Abstract
Purpose
To assess and compare the accuracy of magnitude-based MRI (MRI-M) and complex-based MRI (MRI-C) for estimating hepatic proton density fat fraction (PDFF) in children, using magnetic resonance spectroscopy (MRS) as the reference standard. A secondary aim was to assess the agreement between MRI-M and MRI-C.
Methods
This was a HIPAA-compliant, retrospective analysis of data collected in children enrolled in prospective, IRB-approved studies between 2012 and 2014. Informed consent was obtained from 200 children (ages 8–19 years) who subsequently underwent 3T MR exams that included MRI-M, MRI-C, and T1-independent, T2-corrected, single-voxel STEAM MRS. Both MRI methods acquired 6 echoes at low flip angles. T2*-corrected PDFF parametric maps were generated. PDFF values were recorded from regions of interest (ROIs) drawn on the maps in each of the 9 Couinaud segments and 3 ROIs co-localized to the MRS voxel location. Regression analyses assessing agreement with MRS were performed to evaluate accuracy of each MRI method, and Bland-Altman and ICC analyses were performed to assess agreement between the MRI methods.
Results
MRI-M and MRI-C PDFF were accurate relative to the co-localized MRS reference standard, with regression intercepts of 0.63% and −0.07%, slopes of 0.998 and 0.975, and proportion-of-explained-variance values (R2) of 0.982 and 0.979, respectively. For individual Couinaud segments and for the whole liver averages, Bland-Altman biases between MRI-M and MRI-C were small (ranging from 0.04 to 1.11%) and ICCs were high (≥0.978).
Conclusions
Both MRI-M and MRI-C accurately estimated hepatic PDFF in children, and high inter-method agreement was observed.
Keywords: Magnitude-based, complex-based, pediatric, PDFF, accuracy, spectroscopy
INTRODUCTION
MRI proton density fat fraction (PDFF) is emerging as the leading quantitative imaging biomarker for hepatic steatosis (1–3). PDFF measures the portion of mobile protons attributable to fat, correlates closely with tissue triglyceride concentration (4), accurately predicts the presence and degree of steatosis (4–6), and has been used as an endpoint in clinical trials in non-alcoholic fatty liver disease (NAFLD) (7–9), the most common chronic liver disease in children (10).
PDFF can be measured using one of two general MRI methods, broadly classified as magnitude-based MRI (MRI-M) and complex-based MRI (MRI-C) (2). MRI-M uses only magnitude data and can estimate PDFF from 0 to about 50%, while MRI-C uses both phase and magnitude data and can estimate PDFF from 0–100%. Both methods use low flip angle to minimize T1 weighting effects, acquire multiple echoes to allow for T2* correction, and incorporate a multi-peak spectral model to account for the spectral complexity of fat.
Numerous studies have shown that both the MRI-M (9, 11–15) and MRI-C (16–18) methods accurately quantify PDFF, but gaps in knowledge remain. Until now, the majority of studies have been performed in adults, and the diagnostic accuracy in children has not yet been fully validated(19). Validation in children is necessary because children differ from adults in characteristics relevant to MRI such as ability to tolerate MR exams, breath-hold capacity, and body habitus. These characteristics could influence feasibility and quality of MRI acquisition, thus affecting the diagnostic performance of one or both PDFF quantification methods. Additionally, histological features of NAFLD, including degree and distribution of steatosis, differ between children and adults (20), which potentially could affect MR-based fat quantification.
The purpose of our study was to assess and compare MRI-M and MRI-C in estimating hepatic PDFF in children, using magnetic resonance spectroscopy (MRS) as the reference standard. We chose MRS as our reference because it is widely accepted as the most accurate non-invasive method to measure PDFF (2, 21), it can be performed in the same exam as MRI, and it can be spatially colocalized with MRI reliably. A secondary aim was to assess the agreement of MRI-M and MRI-C for PDFF estimation.
MATERIALS & METHODS
Research Participants and Study Design
This was a single-site, cross-sectional, retrospective analysis of data collected in children enrolled at our institution in prospective cohort studies of pediatric NAFLD. Children in these cohort studies either had known or suspected NAFLD, or they were normal controls with no known liver disease. Inclusion criteria for the current analysis were children aged 8–19 years, who underwent a research MR exam including MRI-M, MRI-C and MRS at our imaging center between January 2012 and December 2014. The study was approved by an Institutional Review Board and was compliant with the Health Insurance Portability and Accountability Act. Children under 18 years signed informed assent, with written informed consent provided by their parent(s)/guardian(s); 18- to 19-year olds signed informed consent. Demographic and anthropometric data were collected on all children.
This study was funded in part by an investigator-initiated research grant from GE Healthcare to CS. None of the authors are employees or consultants for GE. GE had no control over inclusion of any data or other information that might present a conflict of interest.
MR Exam Acquisition
Children were asked to fast for 4 hours before the MR exam and were scanned at 3T (Signa EXCITE HDxt scanner, GE Healthcare, Waukesha WI) in a supine position with an eight-channel torso phased-array coil centered over the liver. A dielectric pad was placed between the coil and the abdomen. Each MR exam included MRS, MRI-M and MRI-C fat quantification acquisitions.
MRS
Using three-plane localizing images, a 20 × 20 × 20mm3 voxel was placed in the right lobe of the liver, avoiding major vasculature, bile ducts, liver edges, and artifacts. After automatic shimming, single-voxel proton MRS was performed in a single breath-hold using stimulated echo acquisition mode (STEAM)(22). Repetition time (TR) was set 3500 ms to minimize effects of T1 weighting. In a single acquisition, five STEAM spectra were collected at TEs of 10, 15, 20, 25 and 30 ms, with a mixing time (TM) of 5 ms. This TM and range of TEs minimizes j-coupling effects, while allowing for T2 correction (22). No water, fat or spatial saturation was applied to ensure a uniform spectral response. An anatomic image illustrating the placement of the MRS voxel was saved. Spectra were transferred offline for analysis.
MRI-M
As previously described (12, 23, 24), two-dimensional, spoiled gradient-recalled-echo axial images of the liver were acquired at six echo times (TEs) at which protons of water and the dominant methylene peak of triglyceride are nominally out of phase or in phase (1.15, 2.30, 3.45, 4.60, 5.75, and 6.90 ms). The six-echo acquisition allows for T2* calculation and correction. A low flip angle (10°) was used with a TR ≥ 120 ms to minimize T1 weighting. Slice thickness was 8 mm, receiver bandwidth was ±125 kHz, and acquisition matrix was 128×192 – 192×224. Externally calibrated parallel imaging was applied with an acceleration factor of 1.25. Rectangular field of view was adjusted depending on body habitus and breath-hold capacity. Complete liver coverage was achieved in one or two breath-holds. A previously described magnitude-data fitting algorithm (25) was applied pixel by pixel to create parametric PDFF maps inline from the source images. The algorithm incorporated a mathematical model that describes the relationship between signal intensity and fat fraction, correcting for T2* exponential decay. The algorithm addresses the spectral complexity of hepatic fat by modeling the composite fat signal as weighted sum of six frequency signals at 0.9, 1.3, 2.1, 2.75, 4.2, and 5.3 ppm (26).
No correction was made for noise bias or eddy currents, as these are considered negligible with magnitude fitting. Source images and PDFF maps were transferred offline for analysis.
MRI-C
As previously described (16, 18, 27), a three-dimensional, complex-based, six-echo, chemical shift-encoded method was used for hepatic fat quantification. The six echoes were acquired in two interleaved shots at TEs optimized for fat water separation using complex fitting (first TE 0.9 – 1.2 ms, ΔTE ~0.8 ms) (28). A low flip angle (3°) was used with a TR ≥ 8.6 ms to minimize T1 weighting (29). Slice thickness was 8 mm, receiver bandwidth was ±125 kHz, and acquisition matrix was 256×160. Auto-calibrating reconstruction (ARC) parallel imaging was used with 2× acceleration in both the x-direction and z-direction. Rectangular field of view was adjusted depending on body habitus and breath-hold capacity. Complete liver coverage was achieved in one breath-hold. A previously described hybrid data-fitting algorithm (25) was applied pixel by pixel to create parametric PDFF maps online from the source images. Similar to the magnitude-data fitting algorithm, this algorithm corrected for T2* exponential decay and spectral complexity of fat (using the same six-peak spectral model) (26, 30). It also corrected for noise (29) and eddy currents (17, 31). Additionally, magnitude water- and fat-only images are reconstructed from the source data. Magnitude images and PDFF maps were transferred offline for analysis.
MR Exam Analysis
MRS
A single blinded MR spectroscopist (GH, 15 years experience) analyzed all spectra. Peak areas were estimated using custom prior knowledge using the Advanced Method for Accurate, Robust and Efficient Spectral fitting of MRS data (AMARES) included in Java-based magnetic resonance user interface software (32, 33). As described previously (26), T2-corrected areas of the water (4–6 ppm) and the fat (0–3 ppm) were estimated as there is insufficient spectral resolution in vivo to accurately characterize the individual fat peaks, or to distinguish water from two nearby fat peaks. The contribution to the water peak from the neighboring fat peaks is corrected using a previous derived fat spectrum post T2 correction, which reassigns these fat peaks from water to the fat signal. (26)
MRI-M and MRI-C
Four analysts, each with a minimum of 2 months of experience, performed all image analysis (Osirix imaging software; Pixmeo, Geneva, Switzerland). For any given child, the same analyst performed all analysis.
Two image analyses were performed. For the first analysis, which focused on accuracy, three circular 1-cm radius regions of interest (ROIs) were placed on the MR images, colocalized to the MRS voxel location, as illustrated in figure 1. For both MRI-M and MRI-C, ROIs were placed on the fifth-echo reconstructed magnitude image. ROIs were colocalized to the superior, middle and inferior portions of the MRS voxel as illustrated on the anatomical image mentioned previously. This was done because the MRS voxel is thicker than the individual MRI slices. For the second image analysis, which focused on inter-MRI-method agreement, one circular 1-cm radius ROI was placed in the center of each of the nine Couinaud liver segments on the fifth echo of the magnitude images. If an artifact covered an entire segment, no ROI was placed in that segment.
Figure 1.
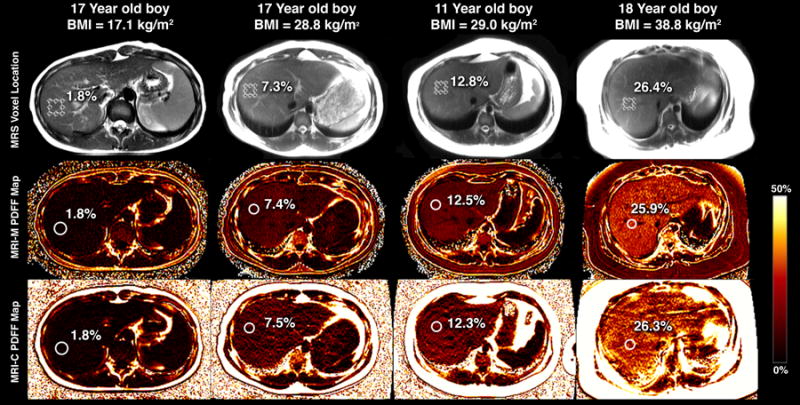
MRS voxel placement (top row) and subsequent ROI colocalization on PDFF maps provided by MRI-M (middle row) and MRI-C (bottom row) for 4 children with varying degrees of steatosis. PDFF quantified by MRS, MRI-M, and MRI-C for each child overlain.
For both analyses, ROIs were copied onto the PDFF maps and mean PDFF values within the ROIs were recorded. ROIs were placed on fifth-echo images initially and subsequently copied onto the PDFF maps for two reasons. Placing ROIs on fifth-echo images prevented the analyst from receiving feedback that may have biased the placement of the ROIs, and the fifth-echo image consistently provided adequate anatomical demarcation for reliable ROI co-localization.
Statistical Analysis
A biostatistical analyst (TW, 20 years experience) performed all statistical analysis under the supervision of a faculty biostatistician, using the R software package (R Foundation for Statistical Computing, Vienna, Austria). Patient characteristics were summarized descriptively. All tests of hypotheses were conducted at significance level of 0.05. Bonferroni’s correction was applied to related sets of tests by aim to ensure a family-wise significance level of 0.05 was established.
Accuracy of MRI-M and MRI-C using MRS as reference
Univariate linear regression analyses were performed, assessing agreement of each of the MRI-based methods with the reference standard (MRS). For these analyses, PDFF values from the three MRI ROIs were averaged to give a single PDFF value, which was compared to the PDFF obtained from MRS. For each regression model, intercept, slope, regression bias and proportion of explained variance (R2) were calculated. Bootstrap-based bias-corrected, accelerated confidence intervals were calculated for all parameters. P-values for comparing slopes to 1 and intercepts to 0 were computed without adjustment, with significance threshold set to 0.05, to allow further investigation of departures from accuracy. A bootstrap-based test was used to compare the two methods on each regression metric. Bonferroni’s correction for multiple comparisons was applied to this set of four comparisons, setting individual significance threshold to 0.0125 to ensure family-wise significance level of 0.05.
Agreement of MRI-M and MRI-C
Bland-Altman and intraclass correlation coefficient (ICC) analyses were performed for the PDFF values obtained from each of the nine Couinaud liver segments, and for the average PDFF values of the nine segments. Bland-Altman bias and its p-value, as well as the 95% limits of agreement were computed as part of the analysis. Bonferroni’s correction was applied to the bias p-values, setting individual significance threshold to 0.005 to ensure family-wise significance level of 0.05. Coefficients of variation (CVs) for paired data with bootstrap confidence intervals were also computed in all cases.
RESULTS
Cohort Characteristics
As summarized in Table 1, the study cohort comprised 200 children (135 boys and 65 girls). Age range was 8 to 19 years (mean 13.6 ± 2.6 years). MRS-PDFF range was 0.9 to 41.9% (mean 13.4 ± 10.5%). BMI range was 14.5 to 49.7 kg/m2 (mean 29.3 ± 6.8 kg/m2).
Table 1.
Cohort Characteristics
| Subjects in study (%) |
Mean Age (years) (range) |
Mean MRS PDFF (%) (range) |
Mean BMI (kg/m2) (range) |
|
|---|---|---|---|---|
| Total | 200 (100) |
13.6 ± 2.6 (8–19) |
13.4 ± 10.5 (0.9 – 41.9) |
29.3 ± 6.8 (14.5 – 49.7) |
| Boys | 135 (67.5) |
13.6 ± 2.5 (8–18) |
14.0 ± 10.7 (0.9 – 41.9) |
28.9 ± 6.5 (15.2 – 48.5) |
| Girls | 65 (32.5) |
13.7 ± 2.9 (8–19) |
12.0 ± 9.8 (1.3 – 33.4) |
30.0 ± 7.3 (14.5 – 49.7) |
Accuracy of MRI-M and MRI-C with MRS as reference
As shown in figure 2, PDFF estimated by both MRI-M and MRI-C agreed closely with PDFF measured by MRS. For both methods, intercepts were less than 1%, slopes were near 1, regression biases were less than 1% and R2 were near 1, all indicating strong agreement with MRS overall. Agreement was not perfect, however, for either method. The intercept of MRI-M (0.63%) was significantly different from 0 (p < 0.001), while the slope of MRI-C (0.975) was significantly different from 1 (p = 0.025), indicating small systematic shifts and resulting in similar regression biases overall. Complete linear regression results and results of MRI-M and MRI-C regression comparisons are found in table 2. As seen in the bottom row of table 2, Bonferroni-adjusted pairwise comparisons showed no significant difference in any regression parameter between the two MRI methods, although the difference in intercepts was borderline.
Figure 2.
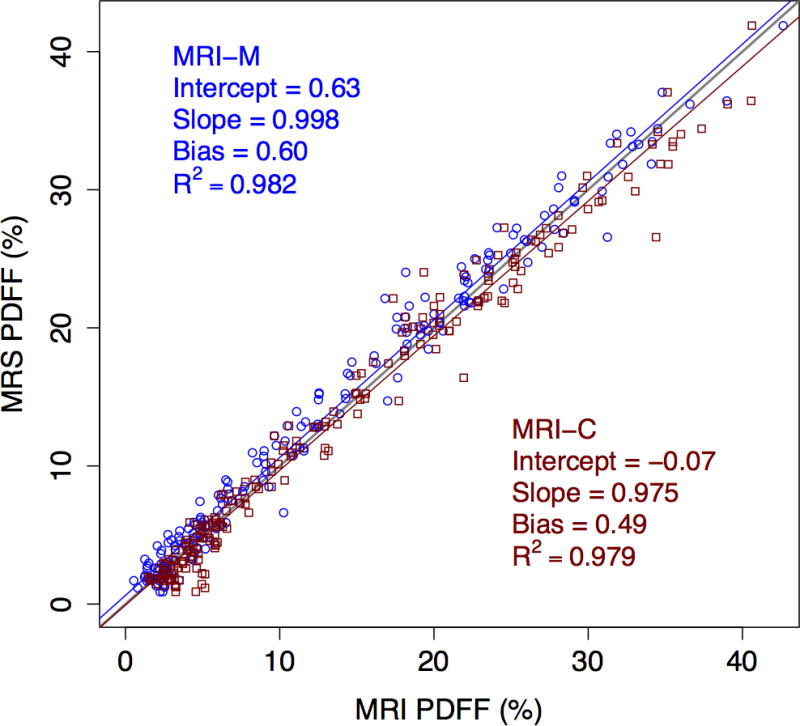
Scatter plot showing accuracy of the two MRI methods for PDFF quantification, using MRS as reference standard. MRI-M regression line and data points are blue circles, and MRI-C regression line and data points are red squares.
Table 2.
Regression Analysis Results for MRI Agreement with MRS
| Intercept (%) (95% CI) |
Slope (95% CI) |
Regression Bias (%) (95% CI) |
R2 (95% CI) |
|
|---|---|---|---|---|
| MRI-M | 0.63 (0.36, 0.90) |
0.998 (0.977, 1.017) |
0.60 (0.41, 0.79) |
0.982 (0.976, 0.986) |
| MRI-C | −0.07 (−0.37, 0.24) |
0.975 (0.950, 0.998) |
0.49 (0.23, 0.79) |
0.979 (0.970, 0.984) |
| p-value | 0.0127 | 0.035 | 0.553 | 0.050 |
Note. p-values are the bootstrap-based p-values for comparing the regression metrics between MRI-M and MRI-C methods. After Bonferroni correction, only p-values of 0.0125 or less are considered to be significant at family-wise 0.05 level. Thus, the difference between intercepts is borderline significant, but there are no other significant differences.
Agreement of MRI-M and MRI-C
As shown in Table 3 (Bland-Altman analyses), MRI-M estimated lower PDFF values than MRI-C at the whole-liver level (Bland-Altman bias of 0.60%), and for every Couinaud segment (Bland-Altman biases < 1.2%); paired comparisons showed that all biases were significant after Bonferroni’s correction (p < 0.001) with the exception of the biases in segments 4a and 4b (p = 0.076, 0.697 respectively). Figure 3 shows the Bland-Altman plot for the whole-liver average PDFF values as estimated by MRI-M and MRI-C.
Table 3.
MRI-C vs MRI-M Bland-Altman Analysis
| Bias | Lower LOA | Upper LOA | SD | p-value | |
|---|---|---|---|---|---|
| 9 segment average | 0.60 | −0.95 | 2.15 | 0.79 | <0.001 |
| 1 | 0.41 | −2.41 | 3.24 | 1.44 | <0.001 |
| 2 | 0.86 | −2.73 | 4.44 | 1.83 | <0.001 |
| 3 | 0.45 | −2.98 | 3.87 | 1.75 | <0.001 |
| 4a | 0.21 | −3.10 | 3.53 | 1.69 | 0.076 |
| 4b | 0.04 | −2.70 | 2.78 | 1.40 | 0.697 |
| 5 | 0.98 | −1.72 | 3.69 | 1.38 | <0.001 |
| 6 | 1.11 | −1.23 | 3.44 | 1.19 | <0.001 |
| 7 | 0.73 | −1.79 | 3.25 | 1.29 | <0.001 |
| 8 | 0.58 | −2.32 | 3.48 | 1.48 | <0.001 |
Note. Bias p-values are obtained via the paired-sample t-test. Except for segments 4a and 4b, all biases are significantly different from 0 after Bonferroni’s correction.
Figure 3.
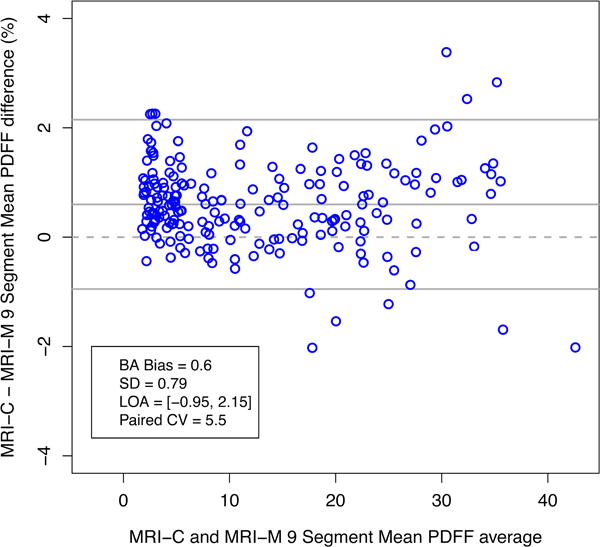
Bland-Altman plot showing agreement of MRI-M with MRI-C for 9-segment average PDFF. The difference between PDFF estimated by MRI-M and MRI-C is plotted on the y-axis, and the average of the PDFF values estimated by the two MRI methods is plotted on the x-axis.
As shown in table 4, the ICCs of the individual segments ranged from 0.978 – 0.991 and the ICC of the nine-segment average was 0.995, (95% CI 0.981, 0.998). The CVs of the individual segments ranged from 7.6 – 12.3%, and the CV of the nine-segment average was 5.5%, (95% CI 4.9%, 6.4%).
Table 4.
ICC and CV Analyses Assessing Agreement of MRI-M and MRI-C
| ICC (95% CI) |
CV (%) (95% CI) |
|
|---|---|---|
| 9 Segment Average | 0.995 (0.981, 0.998) |
5.5 (4.9, 6.4) |
| 1 | 0.989 (0.984, 0.992) |
8.4 (7.3, 9.7) |
| 2 | 0.978 (0.959, 0.987) |
12.3 (10.6, 14.4) |
| 3 | 0.985 (0.979, 0.989) |
10.4 (8.9, 12.3) |
| 4a | 0.984 (0.979, 0.988) |
9.7 (8.4, 11.5) |
| 4b | 0.991 (0.987, 0.993) |
8.0 (6.9, 9.6) |
| 5 | 0.987 (0.955, 0.994) |
9.6 (8.3, 11.3) |
| 6 | 0.988 (0.927, 0.996) |
8.9 (7.9, 10.0) |
| 7 | 0.990 (0.977, 0.995) |
7.6 (6.6, 8.9) |
| 8 | 0.989 (0.982, 0.993) |
8.2 (7.0, 10.4) |
DISCUSSION
This study assessed in children the accuracy of MRI-M and MRI-C PDFF for PDFF quantification, using MRS as the reference standard and the agreement of PDFF as estimated by the two MRI-based methods. Both MRI methods agreed closely with MRS (regression biases 0.49–0.60%), signifying high PDFF quantification accuracy. No accuracy metric was significantly different between methods after the adjustment for multiple comparisons (although difference in regression intercepts reached borderline significance), and in particular, the average regression bias of the two methods was similar, suggesting that the two methods had comparable accuracy overall. Moreover, the two methods agreed closely with each other for each segment (Bland-Altman biases ranging from 0.04 to 1.11%) as well as for the liver as a whole (Bland-Altman bias 0.60%), indicating high inter-method reproducibility.
Although both methods were accurate, neither was perfect. MRI-M underestimated PDFF by a fixed amount (~0.63%) relative to the reference value measured by MRS, while MRI-C overestimated PDFF by a proportional amount (regression slope 0.975). Since MRI-M uses magnitude fitting, it has a Rician noise distribution, which adds spurious signal to the liver at all echo times, reducing the relative signal oscillation and causing PDFF underestimation (34). By comparison, the proportional overestimation of PDFF by MRI-C likely reflects residual T1 related bias, which results in greater overestimation at higher reference PDFF values. Although the use of low flip angles mitigates T1 related bias, it does not entirely eliminate them (29). The degree of residual T1 related bias depends on the relative flip angle and TR used for the acquisition. Based on the gradient echo signal equation, the flip angle/TR combination of the 3D acquisition (≥ 8.6 ms/3 degrees) for MRI-C results in greater T1 bias than that of the interleaved 2D acquisition (≥ 120 ms/10 degrees) used for MRI-M. This likely explains why the slope of the comparison between MRS-PDFF and MRI-C is less than 1.0 while the slope of the comparison between MRS-PDFF and MRI-M is not different from 1.0. Since MRI-C overestimates PDFF slightly relative to MRS and MRI-M underestimates slightly, the observed slight Bland-Altman biases between MRI-M and MRI-C are expected. The small differences between each MRI method and MRS and between the two MRI methods are unlikely to be clinically meaningful, however, since hepatic PDFF in NAFLD ranges from about 0 to 50% (3, 5, 13).
Several factors may potentially confound hepatic steatosis quantification in the pediatric population. Children do not tolerate MR exams as well as adults, and pediatric NAFLD differs from adult NAFLD (20). Our results suggest that these factors did not negatively affect accuracy for either MRI method. The close agreement between MRI-M and MRI-C provided by the two MRI methods also demonstrates that PDFF is reproducible across estimation methods. The accuracy of the two MRI methods in a pediatric cohort, and the observation that PDFF values do not depend on MRI estimation method, are important findings in the continued validation of the MRI PDFF biomarker.
While many studies have been published on the accuracy of MRI PDFF in adult or mixed cohorts, relatively few have focused exclusively on children. Our study corroborates and builds on previously published findings in adults showing high accuracy of MRI-M (11) and MRI-C (16, 18) in quantifying steatosis. Only one study has assessed the reproducibility of PDFF across MRI estimation methods (35), and this study was performed in an obese adult cohort. Our study arrived at similar findings in a pediatric cohort, suggesting that PDFF is reproducible across MRI methods in children as well.
Although commercial versions of MRI-C are increasingly available, continued validation of MRI-M is relevant. MRI-C methods are not standard on all new scanners and may not be available on older platforms. In these situations, MRI-M may be the only option for PDFF estimation. The results of our study suggest that either MRI method may be used to quantify hepatic steatosis in children, giving clinical centers and clinical trials the flexibility of using either method, depending on technical capability.
One study limitation is that all MR exams were performed at a single site with one scanner in one scanning session. Further investigation is needed to confirm these results across scanner manufacturers, field strengths and at different sites. Our cohort included children ages 8–19. Inclusion of children under the age of 8 may be important for future work, as these children tend to have more difficulty tolerating MR exams. Another limitation was that the study did not determine the number or location of ROIs best suited to estimate PDFF over the entire liver; however, this was not an aim of this study.
In conclusion, this study demonstrated that both MRI-M and MRI-C can accurately estimate PDFF in children, and that the two methods agree closely with each other, confirming the reproducibility of PDFF estimation across independent confounder-corrected MRI methods. Our results demonstrate that either MRI method can be used to quantify hepatic PDFF in children.
Acknowledgments
Study supported by NIH grants: DK088925-05, DK088831, DK090350, DK061730, and DK061734-13. Supported in part by a grant from GE Healthcare to C.B.S.
References
- 1.Reeder SB, Hu HH, Sirlin CB. Proton density fat-fraction: a standardized MR-based biomarker of tissue fat concentration. Journal of magnetic resonance imaging. 2012;36(5):1011–4. doi: 10.1002/jmri.23741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reeder SB, Sirlin CB. Quantification of liver fat with magnetic resonance imaging. Magnetic resonance imaging clinics of North America. 2010;18(3):337–57, ix. doi: 10.1016/j.mric.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwimmer JB, Middleton MS, Behling C, et al. Magnetic resonance imaging and liver histology as biomarkers of hepatic steatosis in children with nonalcoholic fatty liver disease. Hepatology. 2015;61(6):1887–95. doi: 10.1002/hep.27666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannas P, Kramer H, Hernando D, et al. Quantitative MR Imaging of Hepatic Steatosis: Validation in Ex Vivo Human Livers. Hepatology. 2015;62(5):1444–55. doi: 10.1002/hep.28012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Permutt Z, Le TA, Peterson MR, et al. Correlation between liver histology and novel magnetic resonance imaging in adult patients with non-alcoholic fatty liver disease - MRI accurately quantifies hepatic steatosis in NAFLD. Alimentary pharmacology & therapeutics. 2012;36(1):22–9. doi: 10.1111/j.1365-2036.2012.05121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang A, Desai A, Hamilton G, et al. Accuracy of MR imaging-estimated proton density fat fraction for classification of dichotomized histologic steatosis grades in nonalcoholic fatty liver disease. Radiology. 2015;274(2):416–25. doi: 10.1148/radiol.14140754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loomba R, Sirlin CB, Ang B, et al. Ezetimibe for the treatment of nonalcoholic steatohepatitis: assessment by novel magnetic resonance imaging and magnetic resonance elastography in a randomized trial (MOZART trial) Hepatology. 2015;61(4):1239–50. doi: 10.1002/hep.27647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le TA, Chen J, Changchien C, et al. Effect of colesevelam on liver fat quantified by magnetic resonance in nonalcoholic steatohepatitis: a randomized controlled trial. Hepatology. 2012;56(3):922–32. doi: 10.1002/hep.25731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noureddin M, Lam J, Peterson MR, et al. Utility of magnetic resonance imaging versus histology for quantifying changes in liver fat in nonalcoholic fatty liver disease trials. Hepatology. 2013;58(6):1930–40. doi: 10.1002/hep.26455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwimmer JB, Deutsch R, Kahen T, Lavine JE, Stanley C, Behling C. Prevalence of fatty liver in children and adolescents. Pediatrics. 2006;118(4):1388–93. doi: 10.1542/peds.2006-1212. [DOI] [PubMed] [Google Scholar]
- 11.Yokoo T, Bydder M, Hamilton G, et al. Nonalcoholic fatty liver disease: diagnostic and fat-grading accuracy of low-flip-angle multiecho gradient-recalled-echo MR imaging at 1.5 T. Radiology. 2009;251(1):67–76. doi: 10.1148/radiol.2511080666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yokoo T, Shiehmorteza M, Hamilton G, et al. Estimation of hepatic proton-density fat fraction by using MR imaging at 3.0 T. Radiology. 2011;258(3):749–59. doi: 10.1148/radiol.10100659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang A, Tan J, Sun M, et al. Nonalcoholic fatty liver disease: MR imaging of liver proton density fat fraction to assess hepatic steatosis. Radiology. 2013;267(2):422–31. doi: 10.1148/radiol.12120896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang GH, Cruite I, Shiehmorteza M, et al. Reproducibility of MRI-determined proton density fat fraction across two different MR scanner platforms. Journal of magnetic resonance imaging. 2011;34(4):928–34. doi: 10.1002/jmri.22701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang BK, Yu ES, Lee SS, et al. Hepatic fat quantification: a prospective comparison of magnetic resonance spectroscopy and analysis methods for chemical-shift gradient echo magnetic resonance imaging with histologic assessment as the reference standard. Investigative radiology. 2012;47(6):368–75. doi: 10.1097/RLI.0b013e31824baff3. [DOI] [PubMed] [Google Scholar]
- 16.Hines CD, Frydrychowicz A, Hamilton G, et al. T(1) independent, T(2) (*) corrected chemical shift based fat-water separation with multi-peak fat spectral modeling is an accurate and precise measure of hepatic steatosis. Journal of magnetic resonance imaging. 2011;33(4):873–81. doi: 10.1002/jmri.22514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu H, Shimakawa A, Hines CD, et al. Combination of complex-based and magnitude-based multiecho water-fat separation for accurate quantification of fat-fraction. Magnetic Resonance in Medicine. 2011;66(1):199–206. doi: 10.1002/mrm.22840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meisamy S, Hines CD, Hamilton G, et al. Quantification of hepatic steatosis with T1-independent, T2-corrected MR imaging with spectral modeling of fat: blinded comparison with MR spectroscopy. Radiology. 2011;258(3):767–75. doi: 10.1148/radiol.10100708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rehm JL, Wolfgram PM, Hernando D, Eickhoff JC, Allen DB, Reeder SB. Proton density fat-fraction is an accurate biomarker of hepatic steatosis in adolescent girls and young women. European radiology. 2015;25(10):2921–30. doi: 10.1007/s00330-015-3724-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwimmer JB, Behling C, Newbury R, et al. Histopathology of pediatric nonalcoholic fatty liver disease. Hepatology. 2005;42(3):641–9. doi: 10.1002/hep.20842. [DOI] [PubMed] [Google Scholar]
- 21.Lindback SM, Gabbert C, Johnson BL, et al. Pediatric nonalcoholic fatty liver disease: a comprehensive review. Advances in pediatrics. 2010;57(1):85–140. doi: 10.1016/j.yapd.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 22.Hamilton G, Middleton MS, Bydder M, et al. Effect of PRESS and STEAM sequences on magnetic resonance spectroscopic liver fat quantification. Journal of magnetic resonance imaging. 2009;30(1):145–52. doi: 10.1002/jmri.21809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zand KA, Shah A, Heba E, et al. Accuracy of multiecho magnitude-based MRI (M-MRI) for estimation of hepatic proton density fat fraction (PDFF) in children. Journal of magnetic resonance imaging. 2015;42(5):1223–32. doi: 10.1002/jmri.24888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson BL, Schroeder ME, Wolfson T, et al. Effect of flip angle on the accuracy and repeatability of hepatic proton density fat fraction estimation by complex data-based, T1-independent, T2*-corrected, spectrum-modeled MRI. Journal of magnetic resonance imaging. 2014;39(2):440–7. doi: 10.1002/jmri.24153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bydder M, Yokoo T, Hamilton G, et al. Relaxation effects in the quantification of fat using gradient echo imaging. Magnetic resonance imaging. 2008;26(3):347–59. doi: 10.1016/j.mri.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamilton G, Yokoo T, Bydder M, et al. In vivo characterization of the liver fat (1)H MR spectrum. NMR in biomedicine. 2011;24(7):784–90. doi: 10.1002/nbm.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reeder SB, McKenzie CA, Pineda AR, et al. Water-fat separation with IDEAL gradient-echo imaging. Journal of magnetic resonance imaging. 2007;25(3):644–52. doi: 10.1002/jmri.20831. [DOI] [PubMed] [Google Scholar]
- 28.Reeder SB, Pineda AR, Wen Z, et al. Iterative decomposition of water and fat with echo asymmetry and least-squares estimation (IDEAL): application with fast spin-echo imaging. Magnetic resonance in medicine. 2005;54(3):636–44. doi: 10.1002/mrm.20624. [DOI] [PubMed] [Google Scholar]
- 29.Liu CY, McKenzie CA, Yu H, Brittain JH, Reeder SB. Fat quantification with IDEAL gradient echo imaging: correction of bias from T(1) and noise. Magnetic Resonance in Medicine. 2007;58(2):354–64. doi: 10.1002/mrm.21301. [DOI] [PubMed] [Google Scholar]
- 30.Reeder SB, Robson PM, Yu H, et al. Quantification of hepatic steatosis with MRI: the effects of accurate fat spectral modeling. Journal of magnetic resonance imaging. 2009;29(6):1332–9. doi: 10.1002/jmri.21751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hernando D, Hines CD, Yu H, Reeder SB. Addressing phase errors in fat-water imaging using a mixed magnitude/complex fitting method. Magnetic resonance in medicine. 2012;67(3):638–44. doi: 10.1002/mrm.23044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naressi A, Couturier C, Castang I, de Beer R, Graveron-Demilly D. Java-based graphical user interface for MRUI, a software package for quantitation of in vivo/medical magnetic resonance spectroscopy signals. Computers in Biology and Medicine. 2001;31(4):269–86. doi: 10.1016/s0010-4825(01)00006-3. [DOI] [PubMed] [Google Scholar]
- 33.Vanhamme L, van den Boogaart A, Van Huffel S. Improved method for accurate and efficient quantification of MRS data with use of prior knowledge. Journal of Magnetic Resonance. 1997;129(1):35–43. doi: 10.1006/jmre.1997.1244. [DOI] [PubMed] [Google Scholar]
- 34.Manning PM, Hamilton G, Wang K, Park C, Hooker JC, Wolfson T, Gamst A, Haufe WM, Schlein AN, Middleton MS, Sirlin CB. Agreement between region-of-interest-and parametric map-based hepatic proton density fat fraction estimation in adults with chronic liver disease. Abdominal Radiology. 2016 doi: 10.1007/s00261-016-0925-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Artz NS, Haufe WM, Hooker CA, et al. Reproducibility of MR-based liver fat quantification across field strength: Same-day comparison between 1.5T and 3T in obese subjects. Journal of magnetic resonance imaging. 2015;42(3):811–7. doi: 10.1002/jmri.24842. [DOI] [PMC free article] [PubMed] [Google Scholar]


