Abstract
Blood-brain barrier (BBB) coverage plays a central role in the homeostasis of the central nervous system (CNS). The BBB is dynamically maintained by astrocytes, pericytes and brain endothelial cells (BECs). Here, we detail methods to assess BBB coverage using single cultures of immortalized human BECs, single cultures of primary mouse BECs, and a humanized triple culture model (BECs, astrocytes and pericytes) of the BBB. To highlight the applicability of the assays to disease states, we describe the effect of oligomeric amyloid-β (oAβ), which is an important contributor to Alzheimer's disease (AD) progression, on BBB coverage. Further, we utilize the epidermal growth factor (EGF) to illuminate the drug screening potential of the techniques. Our results show that single and triple cultured BECs form meshwork-like structures under basal conditions, and that oAβ disrupts this cell meshwork formation and degenerates the preformed mesh structures, but EGF blocks this disruption. Thus, the techniques described are important for dissecting fundamental and disease-relevant processes that modulate BBB coverage.
Keywords: Medicine, Issue 124, Blood-brain barrier, brain endothelial cells, pericytes, astrocytes, cell culture, meshwork formation, meshwork disruption, amyloid beta, epidermal growth factor
Introduction
The blood-brain barrier (BBB) of cerebral capillaries is the largest interface of blood-to-brain contact and plays a central role in the homeostasis of the central nervous system (CNS)1,2. Dynamic processes at the BBB prevent the uptake of unwanted molecules from the blood, remove waste products from the CNS, supply essential nutrients and signaling molecules to the CNS, and modulate neuroinflammation1,2,3,4,5. BBB damage is prevalent during aging and several neurodegenerative disorders including Alzheimer's disease (AD), multiple sclerosis and stroke1,2,3,4,5,6. Therefore, BBB dysfunction may play a key role in neurodegenerative disorders, including as a therapeutic target.
Maintaining vessel coverage is important for the homeostatic functions of the BBB. However, in vivo and in vitro data conflict on whether the processes involved in neurodegenerative disorders cause higher or lower BBB coverage6,7,8,9,10,11,12,13, particularly in AD. Therefore, there is a strong rationale for the development of in vitro models using relevant cell types to assess and more comprehensively understand the dynamics of BBB coverage. Cerebral capillaries are composed of astrocytes, pericytes and brain endothelial cells (BECs)3. All cell types contribute to the function of the BBB through structural support and via the secretion of effector molecules such as angiogenic growth factors, cytokines and chemokines that act in paracrine- and autocrine-like fashion. However, the major effector cells of the BBB are BECs3. In general, the cell culture techniques for assessing BBB function are permeability assays performed on cells grown on filter inserts, or assessing levels of key BEC proteins, both after the addition of stressors14,15,16. Although important, these assays do not focus on the cerebrovascular coverage.
Here, our previous methods17 are detailed to assess BEC coverage and meshwork-like structures using single cultures of immortalized human BECs, single cultures of primary mouse BECs, and a humanized triple culture model (BECs, astrocytes and pericytes) of the BBB. The goal was to demonstrate the detrimental effect of oAβ, which is considered an important contributor to AD progression, on BEC coverage. The protective effect of the epidermal growth factor (EGF) highlights the potential of the technique as a therapeutic screening tool. The technique has several broad applications for basic and applied research including: 1) delineating the role of specific pathways on angiogenesis and vessel coverage, 2) evaluating the effects of disease and aging-relevant factors on angiogenesis and vessel coverage, and 3) identifying pharmacological targets.
Protocol
All experiments follow the University of Illinois, Chicago Institutional Animal Care and Use Committee protocols.
1. General Preparation
NOTE: The brain microvascular endothelial cell line (hCMEC/D3) is an extensively characterized immortalized human BEC line14,15,16,18,19. Culture the hCMEC/D3 cells on tissue culture flasks coated with collagen Type I (calf skin, 1:20 dilution of 0.1% solution in Hank's Balanced Salt Solution (HBSS) containing Ca2+ and Mg2+) in basal Endothelial Growth Basal Medium (EBM-2) containing 2-5% Fetal Bovine Serum (FBS), 10% ascorbic acid, 10% gentamicin sulphate, 25% hydrocortisone and 1/4 of the total volume of the supplied growth factor supplements per 500 mL of media [vascular endothelium growth factor (VEGF), epidermal growth factor (EGF), insulin-like growth factor 1 (IGF-1) and human basic fibroblastic growth factor (bFGF), see Table of Materials]. NOTE: EBM-2 medium with FBS and growth factors is referred to as "EBM-2 complete". EBM-2 without FBS and supplements is referred to as "EBM-2 basal". At full confluence, the hCMEC/D3 cells are ~1 x 105 cells/cm2.
Passage the hCMEC/D3 cells at a ratio of 1:5 by first incubating with HBSS for 3 min without Ca2+ and Mg2+ followed by detachment with 5 mL trypsin/EDTA (0.25%) for 5 min. Neutralize the trypsin using EBM-2 complete (1:1 neutralization), and centrifuge at 290 x g for 5 min at 4 °C; discard the supernatant. Resuspend the pellet in EBM-2 complete and re-plate at 1:5 ratio.
Culture the primary human pericytes and astrocytes according to the supplier's protocol [Table of Materials]. Culture the pericytes in pericyte basal medium with FBS and pericyte growth supplement.
Culture the human astrocytes in astrocyte medium containing astrocyte growth factors. Culture both the pericytes and the astrocytes in tissue culture flasks coated with 3 mL of poly-L-lysine (PLL) for cell adherence and utilize the cells between passages 2 and 5.
Euthanize 7 mice and remove the brain stems and cerebella with forceps; detach the meninges by carefully rolling the brains on gauze. Mince the remaining brain tissue in a Petri dish in Minimal Essential Medium (MEM) with HEPES (5 mL per brain) with a sterile razor blade. Isolate the primary mouse BECs from 2-month-old C57BL/6J mice according to the referenced protocol20.
Transfer the minced brain tissue to a 15 mL conical tube. Centrifuge at 290 x g for 5 min at 4 °C. Remove the supernatant and incubate the tissue in a papain (833.33 µL per brain) and DNase (41.7 µL per brain) solution, in a 37 °C water bath for 1 h, to digest the tissue.
Triturate the homogenate sequentially through 19 and 21 gauge needles, mix at a 1:1 ratio with a 22% Bovine Serum Albumin (BSA) solution, and centrifuge at 1360 x g for 10 min at 4 °C.
Resuspend the resulting pellet in 1 mL EBM-2 complete and centrifuge at 290 x g for 5 min at 4 °C. Resuspend again in 1 mL EBM-2 complete and plate the cells (1 mL/well) on 6-well plates coated with collagen (~1 brain per well).
Replace the media 24 h later with EBM-2 complete containing 4 µg/mL puromycin hydrochloride; replace with EBM-2 complete after 2 days. The primary BECs are cultured as for the hCMEC/D3 cells and are at full confluence at 1 x 105 cells/cm2.
Prepare a 100 µM of oAβ, 24 h before the assay. NOTE: oAβ is considered the disease relevant form of A . For oAβ, use the well-characterized preparations described by Dahlgren et al.21.
Thaw the basement membrane stock solution overnight at 4 °C and aliquot into sterile PCR tube strips (8 tubes) at 140 µL basement membrane per tube. Refreeze each strip and store at -20 °C. Thaw one strip per 96-well plate at 4 °C, 24 h prior to the assay. Pre-cool the pipette tips to avoid solidification. NOTE: All handling of the basement membrane matrix must be carried out on ice to avoid rapid solidification. As 10 µL per well of basement membrane is used on the day of the assay, each strip is sufficient for one 96-well plate.
Incubate fully confluent BECs in EBM-2 basal, 24 h prior to the assay. NOTE: The rationale is: EBM-2 complete contains supplemented factors and FBS with the purpose of promoting optimal cell growth; however, the same molecular pathways that promote cell growth are also important for brain endothelial cell coverage and dynamics, both in the presence and absence of a stressor. We therefore serum and supplement starve the BECs to reduce the confounding effect of residual activation of cellular signaling pathways. Of note, the cells are briefly (5 min) exposed to FBS during the neutralization of trypsinized cells prior to the assay. In contrast to BECs, the pericytes and astrocytes are not serum starved, due to the requirement of different factors for growth and the relative instability of these cell types in response to serum starvation (Tai laboratory, unpublished observations). NOTE: EBM-2 basal is utilized during the assay for the single and triple culture meshwork formation and disruption assays. This is critical to prevent confounding stressor or treatment dependent signaling.
2. Meshwork-like Formation and Disruption Assays
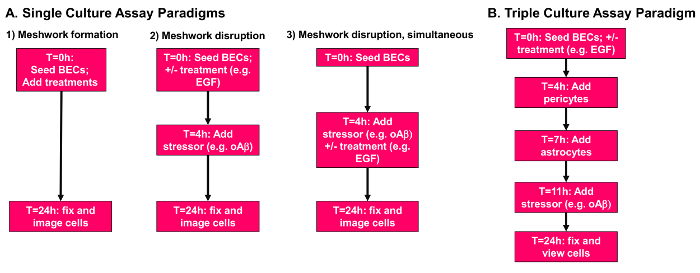
- Single BEC culture assays: NOTE: Three different paradigms for the single cultures of BECs are shown in Figure 1A. Each paradigm is designed to assess the response of BECs in different models of vessel coverage. Paradigm 1 is meshwork formation. This paradigm is designed to examine the effects of stressors and/or treatments on angiogenesis and meshwork formation. BECs, oAβ, and treatments are all added to the plate at the 0 h time point. Paradigm 2 is prevention of meshwork disruption. Cells are plated in the presence of a growth factor or drug and incubated for 4 h to form meshwork-like structures. A stressor, oAβ in this case, is added after 4 h. This paradigm assesses the ability of a treatment to prevent stressor-induced damage to preformed meshwork-like structures. Paradigm 3 is simultaneous treatment of meshwork disruption. Cells are plated and allowed to form meshwork-like structures for 4 h. Treatments and oAβ are then added simultaneously at the 4 h time point, assessing the ability of the preformed cell network to respond to various treatments. All the paradigms follow similar common steps, except with differences in the timing of treatment addition.
- On the day of the assay, pipette the basement membrane matrix at 10 µL/well into the bottom of 96-well plates and allow to set for 1 h at 37 °C.
- Suspend the lyophilized green cell tracking dye (see Table of Materials) in 10 µL DMSO to generate a 10-mM stock solution, and dilute to 10 µM (1:1000) in EBM-2 basal. Remove the media from the fully confluent flask and replace with EBM-2 basal containing the green cell tracking dye (5 mL for a 75 cm2 flask). Preload BECs with the green cell tracking dye, 20 min prior to the start of the assay.
- Incubate the cells for 20-30 min at 37 °C, remove the medium with cell tracking dye, and detach the cells as described in Step 1.1. Neutralize the media with EBM-2 containing 10% FBS and centrifuge at 240 x g for 5 min at 4 °C. Resuspend the pellet in 1 mL of EBM-2 basal.
- Plate the BECs in the 96-well plate at 10,000 cells/well in EBM-2 basal (0 h time point in all paradigms). NOTE: Depending on the paradigm being utilized, treatments, stressors, or vehicle controls are added at the specified time points. In all paradigms, the medium is harvested for subsequent analysis (e.g. ELISA analysis) and cells are fixed at 24 h with freshly prepared 4% paraformaldehyde in Phosphate Buffered Serum (PBS). As an alternative approach to determine the term effects of oAβ on mesh formation, the protocol could be modified to pre-incubate confluent cell culture flasks with oAβ, and then perform the meshwork formation paradigms (+/-oAβ). NOTE: The final volume in each well for all paradigms is 70 µL. All treatments (oAβ, EGF, etc.) are added to their respective wells at a 1:20 dilution i.e. 3.5 µL per treatment. For Paradigm 1, the cells are added in a cell suspension solution at 63 µL/well. Immediately oAβ, desired treatments, and controls are added at 3.5 µL/well each, giving a total of 70 µL. For Paradigm 2, 63 µL of cell suspension and 3.5 µL of treatment (e.g. 100 ng/mL EGF) are added at the 0 h time point. After 4 h incubation, 3.5 µL of an oAβ stock is added (e.g. for 100 nM final oAβ concentration, 3.5 µL of 2 µM oAβ stock). For Paradigm 3, 63 µL of cell suspension is added at the 0 h time point and at 4 h, oAβ and desired treatments are added at 3.5 µL/well each, giving a total of 70 µL. These volumes can be adjusted according to treatments. For example, if only one treatment is required, the cell suspension volume can be adjusted to 66.5 µL (maintaining 10,000 cells/well) and the treatment volume can remain at 3.5 µL.
- Triple culture assay: NOTE: In addition to the single culture assays of BECs, we have developed a triple culture assay paradigm (Figure 1B) to determine the response of BECs, pericytes, and astrocytes within preformed networks to relevant stressors (e.g.oAβ). BECs are plated in the presence of desired treatments at 0 h. At 4 h, pericytes are gently added to the plate. At 7 h, astrocytes are added to the plate, followed by the addition of a stressor at 11 h. Cells are then incubated until the 24 h time point. NOTE: For the triple culture assays, it is important to consider timing to ensure all cells are confluent at the same time. Human astrocytes and pericytes should be cultured 1 week before the assay, whereas the hCMEC/D3 cells need to be plated or passaged 4-5 days before the assay date. Further, it is important to use low passage (2-5) primary cells to avoid phenotypic drift.
- Add the green cell tracking dye working solution to the hCMEC/D3 cells 20 min prior to starting the assay. Plate the hCMEC/D3 cells at 10,000 cells/well in EBM-2 basal (0 h time point). The volumes used are 45 µL of cell suspension and 3.5 µL of treatment (i.e. final EGF concentrations of 50 ng/mL, 100 ng/mL, or 1,000 ng/mL from stocks that are 20 times more concentrated).
- Following the 3.5 h incubation at 37 °C, add the blue cell tracking dye working solution (10 µM cell tracker blue in EBM-2 basal) to the human primary pericytes. At the 4 h time point (~30 min incubation with cell tracking dye), gently add 2,000 pericytes/well to the plate containing the hCMEC/D3 in a volume of 6 µL/well.
- Following an additional 2.5 h incubation (total 6.5 h from 0 h time point), add the orange cell tracking dye working solution (10 µM cell tracker orange in EBM-2 basal) to the human primary astrocytes. At the 7 h time point, gently add 10,000 astrocytes/well to the plate in 12 µL volume. Therefore, the overall cellular ratio for endothelial cells:pericytes:astrocytes is 5:1:5. Further, the volume achieved to this point is 66.5 µL.
- Add oAβ (3.5 µL of 100 µM stock) 4 h later (total 11 h from 0 h time point).
- Fix the cells 13 h later in freshly prepared 4% paraformaldehyde in PBS. Caution: Wear protective clothing, gloves and eyeglasses while handling paraformaldehyde.
3. Quantification
Capture fluorescence images of the whole well of the 96-well plate at 1.6X magnification with a 2 s exposure time at 50% maximum power using a dissecting microscope. NOTE: Equivalent microscopes can be utilized; however, it is critical to capture the entire well for comprehensive analysis.
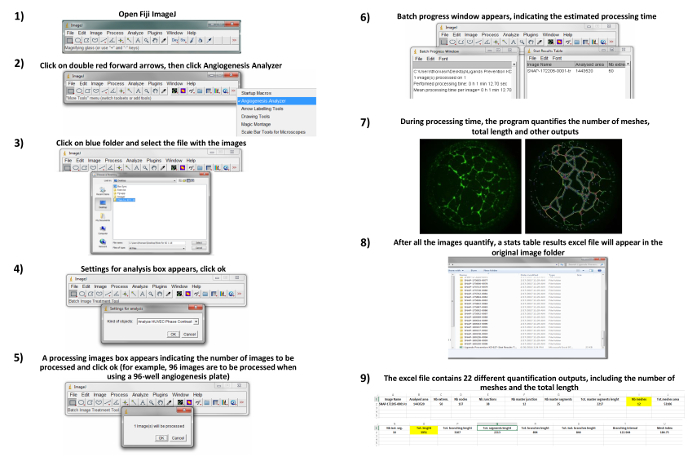
- For quantitative analysis, process the images using the ImageJ angiogenesis analyzer plugin22 to quantify the number of branches, number of meshes, and total cell length (these readouts are automatically tabulated by the software) as described below. A detailed visual protocol of this step is provided in Figure 2.
- Open fiji ImageJ by clicking on the software icon.
- Click on the double red forward arrows located at the end of the toolbar, then click "Angiogenesis Analyzer".
- Select the folder with image files, click "Open".
- When the "settings for analysis" box appears, click "OK".
- When the "processing images" box appears, which indicates the number of images to be processed, click "OK".
- When the "batch progress window" box appears, which indicates the estimated processing time. During this time the program quantifies the outputs including the number of branches, number of meshes, and total cell length. NOTE: Once all the images are quantified, the results file will appear in the original image folder as a spreadsheet document.
- Select the spreadsheet file containing results and compare the number of branches, meshes and total cell length between groups. NOTE: In the triple culture assay ImageJ is further utilized to analyze pericyte/astrocyte coverage and number. Images are thresholded by a blinded experimenter and the "analyze particles" function utilized to generate readouts. Fixed cells and tube-like structures can also be immunostained for Aβ17 or other markers.
Representative Results
In single cultures, both the hCMEC/D3 cells (Figure 3A) and the primary mouse BECs (Figure 3B) form meshwork-like structures throughout the well. The structures are characterized by a meshwork of interlinked nodes (Figure 3). In all the paradigms described (Figure 1), the meshwork-like structures are similar after 24 h in the control groups, forming ~20 meshwork-like structures with a total cellular length of ~10,000 pixels.
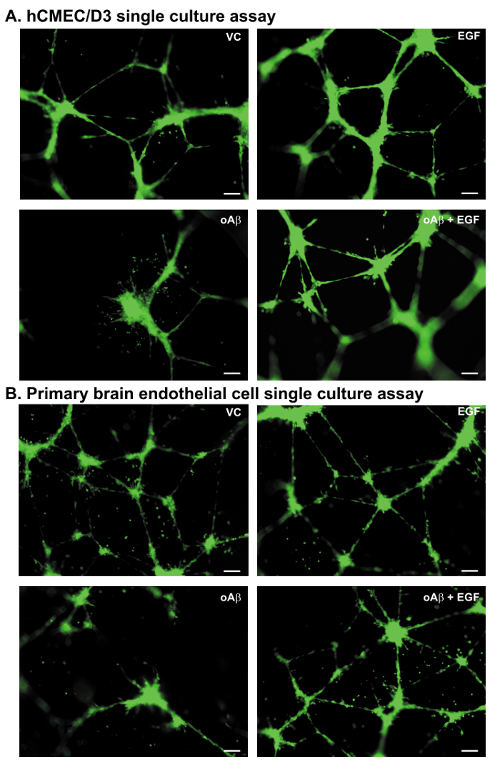
To highlight the applicability of the methods to disease relevant stressors, oAβ was added to the hCMEC/D3 and the primary mouse BECs in two paradigms. In the disruption of cell meshwork formation (Paradigm 1), oAβ and cells are plated at the same time. In the disruption of preformed meshwork (Paradigm 2), oAβ is added 4 h after plating the cells (see Figure 1A). oAβ at 100 nM induces disruption of the meshwork-like formation and degeneration of preformed meshwork (Figure 3A, 3B). For example, using the hCMEC/D3 cells in both paradigms, quantification of the total cell coverage/length is 16-20% lower with 100 nM oAβ17. Further, the number of meshes is reduced by 40% with 100 nM oAβ in both paradigms17. Thus, a disease relevant stressor induces a similar detrimental effect on human and mice BEC coverage.
A key advantage of the cell culture system is the ability to identify factors or treatments that prevent disease-relevant damage, which can then advance to in vivo testing. As a proof of principle experiment, the effects of the main angiogenic growth factors on preventing oAβ-induced changes to vessel coverage were assessed17. The EGF prevented oAβ-induced damage to the hCMEC/D3 cells. Based on those data, EGF was tested in a prevention paradigm using a transgenic mouse model that recapitulates critical aspects of AD-like pathology. EGF treatment prevented the cognitive and BBB deficits, including vessel degeneration23. These data support the predictive potential of the in vitro system for in vivo activity. Currently, we utilize all three developed paradigms for screening: 1) mesh formation, 2) prevention of cell meshwork disruption, and 3) simultaneous treatment of meshwork disruption. As highlighted in Figure 3, EGF can protect against oAβ-induced damage in immortalized and primary mouse BEC cultures.
The BBB consists of BECs, pericytes and astrocytes that collectively contribute to overall cerebrovascular coverage. Therefore, one adaptation of the in vitro system is the incorporation of all three BBB cell types. Our triple culture assay paradigm incorporates the hCMEC/D3 cells, primary human pericytes and primary human astrocytes, which are added sequentially to the basement membrane matrix (Figure 1B). In the triple culture assay paradigm, the hCMEC/D3 cells form a similar meshwork pattern as in the single cultures, but with the pericytes and astrocytes attached to the nodes and connecting branches (Figure 4). The addition of 100 nM oAβ induces meshwork disruption (~10-15%) and a reduction in the number of pericytes contacting the BECs17. In correlation with the data derived from the single culture assay paradigms, oAβ-induced damage is prevented by EGF treatment. Thus, the triple culture BBB model is well suited for studies focused on the interactive effects of astrocytes, pericytes and BECs on vessel dynamics.
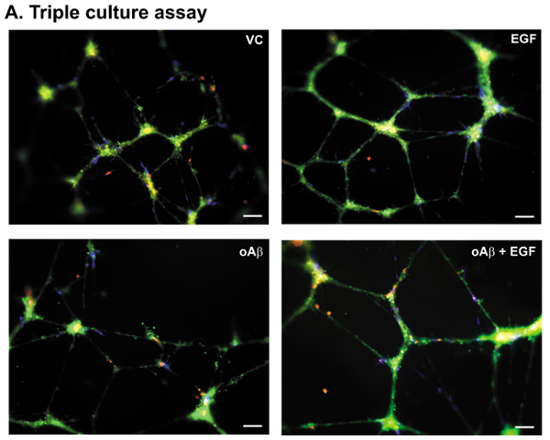
Figure 1: Overview of meshwork formation and disruption assays. (A) Single culture assay paradigms. Paradigm 1, meshwork formation. Cells, stressors and treatments are all added to the plate at the 0 h time point. Paradigm 2, prevention of meshwork disruption. Cells are plated in the presence of desired treatments, incubated for 4 h, and a stressor is added at the 4 h time point. Paradigm 3, simultaneous treatment of meshwork disruption. Cells are plated and allowed to form meshwork-like structures for 4 h before treatments and/or stressors are added simultaneously at the 4 h time point. All paradigms end at the 24 h time point, and cells are fixed with 4% paraformaldehyde. (B) Triple culture assay paradigm. BECs (10,000 cells/well) are plated in the presence of desired treatments at 0 h. At the 4 h time point pericytes (2,000 cells/well) are gently added to the plate. At 7 h astrocytes (10,000 cells/well) are added to the plate, followed by the addition of relevant stressors at 11 h. Cells are then incubated until the 24 h time point and fixed with 4% paraformaldehyde. Please click here to view a larger version of this figure.
Figure 2: Quantitative analysis. All images are opened on Fiji ImageJ and batch-processed using the Angiogenesis Analyzer22. Quantification of total length and number of meshes are utilized. Please click here to view a larger version of this figure.
Figure 3: Meshwork disruption assays: representative images. All images are derived from experiments utilizing Paradigm 2, meshwork disruption.(A) Representative images of the hCMEC/D3 cells plated and treated with vehicle control (VC), EGF (100 nM), oAβ (100 nM), or oAβ (100 nM) + EGF (100 nM). Images at 10X magnification, Scale bar = 100 µm. (B) Representative images of the primary mouse BECs plated and treated with VC, EGF (100 nM), oAβ (100 nM) or oAβ (100 nM) + EGF (100 nM). Images at 10X magnification, green = BECs, Scale bar = 100 µm. Please click here to view a larger version of this figure.
Figure 4. Triple culture assay: representative images. (A) Representative images of the hCMEC/D3 cells, primary human pericytes, and primary human astrocytes plated and treated with VC, EGF (100 nM), oAβ (100 nM), or oAβ (100 nM) + EGF (100 nM) according to the paradigm described in Figure 1B. Images at 10X magnification, green = BECs, blue = pericytes, and red (pseudocolor) = astrocytes. Scale bar = 100 µm. Please click here to view a larger version of this figure.
Discussion
The methods described can be utilized to address several fundamental biological questions surrounding cerebrovascular coverage24. Specifically, they can identify which receptors and signaling pathways play a role in angiogenesis, vessel coverage in cancer tissue, and peripheral endothelial cells relevant to the brain. Examples include angiogenic growth factor receptors, nitric oxide, mitogen activated protein kinase signaling and calcium signaling25,26,27. The hCMEC/D3 and primary BECs are amendable to genetic knockdown approaches to facilitate this effort18. Mechanistic insight can be gained from culturing BECs isolated from mice with complete or endothelial cell-specific knockdown of key proteins with the methods described. Further, the triple culture assays enable in-depth analysis of the astrocyte and pericyte influence on the brain endothelial cell function. For example, pericytes and astrocytes can influence meshwork-like vessel formation through the production of soluble mediators (e.g. angiopoietin, cytokines) and also via structural support24.
The model system can also be applied to disease research. For example, the system can address whether disease- and aging-relevant stressors added exogenously promote angiogenesis and/or meshwork disruption. Stressors can be relevant to a specific disorder, e.g. oAβ, or more generalized, e.g. hydrogen peroxide, cytokines. The triple culture paradigm adds an additional layer of complexity that may improve the understanding of critical, disease-relevant mechanisms that influence BBB dynamics: delineating whether disease relevant stressors activate astrocytes and pericytes to disrupt BEC function. For both single and triple culture assays, the primary cells can be isolated from mouse models that mimic disorders of interest to further delineate functional effects.
There are a number of important considerations for adapting the meshwork-like formation and disruption assays to different cells types. The first is the seeding density. For each new cell line, a critical first step is optimizing the seeding densities in the basement membrane matrix for both the single and triple culture assays. The mesh formation is cell number dependent, as both too high and too low cell density result in a lack of meshwork formation. Further, even for the methods and cells described herein, it is prudent to conduct cell density comparisons to account for any laboratory variations in cell processing prior to conducting larger scale experiments. The second consideration is the temporal sequence of the meshwork formation. Time course experiments should be conducted to identify when meshwork-like structures form and degrade. Typically, a robust meshwork-like formation is observed at ~4 h. The third consideration is the choice of medium prior to seeding and during the experiment. BECs, pericytes and astrocytes are cultured in medium that contains FBS and a plethora of growth factors optimized for cell growth. Although it is preferred to serum and growth factor starve the BECs 24 h prior to the experiment, for certain cell types this may not be feasible. For example, primary cells with specific receptor knockdowns may require additional factors to facilitate growth. These considerations also apply during the experiment, and the choice of medium is dependent on the specific question under investigation. However, when investigating the role of exogenously added stressors and potential treatments, particularly growth factors or related compounds, the use of serum and growth factor free medium during the experiment is ideal. In addition, it may be possible to reduce the length of serum starvation of the BECs, but this is dependent on the signaling pathway under investigation. A fourth consideration is the timing of adding stressors or treatments. As for the choice of medium treatment, the timings are based on the scientific question. The meshwork formation assay is more analogous to angiogenesis, whereas the meshwork disruption assay may be more relevant for a mature BBB. In our experience, once established, the assays produce consistent data between experiments. Consistency issues are often due to seeding density discrepancies between personnel, and importantly, if components of the medium or relevant reagents are unknowingly altered.
There are limitations and areas of further development of the methods described. A strength of this procedure is that low concentrations of stressor (i.e. oAβ) can be utilized to induce meshwork disruption i.e. nM compared to µM, which are more physiologically relevant. However, this strength may also be a limitation. Indeed, higher, but still non-toxic, concentrations of oAβ may be required for drug screening purposes to increase sensitivity, which will apply to other disease relevant stressors. A limitation of the method is that the meshwork of cells is only stable over 24 h in the protocols described. Indeed, after 24 h, the cell meshworks begin to degenerate. Therefore, we add oAβ after a relatively short time (4 h) after the meshwork formation, to enable a total oAβ incubation time of 18 h. Perhaps lower cell seeding densities may enable longer treatment protocols. A further potential limitation is that the in vitro system may not mimic a stable meshwork of cells as would be observed in in vivo. Isolating the cells from aged mice may more closely mimic the in vivo scenario.
Disclosures
The authors have nothing to disclose.
Acknowledgments
Leon Tai is funded by University of Illinois Chicago start-up funds.
References
- Abbott NJ. Blood-brain barrier structure and function and the challenges for CNS drug delivery. J Inherit Metab Dis. 2013;36(3):437–449. doi: 10.1007/s10545-013-9608-0. [DOI] [PubMed] [Google Scholar]
- Engelhardt B, Liebner S. Novel insights into the development and maintenance of the blood-brain barrier. Cell Tissue Res. 2014;355(3):687–699. doi: 10.1007/s00441-014-1811-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbott NJ, Patabendige AA, Dolman DE, Yusof SR, Begley DJ. Structure and function of the blood-brain barrier. Neurobiol Dis. 2010;37(1):13–25. doi: 10.1016/j.nbd.2009.07.030. [DOI] [PubMed] [Google Scholar]
- Zlokovic BV. Neurovascular pathways to neurodegeneration in Alzheimer's disease and other disorders. Nat Rev Neurosci. 2011;12(12):723–738. doi: 10.1038/nrn3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardridge W. Targeted delivery of protein and gene medicines through the blood-brain barrier. Clin Pharmacol Ther. 2014;97(4):347–361. doi: 10.1002/cpt.18. [DOI] [PubMed] [Google Scholar]
- Tai LM, et al. The role of APOE in cerebrovascular dysfunction. Acta Neuropathol. 2016;131(5):709–723. doi: 10.1007/s00401-016-1547-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biron KE, Dickstein DL, Gopaul R, Jefferies WA. Amyloid triggers extensive cerebral angiogenesis causing blood brain barrier permeability and hypervascularity in Alzheimer's disease. PLoS One. 2011;6(8):e23789. doi: 10.1371/journal.pone.0023789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron DJ, et al. Alzheimer's-related peptide amyloid-beta plays a conserved role in angiogenesis. PLoS One. 2012;7(7):e39598. doi: 10.1371/journal.pone.0039598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscolo E, et al. Beta amyloid angiogenic activity in vitro and in vivo. Int J Mol Med. 2007;19(4):581–587. [PubMed] [Google Scholar]
- Paris D, et al. Impaired angiogenesis in a transgenic mouse model of cerebral amyloidosis. Neurosci Lett. 2004;366(1):80–85. doi: 10.1016/j.neulet.2004.05.017. [DOI] [PubMed] [Google Scholar]
- Kitaguchi H, Ihara M, Saiki H, Takahashi R, Tomimoto H. Capillary beds are decreased in Alzheimer's disease, but not in Binswanger's disease. Neurosci Lett. 2007;417(2):128–131. doi: 10.1016/j.neulet.2007.02.021. [DOI] [PubMed] [Google Scholar]
- Jantaratnotai N, Ryu JK, Schwab C, McGeer PL, McLarnon JG. Comparison of Vascular Perturbations in an Abeta-Injected Animal Model and in AD Brain. Int J Alzheimers Dis. 2011;2011:918280. doi: 10.4061/2011/918280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnini S, et al. Abeta peptides accelerate the senescence of endothelial cells in vitro and in vivo, impairing angiogenesis. FASEB J. 2010;24(7):2385–2395. doi: 10.1096/fj.09-146456. [DOI] [PubMed] [Google Scholar]
- Tai LM, Holloway KA, Male DK, Loughlin AJ, Romero IA. Amyloid-beta-induced occludin down-regulation and increased permeability in human brain endothelial cells is mediated by MAPK activation. J Cell Mol Med. 2010;14(5):1101–1112. doi: 10.1111/j.1582-4934.2009.00717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai LM, Loughlin AJ, Male DK, Romero IA. P-glycoprotein and breast cancer resistance protein restrict apical-to-basolateral permeability of human brain endothelium to amyloid-beta. J Cereb Blood Flow Metab. 2009;29(6):1079–1083. doi: 10.1038/jcbfm.2009.42. [DOI] [PubMed] [Google Scholar]
- Tai LM, et al. Polarized P-glycoprotein expression by the immortalised human brain endothelial cell line, hCMEC/D3, restricts apical-to-basolateral permeability to rhodamine 123. Brain Res. 2009;1292:14–24. doi: 10.1016/j.brainres.2009.07.039. [DOI] [PubMed] [Google Scholar]
- Koster KP, Thomas R, Morris AW, Tai LM. Epidermal growth factor prevents oligomeric amyloid-beta induced angiogenesis deficits in vitro. J Cereb Blood Flow Metab. 2016;36(11):1865–1871. doi: 10.1177/0271678X16669956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weksler B, Romero IA, Couraud PO. The hCMEC/D3 cell line as a model of the human blood brain barrier. Fluids Barriers CNS. 2013;10(1):16. doi: 10.1186/2045-8118-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weksler BB, et al. Blood-brain barrier-specific properties of a human adult brain endothelial cell line. FASEB J. 2005;19(13):1872–1874. doi: 10.1096/fj.04-3458fje. [DOI] [PubMed] [Google Scholar]
- Welser-Alves JV, Boroujerdi A, Milner R. Isolation and culture of primary mouse brain endothelial cells. Methods Mol Biol. 2014;1135:345–356. doi: 10.1007/978-1-4939-0320-7_28. [DOI] [PubMed] [Google Scholar]
- Dahlgren KN, et al. Oligomeric and fibrillar species of amyloid-beta peptides differentially affect neuronal viability. J Biol Chem. 2002;277(35):32046–32053. doi: 10.1074/jbc.M201750200. [DOI] [PubMed] [Google Scholar]
- Carpentier G. Angiogenesis Analyzer. ImageJ News. 2012.
- Thomas R, et al. Epidermal growth factor prevents APOE4 and amyloid-beta-induced cognitive and cerebrovascular deficits in female mice. Acta Neuropathol Commun. 2016;4(1):111. doi: 10.1186/s40478-016-0387-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai LM, et al. The role of APOE in cerebrovascular dysfunction. Acta Neuropathol. 2016;131(5):709–723. doi: 10.1007/s00401-016-1547-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose CT. Neuroangiogenesis: a vascular basis for Alzheimer's disease and cognitive decline during aging. J Alzheimers Dis. 2012;32(3):773–788. doi: 10.3233/JAD-2012-120067. [DOI] [PubMed] [Google Scholar]
- Ambrose CT. A therapeutic approach for senile dementias: neuroangiogenesis. J Alzheimers Dis. 2015;43(1):1–17. doi: 10.3233/JAD-140498. [DOI] [PubMed] [Google Scholar]
- Ambrose CT. The Role of Capillaries in the Lesser Ailments of Old Age and in Alzheimer's Disease and Vascular Dementia: The Potential of Pro-Therapeutic Angiogenesis. J Alzheimers Dis. 2016;54(1):31–43. doi: 10.3233/JAD-160303. [DOI] [PubMed] [Google Scholar]


