Abstract
Impressive results have been achieved by adoptively transferring T-cells expressing CD19-specific CARs with binding domains from murine mAbs to treat B-cell malignancies. T-cell mediated immune responses specific for peptides from the murine scFv antigen-binding domain of the CAR can develop in patients and result in premature elimination of CAR-T-cells increasing the risk of tumor relapse. As fully human scFv might reduce immunogenicity, we generated CD19-specific human scFvs with similar binding characteristics as the murine FMC63-derived scFv using human Ab/DNA-libraries. CARs were constructed in various formats from several scFvs and used to transduce primary human T-cells. The resulting CD19-CAR-T-cells were specifically activated by and lysed CD19-positive tumor cell lines and primary CLL cells, and eliminated human lymphoma xenografts in immunodeficient mice. Certain fully human CAR constructs were superior to the FMC63-CAR, which is widely used in clinical trials. Imaging of cell surface distribution of the human CARs revealed no evidence of clustering without target cell engagement, and tonic signaling was not observed. To further reduce potential immunogenicity of the CARs, we also modified the fusion sites between different CAR components. The described fully human CARs for a validated clinical target may reduce immune rejection compared with murine based CARs.
Introduction
Adoptive immunotherapy with gene-modified T-cells expressing a tumor-reactive CAR has rapidly evolved with the most impressive clinical results using autologous T-cells expressing a CD19-specific CAR to treat B-cell malignancies such as acute lymphoblastic leukemia (ALL), chronic lymphocytic leukemia (CLL), and non-Hodgkin’s lymphoma1–9. Tumor regression has correlated with the level of CAR-T-cell proliferation and the duration of their persistence in the blood4–11. The length of time that CAR-T-cells must persist in vivo to attain complete disease eradication has not been established. However, in patients with ALL the loss of CAR-T-cells after an initial expansion phase coincided with the return of normal B-cells and an increased risk of relapse with CD19+ malignancy3. Multiple mechanisms may be responsible for the inability of certain CAR-T-cells to survive in vivo. One such mechanism is the development of an HLA-restricted T-cell mediated immune response against epitopes derived from the murine scFv used as the antigen-binding domain of the CAR. We previously described T-cell responses to foreign transgene products in patients receiving modified T-cells expressing hygromycin-phosphotransferase and herpes-simplex-virus thymidine-kinase12–13, and recently reported that some patients treated with CD19-CAR-T-cells developed an immune response specific for epitopes in the murine scFv and rendered subsequent T-cell infusions ineffective3.
CARs are synthetic proteins consisting of an antigen-binding moiety, usually an scFv derived from non-human mAbs, linked by hinge/spacer and transmembrane sequences to an intracellular signaling module. CARs may contain unique peptide sequences that could be presented by MHC and potentially be immunogenic. Such epitopes could come from a non-human scFv, fusion sites between different human CAR components, and any additional amino acid (aa) modifications to the CAR. In addition to T-cell responses, CAR-specific Abs, including IgE responses that have induced anaphylaxis, may develop after adoptive transfer of CAR-T-cells, particularly those not targeting B-cells, as with CD19-CARs14–16. Reducing immunogenicity of CARs by using humanized17–19 or fully human scFvs20–22 may improve the longevity of CAR-T-cell persistence and enhance their therapeutic efficacy in patients.
All published clinical trials targeting CD19 have utilized scFvs derived either from the murine FMC63- or SJ25C1-mAbs3–5, 7. Here we describe the successful generation and isolation of anti-human CD19 scFvs from human Ab/DNA-libraries with similar binding characteristics as an scFv derived from FMC63. When tested in CAR formats, certain human scFvs showed improved in vitro functions against tumor cell lines and primary CLL and were more efficient in eliminating lymphoma xenografts in immunodeficient mice than the FMC63-CAR. These data indicate that functional fully human CARs against an antigen that has been successfully targeted in patients can be generated to potentially overcome the immunologic barriers that exist with CARs constructed from scFvs that are not fully human in origin.
Materials and Methods
Cells
HEK293T (ATCC_CRL-11268), HEK293 (ATCC_CRL-1573), and CD19-transfected HEK293 (HEK293/CD19) cells were cultured in DMEM, 10% FCS, and 100 U/ml penicillin/streptomycin. K562 (ATCC_CCL-243), K562/CD1923, Raji (ATCC_CCL-86), and Raji/ffluc24 cells were cultured in RPMI-1640, 5% FCS, and 100 U/ml penicillin/streptomycin. Truncated rhesus macaque CD19 (including the extracellular and transmembrane regions) and chimeric rhesus/human versions of truncated CD19 were cloned into the retroviral plasmid pMP7125. K562 cells were transduced with genes encoding rhesus CD19 and rhesus/human CD19 chimeric molecules, and transgene-positive cells enriched by FACS after staining with an anti-CD19 mAb (BD Bioscience, #555415). The absence of mycoplasma was confirmed for all cell lines by monthly testing. T-cells were isolated and cultured as described26. PBMC isolated from blood of CLL patients with high circulating tumor burdens were used as primary CLL samples.
Screening for human anti-CD19 scFvs from human Ab-chain-libraries
DsDNA displayed fusion libraries were purified, and eluted in binding buffer containing 1 mg/ml BSA (Life Technology) and 0.1 mg/ml of ssDNA (Life Technologies) as blocking reagents. Counter-selections were carried out three times with parental HEK293 or K562 cells, each with 107 pre-blocked cells for 1 h at 4°C followed by centrifugation. Unbound library members were transferred to a new tube, incubated with 107 pre-blocked HEK293/CD19 or K562/CD19 cells for 1 h on ice to prevent internalization and then washed with binding buffer. Enriched pools were generated by addition of murine FMC63 IgG to elute binders from the cell surface that recognize overlapping epitopes as the murine reference antibody.
After six rounds of enrichment, the scFv pools were subcloned into the periplasmic expression vector pET-22b (Life Technologies) and transformed into BL21(DE3) bacterial cells for expression. Supernatants from several hundred clones were screened in a flow cytometry based binding assay. ScFvs specifically binding to CD19+ cells were then scaled up for production and purified through a C-terminal His-tag.
Binding and competition flow cytometry assays
2x105 K562/CD19 cells were incubated for 1 h at 4°C with the indicated concentration of His-tagged anti-CD19 scFvs for binding assays and with a mixture of 10 nM myc-tagged FMC63 scFv and the indicated concentration of His-tagged human scFvs for competition binding assays, respectively. Following two washes, cells were stained with an anti-His mAb (Qiagen, #35370) for binding assays or an anti-myc mAb (EMD Millipore, clone 9E10) for competition binding assays. Mean fluorescence intensity was measured using a Guava flow cytometer (EMD Millipore).
Lentiviral vector construction, virus preparation, transduction of T-cells, and in vitro functional assays
The CD19-specific FMC63 CAR was described previously26. Sequences encoding human anti-CD19 scFvs were linked to a modified IgG4 hinge region with a serine to proline substitution at position 10, the CD28-derived transmembrane region, a 4-1BB-derived costimulatory domain, and intracellular CD247. Codon-optimized CAR genes were combined with a sequence endoding truncated epidermal growth factor receptor (EGFRt) by a T2A sequence and cloned into the lentiviral vector epHIV727. To generate CAR-eGFP fusion genes, eGFP was linked in frame to CD247 replacing T2A-EGFRt. To generate 35-G01S HL mutant, one threonine and two serines were mutated to alanine in a predicted O-glycosylation site of 35-G01S by site directed mutagenesis. The new fusion site between CD28 transmembrane and 4-1BB signaling domain that extended the CD28 sequence by two aa was created by overlapping PCRs. Lentivirus was generated by transient transfection of HEK293T-cells28 using psPAX2 and pMD2G packaging plasmids. Primary human T-cells were activated, transduced, enriched, and expanded as described26. Cytotoxicity was analyzed by a standard 4 h chromium release assay, cytokine concentrations were determined by ELISA, and proliferation was measured by carboxyfluorescein succinimidyl ester (CFSE) dilution as described29.
Flow cytometry
Transduced T-cells were stained with biotin-conjugated (EZ-Link Sulfo-NHS-Biotin, Thermo Scientific) anti-EGFR mAb (ImClone Systems Incorporated) and streptavidin-PE (BD Biosciences, #554061). T-cells were stained with anti-PD-1 mAbs and matched isotype control (Biolegend, #329907, #400119). Flow cytometry was performed on a FACSCantoII and data analyzed with FlowJo (Treestar).
Western Blot
5x106 CAR-T-cells were lysed and 10 μg of protein were separated on a 10% polyacrylamide gel. After blotting, membranes were incubated with an anti-CD247 mAb (BD Biosciences, #556366) followed by an HRP-conjugated rabbit anti-mouse secondary Ab (Cell Signaling Technology, #7076S). To remove N-linked glycosylations, protein lysates were incubated with PNGaseF (NEB) for 1 h at 37°C.
Fluorescence microscopy
Live cells were imaged with a DeltaVision microscope (GE Healthcare) and data were analyzed using ImageJ software. Raji and K562 cells were stained with Hoechst33342 (NucBlue® Live reagent, ThermoFisher) for 20 min, washed, and then co-cultured with CAR-T-cells for 10 min at 37°C.
MHC binding prediction
Binding affinities of 9-mer peptides derived from the CAR sequence that are not encoded by the human genome were predicted using NetMHC30.
NOD/SCID/γc−/− (NSG) mouse model
Six to eight week old female NSG mice were engrafted with 5x105 Raji/ffluc cells via tail vein injection. One week later, CAR- or control T-cells that had been expanded with LCL for nine days were injected i.v. Bioluminescence imaging was performed as described29. For experiments where differences between individual mice were expected, at least five mice per experimental group were used for data analysis to provide 81% power to detect an effect size of 1.75, based on a t-test with a 1-sided 0.05 level of significance. The investigator was blinded to group allocation.
Study approval
Blood donors provided written informed consent for research protocols approved by the Institutional Review Board of the Fred Hutchinson Cancer Research Center. The Fred Hutchinson Cancer Research Center Institutional Animal Care and Use Committee approved all mouse experiments.
Results
Selection of human CD19-specific scFvs with similar binding characteristics as FMC63 from human Ab-chain-libraries
Highly diverse human VH- and VL-libraries were constructed by RT-PCR from bone marrow and PBMC samples from twenty donors and displayed in a cell-free system as dsDNA-Ab-libraries31. A VH-library was selected through three rounds for binding to HEK293/CD19 cells, but not to HEK293 cells. The enriched VH-library containing ~104–105 different sequences was then converted to a scFv-library by shuffling with the naïve VL-library in a VH-(G4S)3-VL format. This scFv-library comprising at least 109 sequences was further enriched by two rounds of negative selections on HEK293 or K562 cells followed by positive selections on HEK293/CD19 or K562/CD19 cells. Bound scFvs were displaced off the cell surface by competition with FMC63 with the objective of deriving scFvs that targeted a CD19-epitope overlapping with that recognized by FMC63. The obtained scFv pool was then again negatively selected for members not binding HEK293 and K562 cells, followed by a final positive selection for binding to HEK293/CD19 cells. After this extensive selection, the scFv sequences were cloned into a bacterial vector and crude bacterial supernatants from several hundred randomly picked colonies were screened for scFv binding to CD19/K562 cells.
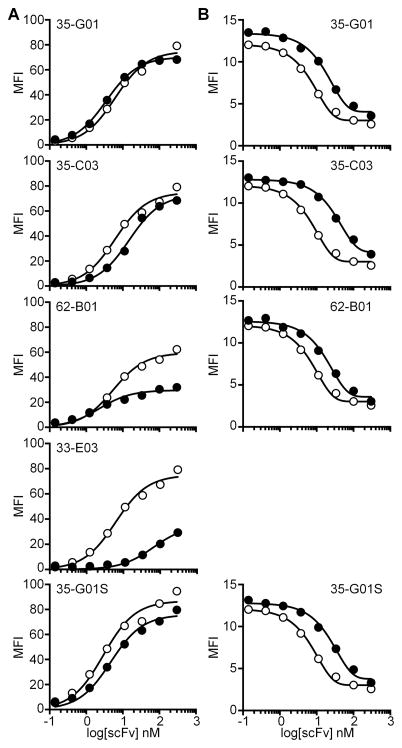
ScFvs from sixty clones that displayed elevated binding to CD19-expressing cells compared to negative cells were purified and characterized for binding affinity (EC50) to CD19 by flow cytometry. We selected three scFvs (35-G01, 35-C03, 62-B01) with a comparable binding affinity to that of the FMC63-scFv (~5 nM), and one scFv (33-E03) with a significantly lower binding activity (Table 1, Fig. 1A). All scFvs included an identical VH paired with different VLs. Analysis of the aa sequence of the 35-G01 scFv identified a cysteine residue in the VL-CDR3 which was not suspected to be involved in target binding as it is the first residue of the VL-CDR3 sequence. This residue represented a potential concern for aggregation due to indiscriminate disulfide bond formation and was therefore converted by site-directed mutagenesis to a serine residue. This derivative scFv (35-G01S) had a similar binding affinity as the original scFv (Table 1, Fig. 1A).
Table 1.
Binding affinity (EC50) and competitive binding activity with FMC63 (IC50) of selected scFvs.
| scFv | EC50 (nM) | IC50 (nM) |
|---|---|---|
| FMC63 | 6.1 ± 0.7 (n=10) | 15.6 ± 2.6 (n=11) |
| 35-G01 | 4.1 ± 0.6 (n=7) | 20.1 ± 5.6 (n=4) |
| 35-G01S | 4.9 ± 0.8 (n=7) | 32.1 ± 2.4 (n=4) |
| 35-C03 | 11.6 ± 1.1 (n=5) | 37.1 ±2.9 (n=3) |
| 62-B01 | 7.0 ± 1.4 (n=5) | 18.2 ± 1.5 (n=2) |
| 33-E03 | >200 (n=4) | not done |
Figure 1. Human CD19-specific scFv with similar characteristics as FMC63 were isolated from a human Ab-chain-library.
(A) ScFv titration to assess binding to K562/CD19 cells. (B) Competition binding assay of each scFv with FMC63. Binding of FMC63 and competition with itself (open symbols) are shown as a comparison in each graph.
An objective was to identify human scFvs that bind CD19 at epitopes shared by FMC63. We therefore analyzed the competitive binding activity (IC50) of individual human scFvs with FMC63. All tested human scFvs displayed 100% competition with FMC63 for binding to K562/CD19 cells with IC50 values in the range of 20–40 nM (Table 1, Fig. 1B). Thus, a number of CD19-specific human scFvs that competed with FMC63 for binding and had a similar affinity as FMC63 for CAR construction were successfully generated.
In vitro function of T-cells expressing CD19-specific CARs constructed with human scFvs
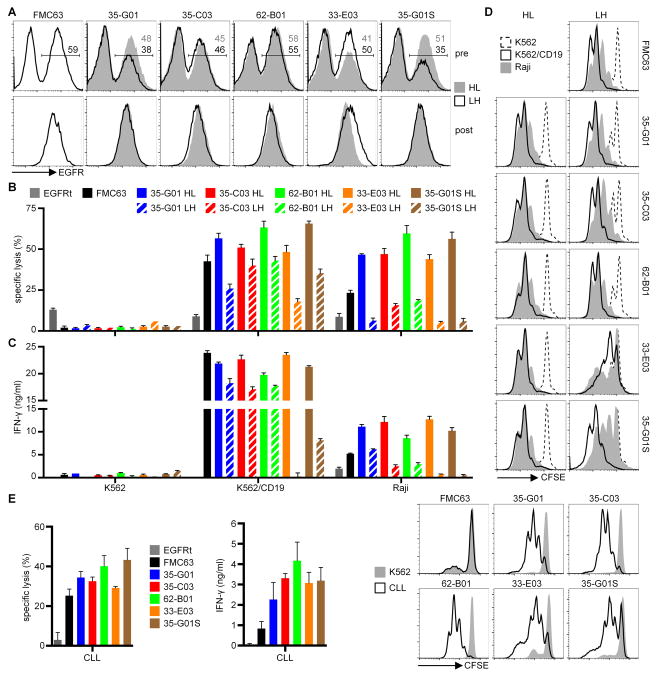
We incorporated each of the five human CD19-specific scFvs shown in Fig. 1 into CARs and compared their ability to redirect T-cell function with that of a CAR containing a murine FMC63-scFv that is currently used in a number of clinical trials3, 5, 7–9. The human VHs and VLs of individual scFvs were linked by a (G4S)3-linker with either the VH at the N-terminus (HL) or the VL at the N-terminus (LH). All CARs contained an extracellular IgG4-hinge region, a CD28 transmembrane region, and an intracellular 4-1BB costimulatory domain linked to the cytoplasmic domain of CD247. Thus, apart from containing human scFv binding regions, the CARs were otherwise comparable to a FMC63-CAR construct used in clinical trials at the FHCRC. The constructs were encoded in lentiviral vectors that contained a truncated epidermal growth factor receptor (EGFRt) to permit purification of transduced T-cells. CD8+ T-cells were isolated from normal donors, transduced with different CAR vectors, and transduction efficiencies were analyzed based on expression of EGFRt. CD8+ T-cells were efficiently transduced with all CAR vectors and transgene-positive T-cells were enriched by sorting based on expression of EGFRt (Fig. 2A). T-cells expressing the FMC63-CAR and each of the human scFv CARs specifically lysed K562 cells transduced with CD19 (K562/CD19) but not control K562 cells, and lysed CD19+ lymphoma cells (Raji) (Fig. 2B). T-cells transduced with human scFv CARs in the HL configuration, which was the original configuration of the scFv-library, were more effective in lysing CD19+ target cells compared to T-cells with LH CARs. These results were consistent with flow cytometry binding assays with purified LH scFvs on K562/CD19 cells that showed reduced or no binding compared to HL scFvs (data not shown).
Figure 2. CARs with human CD19-specific scFvs recognize CD19+ tumor cells.
(A) EGFRt expression on CD8+ T-cells transduced with CD19-CAR-EGFRt constructs before enrichment (pre) and after enrichment and expansion (post). (B) Cytolytic activity of EGFRt+ CD19-CAR-T-cells against CD19+ (K562/CD19, Raji) and control (K562) target cells analyzed by a 4 h chromium release assay at an E:T ratio of 30:1. (C) IFN-γ concentrations in supernatants of CD19-CAR-T-cells after stimulation with K562, K562/CD19, and Raji cells for 24 h analyzed by ELISA. Data in A-C are representative of at least five experiments with T-cells prepared from different donors. Differences between HL and LH constructs are significant (p<0.05) for each CAR. (D) Proliferation of CD19-CAR-T-cells after stimulation with K562, K562/CD19, and Raji cells for 72 h analyzed by a CFSE dilution assay. Data are representative of at least three experiments with T-cells prepared from different donors. (E) Cytolytic activity, IFN-γ production, and proliferation of CD19-CAR-T-cells after co-culture with primary CLL or K562 cells. Data for cytolytic activity and IFN-γ production are shown as means of three CLL samples isolated from different patients. Proliferation data are representative for three CLL samples.
Human HL CAR-T-cells also produced higher cytokine levels and proliferated more after activation with CD19+ cells as compared to LH CAR-T-cells (Fig. 2C,D). When evaluated against Raji lymphoma cells, which express lower levels of CD19 than K562/CD19 cells, we observed superior cytolytic activity, cytokine production and proliferation of T-cells expressing all of the human scFv CARs in the HL configuration compared to T-cells expressing the FMC63-CAR (Fig. 2B–D). The superior effector function of human HL CARs compared with FMC63 observed in this assay was also seen when primary CLL samples were used as target cells (Fig. 2E). Interestingly, T-cells expressing the 33-E03-CAR that contained an scFv with a lower affinity than FMC63 showed significantly more IFN-γ production and proliferation compared to those expressing the FMC63-CAR.
CD19-CARs are expressed at similar levels but differentially glycosylated
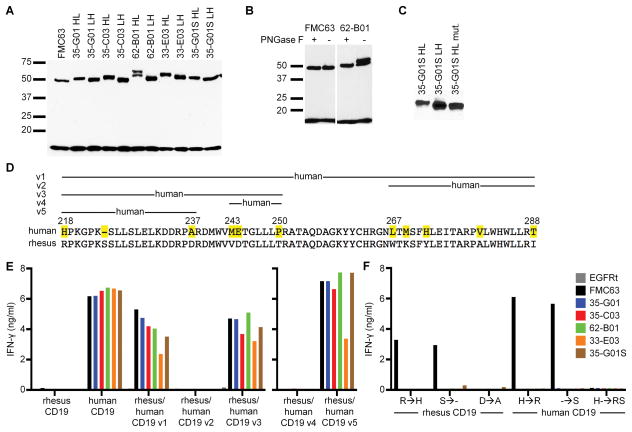
To examine whether the functional differences observed between HL, LH, and FMC63-CARs were due to different expression levels, we analyzed the total amount of CAR-protein. Western blot analysis of cell lysates with a CD247-specific Ab showed that all CARs were expressed at similar levels (Fig. 3A). Although the CARs should have similar molecular weights based on their sequences, Western blot results revealed bands for all HL CARs of a slightly larger molecular weight compared to the LH CARs. In addition, lysates from T-cells expressing the 62-B01 CAR showed two bands. Analysis of the 62-B01 sequence revealed a predicted N-glycosylation site (NetNGlyc 1.0 Server) and treatment of the lysate with PNGaseF removing N-glycosylations resulted in disappearance of the second higher molecular weight band for 62-B01, whereas FMC63 was not affected (Fig. 3B). A potential O-glycosylation site was identified in each of the human HL CARs but not in any of the LH CARs (NetOGlyc 4.0 Server32), and we speculated the size difference in the Western blot results might be due to O-glycosylation at this site. We mutated the site in the 35-G01S HL CAR and showed that the mutated protein had the same apparent size as 35-G01S LH on a Western blot, which suggested that O-glycosylation in HL constructs explained their slightly higher molecular weights (Fig. 3C). However, the additional O-glycosylation did not seem to explain the enhanced function of HL compared to LH CARs, as the in vitro function of 35-G01S HL CARs with and without the glycosylation site were comparable (data not shown).
Figure 3.
(A,B,C) Western Blot analysis of CAR protein (~50 kD) based on CD247 staining. Endogenous CD247 (18 kD) was used as a loading control. Data are representative of at least two experiments. (D) Sequences of human and rhesus macaque CD19 are shown for aa 218–288. Aa that are different are marked in the human sequence. (E) IFN-γ concentrations in supernatants of CD19-CAR-T-cells after co-culture with K562 cells expressing either rhesus macaque, human, or chimeric versions of CD19. (F) IFN-γ concentrations in supernatants of CD19-CAR-T-cells after co-culture with K562 cells expressing either rhesus macaque or human CD19 with single or double mutations to the other species. Data in E and F are representative of at least two experiments.
The fully human CD19-CARs recognize the same region of CD19 as FMC63 but require distinct aa for recognition
The human CD19-CARs in the HL configuration exhibited superior function against CD19+ tumor cells in vitro compared to the FMC63-CAR, despite having similar affinity for CD19. To analyze whether the superior function might be due to differences in the epitope on CD19, we examined the sequences of CD19 that were important for recognition by making human/rhesus chimeric CD19 molecules. The extracellular domains of human and rhesus macaque CD19 are 88% homologous; however, neither FMC63 nor the human CD19-CARs recognized K562 cells expressing rhesus CD19 (Fig. 3D,E). We first replaced aa 218–288 of rhesus CD19 by the human sequence because the epitope for FMC63 is assumed to be located in this region33. CD8+ T-cells expressing either the FMC63- or a human CAR recognized K562 cells transduced with this chimeric CD19 version (rhesus/human CD19_v1), consistent with the requirement for this sequence for recognition (Fig. 3D,E). We next subdivided this region into two parts and constructed CD19 chimeras with the human sequence in either of the two parts (v2: 267–288, v3: 218–250). All CAR-T-cells recognized K562 cells expressing the chimera including aa 218–250 from the human sequence (v3), whereas the other version (v2) was not recognized by any of the CARs (Fig. 3D,E). The region 218–250 was further divided into constructs containing human CD19 sequences corresponding to 243–250 (v4) and 218–237 (v5). All human CARs and FMC63 recognized version 5 but not 4, suggesting that all of the human CARs that we assessed recognize a highly similar region on CD19 as FMC63, a result that is consistent with the results of competition assays performed with the scFvs.
There are only three differences between the human and rhesus sequences within aa 218–237. The residue in each such position in the rhesus sequence was individually mutated to the corresponding aa in the human sequence and these mutant rhesus CD19 molecules were expressed in K562 cells to allow comparison of the fine specificities of the human CARs and the FMC63-CAR. FMC63-CAR-T-cells recognized K562 cells expressing rhesus CD19 with a mutation at position 218 from arginine to histidine or with a deletion of the serine at position 224, but did not recognize rhesus CD19 with only the aspartic acid to alanine substitution (Fig. 3F). None of the human CARs recognized any of the single mutation constructs demonstrating that the fine specificity of these CARs was different than FMC63. To confirm these results, we mutated human CD19 at position 218 from histidine to arginine and/or added a serine at position 224. Here again, the results indicated that each of these CD19 single aa variants was recognized by the FMC63-CAR but not by any of the human CARs (Fig. 3F). A variant containing both arginine and serine substitutions was not recognized by FMC63 or any of the human scFv CARs. Together, these results show that either the presence of histidine at residue 218 or the absence of serine at residue 224 was necessary for FMC63 recognition, but insufficient to restore recognition by the human CD19-CARs. These results demonstrated that the same region encoded by exon 4 of CD19 was required for recognition by both the human CD19-CARs and the FMC63-CAR, but indicated that the particular aa important in this region for recognition by the different CARs were distinct.
Human CD19-CARs lack tonic signaling and cell surface clustering
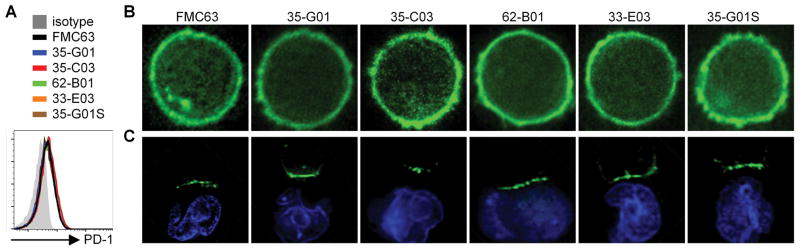
It has been reported that some CARs when expressed in human T-cells result in tonic signaling in the absence of antigen and lead to an exhausted T-cell phenotype, characterized by sustained cell surface expression of PD-1 and loss of function in NSG tumor xenograft models34. We examined the expression of PD-1 on primary T-cells expressing each of the human CD19-CARs ten days after T-cell stimulation and observed only low levels of PD-1, that were comparable to those on T-cells expressing FMC63 (Fig. 4A).
Figure 4.
(A) Flow cytometric analysis of PD-1 expression on CD19-CAR-T-cells ten days after re-stimulation with CD19+ LCL. (B,C) Fluorescence microscopy images of T-cells expressing CD19-CAR-eGFP fusion proteins in the absence (B) or presence (C) of Hoechst33342-labeled (blue) Raji cells. Data in A–C are representative of at least two experiments.
Tonic signaling has also been linked to clustering of CAR molecules on the T-cell surface34, which may preclude their efficient recruitment to synapses with tumor cells. To examine the distribution of CD19-CARs on T-cells, we linked eGFP to the cytoplasmic tail of CD247 and expressed these constructs in CD8+ T-cells. All human CD19-CARs showed a uniform surface distribution on T-cells comparable to the FMC63-CAR, and no evidence of CAR clustering was observed (Fig. 4B). To analyze whether the CARs could cluster in an immunological synapse, we co-cultured the CAR-T-cells with CD19+ Raji cells for 10 min at 37°C before imaging. All CARs concentrated at the interface between the T-cells and target cells, indicating that they could efficiently form an immunological synapse with ligand expressing tumor cells (Fig. 4C). No CAR clustering in synapses was observed when T-cells were co-cultured with CD19− K562 cells (data not shown). In addition, we did not observe constitutive proliferation in the absence of T-cell stimulation as has been reported for c-Met-specific CARs35 (data not shown).
Human CD19-CARs eliminate tumors in vivo
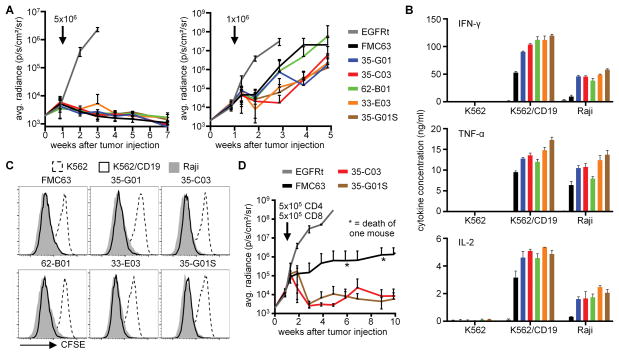
Our in vitro data suggested that the human HL CARs were functionally superior to LH CARs and to the murine FMC63-CAR in tumor recognition assays. To determine if the HL CARs were functional in vivo and to compare their activity to FMC63, NSG mice that had been injected with 5x105 Raji/ffluc cells one week before were treated with 5x106 or 1x106 CD8+ T-cells expressing each of the human scFv CARs or FMC63. At the high dose, T-cells expressing FMC63 and the human CARs eliminated the Raji tumors whereas control T-cells were ineffective, however at the 1x106 cell dose, tumors progressed and progression was more rapid in mice treated with T-cells expressing FMC63 or 62-B01 CARs (Fig. 5A).
Figure 5. CAR-T-cells with a human CD19-specific scFv eradicate tumors in vivo.
(A) Bioluminescence imaging of Raji/ffluc tumor growth in NSG mice that received 5x106 (4 mice per group) and 1x106 (9 mice per group) CD8+ CD19-CAR-T-cells, respectively. EGFRt-T-cells were used as control (4 mice per group). Arrows mark the day of T-cell transfer. (B) Cytokine concentrations in supernatants of CD4+ CD19-CAR-T-cells after stimulation with K562, K562/CD19, and Raji cells for 24 h. (C) Proliferation of CD4+ CD19-CAR-T-cells after stimulation with K562, K562/CD19, and Raji cells for 72 h. Data in B and C are representative of three experiments with T-cells prepared from different donors. (D) Bioluminescence imaging of Raji/ffluc tumor growth in NSG mice (5 mice per group) that received a mixture of 5x105 CD4+ and 5x105 CD8+ CAR-T-cells. EGFRt-T-cells were used as a control (4 mice per group). The arrow marks the day of T-cell transfer.
Our initial functional studies were performed only using transduced CD8+ T-cells, and we have previously shown that CAR-T-cell products containing both CD4+ and CD8+ CAR-T-cells are more potent than either subset alone, and can allow complete tumor eradication at very low cell doses26. To confirm that the human scFv CARs functioned in CD4+ T-cells, we transduced CD4+ T-cells and analyzed their function in vitro. All CD4+ CAR-T-cells released substantial amounts of Th1 cytokines and proliferated after stimulation with CD19+ cells (Fig. 5B,C). Consistent with the results in CD8+ T-cells, CD4+ T-cells expressing the human CARs released significantly more cytokines than CD4+ T-cells expressing the FMC63-CAR.
We then treated Raji/ffluc-bearing NSG mice with 1x106 CAR-T-cells comprised of a 1:1 mixture of CD4+ and CD8+ CAR-T-cells and analyzed tumor growth. These experiments compared the FMC63-CAR currently in the clinic with 35-C03 and 35-G01S, which were potent in the experiments with CD8+ T cells alone. Raji tumors were rejected in mice treated with the mixture of CD4+ and CD8+ CAR-T-cells expressing the human scFvs at this low cell dose, and mice remained tumor free without side effects for >10 weeks. Both human CD19-CARs exhibited better antitumor activity than FMC63 (Fig. 5D). Collectively, the data show that CARs constructed from the fully human CD19-specific scFvs exhibited superior function in vitro and in vivo compared to the FMC63-CAR utilized in clinical trials.
Modifying the CD28/4-1BB fusion site to reduce MHC class I binding peptides
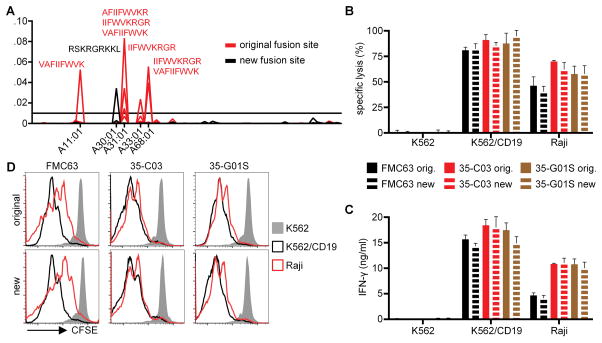
The rationale for replacing murine with human scFvs in CAR design is to reduce the immunogenicity in patients, since it is expected that human T-cells will be tolerant to the peptide sequences of human scFvs, and that Ab responses are less likely to develop. However, even if all individual segments of a CAR are of human origin, fusion sites between these segments could potentially be immunogenic. We have previously shown that CD8+ but not CD4+ T cells are cytotoxic to transgene positive T cells in patients13. Because binding to MHC I molecules is a prerequisite for a peptide to be recognized by cytotoxic T-cells, we analyzed all 9-mer peptides located in the regions spanning each of the fusion sites (H-chain/G4S/L-chain, L-chain/IgG4-hinge, IgG4-hinge/CD28-transmembrane, CD28-transmembrane/4-1BB, 4-1BB/CD247, CD247/T2A) of the CD19-CARs for their potential to bind to human MHC I alleles using the NetMHC prediction algorithm (Fig. 6A and Suppl.Fig. 1A). We identified the fusion site between the CD28 transmembrane and the 4-1BB costimulatory domain as a region of concern that contained seven 9-mer peptide sequences with an affinity to several MHC class I molecules of <100 nM based on MHC binding prediction (Fig. 6A). To reduce the potential of this fusion site to provide immunogenic epitopes, we extended the CD28 sequence by two additional aa (Suppl.Fig. 1B), which resulted in only one predicted 9-mer peptide for HLA-A30:01 with an affinity of <100 nM (Fig. 6A). FMC63, 35-C03, and 35-G01S CARs with this new fusion site were constructed and analyzed for in vitro function. We observed no significant differences between CARs with the original and the new fusion site in cytotoxicity, cytokine release, and proliferation (Fig. 6B,C,D), demonstrating that highly functional CARs with an optimized fusion site between the CD28 transmembrane and the 4-1BB signaling domains can be generated.
Figure 6. The CD28/4-1BB fusion site can be modified to reduce the probability of creating an immunogenic peptide.
(A) Predicted affinities to MHC I of 9-mer peptides derived from the CD28/4-1BB fusion site. Reciprocals of predicted affinities in nM were plotted for all tested MHC types. A predicted affinity of <100 nM (>0.01) was chosen as a threshold. Peptides above the threshold and corresponding HLA types are specified. (B) Cytolytic activity, (C) IFN-γ production, and (D) proliferation of CD8+ T-cells expressing CARs with either the original or the new CD28/4-1BB fusion site after co-culture with K562, K562/CD19, and Raji cells. Data in B–D are representative of at least two experiments with T-cells prepared from different donors.
Discussion
Adoptive transfer of T-cells engineered to express a receptor that re-directs T-cell specificity to a molecule expressed on malignant cells has shown potent antitumor effects in patients with B-cell malignancies and is being investigated for common epithelial tumors19, 36–37. T-cells expressing CARs constructed from scFvs of murine mAbs specific for CD19 have resulted in durable remissions in a subset of patients with relapsed or refractory ALL, CLL, and non-Hodgkin’s lymphoma3–5, 7–8. Relapse with tumor cells that have lost expression of the CD19 epitope targeted by the CAR is a cause of failure38, however some of the patients have relapsed with CD19+ tumor cells after an initial response to therapy, and relapse in these patients has been observed to coincide with a loss of detectable CAR-T-cells in the blood3, 9, 39. The duration that transferred T-cells persist could reflect cell intrinsic properties of the infused T-cells, the costimulatory domain used in the CAR40, inhibitory effects of the patient’s tumor on CAR-T-cell survival, and/or the development of immune responses to the foreign transgene product. Prior work has demonstrated that the transfer of autologous human T-cells expressing foreign proteins, including CARs derived from murine scFvs can elicit cellular and humoral immune responses14–15. In patients treated with CD19-CAR-T-cells, normal B-cells are eliminated thereby compromising humoral immunity, but T-cell responses that primarily targeted peptide epitopes derived from the murine scFv have been detected in patients in whom CAR-T-cells were lost3. Thus, the immunologic barrier to cells expressing foreign proteins represents a challenge for using CAR-T-cells for cancer therapy, where a long duration of persistence may be required to eliminate all sites of metastatic disease. Immune responses to CAR-T-cells may be even more problematic for patients earlier in their disease course when the immune system has not been suppressed by extensive prior chemotherapy, and for targets other than CD19, where Ab responses to the CAR are more likely.
Using a human or humanized scFv in CAR design should reduce the immunogenicity of synthetic CARs, but this approach has not yet been applied successfully to a clinically validated target. Alonso-Camino et al. used lymphocyte display of a lentivirally encoded human scFv-library in a CAR format to screen for human Abs that recognized cell surface molecules on tumor cells22. This approach may be useful as an initial screen to identify candidate human scFvs that recognize cancer cells but requires substantial validation to characterize the target and determine potential expression on normal tissues. Fully human anti-folate-receptor-alpha and anti-mesothelin scFvs were isolated by guided selection and chain shuffling from phage and yeast display human scFv-libraries20–21. These fully human scFvs were functional in vitro and in NSG mouse models20–21; however whether these molecules can be targeted safely or effectively with CAR-T-cells in the clinic remains to be established. The folate-receptor-alpha is expressed on ovarian cancer and on the apical surface of normal epithelial cells, and an initial study of CAR-T-cells derived from murine scFvs was not successful15. Mesothelin is expressed on mesothelioma, lung, ovarian, and pancreatic cancers, but also on normal pericardium and pleura41–42. Analysis of the safety in patients of mesothelin-CARs constructed from murine and human scFvs, respectively, has been initiated by multiple groups16, 43. Both humanized and human scFvs that target EGFR variant III, which is expressed on a subset of glioblastoma, have been described and are effective in animal models in vivo19. However, efforts to target EGFRvIII in clinical trials with CAR-T-cells have thus far been ineffective44.
We focused on developing fully human scFv CARs for CD19 as a target because CARs with murine scFvs are effective in inducing remissions in patients with relapsed and refractory B-cell malignancies. We screened highly diverse human VH- and VL-libraries with the goal of isolating human scFvs that recognized the same epitope of CD19 as FMC63 and with a similar affinity. We identified sixty candidate scFvs and selected five with an identical VH paired with different VL to design CARs with identical spacer, transmembrane, costimulatory, and signaling domains as present in our FMC63-CAR construct3. All scFvs were efficiently expressed as CARs, and in the HL configuration conferred superior recognition of CD19+ tumor cell lines and primary human leukemia compared to the FMC63-CAR. We did not observe evidence of tonic signaling, and cell surface imaging using eGFP fusion constructs showed uniform distribution, similar to FMC63. T-cells expressing the human CD19-CARs were more potent than those expressing FMC63 in eliminating lymphoma xenografts in NSG mice. The superior efficacy of the human scFv CARs was unexpected since they were selected to have similar CD19 binding properties to FMC63. Expression of all CARs was also similar as measured by Western blot and the level of co-expressed eGFP for the fusion constructs. Mapping of sequences involved in recognition of CD19 demonstrated that the same 19 aa region encoded by exon 4 of CD19 was required for recognition by all of the human scFvs and FMC63, but identified a distinct role for specific aa within this region. This difference in fine specificity or slight differences in CAR surface expression may account for the improved function of CARs constructed from the human scFvs.
A comprehensive assessment of potential for immunogenicity of synthetic receptors requires analysis of all foreign peptide sequences that are present, including fusion sites between human components of the CAR. Prior studies have suggested that CD8+ T-cells that recognize peptides from foreign transgene products presented by HLA class I molecules on transduced T-cells are a major mediator of immune mediated elimination12. We used bioinformatics to evaluate the potential for peptides comprising the fusion sites to bind to human HLA class I alleles, and identified candidate immunogenic sequences between CD28 and 4-1BB that could be removed by altering the site of the fusion.
In conclusion, we describe the derivation, characterization, and function of CD19-specific CARs derived from fully human scFvs. These novel CARs can be used for reprogramming T-cells against a validated clinical target. Using fully human CARs may reduce the risk of developing anti-CAR immune responses that have contributed to their premature elimination and a higher risk of relapse in prior trials. A fully human CD19-CAR may also allow repeat infusions enabling dose escalation of T-cells to reduce the severity of cytokine release syndrome and other toxicities of CAR-T-cells.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health CA136551 and CA114536 (S.R.R.) and Juno Therapeutics, Inc.
The authors would like to thank Melissa Comstock and Don Parilla (Shared Resources, Fred Hutchinson Cancer Research Center) for expertise in performing mouse experiments. This work was supported by grants from the National Institutes of Health CA136551 and CA114536 (S.R.R.) and Juno Therapeutics, Inc.
Footnotes
Authorship Contributions:
Contribution: D.S. designed and performed research, analyzed data, and wrote the manuscript; T.H., S.M.S., and A.I.S. designed and performed research and analyzed data; Y.C. and K.M.M. provided expert advice and analyzed data, S.R.R. designed research, analyzed data, and wrote the manuscript.
Conflict of interest statement:
S.R.R. is founder and shareholder of Juno Therapeutics, Inc.
References
- 1.Jensen MC, Riddell SR. Design and implementation of adoptive therapy with chimeric antigen receptor-modified T cells. Immunol Rev. 2014 Jan;257(1):127–144. doi: 10.1111/imr.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kochenderfer JN, Rosenberg SA. Treating B-cell cancer with T cells expressing anti-CD19 chimeric antigen receptors. Nat Rev Clin Oncol. 2013 May;10(5):267–276. doi: 10.1038/nrclinonc.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turtle CJ, Hanafi LA, Berger C, Gooley TA, Cherian S, Hudecek M, et al. CD19 CAR-T cells of defined CD4+:CD8+ composition in adult B cell ALL patients. J Clin Invest. 2016 Jun 1;126(6):2123–2138. doi: 10.1172/JCI85309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davila ML, Riviere I, Wang X, Bartido S, Park J, Curran K, et al. Efficacy and Toxicity Management of 19–28z CAR T Cell Therapy in B Cell Acute Lymphoblastic Leukemia. Sci Transl Med. 2014 Feb 19;6(224):224ra225. doi: 10.1126/scitranslmed.3008226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grupp SA, Kalos M, Barrett D, Aplenc R, Porter DL, Rheingold SR, et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N Engl J Med. 2013 Apr 18;368(16):1509–1518. doi: 10.1056/NEJMoa1215134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalos M, Levine BL, Porter DL, Katz S, Grupp SA, Bagg A, et al. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci Transl Med. 2011 Aug 10;3(95):95ra73. doi: 10.1126/scitranslmed.3002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kochenderfer JN, Dudley ME, Feldman SA, Wilson WH, Spaner DE, Maric I, et al. B-cell depletion and remissions of malignancy along with cytokine-associated toxicity in a clinical trial of anti-CD19 chimeric-antigen-receptor-transduced T cells. Blood. 2012 Mar 22;119(12):2709–2720. doi: 10.1182/blood-2011-10-384388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Porter DL, Levine BL, Kalos M, Bagg A, June CH. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N Engl J Med. 2011 Aug 25;365(8):725–733. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maude SL, Frey N, Shaw PA, Aplenc R, Barrett DM, Bunin NJ, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med. 2014 Oct 16;371(16):1507–1517. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Savoldo B, Ramos CA, Liu E, Mims MP, Keating MJ, Carrum G, et al. CD28 costimulation improves expansion and persistence of chimeric antigen receptor-modified T cells in lymphoma patients. J Clin Invest. 2011 May;121(5):1822–1826. doi: 10.1172/JCI46110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brentjens RJ, Riviere I, Park JH, Davila ML, Wang X, Stefanski J, et al. Safety and persistence of adoptively transferred autologous CD19-targeted T cells in patients with relapsed or chemotherapy refractory B-cell leukemias. Blood. 2011 Nov 3;118(18):4817–4828. doi: 10.1182/blood-2011-04-348540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riddell SR, Elliott M, Lewinsohn DA, Gilbert MJ, Wilson L, Manley SA, et al. T-cell mediated rejection of gene-modified HIV-specific cytotoxic T lymphocytes in HIV-infected patients. Nat Med. 1996 Feb;2(2):216–223. doi: 10.1038/nm0296-216. [DOI] [PubMed] [Google Scholar]
- 13.Berger C, Flowers ME, Warren EH, Riddell SR. Analysis of transgene-specific immune responses that limit the in vivo persistence of adoptively transferred HSV-TK-modified donor T cells after allogeneic hematopoietic cell transplantation. Blood. 2006 Mar 15;107(6):2294–2302. doi: 10.1182/blood-2005-08-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamers CH, Willemsen R, van Elzakker P, van Steenbergen-Langeveld S, Broertjes M, Oosterwijk-Wakka J, et al. Immune responses to transgene and retroviral vector in patients treated with ex vivo-engineered T cells. Blood. 2011 Jan 6;117(1):72–82. doi: 10.1182/blood-2010-07-294520. [DOI] [PubMed] [Google Scholar]
- 15.Kershaw MH, Westwood JA, Parker LL, Wang G, Eshhar Z, Mavroukakis SA, et al. A phase I study on adoptive immunotherapy using gene-modified T cells for ovarian cancer. Clin Cancer Res. 2006 Oct 15;12(20 Pt 1):6106–6115. doi: 10.1158/1078-0432.CCR-06-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maus MV, Haas AR, Beatty GL, Albelda SM, Levine BL, Liu X, et al. T cells expressing chimeric antigen receptors can cause anaphylaxis in humans. Cancer Immunol Res. 2013 Jul;1(1):26–31. doi: 10.1158/2326-6066.CIR-13-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Y, Wang QJ, Yang S, Kochenderfer JN, Zheng Z, Zhong X, et al. A herceptin-based chimeric antigen receptor with modified signaling domains leads to enhanced survival of transduced T lymphocytes and antitumor activity. J Immunol. 2009 Nov 1;183(9):5563–5574. doi: 10.4049/jimmunol.0900447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun M, Shi H, Liu C, Liu J, Liu X, Sun Y. Construction and evaluation of a novel humanized HER2-specific chimeric receptor. Breast Cancer Res. 2014;16(3):R61. doi: 10.1186/bcr3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson LA, Scholler J, Ohkuri T, Kosaka A, Patel PR, McGettigan SE, et al. Rational development and characterization of humanized anti-EGFR variant III chimeric antigen receptor T cells for glioblastoma. Sci Transl Med. 2015 Feb 18;7(275):275ra222. doi: 10.1126/scitranslmed.aaa4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanitis E, Poussin M, Hagemann IS, Coukos G, Sandaltzopoulos R, Scholler N, et al. Redirected antitumor activity of primary human lymphocytes transduced with a fully human anti-mesothelin chimeric receptor. Mol Ther. 2012 Mar;20(3):633–643. doi: 10.1038/mt.2011.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song DG, Ye Q, Poussin M, Liu L, Figini M, Powell DJ., Jr A fully human chimeric antigen receptor with potent activity against cancer cells but reduced risk for off-tumor toxicity. Oncotarget. 2015 Aug 28;6(25):21533–21546. doi: 10.18632/oncotarget.4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alonso-Camino V, Sanchez-Martin D, Compte M, Nunez-Prado N, Diaz RM, Vile R, et al. CARbodies: Human Antibodies Against Cell Surface Tumor Antigens Selected From Repertoires Displayed on T Cell Chimeric Antigen Receptors. Mol Ther Nucleic Acids. 2013;2:e93. doi: 10.1038/mtna.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terakura S, Yamamoto TN, Gardner RA, Turtle CJ, Jensen MC, Riddell SR. Generation of CD19-chimeric antigen receptor modified CD8+ T cells derived from virus-specific central memory T cells. Blood. 2012 Jan 5;119(1):72–82. doi: 10.1182/blood-2011-07-366419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hudecek M, Sommermeyer D, Kosasih PL, Silva-Benedict A, Liu L, Rader C, et al. The nonsignaling extracellular spacer domain of chimeric antigen receptors is decisive for in vivo antitumor activity. Cancer Immunol Res. 2015 Feb;3(2):125–135. doi: 10.1158/2326-6066.CIR-14-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engels B, Cam H, Schuler T, Indraccolo S, Gladow M, Baum C, et al. Retroviral vectors for high-level transgene expression in T lymphocytes. Hum Gene Ther. 2003 Aug 10;14(12):1155–1168. doi: 10.1089/104303403322167993. [DOI] [PubMed] [Google Scholar]
- 26.Sommermeyer D, Hudecek M, Kosasih PL, Gogishvili T, Maloney DG, Turtle CJ, et al. Chimeric antigen receptor-modified T cells derived from defined CD8(+) and CD4(+) subsets confer superior antitumor reactivity in vivo. Leukemia. 2016 Feb;30(2):492–500. doi: 10.1038/leu.2015.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Chang WC, Wong CW, Colcher D, Sherman M, Ostberg JR, et al. A transgene-encoded cell surface polypeptide for selection, in vivo tracking, and ablation of engineered cells. Blood. 2011 Aug 4;118(5):1255–1263. doi: 10.1182/blood-2011-02-337360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leisegang M, Engels B, Meyerhuber P, Kieback E, Sommermeyer D, Xue SA, et al. Enhanced functionality of T cell receptor-redirected T cells is defined by the transgene cassette. J Mol Med. 2008 May;86(5):573–583. doi: 10.1007/s00109-008-0317-3. [DOI] [PubMed] [Google Scholar]
- 29.Hudecek M, Lupo-Stanghellini MT, Kosasih PL, Sommermeyer D, Jensen MC, Rader C, et al. Receptor Affinity and Extracellular Domain Modifications Affect Tumor Recognition by ROR1-Specific Chimeric Antigen Receptor T Cells. Clin Cancer Res. 2013 Jun 15;19(12):3153–3164. doi: 10.1158/1078-0432.CCR-13-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lundegaard C, Lamberth K, Harndahl M, Buus S, Lund O, Nielsen M. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8–11. Nucleic Acids Res. 2008 Jul 1;36(Web Server issue):W509–512. doi: 10.1093/nar/gkn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner RW, Litovchick A, Chen Y. Protein screening methods. US9134304 B2. US Patent. 2015
- 32.Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT, et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013 May 15;32(10):1478–1488. doi: 10.1038/emboj.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Oliveira SN, Wang J, Ryan C, Morrison SL, Kohn DB, Hollis RP. A CD19/Fc fusion protein for detection of anti-CD19 chimeric antigen receptors. J Transl Med. 2013;11:23. doi: 10.1186/1479-5876-11-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Long AH, Haso WM, Shern JF, Wanhainen KM, Murgai M, Ingaramo M, et al. 4-1BB costimulation ameliorates T cell exhaustion induced by tonic signaling of chimeric antigen receptors. Nat Med. 2015 Jun;21(6):581–590. doi: 10.1038/nm.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frigault MJ, Lee J, Basil MC, Carpenito C, Motohashi S, Scholler J, et al. Identification of chimeric antigen receptors that mediate constitutive or inducible proliferation of T cells. Cancer Immunol Res. 2015 Apr;3(4):356–367. doi: 10.1158/2326-6066.CIR-14-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morello A, Sadelain M, Adusumilli PS. Mesothelin-Targeted CARs: Driving T Cells to Solid Tumors. Cancer Discov. 2016 Feb;6(2):133–146. doi: 10.1158/2159-8290.CD-15-0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berger C, Sommermeyer D, Hudecek M, Berger M, Balakrishnan A, Paszkiewicz PJ, et al. Safety of Targeting ROR1 in Primates with Chimeric Antigen Receptor-Modified T Cells. Cancer Immunol Res. 2015 Feb;3(2):206–216. doi: 10.1158/2326-6066.CIR-14-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sotillo E, Barrett DM, Black KL, Bagashev A, Oldridge D, Wu G, et al. Convergence of Acquired Mutations and Alternative Splicing of CD19 Enables Resistance to CART-19 Immunotherapy. Cancer Discov. 2015 Dec;5(12):1282–1295. doi: 10.1158/2159-8290.CD-15-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee DW, Kochenderfer JN, Stetler-Stevenson M, Cui YK, Delbrook C, Feldman SA, et al. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet. 2015 Feb 7;385(9967):517–528. doi: 10.1016/S0140-6736(14)61403-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawalekar OU, O’Connor RS, Fraietta JA, Guo L, McGettigan SE, Posey AD, Jr, et al. Distinct Signaling of Coreceptors Regulates Specific Metabolism Pathways and Impacts Memory Development in CAR T Cells. Immunity. 2016 Feb 16;44(2):380–390. doi: 10.1016/j.immuni.2016.01.021. [DOI] [PubMed] [Google Scholar]
- 41.Lamberts LE, de Groot DJ, Bense RD, de Vries EG, Fehrmann RS. Functional genomic mRNA profiling of a large cancer data base demonstrates mesothelin overexpression in a broad range of tumor types. Oncotarget. 2015 Sep 29;6(29):28164–28172. doi: 10.18632/oncotarget.4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang K, Pastan I. Molecular cloning of mesothelin, a differentiation antigen present on mesothelium, mesotheliomas, and ovarian cancers. Proc Natl Acad Sci U S A. 1996 Jan 9;93(1):136–140. doi: 10.1073/pnas.93.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adusumilli PS, Cherkassky L, Villena-Vargas J, Colovos C, Servais E, Plotkin J, et al. Regional delivery of mesothelin-targeted CAR T cell therapy generates potent and long-lasting CD4-dependent tumor immunity. Sci Transl Med. 2014 Nov 5;6(261):261ra151. doi: 10.1126/scitranslmed.3010162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Rourke D, Desai A, Morrissette J, Martinez-Lage M, Nasrallah M, Brem S, et al. Abstracts from the 20th Annual Scientific Meeting of the Society for Neuro-Oncology, November 19 – 22, 2015, San Antonio, Texas: IMCT-15 PILOT STUDY OF T CELLS REDIRECTED TO EGFRvIII WITH A CHIMERIC ANTIGEN RECEPTOR IN PATIENTS WITH EGFRvIII+ GLIOBLASTOMA. Neuro-Oncology. 2015;17(suppl 5):v110–v111. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.