Abstract
Claudin-low breast cancer (CLBC) is a poor prognosis molecular subtype showing stemness and mesenchymal features. We previously discovered that deletion of a Notch signaling modulator, Lunatic Fringe (Lfng), in the mouse mammary gland induced a subset of tumors resembling CLBC. Here we report that deletion of one copy of p53 on this background not only accelerated mammary tumor development but also led to a complete penetrance of the mesenchymal stem-like phenotype. All mammary tumors examined in the Lfng/p53 compound mutant mice displayed a mesenchymal/spindloid pathology. These tumors showed high level expressions of epithelial-to-mesenchymal transition (EMT) markers including Vimentin, Twist, and PDGFRα, a gene known to be enriched in CLBC. Prior to tumor onset, Lfng/p53 mutant mammary glands exhibited increased levels of Vimentin and E-cadherin, but decreased expressions of cytokeratin 14 and cytokeratin 8, accompanied by elevated basal cell proliferation and an expanded mammary stem cell-enriched population. Lfng/p53 mutant glands displayed increased accumulation of Notch3 intracellular fragment, up-regulation of Hes5 and down-regulation of Hes1. Analysis in human breast cancer datasets found the lowest HES1 and second lowest LFNG expressions in CLBC among molecular subtypes, and low level of LFNG is associated with poor survival. Immunostaining of human breast cancer tissue array found correlation between survival and LFNG immunoreactivity. Finally, patients carrying TP53 mutations express lower LFNG than patients with wild type TP53. Taken together, these data revealed genetic interaction between Lfng and p53 in mammary tumorigenesis, established a new mouse model resembling CLBC, and may suggest targeting strategy for this disease.
Introduction
Breast cancer is a heterogeneous group of molecularly distinct diseases of different cellular origins associated with diverse oncogenic signaling. Based on gene expression profiling, breast cancer was initially subclassified into five groups: luminal A, luminal B, HER2-positive, basal-like, and normal-like breast cancer [1], [2]. Subsequent study found that basal-like breast cancer (BLBC) included another subtype, termed claudin-low breast cancer (CLBC) [3]. In addition, the triple negative breast cancer (TNBC), a collection of loosely defined diseases on the basis of being negative for the expression of estrogen receptor α (ERα), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2), can be subdivided into six subgroups: basal-like 1, basal-like 2, immunomodulatory, mesenchymal, mesenchymal stem-like, and luminal androgen receptor-expressing [4]. The mesenchymal stem-like subgroup is similar to CLBC. BLBC expresses markers of myoepithelial/basal cells and are thought to have originated from mammary bipotent and/or luminal progenitor cells [5], [6]. CLBC shares more features with mammary stem cells and cells that have undergone epithelial-to-mesenchymal transition (EMT), thus may have originated from mammary stem cells [3], [7], [8], [9]. These two subtypes have a high proliferation index and poor cellular differentiation, often harboring recurrent copy number abnormalities and mutations in the TP53 gene [1], [10], [11]. BLBC and CLBC preferentially affect younger patients, are more common in women of African ancestry than women of other ethnic origins [12], [13].
Notch signaling controls mammary stem cell self-renewal and differentiation, and regulates EMT in breast cancer cells [14], [15], [16], [17]. Deregulated Notch activation has been implicated in breast cancer, especially in subtypes with stem-like features and EMT [18]. The Fringe family of β3 N–acetylglucosaminyl-transferases, including Lunatic fringe (Lfng), Manic Fringe (Mfng), and Radical Fringe (Rfng), are known to modify EGF repeats in the extracellular domains of Notch receptors thereby modulating ligand-mediated Notch activation [19], [20], [21]. Interestingly, Lfng expression is restricted to the stem/progenitor cells in the mouse mammary gland. Mammary-specific deletion of Lfng in mice using MMTV-Cre induced mammary tumors with consistent selection of an amplicon including the Met and Cav1, Cav2 genes. Histology, immunohistochemistry, and molecular profiling indicated that two-thirds of these tumors are similar to BLBC and one-third resembles CLBC [22].
TP53 is the most commonly mutated gene in breast cancer and its mutation rate is significantly higher in basal-like compared to other subtypes [11]. Interestingly, loss of p53 function has recently been linked to the induction of EMT and acquisition of stemness properties. Deletion of p53 in mammary epithelial cells resulted in decreased expression of miR-200c and activated EMT program, accompanied by an increase in mammary stem cell population [23]. Thus, p53 deficiency may contribute to the pathogenesis of mesenchymal stem-like breast cancer. Indeed, deletion of p53 in combination with other genetic alterations (e.g. inactivation of Rb or Pten, overexpression of prolactin or Met) resulted in mammary tumors resembling CLBC [24], [25], [26], [27]. A recent study showed that wild type p53 suppressed transcriptional activity of Notch1 through direct interaction with the Notch transcriptional complex [28]. In this study, we generated Lfng and p53 compound deletion mice to determine whether these two genes cooperate in suppressing CLBC development.
Materials and Methods
Mice
Mouse experiments were performed in accordance with a protocol approved by UMMC Institutional Animal Care and Use Committee. All mouse strains have been previously described [22], [29]. Lfngflox/flox, p53flox/flox, and MMTV-Cre strains were interbred and maintained on a mixed FVB/C57BL/6 background.
Mammary Gland Histology and Immunohistochemistry
Human breast tissue array slides HBre-Duc090Sur-01 were purchased from US Biomax. Formalin-fixed paraffin-embedded mouse mammary tissues were processed for histology and immunohistochemistry by standard procedures. Representative images were acquired with a Nikon Eclipse 80i microscope. Primary antibodies used for immunostaining were: Notch1 (Cell Signaling, No. 3608, 1:100), Notch2 (DSHB, University of Iowa, C651.6DbHN, 1:200), Notch3 (ProteinTech, 55,114–1-AP, 1:100), E-cadherin (Cell Signaling, No. 3195, 1:100), Vimentin (Cell Signaling, No. 5741, 1:100), Twist (Santa Cruz, sc-6070, 1:100), PDGFRα (Santa Cruz, sc-338, 1:100), Cytokeratin 8 (Santa Cruz, sc-101,459, 1:200), Cytokeratin 14 (Santa Cruz, sc-53,253, 1:200), Ki67 (Abcam, ab16667, 1:200), and Lfng (Abcam, ab192788, 1:100).
Western Blot Analysis
Mouse mammary tissues were lysed in RIPA buffer (Boston BioProducts) supplemented with protease inhibitor (Roche), and processed for Western blot analyses according to standard methodology. Antibodies for probing specified proteins are as follows: Notch1 (Cell Signaling, No. 3608), Notch2 (DSHB, University of Iowa, C651.6DbHN), Notch3 (ProteinTech, 55,114–1-AP), Notch4 (Millipore, 07–189), Jagged1 (Santa Cruz, sc-6011), ERα (Santa Cruz, sc-542), Vimentin (Cell Signaling, No. 5741), E-cadherin (Cell Signaling, No. 3195), Cytokeratin 8 (Santa Cruz, sc101459), Cytokeratin 14 (Santa Cruz, sc53253), and β-Actin (Santa Cruz, sc-81,178), all with 1:1000 dilution. Western blots were performed three times using tissues from different animals and obtained similar results.
Flow Cytometry and Mammosphere Assay
Mouse mammary tissues were dissociated using Collagenase/Hyaluronidase solution (StemCell Technologies) and single cell suspensions were generated according to manufacturer's instructions. Lineage-depleted mammary epithelial cells were prepared using an EasySep mouse mammary stem cell enrichment kit (StemCell Technologies). Flow cytometry was performed by standard procedures using anti-mouse CD24-PE and anti-CD49f-FITC (StemCell Technologies). Fluorescence was recorded using Gallios Flow Cytometer (Beckman Coulter) and analyzed with Kaluza flow cytometry analysis software. Mammary tissues from three animals per genotype were analyzed with similar results. Percentages of stem cell-enriched cell populations in each genotype were quantitated. Lineage-depleted mammary epithelial cells were cultured for mammosphere formation for 7 days as previously described [30].
Quantitative RT-PCR
Total RNA was extracted from mouse mammary tissue using RNeasy Mini kit (Qiagen), and reverse transcription was performed with iScript cDNA synthesis kit (Bio-rad). PCR was performed using QuantiTect SYBR Green PCR Kits (Qiagen) and quantitated with BioRad CFX96 qPCR System. Primer sequences for mouse Hes1, Hes5, Hey1, Hey2 genes were previously described [31]. Relative abundance of mRNA for each gene to GAPDH was determined using the equation 2-△CT, where ΔCT=CTgene tested – CTGAPDH. Three animals per genotype were used for quantitative RT-PCRs.
Gene Expression Analysis of Human Data Sets
Human breast cancer gene expression data set GSE18229 (n = 372) was downloaded from GEO (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE18229). Data were z-transformed and applied quantile normalization by preprocessCore package from Bioconductor in R software. Gene expressions were summarized by average value if multiple probes mapping to the same gene. Preprocessed METABRIC breast cancer expression data (n = 2509) was downloaded from cBioportal web portal (http://www.cbioportal.org/). Differential expressions of LFNG, NOTCH3, JAG1, HES1 and HEY1 among different breast cancer subtypes were analyzed by one way ANOVA test. p values were calculated by comparing expression means across all subtypes. For the survival analysis related to LFNG expression (probeset 228762_at), we used online tool (http://www.kmplot.com/) to perform univariate Cox regression analysis and Kaplan–Meier survival curves with lower quartile as cutoff. The hazard ratio with 95% confidence and p values were calculated from 1402 patients with overall survival, 3955 patients with recurrence free survival, and 1747 patients with post progression survival information for breast cancer.
Statistics
For mouse experiments, statistical analyses were performed using SigmaPlot (Systat Software) by applying the Log-rank test to compare survival data and the unpaired two-tailed t-test for all other comparisons.
Results
Accelerated Mammary Tumor Development in the Lfngflox/flox;p53flox/+;MMTV-Cre Mice
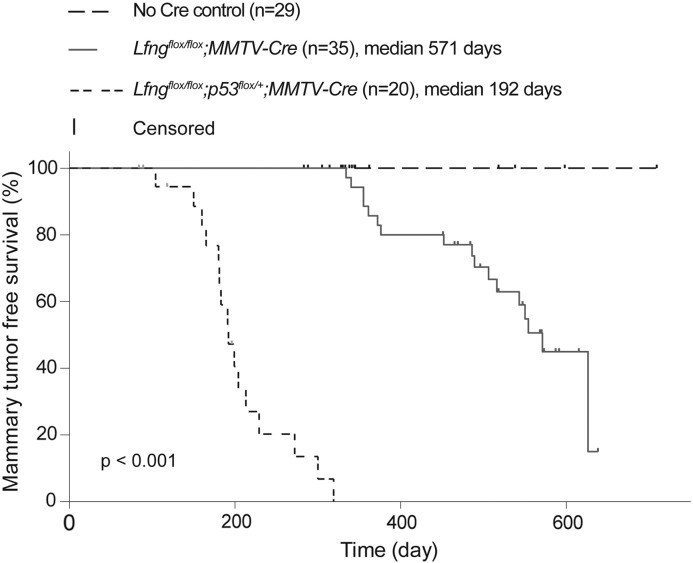
To test the impact of deletion of p53 on BLBC and CLBC development, a conditional knockout allele of p53 was introduced into the Lfngflox/flox;MMTV-Cre mice. All 22 mice of Lfngflox/flox;P53flox/flox;MMTV-Cre (18 females and 4 males) under tumor watch died of thymic lymphomas and metastasis between 3 and 6 months of age (data not shown). Thus, homozygous deletion of both Lfng and p53 using MMTV-Cre was untolerated in the hematopoietic lineage, precluding examination of mammary tumorigenesis in these mice. Of note, a previous study found thymic lymphomas in conditional deletion of DNA polymerase ζ catalytic subunit REV3L using MMTV-Cre, with decreased latency and higher incidence on the p53-null background [32]. Three out of 20 females of Lfngflox/flox;P53flox/+;MMTV-Cre also succumbed to lymphoma prior to mammary tumor formation. The remaining animals developed mammary tumors without lymphoma, and they had drastically shortened median tumor-free survival as compared to the Lfngflox/flox;MMTV-Cre mice (192 and 571 days, respectively, Figure 1). In addition, these animals often developed multiple mammary tumors (1.8 tumors/mouse in average), which was extremely rare in Lfngflox/flox;MMTV-Cre mice. Despite rapid mammary tumor development, no metastasis was found in Lfngflox/flox;P53flox/+;MMTV-Cre mice when animals had to be euthanized due to large-size mammary tumors.
Figure 1.
Drastically accelerated mammary tumor development in the Lfngflox/flox;p53flox/+;MMTV-Cre mice as compared to the Lfngflox/flox;MMTV-Cre mice. Kaplan–Meier analysis was performed for mammary tumor-free survival in female mice of these two genotypes. Wild type, Lfngflox/flox, and Lfngflox/flox;p53flox/+ animals were included in the “No Cre” control.
Lfngflox/flox;p53flox/+;MMTV-Cre Mammary Tumors Exhibit Characteristics of Mesenchymal Stem-Like/Claudin-Low Breast Cancer
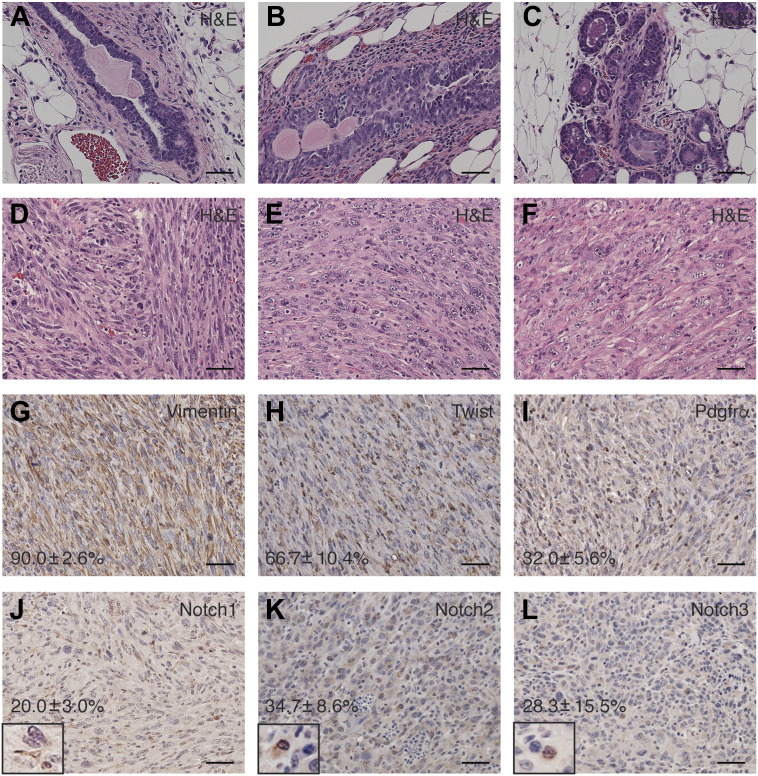
Histological analysis revealed presence of ductal hyperplasia and atypia (Figure 2, B and C) among relatively normal ducts (Figure 2A) in the Lfngflox/flox;p53flox/+;MMTV-Cre mammary gland prior to tumor onset. Interestingly, all the Lfngflox/flox;P53flox/+;MMTV-Cre mammary tumors were poorly differentiated, mainly consisted of mesenchymal/spindle-shaped cells, similar to the claudin-low tumors found in the Lfngflox/flox;MMTV-Cre mice (Figure 2, D–F) [22]. Immunohistochemistry showed robust expressions of Vimentin in approximately 90% of tumor cells, suggesting that these cells have undergone EMT. In addition, these tumors expressed high levels of Twist and PDGFR-α, which are known to be associated with EMT and highly enriched in CLBC (Figure 2, G–I) [4]. Immunoreactivities of Notch receptors, including nuclear staining, were detected in a subset of cells in most tumors, indicating activation of the Notch signaling pathway (Figure 2, J–L). These results suggest that deletion of one copy of p53 on the Lfngflox/flox;MMTV-Cre background caused skewing of mammary tumor subtype from BLBC to CLBC.
Figure 2.
Histological and immunohistochemical analyses of preneoplastic lesions and mammary tumors in the Lfngflox/flox;p53flox/+;MMTV-Cre mice. (A-C) Representative photomicrographs of H&E stained mammary gland sections at 2 months of age. (D-F) Representative photomicrographs of H&E stained mammary tumor sections. (G-L) Representative photomicrographs of immunostaining in mammary tumors. Insets: high-magnification images of Notch nuclear staining. Percentage of positively stained cells are quantitated and presented as mean ± standard deviation. Scale bars: 50 μm.
Increased Basal Cell Proliferation, Decreased Differentiation, and Expanded Stem Cell-Enriched Population in the Lfngflox/flox;p53flox/+;MMTV-Cre Mammary Gland
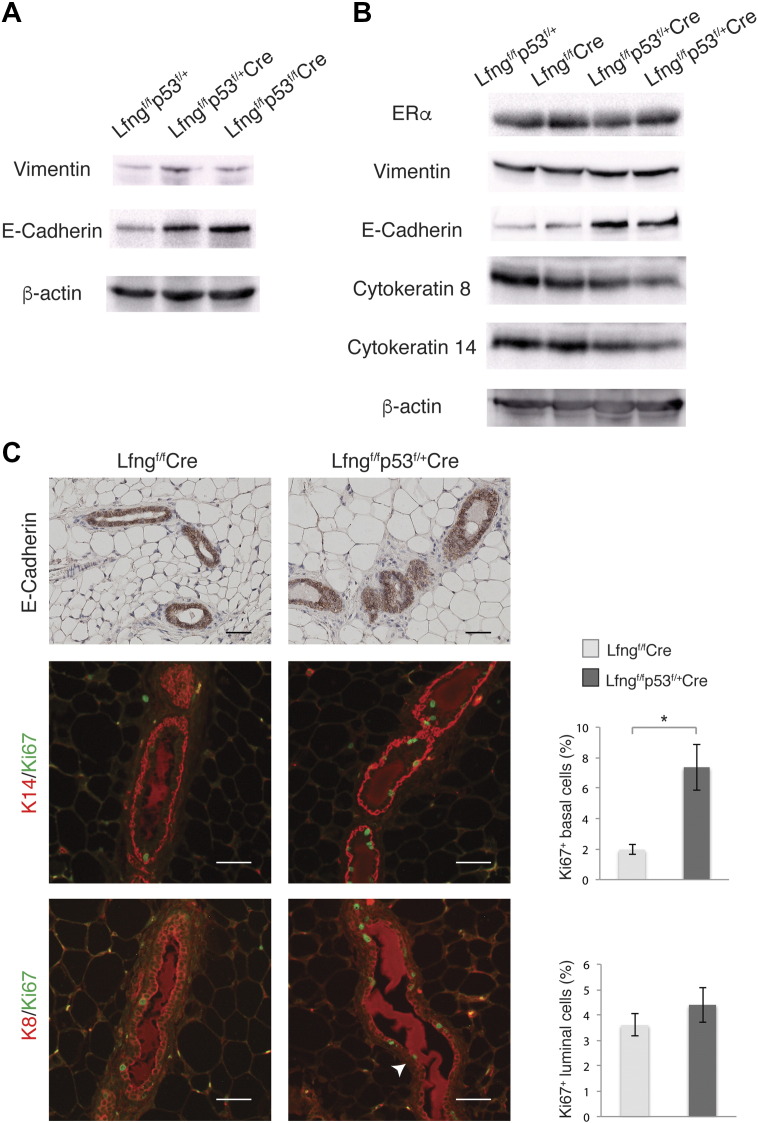
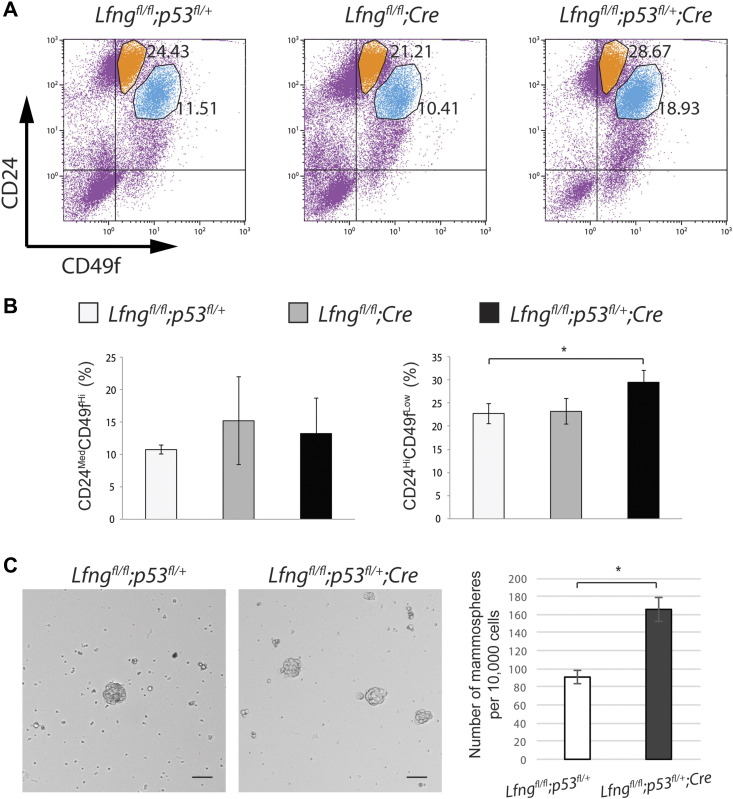
We examined mammary tissues for epithelial cell proliferation and differentiation prior to the tumor onset. Western blot analysis for mammary tissues from littermate mice at 3 months of age showed increased levels of both Vimentin and E-cadherin in the Lfngflox/flox;P53flox/+;MMTV-Cre and Lfngflox/flox;P53flox/flox;MMTV-Cre mice, as compared to the Lfngflox/flox;P53flox/+ control mice (Figure 3A). In another litter including 4 females, both of Lfngflox/flox;P53flox/+;MMTV-Cre mice exhibited significantly increased levels of E-cadherin and slightly increased Vimentin, as compared to the Lfngflox/flox;MMTV-Cre and Lfngflox/flox;P53flox/+ control mice. Interestingly, expressions of Cytokeratin 8 and Cytokeratin 14 were down-regulated in these 2 mice, while their ERα level was similar to the control mice. It is noteworthy that expressions of these proteins in the Lfngflox/flox;MMTV-Cre gland were comparable to those in the Lfngflox/flox;P53flox/+ control at this stage (Figure 3B). Immunohistochemistry showed that E-Cadherin is localized to the ductal epithelial cells, with comparable intensity in Lfngflox/flox;MMTV-Cre and Lfngflox/flox;P53flox/+;MMTV-Cre glands, suggesting that elevated E-Cadherin level shown in Western blot may reflect increased number of epithelial cells. Indeed, Ki67/Cytokeratin 14 double immunofluorescence staining revealed significantly more proliferating basal epithelial cells in the Lfngflox/flox;P53flox/+;MMTV-Cre gland, whereas numbers of proliferating luminal epithelial cells were similar in Lfngflox/flox;P53flox/+;MMTV-Cre and Lfngflox/flox;MMTV-Cre glands (Figure 3C). In agreement with the decreased overall level of Cytokeratin 8 by Western blot, we noted absence of Cytokeratin 8 staining in a small number of luminal cells in the Lfngflox/flox;P53flox/+;MMTV-Cre gland (arrow in Figure 3C), suggesting impaired luminal cell differentiation [33]. Thus, there were increased proliferation and decreased differentiation in the mammary epithelium of Lfngflox/flox;P53flox/+;MMTV-Cre mice. Flow cytometry analysis detected expansion of the stem/progenitor cell-enriched populations (CD24MedCD49fHi and CD24HiCD49fLow) in the mutant mammary gland. Interestingly, the CD24HiCD49fLow population was found expanded in the Lfngflox/flox;P53flox/+;MMTV-Cre gland, not the Lfngflox/flox;MMTV-Cre gland (Figure 4, A and B). It has been reported that while CD24HiCD49fLow cells produced complex mammary outgrowths when implanted into cleared fat pads of nulliparous hosts, CD24MedCD49fHi cells failed to develop ductal structures containing side branches or lobules [34]. Thus, Lfngflox/flox;P53flox/+;MMTV-Cre mammary glands may have accumulated more stem-like cells as compared to Lfngflox/flox;P53flox/+ and Lfngflox/flox;MMTV-Cre glands. We performed mammosphere assay for the assessment of stem cell activity and self-renewal in freshly prepared mammary epithelial cells from 2-month-old Lfngflox/flox;P53flox/+ and Lfngflox/flox;P53flox/+;MMTV-Cre mice. Indeed, the latter exhibited significantly increased mammosphere-forming capability (Figure 4C).
Figure 3.
Altered expressions of differentiation markers and increased basal cell proliferation in the Lfngflox/flox;p53flox/+;MMTV-Cre mammary epithelium. (A and B) Representative Western blots for epithelial and mesenchymal markers in the mammary glands of littermate mice at 3 months of age. (C) Representative photomicrographs of E-Cadherin immunostaining, Ki67/cytokeratin 8 (K8) and Ki67/cytokeratin 14 (K14) immunofluorescence staining in the mammary glands of 2-month-old Lfngflox/flox;MMTV-Cre and Lfngflox/flox;p53flox/+;MMTV-Cre mice. Scale bars: 50 μm. The percentage of Ki67+ cells among K8+ luminal cells and K14+ basal cells was quantitated and presented as mean ± standard deviation. *P < .05.
Figure 4.
Expansion of stem/progenitor cell-enriched population in the Lfngflox/flox;p53flox/+;MMTV-Cre mammary gland. (A) Representative flow cytometry analysis of lineage-depleted mammary cells from littermate mice of indicated genotypes at 2 months of age. (B) Quantitation of CD24MedCD49fHi and CD24HiCD49fLow cell populations from three experiments. Percentages of the populations were presented as mean ± standard deviation. (C) Representative photomicrographs and quantification of mammospheres formed in 3D cultures of the mammary epithelial cells isolated from the Lfngflox/flox;p53flox/+ and Lfngflox/flox;p53flox/+;MMTV-Cre mice at 2 months of age. Scale bars: 60 μm. *P < .05.
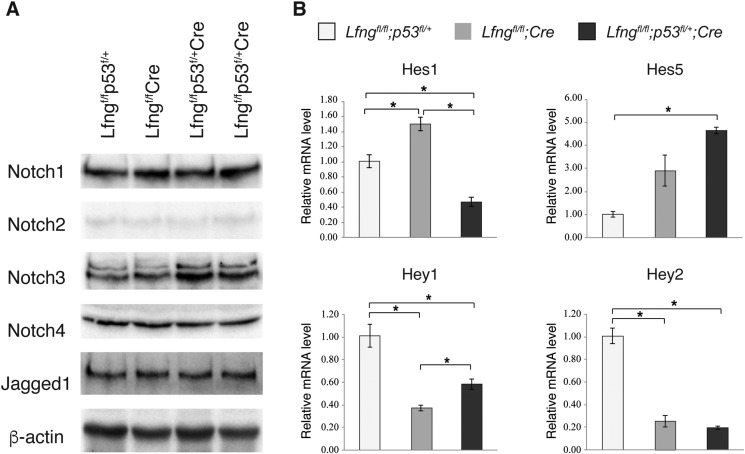
Altered Notch Activation in the Lfngflox/flox;p53flox/+;MMTV-Cre Mammary Gland
To understand the mechanism by which p53 haplodeficiency accelerated mammary tumor development in the Lfngflox/flox;MMTV-Cre mice, we examined Notch activation in these animals. Mammary glands of the Lfngflox/flox;P53flox/+;MMTV-Cre mice at 3 months of age showed increased accumulation of Notch3 intracellular domain (but not of other Notch receptors), compared to the Lfngflox/flox;MMTV-Cre mice. Protein levels of Jagged1 were similar among mice of different genotypes (Figure 5A). Quantitative RT-PCR found significantly decreased Hes1 but increased Hes5 mRNA levels in the Lfngflox/flox;P53flox/+;MMTV-Cre mammary gland, whereas the Lfngflox/flox;MMTV-Cre mammary gland showed increased Hes1 compared to the Lfngflox/flox;P53flox/+ control gland. Hey1 mRNA level was higher in Lfngflox/flox;P53flox/+;MMTV-Cre than that in Lfngflox/flox;MMTV-Cre gland. Both Lfngflox/flox;P53flox/+;MMTV-Cre and Lfngflox/flox;MMTV-Cre glands had down-regulated Hey1 and Hey2 as compared to the control gland (Figure 5B). Taken together, deletion of Lfng in the mammary gland up-regulated Hes1 and Hes5 and down-regulated Hey1 and Hey2, heterozygous deletion of p53 on this background caused a decrease in Hes1 expression but an increase in Hey1 expression.
Figure 5.
Altered Notch signaling in the Lfngflox/flox;p53flox/+;MMTV-Cre mammary gland. (A) Western blot analysis for Notch receptors and Jagged1 ligand in the mammary glands of littermate mice at 3 months of age. (B) Quantitative RT-PCR for Hes1, Hes5, Hey1 and Hey2 in the mammary glands at 3 months of age. *P < .05.
Human CLBC Exhibits Lowest Expression of HES1 and Second Lowest Expression of LFNG Among Six Molecular Subtypes
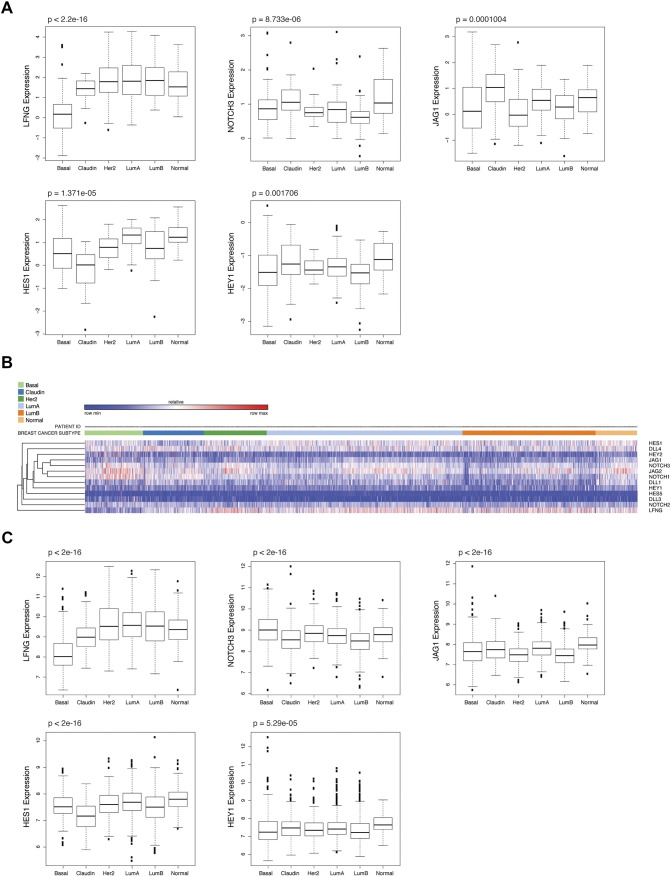
We examined expression levels of the Notch pathway genes in 6 molecular subtypes of human breast cancer using dataset GSE18229 [3]. Notably, the claudin-low subtype exhibits higher expressions of NOTCH3 and JAG1, lower expression of HES1 compared to other subtypes (Figure 6A). Up-regulation of NOTCH3 in the claudin-low subtype is consistent with our result showing accumulation of Notch3 intracellular fragments in the Lfngflox/flox;P53flox/+;MMTV-Cre mice, and may suggest a specific role for Notch3 in the pathogenesis of this subtype. Next, we analyzed expressions of selected Notch pathway genes in human breast cancer using the METABRIC dataset [35], [36]. In agreement with the analysis results in dataset GSE18229, both the heat map and boxplots showed the lowest HES1 expression in claudin-low breast cancer among all subtypes in the METABRIC dataset (Figure 6, B and C). However, elevated NOTCH3 and JAG1 expressions in the claudin-low subtype were not evident in this dataset. Interestingly, expression of LFNG is lower in claudin-low subtype compared to other subtypes except the basal subtype (Figure 6, A and C), suggesting a role for LFNG deficiency in the pathogenesis of basal-like breast cancer and at least a subset of claudin-low breast cancer.
Figure 6.
Human claudin-low breast cancer exhibits low levels of LFNG and HES1 gene expression. (A) Boxplots for expression values of LFNG, NOTCH3, JAG1, HES1 and HEY1 in 6 subtypes of human breast cancer (dataset GSE18229). (B) Heat map for expressions of selected Notch pathways genes in 6 subtypes of human breast cancer (METABRIC dataset). (C) Boxplots for expression values of LFNG, NOTCH3, JAG1, HES1 and HEY1 in 6 subtypes of human breast cancer (METABRIC dataset). Basal: Basal-like. Claudin: Claudin-low. Her2: HER2-positive. LumA: Luminal A. LumB: Luminal B. Normal: Normal breast-like. p values were calculated by comparing expression means across all subtypes.
LFNG Low Expression is Associated with Poor Survival in Human Breast Cancer
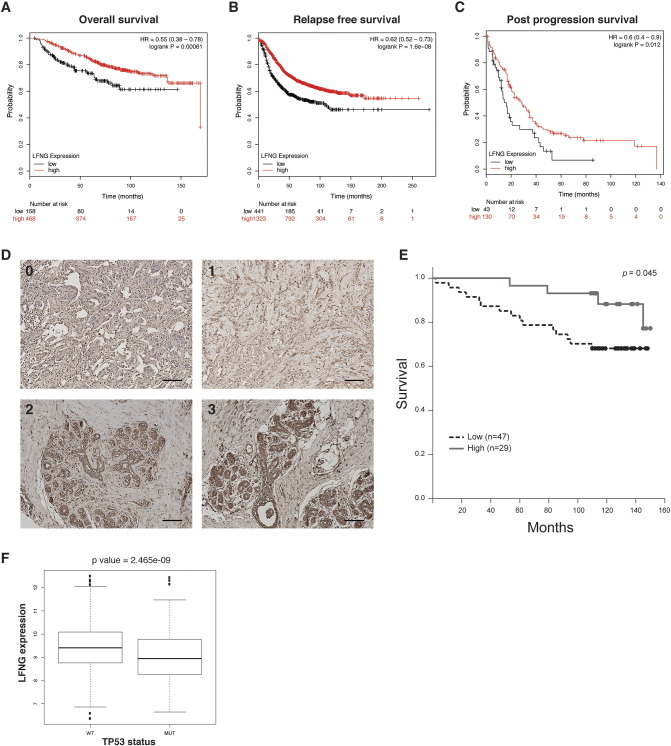
Given that LFNG expression was down-regulated in human BLBC and CLBC (two poor-prognosis breast cancer subtypes) and deletion of Lfng in mice induced mammary tumors resembling BLBC and CLBC [22], low expression of LFNG may predict poor outcome. Indeed, analysis using the online tool (http://www.kmplot.com/) found significantly lower survival rate in patients with low expression of LFNG compared to those showing high LFNG expression in all three categories (overall survival, relapse-free survival, and post-progression survival, Figure 7, A–C). We also performed LFNG immunohistochemistry on human breast cancer tissue array and compared survival of patients showing high or low LFNG immunoreactivity. The low LFNG group had significantly shorter survival compared to the high LFNG group (Figure 7, D and E). Finally, low expression of LFNG was found more prevalent among patients carrying TP53 mutation (Figure 7F), corroborating our study in mice showing genetic interaction between Lfng and p53 in preventing mesenchymal stem-like breast cancer.
Figure 7.
Low level of LFNG expression is associated with poor survival among breast cancer patients and is enriched in patients carrying TP53 mutation. (A-C) Survival analyses in breast cancer patients using univariate Cox regression and Kaplan–Meier methods. Overall survival rate (A), relapse-free survival rate (B), and post-progression survival rate (C) in patients with high LFNG expression (red line) were all significantly higher than that in patients with low LFNG expression (black line). (D) Representative photomicrographs of LFNG immunostaining on a human breast cancer tissue array. Staining intensity scores are: 0 (negative), 1 (weak), 2 (moderate), and 3 (strong). Scale bars: 100 μm. (E) Kaplan–Meier survival analysis for patients with low LFNG expression (staining score 0 or 1) and high LFNG expression (staining score 2 or 3). (F) Boxplot for LFNG expression levels in patients with wild type or mutant TP53 gene.
Discussion
Down-regulation of LFNG and mutations in TP53 are common in human CLBC. In this study we found that deletion of one copy of p53 on the Lfngflox/flox;MMTV-Cre background caused increased basal cell proliferation, impaired luminal cell differentiation, expanded stem-like cell population, as well as EMT in the mammary epithelium, ultimately leading to mesenchymal stem-like mammary tumor formation. Haplodeficiency of p53 drastically shortened tumor latency in these animals, suggesting that Lfng and p53 cooperate to suppress mesenchymal stem-like/claudin-low breast cancer. A recent study showed that p53 negatively regulated activity of the Notch1 transcriptional complex in MCF-7 breast cancer cells [28]. Our previous study suggests that Lfng may exert tumor-suppressive function through inhibition of Jagged1-mediated Notch activation [22]. Both Lfng and p53 negatively regulate Notch signaling in this context. Interestingly, Lfngflox/flox;P53flox/+;MMTV-Cre mammary gland showed increased accumulation of Notch3 intracellular domain compared to the Lfngflox/flox;MMTV-Cre gland. In addition, NOTCH3 was found up-regulated specifically in CLBC subtype in one of the human breast cancer datasets analyzed. Previous studies showed that Notch3 controls self-renewal and hypoxia survival in human mammary stem/progenitor cells [37], [38]. Down-regulation of Notch3, but not Notch1, significantly suppressed proliferation and promoted apoptosis of HER2-negative human breast cancer cells [39]. Recently Notch3 was found to regulate endocrine resistance in metastatic breast cancer [40]. Our present study suggests that signaling through Notch3 may be important in CLBC pathogenesis. CLBC are thought to originate from mammary stem cells. However, Morel et al. have proposed that CLBC could arise from cells committed to luminal differentiation, through a process driven by EMT inducers and combining malignant transformation and transdifferentiation [41]. Given that Notch3 is expressed in a highly clonogenic and transiently quiescent luminal progenitor population [42], it is plausible that dysregulation of Notch3 in these luminal progenitor cells contributes to the initiation and/or progression of CLBC.
Lfngflox/flox;P53flox/+;MMTV-Cre mammary gland showed up-regulation of Hes5 but down-regulation of Hes1 as compared to the Lfngflox/flox;MMTV-Cre gland, suggesting differential regulation of Notch downstream target genes by p53. HES1 expression was also found significantly lower in claudin-low subtype as compared to other subtypes of human breast cancer in both GSE18229 and METABRIC datasets. Notch signaling promotes breast cancer stem cell self-renewal through up-regulation of Hes5 [43]. Interestingly, overexpression of Hes1 was recently shown to reduce CD44+CD24−/low tumor-initiating subpopulation in basal-like breast cancer [44]. In the pancreas, Hes1 controls exocrine cell plasticity and restricts development of pancreatic ductal adenocarcinoma [45]. In this regard, Notch signaling through Hes1 could be tumor-suppressive. Indeed, Notch signaling was reported to cooperate with p53 in restricting cell proliferation and tumor growth in mouse models of human brain tumors, where simultaneous inactivation of the Notch transcriptional effector RBPJκ and p53 induced primitive neuroectodermal-like tumors [46]. Thus, interactions between Notch signaling and p53 may play complex roles in the tumorigenesis of various tissues.
Martin et al. recently reported a novel mouse model of spindle cell metaplastic carcinomas resembling human claudin-low tumors [47]. Our analysis in human breast cancer datasets found association of LFNG low expression with poor survival, as well as the enrichment of LFNG low expression among patients carrying TP53 mutation. Future experiment including gene expression profiling will be needed to confirm the CLBC subtype for mammary tumors developed in the Lfngflox/flox;P53flox/+;MMTV-Cre mice. Nonetheless, these mice may represent a clinically-relevant CLBC model with early tumor onset and complete penetrance, which can be used for the identification of new tumor or tumor microenvironment molecular targets as well as testing of therapeutic strategies.
Conflict of Interest Statement
The authors declare no conflict of interest.
Acknowledgments
Acknowledgements
This work was supported by the National Institutes of Health (grant number R21CA175136) to KX. The authors would like to thank Dr. Sean Egan for providing mouse strain and Dr. Sibali Bandyopadhyay for assistance in flow cytometry analysis.
References
- 1.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheang MC, Voduc D, Bajdik C, Leung S, McKinney S, Chia SK, Perou CM, Nielsen TO. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res. 2008;14:1368–1376. doi: 10.1158/1078-0432.CCR-07-1658. [DOI] [PubMed] [Google Scholar]
- 6.Perou CM, Borresen-Dale AL. Systems biology and genomics of breast cancer. Cold Spring Harb Perspect Biol. 2011:3. doi: 10.1101/cshperspect.a003293. a003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Creighton CJ, Chang JC, Rosen JM. Epithelial-mesenchymal transition (EMT) in tumor-initiating cells and its clinical implications in breast cancer. J Mammary Gland Biol Neoplasia. 2010;15:253–260. doi: 10.1007/s10911-010-9173-1. [DOI] [PubMed] [Google Scholar]
- 8.Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, Lee JS, Fridlyand J, Sahin A, Agarwal R, Joy C. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009;69:4116–4124. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson PA, Brewster AM, Kim-Anh D, Baladandayuthapani V, Broom BM, Edgerton ME, Hahn KM, Murray JL, Sahin A, Tsavachidis S. Selective genomic copy number imbalances and probability of recurrence in early-stage breast cancer. PLoS One. 2011;6 doi: 10.1371/journal.pone.0023543. e23543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas N Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer. 2007;109:1721–1728. doi: 10.1002/cncr.22618. [DOI] [PubMed] [Google Scholar]
- 13.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 14.Bouras T, Pal B, Vaillant F, Harburg G, Asselin-Labat ML, Oakes SR, Lindeman GJ, Visvader JE. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell. 2008;3:429–441. doi: 10.1016/j.stem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 15.Lombardo Y, Faronato M, Filipovic A, Vircillo V, Magnani L, Coombes RC. Nicastrin and Notch4 drive endocrine therapy resistance and epithelial-mesenchymal transition in MCF7 breast cancer cells. Breast Cancer Res. 2014;16 doi: 10.1186/bcr3675. R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raouf A, Zhao Y, To K, Stingl J, Delaney A, Barbara M, Iscove N, Jones S, McKinney S, Emerman J. Transcriptome analysis of the normal human mammary cell commitment and differentiation process. Cell Stem Cell. 2008;3:109–118. doi: 10.1016/j.stem.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 17.Leong KG, Niessen K, Kulic I, Raouf A, Eaves C, Pollet I, Karsan A. Jagged1-mediated Notch activation induces epithelial-to-mesenchymal transition through Slug-induced repression of E-cadherin. J Exp Med. 2007;204:2935–2948. doi: 10.1084/jem.20071082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrison H, Farnie G, Brennan KR, Clarke RB. Breast cancer stem cells: something out of notching? Cancer Res. 2010;70:8973–8976. doi: 10.1158/0008-5472.CAN-10-1559. [DOI] [PubMed] [Google Scholar]
- 19.Cohen B, Bashirullah A, Dagnino L, Campbell C, Fisher WW, Leow CC, Whiting E, Ryan D, Zinyk D, Boulianne G. Fringe boundaries coincide with Notch-dependent patterning centres in mammals and alter Notch-dependent development in Drosophila. Nat Genet. 1997;16:283–288. doi: 10.1038/ng0797-283. [DOI] [PubMed] [Google Scholar]
- 20.Haines N, Irvine KD. Glycosylation regulates Notch signalling. Nat Rev Mol Cell Biol. 2003;4:786–797. doi: 10.1038/nrm1228. [DOI] [PubMed] [Google Scholar]
- 21.Johnston SH, Rauskolb C, Wilson R, Prabhakaran B, Irvine KD, Vogt TF. A family of mammalian Fringe genes implicated in boundary determination and the Notch pathway. Development. 1997;124:2245–2254. doi: 10.1242/dev.124.11.2245. [DOI] [PubMed] [Google Scholar]
- 22.Xu K, Usary J, Kousis PC, Prat A, Wang DY, Adams JR, Wang W, Loch AJ, Deng T, Zhao W. Lunatic fringe deficiency cooperates with the Met/Caveolin gene amplicon to induce basal-like breast cancer. Cancer Cell. 2012;21:626–641. doi: 10.1016/j.ccr.2012.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang CJ, Chao CH, Xia W, Yang JY, Xiong Y, Li CW, Yu WH, Rehman SK, Hsu JL, Lee HH. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat Cell Biol. 2011;13:317–323. doi: 10.1038/ncb2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu JC, Voisin V, Wang S, Wang DY, Jones RA, Datti A, Uehling D, Al-awar R, Egan SE, Bader GD. Combined deletion of Pten and p53 in mammary epithelium accelerates triple-negative breast cancer with dependency on eEF2K. EMBO Mol Med. 2014;6:1542–1560. doi: 10.15252/emmm.201404402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang Z, Deng T, Jones R, Li H, Herschkowitz JI, Liu JC, Weigman VJ, Tsao MS, Lane TF, Perou CM. Rb deletion in mouse mammary progenitors induces luminal-B or basal-like/EMT tumor subtypes depending on p53 status. J Clin Invest. 2010;120:3296–3309. doi: 10.1172/JCI41490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knight JF, Lesurf R, Zhao H, Pinnaduwage D, Davis RR, Saleh SM, Zuo D, Naujokas MA, Chughtai N, Herschkowitz JI. Met synergizes with p53 loss to induce mammary tumors that possess features of claudin-low breast cancer. Proc Natl Acad Sci U S A. 2013;110:E1301–1310. doi: 10.1073/pnas.1210353110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Leary KA, Rugowski DE, Sullivan R, Schuler LA. Prolactin cooperates with loss of p53 to promote claudin-low mammary carcinomas. Oncogene. 2014;33:3075–3082. doi: 10.1038/onc.2013.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yun J, Espinoza I, Pannuti A, Romero D, Martinez L, Caskey M, Stanculescu A, Bocchetta M, Rizzo P, Band V. p53 Modulates Notch Signaling in MCF-7 Breast Cancer Cells by Associating With the Notch Transcriptional Complex Via MAML1. J Cell Physiol. 2015;230:3115–3127. doi: 10.1002/jcp.25052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marino S, Vooijs M, van Der Gulden H, Jonkers J, Berns A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 2000;14:994–1004. [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang S, Chung WC, Miele L, Xu K. Targeting Met and Notch in the -deficient, -amplified triple-negative breast cancer. Cancer Biol Ther. 2014;15:633–642. doi: 10.4161/cbt.28180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsao PN, Wei SC, Wu MF, Huang MT, Lin HY, Lee MC, Lin KM, Wang IJ, Kaartinen V, Yang LT. Notch signaling prevents mucous metaplasia in mouse conducting airways during postnatal development. Development. 2011;138:3533–3543. doi: 10.1242/dev.063727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wittschieben JP, Patil V, Glushets V, Robinson LJ, Kusewitt DF, Wood RD. Loss of DNA polymerase zeta enhances spontaneous tumorigenesis. Cancer Res. 2010;70:2770–2778. doi: 10.1158/0008-5472.CAN-09-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dravis C, Spike BT, Harrell JC, Johns C, Trejo CL, Southard-Smith EM, Perou CM, Wahl GM. Sox10 regulates stem/progenitor and mesenchymal cell states in mammary epithelial cells. Cell Rep. 2015;12:2035–2048. doi: 10.1016/j.celrep.2015.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeselsohn R, Brown NE, Arendt L, Klebba I, Hu MG, Kuperwasser C, Hinds PW. Cyclin D1 kinase activity is required for the self-renewal of mammary stem and progenitor cells that are targets of MMTV-ErbB2 tumorigenesis. Cancer Cell. 2010;17:65–76. doi: 10.1016/j.ccr.2009.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, Pugh M, Jones L, Russell R, Sammut SJ. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7 doi: 10.1038/ncomms11479. 11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sansone P, Storci G, Giovannini C, Pandolfi S, Pianetti S, Taffurelli M, Santini D, Ceccarelli C, Chieco P, Bonafe M. p66Shc/Notch-3 interplay controls self-renewal and hypoxia survival in human stem/progenitor cells of the mammary gland expanded in vitro as mammospheres. Stem Cells. 2007;25:807–815. doi: 10.1634/stemcells.2006-0442. [DOI] [PubMed] [Google Scholar]
- 38.Sansone P, Storci G, Tavolari S, Guarnieri T, Giovannini C, Taffurelli M, Ceccarelli C, Santini D, Paterini P, Marcu KB. IL-6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland. J Clin Invest. 2007;117:3988–4002. doi: 10.1172/JCI32533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamaguchi N, Oyama T, Ito E, Satoh H, Azuma S, Hayashi M, Shimizu K, Honma R, Yanagisawa Y, Nishikawa A. NOTCH3 signaling pathway plays crucial roles in the proliferation of ErbB2-negative human breast cancer cells. Cancer Res. 2008;68:1881–1888. doi: 10.1158/0008-5472.CAN-07-1597. [DOI] [PubMed] [Google Scholar]
- 40.Sansone P, Ceccarelli C, Berishaj M, Chang Q, Rajasekhar VK, Perna F, Bowman RL, Vidone M, Daly L, Nnoli J. Self-renewal of CD133(hi) cells by IL6/Notch3 signalling regulates endocrine resistance in metastatic breast cancer. Nat Commun. 2016;7 doi: 10.1038/ncomms10442. 10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morel AP, Hinkal GW, Thomas C, Fauvet F, Courtois-Cox S, Wierinckx A, Devouassoux-Shisheboran M, Treilleux I, Tissier A, Gras B. EMT inducers catalyze malignant transformation of mammary epithelial cells and drive tumorigenesis towards claudin-low tumors in transgenic mice. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002723. e1002723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lafkas D, Rodilla V, Huyghe M, Mourao L, Kiaris H, Fre S. Notch3 marks clonogenic mammary luminal progenitor cells in vivo. J Cell Biol. 2013;203:47–56. doi: 10.1083/jcb.201307046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xing F, Kobayashi A, Okuda H, Watabe M, Pai SK, Pandey PR, Hirota S, Wilber A, Mo YY, Moore BE. Reactive astrocytes promote the metastatic growth of breast cancer stem-like cells by activating Notch signalling in brain. EMBO Mol Med. 2013;5:384–396. doi: 10.1002/emmm.201201623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.So JY, Wahler J, Das Gupta S, Salerno DM, Maehr H, Uskokovic M, Suh N. HES1-mediated inhibition of Notch1 signaling by a Gemini vitamin D analog leads to decreased CD44(+)/CD24(−/low) tumor-initiating subpopulation in basal-like breast cancer. J Steroid Biochem Mol Biol. 2015;148:111–121. doi: 10.1016/j.jsbmb.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hidalgo-Sastre A, Brodylo RL, Lubeseder-Martellato C, Sipos B, Steiger K, Lee M, von Figura G, Grunwald B, Zhong S, Trajkovic-Arsic M. Hes1 Controls Exocrine Cell Plasticity and Restricts Development of Pancreatic Ductal Adenocarcinoma in a Mouse Model. Am J Pathol. 2016;186:2934–2944. doi: 10.1016/j.ajpath.2016.07.025. [DOI] [PubMed] [Google Scholar]
- 46.Giachino C, Boulay JL, Ivanek R, Alvarado A, Tostado C, Lugert S, Tchorz J, Coban M, Mariani L, Bettler B. A tumor suppressor function for notch signaling in forebrain tumor subtypes. Cancer Cell. 2015;28:730–742. doi: 10.1016/j.ccell.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 47.Martin EE, Huang W, Anwar T, Arellano-Garcia C, Burman B, Guan JL, Gonzalez ME, Kleer CG. MMTV-cre;Ccn6 knockout mice develop tumors recapitulating human metaplastic breast carcinomas. Oncogene. 2017;36:2275–2285. doi: 10.1038/onc.2016.381. [DOI] [PMC free article] [PubMed] [Google Scholar]