27 type) | (Cs23 type) | ||
| 24 | 42 | 4 | ||
| Outcross 3 | T10-P102* (HR;SS) × Cs27 (SS) | |||
| Parental type | Recombinant type | |||
| HR;SS | SS | HR;SF | ||
| (T10 type) | (Cs27 type) | (Cs23 type) | ||
| 15 | 44 | 4 | ||
| Outcross 4 | T10-P48-4** (HR;SS) × TC27G-1*** (GR;SS) | |||
| Parental type | Recombinant type | |||
| HR;SS | GR;SS | GR;SF | HR;GR;SF | |
| (T10 type) | (Cs27 type) | (Cs23 type) | (Cs23 type) | |
| 47 | 59 | 4 | 4 | |
HR: resistant to hygromycin B, GR: resistant to geneticin, SS: self-sterile, SF: self-fertile
* T10-P48 and T10-P102 are T10-type (HR;SS) progeny obtained from outcross 1.
** T10-P48-4 is a T10-type (HR;SS) progeny from outcross 2.
*** TC27G-1 is a transgenic Cs27 strain carrying the gen marker in the 3'flank of MAT1-1-3.
Unlike T10 derived by deleting DR2 from Cs23, all transgenic strains generated by insertion of MAT1-2-1 by homologous recombination into the MAT region of Cs27, were mitotically unstable. For example, hygBR strain T27M12b-H3 (Fig 5C) completely lost resistance to hygromycin B after three successive transfers on PDA without the drug (S4 Fig). In the first transfer, these strains formed brownish stroma-like structures, similar in morphology to those formed by self-fertile Cs23, but no perithecia. In subsequent transfers, stroma-like structures no longer developed, although white hyphal aggregates, similar to those formed by T10 (Fig 4A top), were produced occasionally. T27M12a-H1 (Fig 5A) showed phenotypes similar to those of the T27M12b-H3 strain. In summary, all Cs27 transformants generated by homologous recombination, were mitotically unstable for hygB and did not produce normal-looking stroma (S4 Fig). We consider it likely that these are unstable because our introduced constructs created nearby repeated regions upon homologous integration.
In contrast, all of Cs27 transgenic strains examined (Fig 5B, lanes 1–4) carrying an intact copy of MAT1-2-1 at an ectopic position (e.g. T27M12a-E5 in Fig 4A, Fig 5B, lane 1, asterisk) were mitotically stable, based on resistance to hygromycin B, and produced normal-looking stroma similar to stroma produced by self-fertile Cs23 (Fig 4A, bottom). However, none of these strains developed fertile perithecia on the stroma. One of these strains (T27M12a-E25, Fig 4, Fig 5B, lane 2, asterisk), produced black perithecia partially immersed in the stroma (Fig 4A, bottom), but no asci/ascospores (Fig 4B, bottom).
Thus, overall, all hygB stable transgenic C. spinulosa strains examined, whether from Cs23 or Cs27, were self-sterile. In the following, we focus on analyses of T10, the isogenic strain of Cs23 lacking DR2, and also on progeny from crosses in which T10 was a parent.
Mating ability of strain T10 and progeny from outcrosses to Cs27
Although strain T10 carrying both MAT1-1 and MAT1-2 was self-sterile, when out-crossed to self-sterile strain Cs27 carrying only MAT1-1, sexual progeny were produced (Fig 6). Four out-crosses were set up using the original T10 strain or T10-type progeny as one parent and Cs27 or geneticin-resistant (genR) Cs27 strain TC27G-1 as the other parent (Table 1). The first cross was between the original T10 strain and Cs27, the second and third were between T10-type progeny obtained from the first out-cross, and Cs27, and the fourth was between T10-type progeny from the second out-cross and the genR Cs27 strain TC27G-1 carrying gen in the 3’ flank of MAT1-1-3 (Table 1). As all self-sterile progeny were similar in both sexual development and colony morphology to the original parental T10 (hygBR, some hyphal aggregates) or Cs27 (hygBS, no stroma) strains, while the self-fertile progeny were identical to Cs23 (hygBS, perithecia), we designated these progeny T10-type, Cs27-type, and Cs23 type, respectively. The first out-cross produced progeny with phenotypes that were of four types. These included the parental types- hygBR and self-sterile, like T10, and hygBS and self-sterile, like Cs27, and two recombinant types- hygBR and self-fertile and hygBS and self-fertile. The fourth out-cross also produced progeny with four phenotypes. These included the parental types- hygBR and self-sterile, like T10, and genR and self-sterile, like Cs27-G (Cs27), and two recombinant types- genR and self-fertile (Cs23 type) and hygBRgenR and self-fertile (Cs23 type). The second and third crosses produced progeny with three phenotypes-hygBR and self-sterile, like T10, hygBS and self-sterile (like Cs27), hygBR and self-fertile. Note that in all four crosses between self-sterile parents a low percentage of self-fertile progeny were recovered. No self-fertile progeny would be expected from a typical cross between self-sterile heterothallic parents.
Fig 6. Outcrosses of self-sterile strain T10 to self-sterile Cs27 are fertile.
Top row: Cross plates showing selfs of control Cs23 and Cs27 strains and a cross between them. Note stroma with perithecia formed on the Cs23 and the cross plate, as in Fig 4. Bottom row: Cross plates of a self of self-sterile T10 and a cross between T10 and self-sterile Cs27. Note formation of stroma bearing fertile perithecia on the cross plate.
The ratio of hygBR to hygBS progeny varied but was approximately 2:1, 1:2, and 1:2, in crosses 1–3, respectively. Self-fertile (Cs23 type) progeny were produced at frequencies of 21%, 5.7%, and 6.3% (Table 1). In the fourth out-cross, each parental phenotype (hygBR;self-sterile, and genR;self-sterile) segregated approximately 1:1 (47:59), and recombinant phenotypes (hygBR;genR;self-fertile and genR;self-fertile) occurred with a frequency of 7%. In addition to the outcrosses described above, selfs of two hygBR Cs23 type progeny obtained from outcross 1 produced both self-sterile (belonging to either the T10- or Cs27-type) and self-fertile progeny (Cs23 type). Self-fertile progeny were recovered at a much lower frequency (4/30 = 13.3%), than in selfs of the original Cs23 strain (~50% self-fertile and 50% self-sterile) in one cross only (Table 2). In addition, overall ascospore numbers were significantly lower than from selfs of the wild-type (WT) strain (~ 10% of Cs23).
Table 2. Phenotype of progeny from a self of the Cs23 type progeny derived from outcross 1 (Table 1).
| Self 1 | P28 (HR;SF) | ||
| HR;SS (T10 type) | SS (Cs27 type) | HR;SF (Cs23 type) | |
| 17 | 9 | 4 | |
| Self 2 | P26 (HR:SF) | ||
| HR;SS (T10 type) | SS (Cs27 type) | HR;SF (Cs23 type) | |
| 5 | 7 | 0 | |
HR: resistant to hygromycin B, SS: self-sterile, SF: self-fertile
Finally, unlike the T10-derived strains, none of the Cs27-derived transformants that carried MAT1-2-1 at the MAT1-1 locus or an ectopic position in the genome was able to produce fertile perithecia in outcrosses to Cs27. The genetic event(s) responsible for the occurrence of self-fertile progeny in any of the above outcrosses remains unclear.
Molecular structure of MAT in progeny with different mating phenotypes
We analyzed the structure of MAT loci in progeny of out-crosses described above, using DNA gel blots (S5 Fig) and quantitative real-time PCR (qPCR) (Figs 7 and 8). The appearance of a single hybridizing 2.6 kb-SacI-digested band (‘b’ in S5 Fig) in the DNA of T10-type progeny, probed with MAT1-1-1 or a DR region, demonstrated that all T10-type progeny examined carried MAT1-1-1L and one DR near MAT1-1-1L as in the original T10 strain (S5A–S5C Fig). Additionally, the 6.1 kb-MAT1-2-1-hybridizing band (‘a’ in S5D Fig) showed intensities in T10-type progeny similar to those hybridizing to MAT1-1-1L (S4A and S4C Fig), supporting the genomic architecture of the MAT loci depicted in S5A Fig (top panel). Note that the 2.6 and 6.1 kb bands were not visible in progenitor Cs23 DNA, again supporting the notion that nuclei containing MAT1-2-1 are in the minority in this strain. The 3.3-kb hybridizing band (‘c’ in S5B and S5C Fig) confirmed the presence of only MAT1-1 in Cs27-type progeny, as in the original Cs27 strain. This band is also visible in Cs23 as expected.
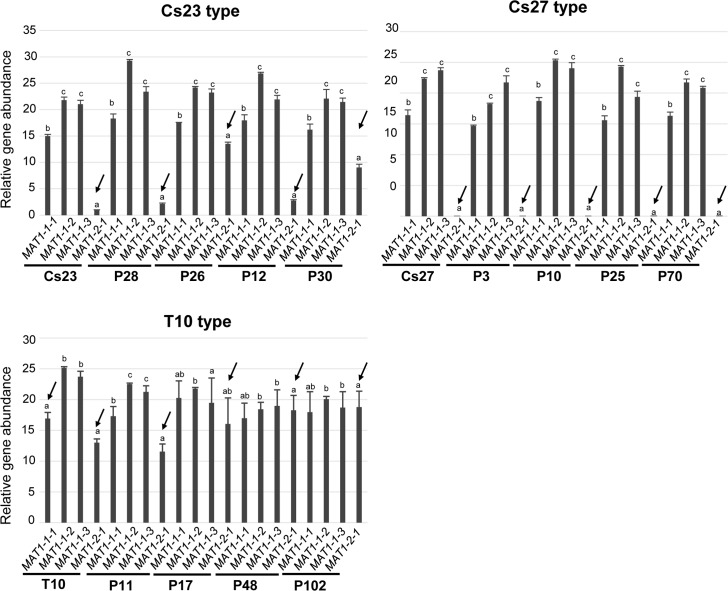
Fig 7. Relative amounts of individual MAT genes determined by qPCR using genomic DNA.
Amplification level (amount) of MAT1-2-1 is indicated by arrows, with MAT1-2-1 level in Cs23 as a reference. The same letter above bars indicates no significant difference. The three panels represent Cs23 and Cs23-type progeny, C27 and Cs27-type progeny and T10 and T10-type progeny from the T10 by Cs27 crosses. Primer pairs used: for MAT1-1-1: qCsmat1-1for7 & qCsmat1-1rev7, for MAT1-1-2: qCsmat1-2for6 & qCsmat1-2rev6, for MAT1-1-3: qCsmat1-3for4 & qCsmat1-3rev4, for MAT1-2-1: qCsmat2for4 & qCsmat2rev4.
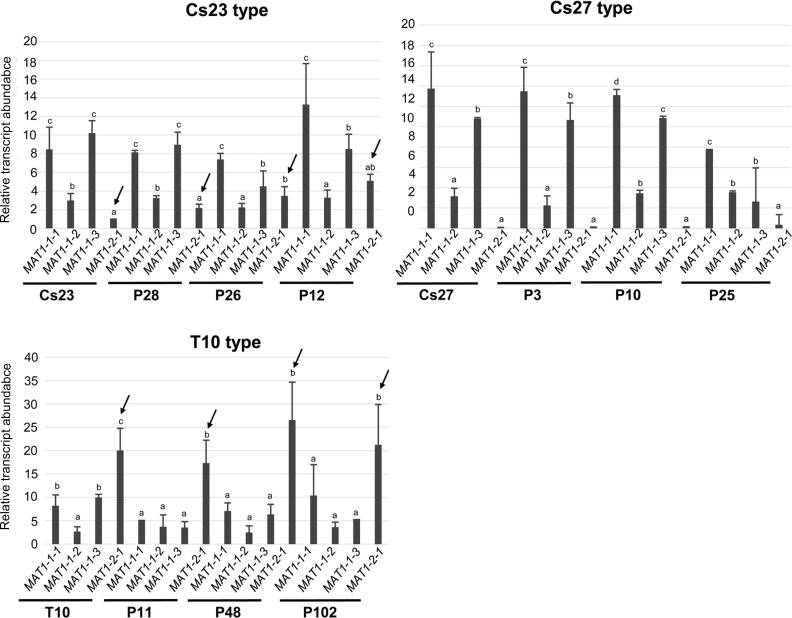
Fig 8. Relative transcript levels for individual MAT genes determined by qPCR using total RNA.
Amplification level (amount) of MAT1-2-1 is indicated by arrows, with MAT1-2-1 level in Cs23 as a reference (set to 1). The same letter above bars indicates no significant difference. The three panels represent Cs23 and Cs23-type progeny, C27 and Cs27-type progeny and T10 and T10-type progeny from the T10 by Cs27 crosses, as in Fig 7. Primer pairs used as in Fig 7.
In contrast, in some Cs23-type self-fertile progeny (P12, P26, P30, and P60), two SacI bands (3.3 and 2.6 kb) were visible (S5B and S5C Fig) demonstrating that these progeny carried both versions of MAT (S5A Fig, middle). The ratio of intensities of the two hybridizing bands varied, suggestive of unequal numbers of nuclei carrying each MAT structure, as proposed for the original Cs23 strain (Fig 2).
We performed qPCR on genomic DNA to determine copy number of individual MAT genes in the progeny that we examined by Southern hybridization (Fig 7). In Cs23 and Cs23-type progeny, the relative amounts of MAT1-1-2 and MAT1-1-3 did not differ significantly from each other in any of the strains examined. MAT1-1-1 was less abundant than these two MAT1-1 genes, but the largest fold-changes among the three MAT1-1 genes in the same strain were less than ~1.7 (e.g., Cs23 type progeny P28 and P26, Fig 7). In contrast, they were ~15- to ~25-fold-higher than the relative amount (1.0) of MAT1-2-1 in several Cs23 types (e.g., Cs23, P12 and P28). The relative amounts of MAT1-2-1 in T10 and T10-type progeny did not differ significantly (less than a two-fold change) from those of the MAT1-1 genes in the same strain. In particular, T10 and progeny P17, P48, and P102 showed MAT1-2-1 levels similar to those of the MAT1-1 genes within the same strain (e.g., ~0.8- to ~1.1-fold changes compared with MAT1-1-3). This is in marked contrast to the MAT1-2-1 levels in Cs23 type progeny which were much lower than those of the MAT1-1 genes, ranging from ~0.04- to ~0.4-fold compared with MAT1-2-1 in the same progeny. No MAT1-2-1 signal was detected in Cs27 and Cs27 type progeny and the profiles of all progeny mirrored those of Cs27 (Fig 7).
Expression of MAT genes in parents and progeny
We examined expression patterns of each MAT gene in a subset of the progeny shown in Fig 7 and S5 Fig by qPCR using total RNAs from progeny grown on PDA (Fig 8). In all strains examined, the MAT1-1-2 transcript accumulated at the lowest levels among the three MAT1-1 transcripts at the MAT1-1 locus, which is consistent with the expression pattern of the three MAT1-1 transcripts in self-fertile F. graminearum [25]. The levels of the MAT1-2-1 transcript varied among the strains examined. In Cs23 and Cs23-type progeny the signal was lower than those of the MAT1-1 genes (lower than those of all three MAT1-1 transcripts in Cs23 and P28 strains, and lower than those of the MAT1-1-1 and MAT1-1-3 transcripts in P12 and P26 strains). In contrast, MAT1-2-1 transcripts in the T10-type progeny showed the highest accumulation levels among all MAT transcripts, ranging from ~4- to ~13-fold higher than MAT1-1-2 transcripts. The expression pattern of MAT1-2-1 in the T10-type progeny was consistent with that of MAT1-2-1 in F. graminearum, where the transcript level of MAT1-2-1 was the highest of all MAT genes [25]. In contrast, no significant MAT1-2-1 expression was detected in Cs27 and Cs27-type strains.
To determine whether or not MAT1-1-1L was expressed, we used qPCR analysis of total RNA with PCR primers (S6 Fig, S2 Table) that could distinguish MAT1-1-1L from intact MAT1-1-1 (S7 Fig). These experiments revealed that the MAT1-1-1L fragment was indeed expressed in Cs23 and Cs23-type progeny (MAT1-2-1 was set to as reference) because the expression level of MAT1-1-1L was almost same as that of MAT1-2-1. In contrast, MAT1-1-1L transcript levels in T10 or T10-type progeny were much higher than those in Cs23 strains (S7A Fig).
qPCR on genomic DNA determined that copy number of MAT1-1-1L was less than MAT1-1-1. MAT1-1-1L signals from genomic DNA were not significantly different from those of MAT1-2-1 (set to 1 as reference) in Cs23 and Cs23-type progeny, whereas MAT1-1-1L signals in T10 strains were higher but also similar to those of MAT1-2-1 (S7B Fig).
Test of MAT1-1-1L protein function by heterologous expression in a MAT1-1-1 deletion strain of F. graminearum
To determine if Cs23 MAT1-1-1L is functional, we inserted it and MAT1-1-1 (as a positive control), separately, into a F. graminearum ΔMAT1-1-1 strain [25] (S8A Fig). PCR amplification analysis revealed that all candidate hygBR, genR strains, transformed with C. spinulosa MAT1-1-1 (designated FgΔMAT1-1-1::CsMAT1-1-1), MAT1-1-1L (FgΔMAT1-1-1::CsMAT1-1-1L) or F. graminearum MAT1-1-1 carried the transgenes at ectopic positions (S8A Fig). Wild-type self-fertile F. graminearum positive control strain Z3643 and all transgenic (FgΔMAT1-1-1::FgMAT1-1-1 strains, began to form protoperithecia at 3 dai (days after inoculation) and were fully fertile after 6–7 dai, containing asci with eight ascospores. The negative control strain, F. graminearum ΔMAT1-1-1 formed perithecia that were 4~5 times smaller than WT perithecia and barren even four weeks after perithecial induction (S8B and S8C Fig). Transgenic FgΔMAT1-1-1::CsMAT1-1-1 and FgΔMAT1-1-1::CsMAT1-1-1L strains also produced small, barren perithecia, but these were at least two times bigger than those of the negative control, and were not significantly different from each other (S8B and S8C Fig). Although they could not completely complement function, both C. spinulosa MAT1-1-1L and MAT1-1-1 performed similarly, thus we conclude that both are able of promoting perithecial development in F. graminearum and that MAT1-1-1L is functional.
Staining of ascospore nuclei with DAPI
We attempted to determine whether nuclei in large versus small spores could be distinguished using DAPI staining. Ascospores generated by selfing Cs23 revealed that only immature asci in which ascospores could not be seen clearly or were just starting to be visible were stained well; mature tetrads (carrying 16 ascospores of two sizes) were barely stained (S9A and S9B Fig). Immature asci carrying nuclei in various stages of division (ranging from nuclei in the diploid zygote to those in complete tetrads) were distinguishable (S9A andS9B Fig). The presence of several types of ascospore arrangement within an ascus (8L:8S, 4L:8S:4L, 4S:8L:4S, 4L:4S:4L:4S; large = L, small = S) indicates that second, as well as first meiotic division segregations occurred frequently. At least 2 to 3 nuclei were present per single ascospore, though the exact number was unclear (S10 Fig). As they matured, nuclei in small ascospores were less likely to be stained than in large (S10 C, D, leftmost ascus), which may suggest that small ascospores mature earlier than large ones. Nevertheless, no significant difference between large and small ascospores within as ascus (number/or intensity of nuclei) was found.
Asci from an outcross between T10 and Cs27 were not significantly different from those from the Cs23 self in terms of spore number and size segregation (S9C and S9D Fig). However, the frequency of asci segregating for size in the first meiotic division was different between the Cs23 self (22.9%, 8 out of 35 asci) and the outcross (60.5%, 17 out of 25 asci).
Discussion
As described above, WT self-fertile strain Cs23 (Figs 1 and 2, S1 Fig) carries two versions of MAT, one with MAT1-2 closely linked to MAT1-1 (MAT1-1;MAT1-2), the other with only MAT1-1. The architecture of the former is similar to MAT structure in several other homothallic Sordariomycetes, including F. graminearum, in that it includes genes encoded at both MAT1-1 and MAT1-2 counterparts of heterothallic species [27]. Cs23 MAT1-1;MAT1-2, however, has several noteworthy structural variations. Direct repeats are found 5’ and 3’ of MAT1-2-1, the MAT1-1-1 ORF is interrupted by MAT1-2-1 and both fragments of MAT1-1-1 are bordered by a repeat. In contrast, the WT self-sterile strain Cs27 (S10 Fig) carries only MAT1-1 with three complete genes (Figs 2 and 9). As noted above, this form of MAT is also found in homothallic Cs23.
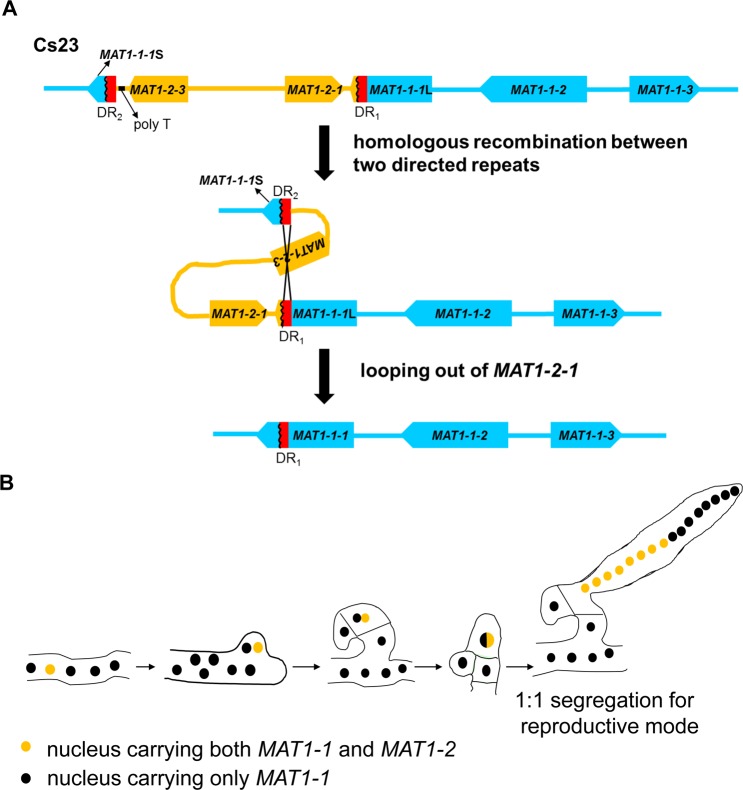
Fig 9. Proposed model for elimination of MAT1-2 and acquisition of concomitant homothallic capability involving recognition between two types of nuclei in the same individual.
(A). Looping out of the MAT1-2 region from self-fertile C. spinulosa strain 23 by homologous recombination between the 115-bp direct repeats. (B). Model for C. spinulosa ascus development. The two types of nuclei in the ascogenous hyphae are present in unequal proportions, with nuclei carrying MAT1-1;MAT1-2 in the minority. A nucleus carrying MAT1-1;MAT1-2 (yellow) recognizes and pairs with a nucleus carrying MAT1-1 only (black) and migrates into the developing crozier. The nuclear pair are partitioned in the penultimate cell of the crozier where they undergo karyogamy, meiosis, and ascus development. The diploid nucleus undergoes meiosis followed by two rounds of mitosis, yielding 16 ascospores segregating 1:1 ratio for self-fertility and large size versus self-sterility and small size in a single ascus.
Development of several different types of MAT-manipulated strain provided insight into mechanisms of homothallism and unidirectional mating-type alteration in C. spinulosa. One type of transgenic strain, derived from Cs23, carried the MAT1-1;MAT1-2 organization but lacked one of the DRs. The poly T tract and MAT1-2-3 were intact. These manipulations provide robust evidence that the DRs, but not the poly T tract, or the MAT1-2-3 protein are key to homothallic behavior. Transgenic strains generated from Cs27, carried a copy of MAT1-2-1 introduced adjacent to the MAT1-1 locus by homologous recombination or integrated at an ectopic position. These strains have the two repeats and an intact MAT1-2 and MAT1-1. One version typified by homologous integrant T27M12a-H1 (Fig 5) also carried the poly T tract and MAT1-2-3, while the other version, typified by homologous integrant T27M12b-H3 lacked poly T and MAT1-2-3 and had a duplication of most of the MAT1-1 region (Fig 5). We consider it likely that the mitotic instability we detected for both types of transgenic strain reflects an inherent instability trait of the MAT region that is tied to the mechanism of homothallism. Naturally occurring Cs23 is mitotically unstable due to naturally occurring repeated regions, while our engineered Cs27 strains are mitotically unstable because we introduced repeated regions at MAT. This notion is supported by the fact that Cs27 transgenic strains carrying an ectopic copy of MAT1-2 were mitotically stable. Mating assays with these strains, along with qPCR of MAT DNA and RNA, provided data crucial for addressing the following questions.
Are the direct repeats associated with unidirectional mating-type alteration in C. spinulosa?
The presence of 115-bp direct repeats bordering the MAT1-2 locus in Cs23 suggested a possible mechanism for elimination of MAT1-2-1, based on homologous recombination between the DRs, as pointed out by us in preliminary work [22]. Such an event would loop out MAT1-2-1, leaving only the MAT1-1 locus, now with an intact MAT1-1-1 ORF. Importantly, this would result in two types of nuclei in a common cytoplasm, one type carrying MAT1-1;MAT1-2 and the other type carrying MAT1-1 only, as is actually found in homothallic Cs23 (Fig 9).
The weak intensity of the band hybridizing to MAT1-2-1 compared with the band hybridizing to MAT1-1-1 on DNA gel blots of Cs23 (Fig 2) supports a model in which a minority of nuclei carry both MAT versions during mitotic growth (Fig 9B, S10 Fig). That the DRs are important in elimination of MAT1-2-1 and subsequent unevenness of nuclei carrying this MAT gene was confirmed by comparing DNA blots of WT Cs23 to those of transgenic strains derived from it (T10 and T10-type progeny) that lack DR1 and are self-sterile. Unlike Cs23, which shows two hybridizing bands when hybridized with MAT1-1-1, one of which also hybridizes to MAT1-2-1, T10 strains showed only one, and the same, band, when hybridized separately with MAT1-1-1 and MAT1-2-1 (Fig 3). Furthermore, in contrast to WT Cs23, the intensities of bands hybridizing to MAT1-1-1 or MAT1-2-1 in T10 were similar (Fig 3, S4 Fig). This indicates that the MAT1-1;MAT1-2 organization in T10 was stably maintained during vegetative growth and sexual development. T10 strains were no longer self-fertile, which bolsters our hypothesis that removal of one repeat from Cs23 would prevent looping out of MAT1-2-1, and abolish capability for unidirectional mating-type alteration and self-mating.
Although MAT1-1-1 at the MAT1-1;MAT1-2 locus is split to a large (MAT1-1-1L) and small (MAT1-1-1S) fragment, this may not lead to functional disruption. MAT1-1-1L, which consists of ~83% of MAT1-1-1 including the alpha-box (S3 Fig), is expressed (S7 Fig) and capable of promoting perithecium formation as well as the intact CsMAT1-1-1 in a MAT1-1-1-deletion strain of F. graminearum (S8 Fig). Therefore, it is likely that Cs23 carries two different types of nuclei, one expressing three functional MAT1-1 genes (including MAT1-1-1L) and MAT1-2-1, and the other expressing only MAT1-1 genes (including MAT1-1-1), and that karyogamy between these two different nuclei is necessary for sexual development (see details below).
To date, the involvement of DNA rearrangement in mating-type switching has been elucidated in depth for only a few yeast species that show bi-directional switching. S. cerevisiae, which carries three MAT loci (an active MAT locus and two silent MAT loci) on the same chromosome, can switch mating type through a mitotic recombination-dependent gene conversion between the silent and active copies [7]. The heavily studied yeast model, S. pombe operates similarly [28]. Beyond the Saccharomycetaceae with three MAT loci [29,30], methylotropic yeasts such as H. polymorpha and P. pastoris, which carry two linked MAT loci, undergo mating-type inversion mediated by inverted repeats [8,9], that silences one of the two MAT loci and thus changes mating type [10]. Our in depth probing of MAT alteration with the filamentous Sordariomycete C. spinulosa supports our earlier hypotheses [22] and has parallels with recently described features of mating behavior of the Leotiomycete, S. trifoliorum [21] and the Sordariomycete, C. fimbriata [20] Although the gene complement at MAT in homothallic strains of these species varies, both carry MAT1-1 and MAT1-2, and have structural features (two direct repeats flanking MAT1-2) that testify to a mechanism by which the MAT1-2 region is eliminated via homologous recombination between repeats (Fig 9A). S3 Table summarizes the similarities and differences among C. spinulosa, S. trifoliorum, and C. fimbriata MAT features. To our knowledge no functional analyses by gene manipulation have been reported for S. trifoliorum or C. fimbriata. Through DR and MAT-manipulation, crossing experiments with transgenic strains, and examination of MAT gene copy number and expression levels in WT strains and progeny, we provide experimental evidence for the looping mechanism and demonstrate that the MAT1-1;MAT1-2 region with associated DRs is unstable during vegetative growth. This feature is central to the mechanism of homothallism. Crucially, this process also leaves two types of nuclei, in unequal numbers, in a common cytoplasm.
Is maintenance of opposite mating-type loci in a single nucleus sufficient for homothallism in C. spinulosa?
An unexpected result of this study was that transgenic C. spinulosa strains carrying both MAT1-1 and MAT1-2 loci in a single nucleus were self-sterile. Transgenic T10 and T10-type progeny, with MAT1-1;MAT1-2 architecture identical to that of Cs23 except for the absence of DR1 (replaced with hygB) were unable even to produce stroma, although all four MAT genes were transcribed to levels similar to levels in self-fertile Cs23 on PDA medium. Transgenic Cs27 strains carrying an intact MAT1-2-1 gene at an ectopic position produced normal-looking stroma with occasional perithecia, however all were barren. These results strongly suggest that presence of opposite forms of MAT in a single nucleus is not sufficient for self-fertility in C. spinulosa, even though this appears to be sufficient for many fungi that self successfully [22,27,31,32]. Supporting our findings are several reports that heterothallic species carrying both the native MAT locus and an introduced MAT locus of opposite mating type are not fertile [33–36].
What is the molecular mechanism for homothallism during unidirectional mating-type switching in C. spinulosa?
Based on the unequal intensity of MAT1-1-1- and MAT1-2-1-hybridizing bands on DNA gel blots of Cs23, we suggest that Cs23 hyphae carry two different types of nuclei in unequal numbers. One type contains the MAT1-1;MAT1-2 structure and the other, MAT1-1 only (Fig 9, S10 Fig). We propose that this unusual MAT organization is achieved by deletion of MAT1-2-1 from a majority of the nuclei within a common cytoplasm during mitotic growth, via a DR-mediated looping out mechanism that reconstructs the full MAT1-1-1 ORF and leaves the MAT1-1 locus only. Because the MAT1-1-1L fragment is expressed and appears to be as functional as the intact MAT1-1-1 ORF in a MAT1-1-1-deletion strain of F. graminearum, we hypothesize that this loop out step is required for subsequent karyogamy. Given that T10-type strains, constructed in the Cs23 genetic background, but carrying only one version of MAT with both MAT1-1 and MAT1-2, were completely self-sterile, we propose that karyogamy in WT Cs23 occurs only between the two types of nuclei during sexual development (Fig 9B). Subsequent meiosis would result in production of progeny segregating 1:1 for parental type nuclei (Fig 9B). Specifically, 50% of these progeny would carry both MAT loci and be self-fertile and large [11], while the other 50% would carry only MAT1-1 and be self-sterile and small. When self-fertile progeny are selfed, the same genetic event (i.e., the loss of MAT1-2-1 during mitotic growth) is repeated.
It remains unclear, and a subject of further study, how the two types of nuclei recognize each other. Do they behave as functionally heterothallic, perhaps by a mechanism that allows for activation of MAT1-2-1 expression and repression of expression of the three MAT1-1 genes at a critical stage in nuclei with MAT1-1;MAT1-2? Differential expression and epigenetic inactivation of MAT have been hypothesized [1,2,37]. Also unknown are the particulars of the molecular mechanism underlying the DR-mediated homologous recombination in C. spinulosa. MAT1-2-1 deletion occurs at high frequency in Cs23 during mitotic growth (most nuclei do not carry it, Fig 9, S10 Fig). Given that DR1 and DR2 are relatively short (115-bp) compared with in the size of inverted repeats (> 2 kb) in methylotropic yeasts [8,9], it is likely that additional genetic element(s) (e.g., site-specific recombinases or transposases) are required for enhancing this recombination event.
In addition, it is difficult at this point, to explain the low frequency of self-fertile progeny in out-crosses between T10 (or T10-type progeny) and Cs27 (Table 1). When selfed (Table 2), one of these self-fertile progeny (P28) produced both self-fertile and self-sterile progeny, but the other (P26) did not. One clue might be that the copy number of MAT1-2-1 in P28 matched that of Cs23, while in P26, MAT1-2-1 copy number was higher (Table 2, Fig 7). Note, however, that the ratio of self-sterile to self-fertile progeny in the P28 self was not 1:1 as in self-fertile WT control Cs23 (Table 2). The connection between ascospore size and MAT genotype remains a mystery.
How does the MAT chromosome evolve in C. spinulosa?
Although the molecular mechanism underpinning the evolutionary origin of C. spinulosa DRs located at the Cs23 MAT locus remains unclear, the availability of MAT sequences of several closely related Trichoderma species allows us to propose a mechanism for evolution of C. spinulosa MAT. Our starting point is the MAT1-1-1/MAT1-2-3 fusion adjacent to MAT1-2 in T. reesei strain QM6a (Fig 1). This strain is capable of mating with H. jercorina MAT1-1 field isolate CBS999.97 [26]. We speculate that the fused ORF resulted from a misalignment of parental MAT chromosomes (i.e., between regions at the 3’ end of MAT1-2-3 and within MAT1-1-1) during meiosis (S11A Fig). Indeed, a putative recombination site can be identified for this event (S11B Fig).
To achieve the C. spinulosa Cs23 MAT1-1;MAT1-2 structure, we need to postulate that at least three different crossover events occurred in crosses between the putative ancestors of C. spinulosa (S12 Fig). In the first (S12A Fig), an unequal crossover would occur as described above, resulting in a MAT1-2 progeny carrying a fused MAT1-1-1/MAT1-2-3 gene (S12A Fig, asterisk) and a MAT1-1-1 progeny, as described in S11A Fig for Trichoderma. In this cross, we assume that DR1 is included in the partial MAT1-1-1 sequence. In the second crossover (S12B Fig), we postulate a similar event on the other side of DR1, resulting in a MAT1-1 progeny (S12B Fig, asterisk). The third crossover would be between progeny of the first and second events (S12A and S12B Fig, asterisks) and would yield a progeny carrying the MAT1-1;MAT1-2 structure present in C. spinulosa Cs23 (S12C Fig). Considering the high frequency of recombination (in the form of gene conversion) within the unusually large MAT locus (>100 kb) in the human pathogenic fungus Cryptococcus neoformans [38,39], this inference warrants further investigation.
Conclusions
Our data clearly support a mechanism in which a DR-mediated looping out of MAT1-2-1 occurs during mitotic growth (i.e., premeiotic) resulting in a strain with two different types of nuclei (genotypes MAT1-1;MAT1-2 and MAT1-1) within a common cytoplasm in self-fertile C. spinulosa. Subsequently, in sexual reproduction, recognition and karyogamy occur between these two types of nuclei, and meiotic progeny are produced that segregate 1:1 for parental nucleus type [11]. Those with MAT1-1;MAT1-2 are self-fertile and large, while those with MAT1-1 only are self-sterile and smaller. We argue, therefore, that C. spinulosa is not primarily homothallic as are true homothallic species (e.g., F. graminearum), but employs a heterothallic mating strategy at the level of the nucleus (not cell), as previously suggested [19]. The MAT1-2 deletion and imbalance of nuclear types are key to unidirectional mating-mode alteration. This hypothesis can also be applied to explain unidirectional mating-type alteration in C. fimbriata and S. trifoliorum [20,21]. Our study is the first to confirm, by MAT manipulation, that a chromosomal looping out-based mechanism underpins irreversible unidirectional mating-type alteration in filamentous fungi. Additionally, our results expand the repertoire of molecular mechanisms underlying homothallism and evolution of MAT loci in fungi.
Methods
Fungal strains, culture conditions, and staining of nuclei
C. spinulosa strains Cs23 and Cs27, which are self-fertile and self-sterile respectively, were kindly provided by Dr. John R. S. Fincham (S1 Fig) in 1997 and were used as the WT strains in this study. Transgenic strains derived from Cs23 and Cs27 mentioned in this study are listed in S1 Table. The WT and transgenic strains were maintained on potato dextrose agar (PDA; Difco Laboratories, Detroit, MI, USA) and stored in 20% glycerol at –70°C. Sexual development was induced on cornmeal agar (CMA, Difco) medium, as described previously [40]. Because of mitotic instability of Cs27-derived transformants carrying MAT1-2-1 at the MAT1-1 locus, ability to self was assayed with mycelia taken directly from transformation plates. For genomic DNA extraction, each strain was grown in 50 mL PD broth at 25°C for 72 h on a rotary shaker (150 rpm).The self-fertile F. graminearum WT strain Z3643 and its self-sterile transgenic MAT1-1-1-deletion strain [25], were maintained on PDA. Sexual development was induced on carrot agar, as previously described [25,41]. Fungal nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen, Carlsbad, CA) and examined as previously described [42].
Nucleic acid manipulations, primers, and qPCR
To isolate genomic DNA, fungal strains grown in PD broth medium for 4 days at 25°C were harvested and lyophilized, as described previously [43]. DNA gel blots were hybridized with biotinylated DNA probes prepared using the BioPrime DNA labeling system (Invitrogen, Carlsbad, CA, USA) and developed using the BrightStar BioDetect Kit (Ambion, Austin, TX, USA). Other general procedures for nucleic acid manipulation were performed as described previously [44]. Total RNA was extracted using the Easy-Spin Total RNA Extraction Kit (iNtRON Biotechnology, Seongnam, Korea) and first-strand cDNA was synthesized from total RNA using ReverTra Ace qPCR RT Master mix (Toyobo, Osaka, Japan). All PCR primers (S2 Table) used in this study were synthesized by the Bioneer Corporation (Chungwon, Korea), diluted to 100 μM in sterilized water, and stored at −20°C. Quantitative real-time PCR (qPCR) was performed with SYBR Green Super Mix (Bio-Rad, Hercules, CA, USA) using first-strand cDNA synthesized from total RNA or genomic DNA from strains grown on PDA for 5 days. Amplification efficiencies of all genes were determined as described previously [45]. Gene expression was measured in three biological replicates from each time point. Statistical analysis was performed by ANOVA by Duncan's multiple range test. The C. spinulosa EF1A gene was used as endogenous control for data normalization [45].
Construction of transforming DNA or plasmids
The DNA construct for deletion of DR1 from strain Cs23 was created using the double-joint (DJ) PCR procedure, as described previously [46]. To delete the DR1 region, the 5′- and 3′-flanking regions of the DR1 sequence (Fig 3) were amplified using the primer pairs CoHo5F/CoHo5RT and CoHo3FT/CoHo3R, respectively, and these were fused to a hygromycin B resistance gene cassette (hygB) amplified from pBCATPH [47] using primers hygB-For and hygB-Rev. The resulting PCR products were used as template for the final PCR to generate the gene deletion, using the primers CoHo5N/pUH-BC/H3 and CoHo3N/pUH-H2. Similarly, the DNA constructs for the generation of the transgenic strain TC27G-1 (S1 Table) were created as described above. Two DNA regions at the 3’ end of MAT1-1-3 corresponding to nucleotide positions 11,702 in the Cs27 MAT and 15,291 in the Cs23 MAT locus, respectively, which start at the 3’ end of MAT1-1-3, were amplified by primer sets CoHoG5F/CoHoG5RT and CoHoG3FT/CoHoG3R, respectively, and fused to the geneticin resistance gene (gen) cassette using the primers CoHoG5N/GenForN and CoHoG3N/GenRenN.
For insertion of MAT1-2 into Cs27, two different plasmid DNAs (pMAT2 and pM2M1, Fig 5), both carrying MAT1-2-1, were constructed. For pMAT2, the 5.3 kb-region carrying the entire MAT1-2-1 ORF and its 5’ flank (1.0 kb) was amplified from Cs23 and put into the pGEMT vector, followed by hygB insertion at a SalI site in pGEMT, as described in pM2M1. The 8.4-kb DNA region carrying MAT1-2-1 and all three MAT1-1 genes, which was amplified from Cs23, was inserted into the pGEMT vector (Promega), followed by the insertion of the hygB cassette at a SalI site in pGEMT, resulting in pM2M1. For heterologous expression of Cs23 MAT1-1-1 in a MAT1-1-1-deletion strain of F. graminearum, the entire ORF of each MAT1-1-1 version amplified from Cs23 [MAT1-1-1L and MAT1-1-1 with primer pairs Cs27forE/Cs23revE and Cs27forE/Cs27revE2 (S2 Table), respectively] was fused to DNA (~ 1 kb) both 5' and 3' of the F. graminearum MAT1-1-1 (annotated as FGSG_08892.3 in the F. graminearum genome database) as described above [46]. The 5′- and 3′-flanking regions of the F. graminearum MAT1-1-1 sequence were amplified using the primer pairs FgM1-1-1F5/FgM1-1-1Rt5Cs27 and Fgmat1-1-1Rt5Cs23 or FgM1-1-1Ft3Cs27 /FgM1-1-1R3Cs27 (S2 Table), respectively, and these were fused to either Cs23 MAT1-1-1L or MAT1-1-1, followed by the final PCR amplification using nested primers FgM1-1-1FNCs27 and FgM1-1-1RNCs27 (S2 Table).
Sequencing of MAT loci
Nucleotide sequences of Cs23 and Cs27 MAT loci (Fig 1) were obtained using conventional PCR amplification followed by a combination of TAIL-PCR and inverse PCR amplification as chromosome walking strategies (S2 Fig). First, a 270-bp fragment of the High Mobility Group (HMG) box of MAT1-2-1 was amplified from genomic DNA of Cs23 using degenerate HMG primers (NcHMG1 and NcHMG2 [48], designated P1 and P2 in S2 Table, S2 Fig), after which TAIL-PCR [49] was performed to obtain sequence beyond the HMG box using combinations of arbitrary and specific primers, such as P4 [49]/P3 for the 3’ flank and P5 [49]/P6 for the 5’ flank. To recover the rest of the MAT region, we employed an inverse PCR strategy, as described previously [31]. Genomic DNA from Cs23 was digested with BamHI, SphI, ClaI, NheI, and SacII, self-ligated, and used as a template with appropriate primer pairs as shown in S2 Fig. Subsequently, sequencing was extended using primers corresponding to previously determined sequences. Sequence assembly (21.9 kb) revealed that the C. spinulosa Cs23 MAT chromosome included MAT1-2-1, three MAT1-1 genes and additional ORFs (S2 Fig, Fig 1). Similarly, a total of 18.3 kb of the MAT chromosomal region was recovered from Cs27 using the inverse PCR strategy and conventional PCR with the primers derived from the Cs23 MAT region, as shown in S2 Fig. Nucleotide sequences were assembled (Cs23, Cs27: GenBank accessions # KY624604 and # KY624603) and analyzed using the DNASTAR software package (DNAStar Inc., Madison, WI, USA). BLAST [50] searches were performed against the NCBI/GenBank databases.
Fungal transformation
The C. spinulosa Cs23, Cs27, and T10 strains were transformed using a polyethylene glycol (PEG)-mediated transformation procedure newly developed in this study. For preparation of young mycelia for protoplasting, approximately 50 agar blocks (3 × 3 mm) from a 7-day old PDA culture of a strain were inoculated into 100 ml PD broth and incubated for 24 h on a shaker (200 rpm). Mycelia, harvested by centrifugation, were re-suspended in 100 ml fresh PD broth in a 500 ml flask and incubated for an additional 24 h under the same conditions. Note that C. spinulosa was unable to produce asexual spores (conidia) under all growth conditions examined. Young mycelia were recovered by centrifugation and suspended in 25 ml osmoticum (0.7 M KCl) containing 800 mg of lysing enzyme from Trichoderma harzianum and 8 mg of cellulase from Trichoderma sp. (Sigma). The suspension was incubated for 3 h at 30°C on a shaker (50 rpm). Protoplasts, which were collected by filtering through three layers of cheesecloth followed by centrifugation, were suspended in 10 ml STC (1.2 M Sorbitol, 10 mM Tris-HCl pH 8, and 50 mM CaCl2). All other transformation steps using protoplasts were performed as described previously [43]. Two genes conferring resistance to hygromycin B (hygB) or geneticin (gen) were used as selectable markers for fungal transformation in this study.
To determine if Cs23 MAT1-1-1 L is functional, we inserted it and MAT1-1-1, separately, into a F. graminearum ΔMAT1-1-1 strain [25]. PCR constructs were directly added to protoplasts of the F. graminearum ΔMAT1-1-1 strain along with the plasmid DNA pSSK660 carrying a geneticin resistance gene (gen) [51] (S8A Fig). As a positive control, the F. graminearum MAT1-1-1 region (carrying the entire MAT1-1-1 along with its 5' and 3' flanking regions) was added using the same strategy.
Supporting information
Strain name, a brief description, and MAT genotype and phenotype are described.
(DOCX)
Primer names, sequence, position in overall sequence and references are listed.
(DOCX)
C. spinulosa, S. trifoliorum, and C. fimbriata MAT structural features are compared.
(DOCX)
The note traces the origin of the Cs23 and Cs27 strains and mentions a 1952 paper [11] describing mating phenomena in this fungus. JF subsequently sent the strains to BGT.
(TIF)
All primers indicated are listed in S2 Table. See Fig 1 legend for gene explanations.
(TIF)
Numbers refer to positions in the 21.9 kb of Cs23 sequence generated (S2 Fig). Note that the 3’ end of the MAT1-1-1L fragment (green box), is lost in the recombination event.
(PPTX)
Control Cs23 and Cs27 in the first rows were grown on PDA without hygromycin B. Typical candidates generated by integration of MAT1-2-1 (see Fig 5), were grown on PDA or PDA amended with hygromycin B. (A). Shows a T27M1M2a-type transformant after three cycles. (B). Shows a T27M1M2b-type transformant after three cycles. Note in both cases, ability to grow on hygromycin medium is lost by cycle 3.
(TIF)
(A). MAT structure in T10, Cs23 and Cs27-type strains. (B). Gel blots of SacI-digested genomic DNAs from strains of all categories represented in (A). Hybridization with MAT1-1-1. (C and D). As in (B), but hybridized with DR and MAT1-2-1 probes, respectively. From left, Cs23, Cs27, T10, TCs27Gen1, Cs23-type progeny (12, 26, 28, 30, and 60), T10-type progeny (48 and 102), and Cs27-type progeny (10, 25, and 70). All strains are listed in S1 Table; progeny are prefixed with “P”. Sizes (in kb) are indicated to the left of the gel.
(TIF)
PCR primer sequences are shown in S2 Table, results in S7 Fig. Primer pair qCs27M1-1-1R/qCs23M1-1-1F, is specific to MAT1-1-1L, while primer pair qCs27M1-1-1R/qCs27M1-1-1F amplifies MAT1-1-1 from strains carrying either MAT1-1-1 or MAT1-1-1L.
(TIF)
(A). Relative expression levels of individual MAT transcripts determined by qPCR on total RNA (A). Amplification level of MAT1-2-1 in Cs23 was used as a reference (set to 1). The same letter above bars indicates no significant difference. Primer pair qCs27M1-1-1R/qCs23M1-1-1F, specific to MAT1-1-1L, amplified the predicted 138 bp fragment from both Cs23 and T10, while the primer pair qCs27M1-1-1R/ qCs27M1-1-1F, amplified the predicted 106 bp fragment from both Cs23 and Cs27. Note that the 106 bp fragment was not amplified from RNA of strain T10 because one of two repeats had been deleted. See S6 Fig for primer positions. Other primer pairs were as described in Fig 7. (B). Relative amounts of individual MAT genes determined by qPCR on genomic DNA.Amplification level of MAT1-2-1 in Cs23 was set to 1 as reference. See (A) for primer details.
(TIF)
(A). Scheme for the insertion of MAT1-1-1L or MAT1-1-1 from Cs23 (designated CsMAT1-1-1) into the FgΔMAT1-1-1 strain by co-transformation with pSSK660 carrying the geneticin resistance gene (gen). The transgenic FgΔMAT1-1-1 strains carrying either CsMAT1-1-1 or CsMAT1-1-1L at an ectopic position are designated as FgΔMAT1-1-1::CsMAT1-1-1 and FgΔMAT1-1-1::CsMAT1-1-1L, respectively. Drug resistance phenotypes are in parentheses. (B). Perithecium formation.: F. graminearum self-fertile WT Z3643 strain, FgΔMAT1-1-1: a MAT1-1-1-deletion strain of Z3643, FgΔMAT1-1-1::FgMAT1-1-1: a control add-back strain of FgΔMAT1-1 carrying an intact copy of FgMAT1-1-1 at an ectopic position, FgΔMAT1-1-1::CsMAT1-1-1-16: a FgΔMAT1-1-1::CsMAT1-1-1 strain, and FgΔMAT1-1-1::CsMAT1-1-1L-35: a FgΔMAT1-1-1::CsMAT1-1-1L strain. Scale-bar = 500 μm. (C). Average diameter of perithecia formed on carrot agar cultures. Perithecial sizes from two independent transformants carrying C. spinulosa MAT1-1-1 (6, 13) or MAT1-1-1L (35, 29) are shown. The same letters on each bar represent no significant difference.
(TIF)
DAPI staining of nuclei in ascospores from a self of strain Cs23 (A-B), and an outcross between strains T10 and Cs27 (C-D). DIC images corresponding to DAPI fluorescence images are shown in B and D, respectively. Large and small ascospores within an ascus are designated with L and S, respectively. Asci containing different numbers of nuclei (ranging from those in the diploid zygote to those in complete tetrads with 16 nuclei) are indicated by arrows. Scale bars = 50μm.
(TIF)
High magnification of DAPI stained nuclei in ascospores from a self of Cs23 (A and C). Corresponding DIC images are shown in B and D, respectively. Large and small ascospores are designated with L and S, respectively. Several ascospores each containing 2–3 nuclei are indicated by arrows. Nuclei in small ascospores were difficult to stain in mature spores (C, D). Scale bars = 20 μm.
(TIF)
Self-fertile Cs23 and Cs23-type progeny carry two different versions of MAT (MAT1-1 only and MAT1-1;MAT1-2) in a common cytoplasm. The latter are in the minority. Self-sterile strains (Cs27, T10, and T10-type progeny) carry nuclei containing a single version of MAT but architecture can vary. Self-sterile T27M12a-E5 carries both MAT1-1 and MAT1-2, but likely not closely linked to each other.
(TIF)
(A). A model for the evolution of the fused MAT1-1-1:MAT1-2-3 gene. We propose that a recombination event occurred via misalignment of paired MAT chromosomes in T. reesei ancestors [represented by T. virens Gv29-8 (a MAT1-2 parent) and H. jecorina (a MAT1-1 parent), respectively] resulting in the fused gene in extant strain QM6a. (B). A possible crossover point for the model shown in (A) in the actual nucleotide sequences of T. virens Gv29-8 and H. jecorina CBS 999.97 strains. Nucleotide sequences conserved between strains are indicated by the same colors.
(TIF)
To achieve the MAT1-1;MAT1-2 structure present in C. spinulosa Cs23, we assume at least three different unequal crossing over events occurred in putative heterothallic ancestors (as in S12 Fig). In the first cross (A), a crossover would occur in a manner similar to that described in S12 Fig, resulting in a MAT1-2 progeny (asterisk) carrying a fused gene consisting of a 3’ portion of MAT1-1-1 and MAT1-2-3. In this case, a possible crossover point would be to the right of the DR (red bar) in the MAT1-1 parent chromosome, leaving the DR1 associated with the MAT1-1-1 fragment in the fused protein. In the second case (B), a similar event might occur, but with a possible crossover site to the left of the DR in the MAT1-1 parent chromosome. This would result in a MAT1-1 progeny (asterisk) carrying a truncated MAT1-1-1 gene that includes the DR sequence at its 3’ end. The third crossover (C) would occur between the MAT1-2 and MAT1-1 progeny generated from the (A) and (B). If the crossover site were between a region 3’ of MAT1-2-1 3’ and a region 3’ of MAT1-1-1, this would yield a progeny carrying MAT organization of C. spinulosa Cs23 (Figs 1 and 2). Refer to Fig 1 for gene organization on the MAT chromosome.
(TIF)
Acknowledgments
This study was begun in 1997 at the prompting of the late John R.S. Fincham, FRS FRSE, as per S1 Fig. We are grateful to him and JM Mathieson (Mayo) for the original cultures.
Data Availability
All data are available from GenBank (accession numbers KY624604 and KY624603).
Funding Statement
This work was supported by Strategic Initiative for Microbiomes in Agriculture and Food, Ministry of Agriculture, Food and Rural Affairs, Republic of Korea (SHY) and Next-Generation Bio Green21 Program (no. PJ0111802017), the Rural Development Administration, Republic of Korea (SHY). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Coppin E, Debuchy R, Arnaise S, Picard M (1997) Mating types and sexual development in filamentous ascomycetes. Microbiol Mol Biol Rev 61: 411–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Metzenberg RL, Glass NL (1990) Mating type and mating strategies in Neurospora. Bioessays 12: 53–59. doi: 10.1002/bies.950120202 [DOI] [PubMed] [Google Scholar]
- 3.Martin T, Lu SW, van Tilbeurgh H, Ripoll DR, Dixelius C, et al. (2010) Tracing the origin of the fungal alpha1 domain places its ancestor in the HMG-box superfamily: implication for fungal mating-type evolution. PLoS One 5: e15199 doi: 10.1371/journal.pone.0015199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelson MA (1996) Mating systems in ascomycetes: a romp in the sac. Trends Genet 12: 69–74. [DOI] [PubMed] [Google Scholar]
- 5.Turgeon BG (1998) Application of mating type gene technology to problems in fungal biology. Annu Rev Phytopathol 36: 115–137. doi: 10.1146/annurev.phyto.36.1.115 [DOI] [PubMed] [Google Scholar]
- 6.Herskowitz I (1988) Life cycle of the budding yeast Saccharomyces cerevisiae. Microbiol Rev 52: 536–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haber JE (2012) Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics 191: 33–64. doi: 10.1534/genetics.111.134577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maekawa H, Kaneko Y (2014) Inversion of the chromosomal region between two mating type loci switches the mating type in Hansenula polymorpha. PLoS Genet 10: e1004796 doi: 10.1371/journal.pgen.1004796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanson SJ, Byrne KP, Wolfe KH (2014) Mating-type switching by chromosomal inversion in methylotrophic yeasts suggests an origin for the three-locus Saccharomyces cerevisiae system. Proc Natl Acad Sci U S A 111: E4851–4858. doi: 10.1073/pnas.1416014111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nieuwenhuis BP, Immler S (2016) The evolution of mating-type switching for reproductive assurance. Bioessays 38: 1141–1149. doi: 10.1002/bies.201600139 [DOI] [PubMed] [Google Scholar]
- 11.Mathieson M (1952) Ascospore dimorphism and mating type in Chromocrea spinulosa (Fuckel) Petch n. comb. Annals of Botany 16: 449–468. [Google Scholar]
- 12.Wheeler HE (1950) Genetics of Glomerella VIII. A genetic basis for the occurrence of minus mutants. Am J Bot 37: 304–312. [Google Scholar]
- 13.Harrington TC, McNew DL (1997) Self-fertility and uni-directional mating-type switching in Ceratocystis coerulescens, a filamentous ascomycete. Curr Genet 32: 52–59. [DOI] [PubMed] [Google Scholar]
- 14.Witthuhn RC, Harrington TC, Steimel JP, Wingfield BD, Wingfield MJ (2000) Comparison of isozymes, rDNA spacer regions, and MAT-2 DNA sequences as phylogenetic characters in the analysis of the Ceratocystis coerulescens complex. Mycologia 92: 447–545. [Google Scholar]
- 15.Uhm JY, Fuji H (1983) Ascospore dimorphism in Sclerotinia trifoliorum and cultural characters of strains from different-sized spores. Phytopathology 73: 565–569. [Google Scholar]
- 16.Leslie JF, Klein KK (1986) Female fertility and mating type effects on effective population size and evolution in filamentous fungi. Genetics 144: 557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faretra F, Antonacci E, Pollastro S (1988) Sexaul behaviour and mating system of Botryotinia fukeliana, teleomorph of Botrytis cinerea. J Gen Microbiol 134: 2543–2550. [Google Scholar]
- 18.Wilson AM, Wilken PM, van der Nest MA, Steenkamp ET, Wingfield MJ, et al. (2015) Homothallism: an umbrella term for describing diverse sexual behaviours. IMA Fungus 6: 207–214. doi: 10.5598/imafungus.2015.06.01.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perkins DD (1987) Mating-type switching in filamentous ascomycetes. Genetics 115: 215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilken PM, Steenkamp ET, Wingfield MJ, de Beer ZW, Wingfield BD (2014) DNA loss at the Ceratocystis fimbriata mating locus results in self-sterility. PLoS One 9: e92180 doi: 10.1371/journal.pone.0092180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu L, Jardini TM, Chen W (2016) Direct repeat-mediated DNA deletion of the mating type MAT1-2 genes results in unidirectional mating type switching in Sclerotinia trifoliorum. Sci Rep 6: 27083 doi: 10.1038/srep27083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Debuchy R, Turgeon BG (2006) Mating-type structure, evolution, and function in Euascomycetes In: Kues U, Fischer R, editors. The Mycota. Berlin, Germany: Springer; pp. 293–323. [Google Scholar]
- 23.Yun SH, Kim HK, Lee T, Turgeon BG (2015) Molecular analysis of unidirectional mating type switching in Chromocrea spinulosa. Fungal Genet Rep 61S: Abstract 178. [Google Scholar]
- 24.Martin SH, Wingfield BD, Wingfield MJ, Steenkamp ET (2011) Structure and evolution of the Fusarium mating type locus: new insights from the Gibberella fujikuroi complex. Fungal Genet Biol 48: 731–740. doi: 10.1016/j.fgb.2011.03.005 [DOI] [PubMed] [Google Scholar]
- 25.Kim HK, Cho EJ, Lee S, Lee YS, Yun SH (2012) Functional analyses of individual mating-type transcripts at MAT loci in Fusarium graminearum and Fusarium asiaticum. FEMS Microbiol Lett 337: 89–96. doi: 10.1111/1574-6968.12012 [DOI] [PubMed] [Google Scholar]
- 26.Seidl V, Seibel C, Kubicek CP, Schmoll M (2009) Sexual development in the industrial workhorse Trichoderma reesei. Proc Natl Acad Sci U S A 106: 13909–13914. doi: 10.1073/pnas.0904936106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yun SH, Arie T, Kaneko I, Yoder OC, Turgeon BG (2000) Molecular organization of mating type loci in heterothallic, homothallic, and asexual Gibberella/Fusarium species. Fungal Genet Biol 31: 7–20. doi: 10.1006/fgbi.2000.1226 [DOI] [PubMed] [Google Scholar]
- 28.Beach DH (1983) Cell type switching by DNA transposition in fission yeast. Nature 305: 682–688. [Google Scholar]
- 29.Dujon B (2010) Yeast evolutionary genomics. Nat Rev Genet 11: 512–524. doi: 10.1038/nrg2811 [DOI] [PubMed] [Google Scholar]
- 30.Gordon JL, Armisen D, Proux-Wera E, OhEigeartaigh SS, Byrne KP, et al. (2011) Evolutionary erosion of yeast sex chromosomes by mating-type switching accidents. Proc Natl Acad Sci U S A 108: 20024–20029. doi: 10.1073/pnas.1112808108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yun SH, Berbee ML, Yoder OC, Turgeon BG (1999) Evolution of the fungal self-fertile reproductive life style from self-sterile ancestors. Proc Natl Acad Sci U S A 96: 5592–5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poggeler S, Risch S, Kuck U, Osiewacz HD (1997) Mating-type genes from the homothallic fungus Sordaria macrospora are functionally expressed in a heterothallic ascomycete. Genetics 147: 567–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Picard M, Debuchy R, Coppin E (1991) Cloning the mating types of the heterothallic fungus Podospora anserina: developmental features of haploid transformants carrying both mating types. Genetics 128: 539–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turgeon BG, Bohlmann H, Ciuffetti LM, Christiansen SK, Yang G, et al. (1993) Cloning and analysis of the mating type genes from Cochliobolus heterostrophus. Mol Gen Genet 238: 270–284. [DOI] [PubMed] [Google Scholar]
- 35.Turgeon BG, Sharon A, Wirsel S, Yamaguchi K, Christiansen SK, et al. (1995) Structure and function of mating type genes in Cochliobolus spp. and asexual fungi. Can J Botany 73(S1): 778–783. [Google Scholar]
- 36.Wirsel S, Turgeon BG, Yoder OC (1996) Deletion of the Cochliobolus heterostrophus mating-type (MAT) locus promotes the function of MAT transgenes. Curr Genet 29: 241–249. [PubMed] [Google Scholar]
- 37.Scazzocchio C (2006) Aspergillus genomes: secret sex and the secrets of sex. Trends Genet 22: 521–525. doi: 10.1016/j.tig.2006.08.004 [DOI] [PubMed] [Google Scholar]
- 38.Hsueh YP, Idnurm A, Heitman J (2006) Recombination hotspots flank the Cryptococcus mating-type locus: implications for the evolution of a fungal sex chromosome. PLoS Genet 2: e184 doi: 10.1371/journal.pgen.0020184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun S, Hsueh YP, Heitman J (2012) Gene conversion occurs within the mating-type locus of Cryptococcus neoformans during sexual reproduction. PLoS Genet 8: e1002810 doi: 10.1371/journal.pgen.1002810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chaverri P, Samuels GJ (2003) Hypocrea/Trichoderma (Ascomycota, Hypocreales, Hypocreaceae): Species with green ascospores. Stud Mycol 48: 1–116. [Google Scholar]
- 41.Kim HK, Jo SM, Kim GY, Kim DW, Kim YK, et al. (2015) A large-scale functional analysis of putative target genes of mating-type loci provides insight into the regulation of sexual development of the cereal pathogen Fusarium graminearum. PLoS Genet 11: e1005486 doi: 10.1371/journal.pgen.1005486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Min K, Son H, Lim JY, Choi GJ, Kim JC, et al. (2014) Transcription factor RFX1 is crucial for maintenance of genome integrity in Fusarium graminearum. Eukaryot Cell 13: 427–436. doi: 10.1128/EC.00293-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim H, Kim HK, Lee S, Yun SH (2015) The white collar complex is involved in sexual development of Fusarium graminearum. PLoS One 10: e0120293 doi: 10.1371/journal.pone.0120293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual Plainview, U.S.A.: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 45.Kim HK, Yun SH (2011) Evaluation of potential reference genes for quantitative RT-PCR analysis in Fusarium graminearum under different culture conditions. Plant Pathol J 27: 301–309. [Google Scholar]
- 46.Yu JH, Hamari Z, Han KH, Seo JA, Reyes-Dominguez Y, et al. (2004) Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol 41: 973–981. doi: 10.1016/j.fgb.2004.08.001 [DOI] [PubMed] [Google Scholar]
- 47.Lee J, Lee T, Lee YW, Yun SH, Turgeon BG (2003) Shifting fungal reproductive mode by manipulation of mating type genes: obligatory heterothallism of Gibberella zeae. Mol Microbiol 50: 145–152. [DOI] [PubMed] [Google Scholar]
- 48.Arie T, Christiansen SK, Yoder OC, Turgeon BG (1997) Efficient cloning of ascomycete mating type genes by PCR amplification of the conserved MAT HMG Box. Fungal Genet Biol 21: 118–130. [PubMed] [Google Scholar]
- 49.Liu YG, Whittier RF (1995) Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25: 674–681. [DOI] [PubMed] [Google Scholar]
- 50.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 51.Kim JE, Kim JC, Jin J, Yun SH, Lee YW (2008) Functional characterization of genes located at the aurofusarin biosynthesis gene cluster in Gibberella zeae. Plant Pathol J 24: 8–16. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Strain name, a brief description, and MAT genotype and phenotype are described.
(DOCX)
Primer names, sequence, position in overall sequence and references are listed.
(DOCX)
C. spinulosa, S. trifoliorum, and C. fimbriata MAT structural features are compared.
(DOCX)
The note traces the origin of the Cs23 and Cs27 strains and mentions a 1952 paper [11] describing mating phenomena in this fungus. JF subsequently sent the strains to BGT.
(TIF)
All primers indicated are listed in S2 Table. See Fig 1 legend for gene explanations.
(TIF)
Numbers refer to positions in the 21.9 kb of Cs23 sequence generated (S2 Fig). Note that the 3’ end of the MAT1-1-1L fragment (green box), is lost in the recombination event.
(PPTX)
Control Cs23 and Cs27 in the first rows were grown on PDA without hygromycin B. Typical candidates generated by integration of MAT1-2-1 (see Fig 5), were grown on PDA or PDA amended with hygromycin B. (A). Shows a T27M1M2a-type transformant after three cycles. (B). Shows a T27M1M2b-type transformant after three cycles. Note in both cases, ability to grow on hygromycin medium is lost by cycle 3.
(TIF)
(A). MAT structure in T10, Cs23 and Cs27-type strains. (B). Gel blots of SacI-digested genomic DNAs from strains of all categories represented in (A). Hybridization with MAT1-1-1. (C and D). As in (B), but hybridized with DR and MAT1-2-1 probes, respectively. From left, Cs23, Cs27, T10, TCs27Gen1, Cs23-type progeny (12, 26, 28, 30, and 60), T10-type progeny (48 and 102), and Cs27-type progeny (10, 25, and 70). All strains are listed in S1 Table; progeny are prefixed with “P”. Sizes (in kb) are indicated to the left of the gel.
(TIF)
PCR primer sequences are shown in S2 Table, results in S7 Fig. Primer pair qCs27M1-1-1R/qCs23M1-1-1F, is specific to MAT1-1-1L, while primer pair qCs27M1-1-1R/qCs27M1-1-1F amplifies MAT1-1-1 from strains carrying either MAT1-1-1 or MAT1-1-1L.
(TIF)
(A). Relative expression levels of individual MAT transcripts determined by qPCR on total RNA (A). Amplification level of MAT1-2-1 in Cs23 was used as a reference (set to 1). The same letter above bars indicates no significant difference. Primer pair qCs27M1-1-1R/qCs23M1-1-1F, specific to MAT1-1-1L, amplified the predicted 138 bp fragment from both Cs23 and T10, while the primer pair qCs27M1-1-1R/ qCs27M1-1-1F, amplified the predicted 106 bp fragment from both Cs23 and Cs27. Note that the 106 bp fragment was not amplified from RNA of strain T10 because one of two repeats had been deleted. See S6 Fig for primer positions. Other primer pairs were as described in Fig 7. (B). Relative amounts of individual MAT genes determined by qPCR on genomic DNA.Amplification level of MAT1-2-1 in Cs23 was set to 1 as reference. See (A) for primer details.
(TIF)
(A). Scheme for the insertion of MAT1-1-1L or MAT1-1-1 from Cs23 (designated CsMAT1-1-1) into the FgΔMAT1-1-1 strain by co-transformation with pSSK660 carrying the geneticin resistance gene (gen). The transgenic FgΔMAT1-1-1 strains carrying either CsMAT1-1-1 or CsMAT1-1-1L at an ectopic position are designated as FgΔMAT1-1-1::CsMAT1-1-1 and FgΔMAT1-1-1::CsMAT1-1-1L, respectively. Drug resistance phenotypes are in parentheses. (B). Perithecium formation.: F. graminearum self-fertile WT Z3643 strain, FgΔMAT1-1-1: a MAT1-1-1-deletion strain of Z3643, FgΔMAT1-1-1::FgMAT1-1-1: a control add-back strain of FgΔMAT1-1 carrying an intact copy of FgMAT1-1-1 at an ectopic position, FgΔMAT1-1-1::CsMAT1-1-1-16: a FgΔMAT1-1-1::CsMAT1-1-1 strain, and FgΔMAT1-1-1::CsMAT1-1-1L-35: a FgΔMAT1-1-1::CsMAT1-1-1L strain. Scale-bar = 500 μm. (C). Average diameter of perithecia formed on carrot agar cultures. Perithecial sizes from two independent transformants carrying C. spinulosa MAT1-1-1 (6, 13) or MAT1-1-1L (35, 29) are shown. The same letters on each bar represent no significant difference.
(TIF)
DAPI staining of nuclei in ascospores from a self of strain Cs23 (A-B), and an outcross between strains T10 and Cs27 (C-D). DIC images corresponding to DAPI fluorescence images are shown in B and D, respectively. Large and small ascospores within an ascus are designated with L and S, respectively. Asci containing different numbers of nuclei (ranging from those in the diploid zygote to those in complete tetrads with 16 nuclei) are indicated by arrows. Scale bars = 50μm.
(TIF)
High magnification of DAPI stained nuclei in ascospores from a self of Cs23 (A and C). Corresponding DIC images are shown in B and D, respectively. Large and small ascospores are designated with L and S, respectively. Several ascospores each containing 2–3 nuclei are indicated by arrows. Nuclei in small ascospores were difficult to stain in mature spores (C, D). Scale bars = 20 μm.
(TIF)
Self-fertile Cs23 and Cs23-type progeny carry two different versions of MAT (MAT1-1 only and MAT1-1;MAT1-2) in a common cytoplasm. The latter are in the minority. Self-sterile strains (Cs27, T10, and T10-type progeny) carry nuclei containing a single version of MAT but architecture can vary. Self-sterile T27M12a-E5 carries both MAT1-1 and MAT1-2, but likely not closely linked to each other.
(TIF)
(A). A model for the evolution of the fused MAT1-1-1:MAT1-2-3 gene. We propose that a recombination event occurred via misalignment of paired MAT chromosomes in T. reesei ancestors [represented by T. virens Gv29-8 (a MAT1-2 parent) and H. jecorina (a MAT1-1 parent), respectively] resulting in the fused gene in extant strain QM6a. (B). A possible crossover point for the model shown in (A) in the actual nucleotide sequences of T. virens Gv29-8 and H. jecorina CBS 999.97 strains. Nucleotide sequences conserved between strains are indicated by the same colors.
(TIF)
To achieve the MAT1-1;MAT1-2 structure present in C. spinulosa Cs23, we assume at least three different unequal crossing over events occurred in putative heterothallic ancestors (as in S12 Fig). In the first cross (A), a crossover would occur in a manner similar to that described in S12 Fig, resulting in a MAT1-2 progeny (asterisk) carrying a fused gene consisting of a 3’ portion of MAT1-1-1 and MAT1-2-3. In this case, a possible crossover point would be to the right of the DR (red bar) in the MAT1-1 parent chromosome, leaving the DR1 associated with the MAT1-1-1 fragment in the fused protein. In the second case (B), a similar event might occur, but with a possible crossover site to the left of the DR in the MAT1-1 parent chromosome. This would result in a MAT1-1 progeny (asterisk) carrying a truncated MAT1-1-1 gene that includes the DR sequence at its 3’ end. The third crossover (C) would occur between the MAT1-2 and MAT1-1 progeny generated from the (A) and (B). If the crossover site were between a region 3’ of MAT1-2-1 3’ and a region 3’ of MAT1-1-1, this would yield a progeny carrying MAT organization of C. spinulosa Cs23 (Figs 1 and 2). Refer to Fig 1 for gene organization on the MAT chromosome.
(TIF)
Data Availability Statement
All data are available from GenBank (accession numbers KY624604 and KY624603).