Abstract
Brain-derived neurotrophic factor (BDNF) is an activity-dependent neurotrophin critical for neuronal plasticity in the hippocampus. BDNF is encoded by multiple transcripts with alternative 5′ untranslated regions (5′UTRS) that display activity-induced targeting to distinct subcellular compartments. While individual Bdnf 5′UTR transcripts influence dendrite morphology in cultured hippocampal neurons, it is unknown whether Bdnf splice variants impact dendrite arborization in functional classes of neurons in the intact hippocampus. Moreover, the contribution of Bdnf 5′UTR splice variants to dendritic spine density and shape has not been explored. We analyzed the structure of CA1 and CA3 dendrite arbors in transgenic mice lacking BDNF production from exon (Ex) 1, 2, 4, or 6 splice variants (Bdnf-e1, -e2, -e4, and -e6 −/− mice) and found that loss of BDNF from individual Bdnf mRNA variants differentially impacts the complexity of apical and basal arbors in vivo. Consistent with subcellular localization studies, Bdnf Ex2 and Ex6 transcripts significantly contributed to dendrite morphology in both CA1 and CA3 neurons. While Bdnf-e2 −/− mice showed increased branching proximal to the soma in CA1 and CA3 apical arbors, Bdnf-e6 −/− mice showed decreased apical and basal dendrite complexity. Analysis of spine morphology on Bdnf-e6 −/− CA1 dendrites revealed changes in the percentage of differently sized spines on apical, but not basal, branches. These results provide further evidence that Bdnf splice variants generate a spatial code that mediates the local actions of BDNF in distinct dendritic compartments on structural and functional plasticity.
Keywords: BDNF transcripts, hippocampus, dendrite branching, spine morphology, spatial code
Introduction
Brain-derived neurotrophic factor (BDNF) is an activity-dependent neurotrophin that mediates diverse brain functions including neuronal survival, neurite morphology, and synaptic plasticity (Cohen-Cory et al. 2010; Huang and Reichardt 2001; McAllister et al. 1999). In the cortex and hippocampus, BDNF regulates local dendritic growth and branching in pyramidal neurons (Horch and Katz 2002; Ji et al. 2005; McAllister et al. 1997; McAllister et al. 1995) as well as granule cells in the dentate gyrus (Gao et al. 2009; Tolwani et al. 2002). The role of BDNF in dendrite morphology is complex as it can enhance branch length and number (Horch and Katz 2002; McAllister et al. 1995; Wirth et al. 2003), but can also induce dendritic instability and limit branch arborization (Horch et al. 1999; Lom and Cohen-Cory 1999; McAllister et al. 1997). Furthermore, BDNF can have opposing effects on apical and basal dendrite arbors (McAllister et al. 1997; McAllister et al. 1995). The molecular mechanisms mediating these diverse and sometimes opposite effects of BDNF on dendrite morphology have been relatively unexplored.
In addition to regulating dendrite formation and maintenance, BDNF signaling plays a pivotal role in modifying the shape and number of dendritic spines in many cell types, including pyramidal neurons (Chakravarthy et al. 2006; Matsutani and Yamamoto 2004; Murphy et al. 1998; Shimada et al. 1998). While disrupting BDNF expression in adult mice decreases spine density on cortical pyramidal neurons (Vigers et al. 2012), exogenous application of BDNF increases spine density on CA1 apical branches in an ERK1/2 dependent manner (Alonso et al. 2004). Interestingly, alternative polyadenylation of Bdnf mRNA transcripts differentially impacts spine morphology and synaptic plasticity in CA1 hippocampal neurons (An et al. 2008). While it is established that BDNF is expressed in hippocampal dendrites and undergoes activity-induced local translation (Baj et al. 2011; Tongiorgi et al. 2004), it has only recently been demonstrated that BDNF engages in autocrine signaling in dendritic spines to locally alter structural and functional plasticity of CA1 neurons (Harward et al. 2016; Hedrick et al. 2016). It is hypothesized that tight regulation of BDNF expression in distinct compartments accounts for local effects of BDNF on dendrite and spine structure.
Production of multiple transcripts is one mechanism by which BDNF expression is tightly controlled. The Bdnf gene has 9 unique promoters that drive transcription of at least 20 different Bdnf transcripts that encode an identical BDNF protein (Aid et al. 2007; Liu et al. 2006; Pruunsild et al. 2007; Timmusk et al. 1993). Each splice variant consists of a 5′ untranslated region (UTR) exon that is alternatively spliced to a downstream common coding exon with a 3′UTR containing 2 different polyadenylation signals (Fig. 1a). The existence of multiple Bdnf splice variants has led to the spatial code hypothesis, which posits that differential expression of 5′UTR transcripts enables local spatial, temporal, and stimulus-specific BDNF production (Tongiorgi 2008). Indeed, Bdnf mRNA variants are differentially expressed across brain regions and show activity-dependent targeting to dendrites, especially in the hippocampus. (Sathanoori et al. 2004; Timmusk et al. 1993; Tongiorgi et al. 2004; Tongiorgi et al. 1997). Upon activation of both cortical and hippocampal neurons, Bdnf Ex1 and Ex4 transcripts are localized to the cell body and proximal dendrites, while Ex2 and Ex6 transcripts are found in distal dendrites (Baj et al. 2011; Chiaruttini et al. 2008; Pattabiraman et al. 2005). Stimuli that enhance Bdnf expression, including antidepressants and physical exercise, also result in targeting of specific 5′UTR transcripts (Baj et al. 2011). In line with these findings, decreasing Ex1 and Ex4 transcripts in cultured hippocampal neurons reduces proximal dendrite number, while decreasing Ex2 and Ex6 transcripts alters distal dendrites (Baj et al 2011). In vivo localization studies and in vitro functional studies provide evidence supporting the notion of unique roles for Bdnf 5′UTR splice variants, but how loss of individual variants impacts dendrite arborization and spine formation in distinct populations of pyramidal neurons in the intact hippocampus has not been examined.
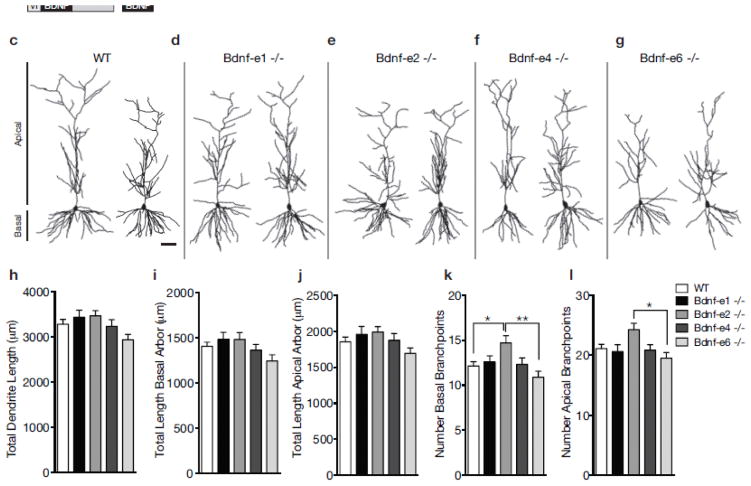
Fig. 1.
Bdnf splice variants impact branching, but not length, of CA1 dendrite arbors. a Schematic of Bdnf gene transcription. Transcription is initiated from promoters upstream of unique 5′-untranslated regions (UTRs) and spliced to the common coding exon IX. Each transcript produces the same BDNF protein. b Design of Bdnf −/− mice using Bdnf-e1 −/− as a representative example. Targeting vectors were designed to insert an enhanced green fluorescent protein (eGFP) upstream of the exon’s splice donor site. Bdnf-e1 mice express a Bdnf-I-eGFP-STOP-Bdnf IX transcript leading to the production of GFP in lieu of BDNF. c–g Representative neurolucida tracings of CA1 pyramidal neuron apical and basal dendrites for WT (c) and Bdnf -e1 (d), -e2 (e), -e4 (f), and -e6 (g) −/− mice. h–l Morphometric analyses of total dendritic length (h), total basal arbor length (i), total apical arbor length (j), number of basal branchpoints (k) and number of apical branchpoints (l) in WT and Bdnf-e1, -e2, -e4, and -e6 −/−CA1 neurons. Bdnf-e2 −/− have significantly more basal branchpoints compared to WT and Bdnf-e6 −/−. Bdnf-e2 −/− also have significantly more apical branchpoints compared to Bdnf-e6 −/−. Data are analyzed using one-way ANOVA followed by Tukey posthoc tests and presented as mean ± SEM, *p<0.05, **p<0.01. Scale bar 50μm. Basal and apical arbors are denoted in (c).
To assess the role of individual Bdnf 5′UTR transcripts on cell morphology, we reconstructed CA1 and CA3 apical and basal dendrite arbors from the brains of mice with selective disruption of BDNF production from promoters I, II, IV, or VI [Bdnf-e1, Bdnf-e2, Bdnf-e4, or Bdnf-e6 mice (Maynard et al. 2016)]. We focused on transcripts derived from these promoters as they represent the majority of Bdnf mRNAs in the brain (Aid et al. 2007; Pruunsild et al. 2007) and disruption of BDNF from these promoters leads to distinct changes in hippocampal BDNF protein expression and unique neurobehavioral deficits (Hill et al. 2016; Maynard et al. 2016). We also analyzed dendritic spine density and morphology on CA1 apical and basal dendrite branches in Bdnf-e6 mutants. We demonstrate that BDNF derived from Ex2 and Ex6 transcripts have opposing effects on both CA1 and CA3 dendrite arbors and provide evidence that Bdnf Ex6 transcripts influence the shape of dendritic spines in distinct dendritic compartments. Our results support the BDNF spatial code hypothesis by demonstrating that individual Bdnf splice variants contribute to structural plasticity in different populations of pyramidal neurons in the intact hippocampus.
Methods
Mouse Generation
Mice with disruption of BDNF production from promoters I, II, IV or VI were generated as previously described [Bdnf-e1, -e2, -e4, and -e6 mice; (Maynard et al. 2016)]. Briefly, an enhanced green fluorescent protein (eGFP)-STOP cassette was inserted upstream of the respective 5′UTR splice donor site of the targeted exon. In Bdnf-e1, -e2, -e4 and -e6 −/− mice, transcription is initiated from promoter I, II, IV, or VI, producing a 5′UTR-eGFP-STOP-Bdnf IX transcript, which leads to GFP production in lieu of BDNF from the targeted promoter (Fig. 1b). While GFP is detected by quantitative PCR and Western blot in all lines, it cannot be visualized by immunofluorescence (Maynard et al. 2016). To facilitate morphological analyses of CA1 and CA3 neurons, a Thy1-M/GFP reporter mouse (Jackson Laboratory Stock #007788, Bar Harbor, Maine, USA), which expresses GFP in a subset of CA1 and CA3 hippocampal neurons under control of the Thy1 promoter (Feng et al. 2000), was backcrossed to C57BL/6J at least 12x and subsequently crossed to each Bdnf mutant line creating Bdnf-e1/ Thy1M, Bdnf-e2/ Thy1M, Bdnf-e4/ Thy1M, and Bdnf-e6/ Thy1M lines. Male WT and Bdnf −/− mice between the ages of 13–15 weeks were used for all experiments. The WT group was mixed and consisted of littermates from Bdnf-e1, -e2, -e4, and -e6 −/− crosses. For each line, WT littermates were housed in the same conditions as corresponding Bdnf mutants (Maynard et al. 2016) with a 12-hour light/ dark cycle and free access to food and water. All animal experiments were approved by the Johns Hopkins University Institutional Animal Care and Use Committee.
Histology
Mice were anesthetized with isoflurane and then transcardially perfused with 4% paraformaldehyde (PFA) in phosphate buffered saline (PBS) pH 7.4. Brains were removed from the skull, post-fixed overnight at 4°C in 4% PFA, cryoprotected in 30% sucrose for at least 48 hours, and coronally sectioned at 150μm on a microtome equipped with a freezing stage. Free-floating sections of hippocampus were stained with 0.05% Sudan Black (Sigma, St. Louis, MO, USA) in 70% ethanol to decrease autofluorescence for optimal imaging of GFP.
Confocal Imaging
GFP-positive CA1 and CA3 apical and basal dendrite arbors were imaged in z-series at 20x magnification using a Zeiss Axioplan Apotome or a Zeiss LSM 700 microscope (Carl Zeiss, Oberkochen, Germany) with the experimenter blinded to genotype. CA1 and CA3 neurons were differentiated by location of cell body within hippocampal anatomy. CA1 neurons were imaged dorsal to the dentate gyrus and CA3 neurons were imaged ventral to the dentate gyrus (Supplementary Figure 1a). Images were montaged in Neurolucida (MicroBright Field Biosciences, Williston, VT, USA) using the Image Montage module. CA1 and CA3 neurons were selected for imaging based on the following criteria: (1) adequate brightness and isolation to allow for unambiguous tracing of dendrite arbors; (2) apical and basal branches were complete and clearly distinguishable from those of neighboring cells; (3) close proximity to the coverslip to allow for capture of all branches in the z-dimension. A representative confocal image of a CA1 neuron selected for reconstruction is depicted in Supplementary Figure 1b. For CA1 dendrites, we imaged 80 neurons from 10 brains for WT, 35 neurons from 8 brains for Bdnf-e1 −/−, 32 neurons from 8 brains for Bdnf-e2 −/−, 46 neurons from 7 brains for Bdnf-e4 −/−, and 43 neurons from 7 brains for Bdnf-e6 −/−. For CA3 dendrites, we imaged 44 neurons from 11 brains for WT, 35 neurons from 11 brains for Bdnf-e1 −/−, 36 neurons from 12 brains for Bdnf-e2 −/−, 31 neurons from 6 brains for Bdnf-e4 −/−, and 35 neurons from 8 brains for Bdnf-e6 −/−.
For Bdnf-e6 −/− dendritic spine studies, isolated oblique apical dendrites or tertiary basal dendrites on CA1 neurons were imaged at 100x magnification on a Zeiss LSM 510 confocal microscope with the experimenter blinded to genotype. Individual branches were selected based on the following criteria: (1) adequate brightness and isolation to allow for clear marking of spine density and morphology (2) unambiguous identification of branch order (3) one branch per neuron and (3) clear attachment to cell body (i.e. we did not image “floating” branches in which cell bodies were in another plane or different section). For CA1 oblique apical dendrites, we imaged 46 branches from 4 brains for WT and 45 neurons from 5 brains for Bdnf-e6 −/−. For CA1 basal dendrites, we imaged 43 branches from 4 brains for WT and 44 neurons from 5 brains for Bdnf-e6 −/−.
Neuronal Reconstruction and Morphometric Analysis
Individual neurons were manually traced from confocal z-stacks using Neurolucida software with experimenter blinded to genotype. For apical and basal arbors, dendritic length and branchpoint number were calculated using NeuroExplorer software (MicroBrightField Biosciences, Williston, VT, USA) and analyzed by one-way analysis of variance (ANOVA) followed by Tukey multiple comparison tests (Supplementary Table 1). For Sholl analysis, concentric three-dimensional shells were centered around the cell body (basal spheres were set every 25μm; apical spheres were set every 50μm) and the number of intersections between the dendrite arbor and a given shell was plotted over distance from the cell body. Concentric circle sizes were selected based on previous studies describing in vivo Bdnf mRNA localization in dendrites (Baj et al. 2012; Chiaruttini et al. 2008; Tongiorgi et al. 2004) as well as pyramidal neuron morphology in the cortex and hippocampus (Chowdhury et al. 2014; Gourley et al. 2013; Maynard and Stein 2012). For CA1 and CA3 apical and basal arbors, four separate repeated measures two-way ANOVAs were used to analyze differences in branching complexity between WT and Bdnf mutants at increasing distances from the cell body (Supplementary Tables 2–4). Bonferroni multiple comparison tests were conducted between WT and each Bdnf mutant. For quantification of spine density (spines per micrometer) and morphology, Neurolucida and Neuroexplorer software were used to calculate the number, height, and width of spines on defined segments of dendrite branches. Student’s t-tests were used to analyze spine density and ordinary two-way ANOVAs followed by Bonferroni posthoc tests were used to examine differences between the percentage of differently sized spines on WT and Bdnf-e6 −/− apical and basal branches (Supplementary Table 5).
Statistical Analysis
Quantitative data are presented as means ± standard error of the mean (SEM). The significance for comparisons was calculated using one-way ANOVAs with Tukey posthoc tests (Figs. 2h–l and 3f–j; Supplementary Table 1) or two-way ANOVAs with Bonferroni posthoc tests where appropriate (Figs. 2e–l, 4e–l, 5d–e, 6d–e; Supplementary Tables 2–5). Statistical analyses were conducted using GraphPad Prism version 7.0a (GraphPad Software, La Jolla, CA, USA). Statistical significance was set at *p<0.05, **p<0.01, &p<0.001, #p<0.0001.
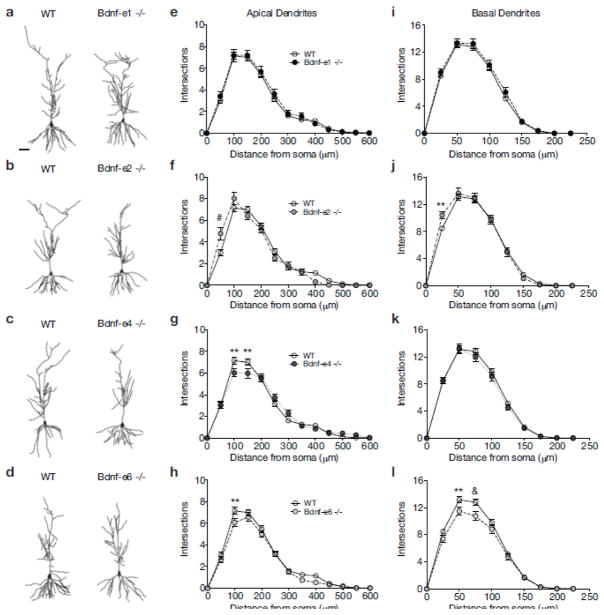
Fig. 2.
BDNF derived from Ex2 and Ex6 transcripts has opposing roles in CA1 dendrite complexity. a–d Illustrative comparisons of WT and Bdnf -e1 (a), -e2 (b), -e4 (c), and -e6 (d) − /− CA1 neurolucida tracings. e–h Sholl analyses of CA1 apical arbors in Bdnf-e1 (e), -e2 (f), -e4 (g) and -e6 (h) −/−mice measuring number of dendritic intersections in 50μm increments from the soma. i–l Sholl analyses of CA1 basal arbors in Bdnf-e1 (i), -e2 (j), -e4 (k) and -e6 (l) −/− mice measuring number of dendritic intersections in 25μm increments from the soma. Bdnf-e2 and -e4 −/− show increased branching compared to WT at particular distances from the soma, while Bdnf-e6 −/− show decreased branching. Data are analyzed using repeated measures two-way ANOVAs followed by Bonferroni posthoc tests and presented as the mean ± SEM, *p<0.05, **p<0.01, &p<0.001, #p<0.0001. Scale bar 50μm. WT group is same for e–h and i–l.
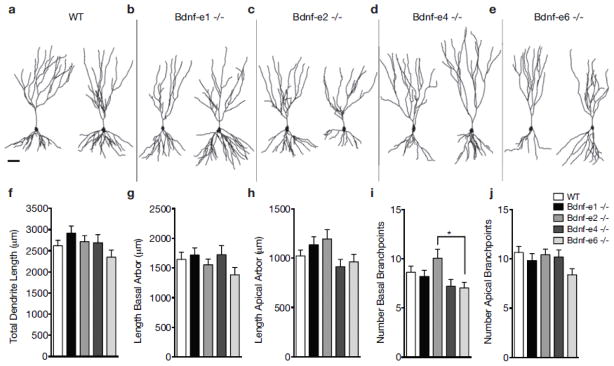
Fig. 3.
Bdnf splice variants influence branching of CA3 dendrite arbors. a–e Representative neurolucida tracings of CA3 pyramidal neuron apical and basal dendrites for WT (a) and Bdnf -e1 (b), -e2 (c), -e4 (d), and -e6 (e) −/− mice. f–j Morphometric analyses of total dendritic length (f), total basal arbor length (g), total apical arbor length (h), number basal branchpoints (i) and number of apical branchpoints (j) in WT and Bdnf-e1, -e2, -e4, and -e6 −/− CA3 neurons. Consistent with findings in CA1 neurons, Bdnf-e2 −/− show increased basal branchpoints compared to Bdnf-6 −/−. Data are analyzed using one-way ANOVA followed by Tukey posthoc tests and presented as mean ± SEM, *p<0.05. Scale bar 50μm.
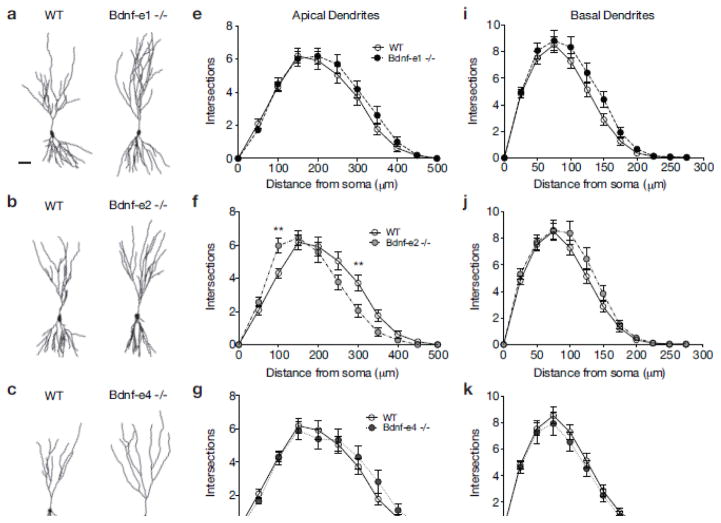
Fig. 4.
Bdnf Ex2 transcripts also influence dendrite complexity in CA3 neurons. a–d Illustrative comparisons of WT and Bdnf -e1 (a), -e2 (b), -e4 (c), and -e6 (d) - /- CA3 neurolucida tracings. eh Sholl analyses of CA3 apical arbors in Bdnf-e1 (e), -e2 (f), -e4 (g) and -e6 (h) −/− mice measuring number of dendritic intersections at 50μm increments from the soma. i–l Sholl analyses of CA3 basal arbors in Bdnf-e1 (i), -e2 (j), -e4 (k) and -e6 (l) −/− mice measuring number of dendritic intersections at 25μm increments from the soma. Bdnf-e2 −/− show a shifted branching curve compared to WT with increased intersections proximal to the soma and decreased intersections distal from the soma. Data are analyzed using repeated measures two-way ANOVAs followed by Bonferroni posthoc tests and presented as the mean ± SEM, **p<0.01. Scale bar 50μm. WT group is same for e–h and i–l.
Fig. 5.
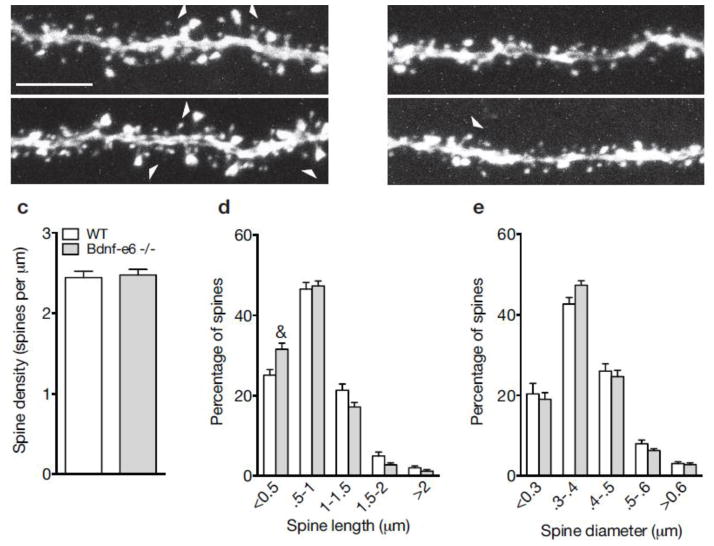
Loss of BDNF derived from Ex6 transcripts impairs spine morphology on CA1 apical dendrites. a–b Confocal z-projections of representative WT (a) and Bdnf-e6 −/− (b) apical branches on GFP-labeled CA1 neurons. c Quantification of spine density (spines per μm) on WT and Bdnf-e6 −/− CA1 apical branches (Student’s t-test). d Frequency histogram of spine height size on WT and Bdnf-e6 −/− CA1 apical branches. Bdnf-e6 −/− show an increase in the percentage of shorter spines compared to WT. e Frequency histogram of spine head width on WT and Bdnf-e6 −/− CA1 apical branches. Bdnf-e6 −/− display strong trends for increases in smaller spine populations and decreases in larger spine populations. Data are analyzed using ordinary two-way ANOVAs followed by Bonferroni posthoc tests and presented as the mean ± SEM, &p<0.001. Scale bar 5μm.
Fig. 6.
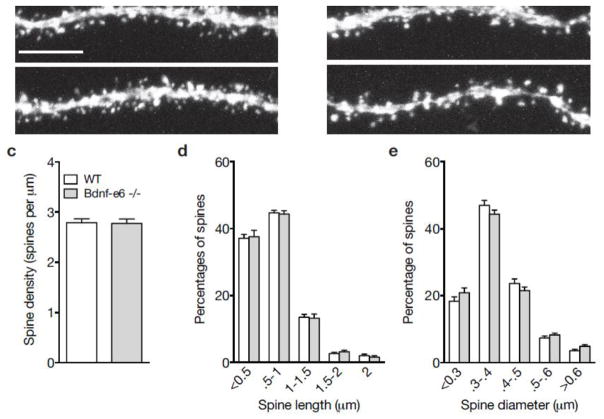
Bdnf-e6 −/− show normal spine density and morphology on CA1 basal branches. a–b Confocal z-projections of representative WT (a) and Bdnf-e6 −/− (b) basal branches on GFP-labeled CA1 neurons. c Quantification of spine density (spines per μm) on WT and Bdnf-e6 −/− CA1 basal branches (Student’s t-test). d Frequency histogram of spine height size on WT and Bdnf-e6 −/− CA1 basal branches. e Frequency histogram of spine head width on WT and Bdnf-e6 −/− CA1 basal branches. Data are analyzed using ordinary two-way ANOVAs followed by Bonferroni posthoc tests and presented as the mean ± SEM. Scale bar 5μm.
Results
Bdnf splice variants differentially impact CA1 dendrite arborization
We generated morphological reconstructions of CA1 pyramidal neurons from WT and Bdnf-e1, -e2, -e4, and -e6 −/− mice (representative reconstructions in Fig. 1c–g). To determine if there were any significant changes in CA1 dendrite growth, we first quantified dendritic arbor length. There were no significant differences in the total length of CA1 dendritic arbors between WT and any Bdnf −/− line (F4,231 = 2.252, p = 0.0643; Fig. 1h). Given that Bdnf 5′UTR variants localize to distinct dendritic compartments, we also analyzed the dendritic length of basal (Fig. 1i) and apical (Fig. 1j) dendritic arbors separately. There were also no significant differences in the total length of basal or apical arbors between WT and any Bdnf −/− line (F4,231 = 2.039, p = 0.0897 and F4,230 = 1.531, p = 0.1940, respectively). However, there was a strong trend for reduced length of basal arbors in Bdnf-e6 −/− mice, which was not observed in Bdnf-e1, -e2, and -e4 mutants. In addition to examining the length of apical and basal arbors, we also analyzed the number of basal and apical branchpoints in CA1 neurons (Figs. 1k and 1l, respectively). There were significant differences in branchpoint number for both basal and apical arbors (F4,231 = 3.502, p = 0.0085 and F4,230 = 2.724, p = 0.0303, respectively; Supplementary Table 1). Posthoc multiple comparison tests revealed that Bdnf-e2 −/− show a 21% increase in basal branchpoints compared to WT and a 35% increase in basal branchpoints compared to Bdnf-e6 −/−. Bdnf-e2 −/− also show a 24% increase in apical branchpoint number compared to Bdnf-e6 −/− and strong trend for increased apical branchpoint number compared to WT. Taken together, morphometric analyses of hippocampal CA1 pyramidal neurons in Bdnf-e1, -e2, -e4, and -e6 −/− mice suggest that Bdnf Ex2 and Ex6 transcripts play an important, and potentially opposing, role in dendrite branching.
To explore how individual Bdnf variants influence apical and basal dendrite complexity in hippocampal CA1 pyramidal neurons, we conducted Sholl analysis, which gives a measure of dendrite arborization by calculating the number of dendritic intersections at increasing distances from the soma. Repeated measures two-way ANOVA revealed a significant interaction between genotype and distance from the soma for apical dendrite intersections (F 52, 3003 = 2.343, p < 0.0001; Supplementary Table 2). Posthoc comparison tests showed that for Bdnf-e1 −/−, apical dendrite arborization was normal and there were no significant differences in the number of intersections at any distance from the soma compared to WT (Fig. 2a and e). For Bdnf-e2 −/− CA1 neurons (Fig. 2b), posthoc comparison tests revealed a significant increase in apical dendrite intersections at 50μm from the soma compared to WT neurons (Fig. 2f). In contrast, Bdnf-e4 −/− CA1 neurons (Fig. 2c) showed significantly fewer intersections at both 100μm and 150μm from the soma compared to WT CA1 neurons (Fig. 2g). Similarly, for Bdnf-e6 −/− CA1 neurons (Fig. 2d), posthoc comparisons indicated significantly fewer apical intersections at 100μm from the soma compared to WT (Fig. 2h).
Sholl analysis of basal dendrite arbors analyzed by repeated measures two-way ANOVA revealed a significant interaction between genotype and distance from the soma for basal dendrite intersections (F36, 2079 = 1.91, p = 0.0009; Supplementary Table 3). While Bdnf-e1 and -e4 −/− CA1 neurons showed no significant differences in basal intersections compared to WT (Fig. 2i and k), Bdnf-e2 −/− showed increased intersections at 25μm from the soma (Fig. 2j) and Bdnf-e6 −/− showed decreased intersections at 50μm and 75μm from the soma compared to WT (Fig. 2l). In summary, loss of BDNF derived from Ex1 transcripts has no effect on CA1 dendrite complexity, while loss of BDNF derived from Ex2, Ex4, and Ex6 transcripts differentially impacts CA1 apical and basal arborization. Taken together, these data suggest that BDNF produced from Ex2 transcripts may restrict dendrite complexity in apical dendrites proximal to the soma, while BDNF produced from Ex4 and Ex6 transcripts may promote more distal branching in both dendritic arbors.
Bdnf splice variants distinctly contribute to CA3 dendrite architecture
We also generated morphological reconstructions of CA3 neurons from each Bdnf −/− line (representative reconstructions in Fig. 3a–e). To quantify the global contribution of Bdnf splice variants to CA3 dendrite morphology, we first measured the total dendritic length of CA3 pyramidal neurons from Bdnf-e1, -e2, -4, and -e6 −/−mice (Fig. 3f). Similar to CA1 neurons, CA3 neurons showed no significant differences in total dendrite length between WT and any Bdnf −/− line (F4, 176 = 1.633, p = 0.1679). Dendritic lengths of CA3 basal (Fig. 3g) and apical (Fig. 3h) arbors were also not significantly different between WT and mutants (F4, 174 = 2.2, p = 0.0710 and F4, 177 = 1.24, p = 0.2955, respectively). In addition to examining dendritic length, we also analyzed the number of apical and basal branchpoints in CA3 neurons (Figs. 3i and 3j, respectively). One-way ANOVA revealed significant differences in basal, but not apical, branchpoints between groups (F4, 175 = 2.958, p = 0.0214 and F4, 177 = 1.947, p = 0.1046, respectively; Supplementary Table 1). Consistent with increased dendritic complexity in Bdnf-e2 −/− and decreased dendritic complexity in Bdnf-e6 −/− CA1 neurons, we found that Bdnf-e2 −/− CA3 neurons showed a 43% increase in basal branchpoint number compared to Bdnf-e6 −/− CA3 neurons. Taken together, morphometric analyses of hippocampal CA3 neurons in Bdnf-e1, -e2, -e4, and -e6 −/− mice suggest that no single Bdnf 5′UTR variant substantially influences dendritic length of apical or basal arbors; however, BDNF derived from Ex2 transcripts may play a unique role in regulating branchpoint number in basal arbors.
To more closely explore how individual Bdnf splice variants influence local apical and basal dendrite arborization in CA3 neurons, we conducted Sholl analysis on reconstructed CA3 neurons from each line. Repeated measures two-way ANOVA revealed a significant interaction between genotype and distance from the soma for apical dendrite intersections (F40, 1770 = 2.646, p < 0.0001; Supplementary Table 4). Posthoc comparison tests showed that for Bdnf-e1 −/−, apical dendrite arborization was normal and there were no significant differences in the number of intersections at any distance from the soma compared to WT (Fig. 4a and e). For Bdnf-e2 −/− CA3 neurons (Fig. 2b), posthoc comparison tests revealed a significant increase in apical dendrite intersections at 100μm from the soma, but a significant decrease in intersections at 300μm from the soma compared to WT neurons (Fig. 4f). In contrast to Bdnf-e4 −/− CA1 neurons, Bdnf-e4 −/− CA3 neurons (Fig. 4c) showed no significant differences in dendrite complexity compared to WT (Fig. 4g). Similarly, for Bdnf-e6 −/− CA3 neurons (Fig. 4d), posthoc comparisons indicated only strong trends for reduced apical intersections at multiple distances from the soma (Fig. 4h).
For basal dendrite Sholl analysis, repeated measures two-way ANOVA of CA3 basal intersections revealed no significant effect of genotype and no interaction between genotype and distance from the soma (Fig. 4i, j, k, l). However, consistent with morphometric analysis of total basal branchpoints (Fig. 3i), Bdnf-e2 −/− CA3 basal arbors showed strong trends for increased basal dendrite complexity (Fig. 4j), while Bdnf-e6 −/− CA3 basal dendrites showed a reduction in intersections at multiple distances from the soma compared to WT (Fig. 4l). In summary, loss of BDNF production from Ex1 and Ex4 transcripts has no effect on CA3 dendrite complexity, while loss of BDNF from Ex2 and Ex6 transcripts differentially impacts CA3 apical arborization. Taken together, these data suggest that Bdnf Ex2 and Ex6 splice variants may serve similar roles in CA1 and CA3 neurons.
Loss of BDNF from Ex6 transcripts impairs aspects of spine morphology
Given that Bdnf-e6 mutants showed the most dramatic changes in CA1 apical and basal dendrite arborization (Figs. 1 and 2), we asked whether loss of BDNF derived from Ex6 transcripts alters the density and morphology of spines on CA1 dendrites in the intact hippocampus. Examination of spine density on CA1 oblique apical dendrites of Bdnf-e6 −/− mice revealed no significant differences compared to WT mice (Fig. 5a–c). Spine distribution also appeared normal with uniform localization along the dendritic shaft. To characterize the structure of dendritic spines, we measured both the height and width of spine heads on WT and Bdnf-e6 −/− apical dendrite branches. For both parameters, spines were classified by size and binned to generate frequency histograms depicting the percentage of differently sized spine populations along a defined branch (Belichenko et al. 2009). Analysis of spine morphology on oblique apical branches by two-way ANOVA followed by Bonferroni posthoc tests showed that Bdnf-e6 −/− branches had a significant 25% increase in the percentage of shorter spines (<0.5μm) and strong trends for decreases in the percentage of longer spines (1–2μm) compared with WT branches (F4, 445 = 5.906, p=0.0001, Fig. 5d; Supplementary Table 5). This reduction in spine height was not accompanied by significant changes in spine head size (Fig. 5e), although Bdnf-e6 −/− apical branches showed trends for increased frequency of spine heads between 0.3μm and 0.4μm in width and decreased frequency of spines heads between 0.5μm and 0.6μm in width compared to WT apical branches. Thus, frequency diagrams of CA1 apical branches suggest an increase in the percentage of short spines and a decrease in the percentage of large spines following loss of BDNF produced from Ex6 transcripts.
To determine if spine changes on Bdnf-e6 −/− CA1 dendrites were selective to apical arbors, we also examined spine morphology on basal dendrites. We focused on tertiary basal dendrites as their branch order was more easily identified and segments were isolated enough for clear confocal imaging. Examination of spine density on Bdnf-6 −/− basal dendrites revealed no significant differences compared to WT dendrites (Fig. 6a–c). Unlike apical branches, Bdnf-e6 −/− basal branches showed no significant changes in the frequency of differently sized spines (Fig. 6d–e). Thus, structural deficits in Bdnf-e6 −/− CA1 basal arbors were specific to dendrites (Fig. 2l) and did not include alterations in dendritic spine shape. Taken together, morphological analysis of CA1 dendritic spines indicates that loss of BDNF from Ex6 transcripts selectively affects spine structure on apical, but not basal dendrites.
Discussion
The existence of multiple Bdnf splice variants with distinct subcellular localizations has led to the hypothesis that different Bdnf 5′UTR transcripts provide a spatial code for the delivery and/or local production of BDNF at specific dendritic locations to mediate structural and functional plasticity events. Here we provide further evidence in support of this hypothesis by demonstrating that loss of BDNF from individual Bdnf 5′UTR splice variants leads to local deficits in CA1 and CA3 apical and basal arbors in the intact hippocampus. Furthermore, we provide new evidence that Bdnf Ex6 transcripts influence dendritic spine structure. Our in vivo morphological analysis of single neurons in Bdnf-e1, -e2, -e4, and -e6 mutant mice is consistent with a cell-autonomous function for BDNF as recently demonstrated (Harward et al. 2016), and suggests a mechanism by which BDNF may differentially impact the development and maintenance of specific dendritic arbors and spines in response to diverse environmental stimuli and experiences.
Bdnf 5′UTR splice variants impact local dendrite complexity of CA1 and CA3 neurons
A large body of evidence has established that Bdnf 5′UTR splice variants are localized to different subcellular compartments in the cortex and hippocampus during both development and adulthood (Pattabiraman et al. 2005; Sathanoori et al. 2004; Tongiorgi 2008). In dissociated hippocampal pyramidal neurons, Bdnf Ex1 and Ex4 transcripts are restricted to the cell soma and proximal dendrites, while Bdnf Ex2c and Ex6 transcripts are expressed in distal dendrites (Baj et al. 2011). These results are supported by in situ hybridization studies in hippocampal sections showing that neuronal activity and epileptogenesis enhance the subcellular localization of Bdnf Ex2c and Ex6 transcripts into distinct dendritic laminae (Baj et al. 2012; Chiaruttini et al. 2008; Tongiorgi et al. 2004). While these studies have provided a strong foundation for the role of individual Bdnf splice variants in regulating hippocampal dendrite morphology, the present study is the first to directly examine the contribution of BDNF derived from distinct 5′UTR Bdnf transcripts to CA1 and CA3 apical and basal dendrite arborization in vivo.
Consistent with previous studies (Baj et al. 2011; Chiaruttini et al. 2008), our results show that loss of BDNF from Ex2 and Ex6 transcripts in Bdnf-e2 and -e6 −/− mice, respectively, significantly impacts CA1 and CA3 dendrite morphology (Figs. 1 and 3). Bdnf-e4 −/− also show mild impairments in distal CA1 dendrite architecture, suggesting that Ex4 transcripts may play an important role in activity-induced dendrite branching despite evidence of restricted localization to the soma and proximal dendrites. While siRNA knockdown of Ex1 and Ex4 transcripts in mixed hippocampal cultures did not significantly alter distal dendrite arborization (Baj et al. 2011), it is possible that subtle branching changes may have been masked in vitro as (1) dissociated neurons are removed from their normal synaptic inputs and (2) CA1 and CA3 neurons are not readily distinguishable making it difficult to detect changes that are unique to each pyramidal neuron class. One advantage to our study is that we reconstruct dendritic morphology in the context of the highly organized tri-synaptic circuit in the intact hippocampus. Furthermore, we are able to differentiate between CA1 and CA3 hippocampal pyramidal neurons, which are functionally distinct and receive inputs from different cell populations. Therefore, the present study may have captured novel roles for Bdnf Ex4 transcripts in distal dendrite branching that were previously difficult to observe in mixed hippocampal cultures. However, it should be noted that we examined neuronal structure at a baseline condition in which mice were not exposed to specific stimuli that enhance neural activity. Future studies should examine whether Bdnf-e1, -e2, -e4, and -e6 −/− mice show impairments in activity-induced remodeling following robust neuronal stimulation, such as kindling or electroconvulsive seizures. As suggested by in vivo localization studies (Chiaruttini et al. 2008; Sathanoori et al. 2004), it is likely that differences between Bdnf splice variants in shaping dendritic architecture would be more dramatic following induction of widespread activity.
Unique roles for Bdnf Ex2 and Ex6 transcripts in dendrite arborization
In addition to implicating Bdnf Ex4 transcripts in dendrite arborization, our in vivo morphological reconstructions provide evidence that Ex2 and Ex6 transcripts are not entirely redundant. In fact, while Ex2 and Ex6 transcripts both affect dendritic morphology, they appear to exert opposing effects in both pyramidal cell types. Loss of BDNF from Ex6 transcripts impairs both apical and basal dendritic architecture at multiple distances from the soma in CA1 neurons (Fig. 2h,l), and there are strong trends for similar phenotypes in CA3 neurons (Figs. 4h,l). On the other hand, loss of BDNF derived from Ex2 transcripts increases apical dendrite complexity proximal to the soma in the stratum lucidum region of the hippocampus in both CA1 and CA3 neurons (Figs. 2f and 4b). In CA3, the stratum lucidum contains the terminal fields of granule cell mossy fibers (Tongiorgi et al. 2004), suggesting that BDNF from Ex2 transcripts may be playing an important role in pruning these synaptic connections. This would be consistent with observations in cortical layer VI neurons, where BDNF has been shown to restrict basal dendrite arborization (McAllister et al. 1997; McAllister et al. 1995). As distal dendrite branching is reduced in Bdnf-e2 −/− CA3 neurons (Fig. 4b), it is possible that BDNF from Ex2 transcripts supports proximal pruning in favor of extending more distal dendrites into other laminae of the hippocampus. Taken together, these results suggest that BDNF synthesized from Ex2 and Ex6 transcripts may have different, possibly opposing, roles in shaping dendritic arbors. Further studies are needed to determine whether these transcripts show differences in activity-induced translational regulation and whether loss of one transcript causes compensatory changes in other Bdnf splice variants. Future developmental studies would also provide important insights into the role of Bdnf Ex2 and Ex6 transcripts in dendrite formation, maintenance, and pruning during critical periods that are especially sensitive to neuronal activity.
Bdnf Ex6 transcripts influence CA1 dendrite architecture
Our results extend previous studies identifying that Bdnf Ex6 transcripts are important for CA3 dendrite morphology by showing that they are also critical for CA1 dendrite morphology. Baj et al. (2012) show that physical exercise and antidepressants induce targeting of Bdnf Ex6 transcripts to CA3 apical dendrites in vivo, suggesting a key role for promoter VI-derived BDNF at synapses between granule cell mossy fibers and CA3 pyramidal neuron dendrites. We demonstrate that BDNF produced from Ex6 transcripts also promotes dendrite growth and branching in CA1 apical and basal dendrites (Figs. 1 and 2), indicating a key role for promoter VI-derived BDNF at synapses between CA1 and CA3 neurons. Indeed, Bdnf-e6 −/− CA1 neurons show significant decreases in apical dendrite complexity at 100μm from the soma in the stratum radiatum region, which is a functionally distinct compartment that receives large numbers of inputs from CA3 via Schaffer collaterals. We also show an important function for BDNF from Ex6 transcripts in CA1 basal dendrite branching, which has been previously unexplored both in vitro and in vivo. Although in situ hybridization data suggest that activity does not induce localization of Bdnf mRNA into the stratum oriens region of the hippocampus, a compartment which harbors the basal dendrites of CA1 and CA3 neurons (Tongiorgi et al. 2004), our results demonstrate that Bdnf Ex6 transcripts may still have a role in shaping these arbors. Dynamic changes in local translation of Bdnf Ex6 transcripts following activity might explain a unique role for this transcript in activity-dependent structural plasticity despite static mRNA localization.
Bdnf splice variants and CA1 dendritic spine morphology
In light of new evidence that BDNF is released from CA1 dendrites and engages in autocrine signaling within individual dendritic spines (Harward et al. 2016; Hedrick et al. 2016), it is timely to consider the contribution of Bdnf 5′UTR splice variants to spine formation and maintenance. Previous studies have shown that alternative Bdnf 3′UTRs differentially contribute to spine morphology in CA1 pyramidal neurons (An et al. 2008). In particular, long 3′ UTR mRNAs are expressed in dendrites and disruption of these transcripts leads to enlargement of spines and impairments in spine pruning. However, the contribution of individual 5′UTR splice variants to spine density and shape has not been previously examined.
Here we provide evidence that loss of BDNF from Ex6 transcripts in Bdnf-e6 −/− mice is associated with shifts in the size of differently sized spine populations on CA1 apical dendrites (Fig 5d). These results are consistent with morphological studies in mice with conditional deletion of BDNF in post-mitotic neurons showing a decrease in mature, mushroom-type spines and an increase in immature thin-type spines on CA1 mid-distal apical dendrites (Rauskolb et al. 2010). Decreases in spine length in Bdnf-e6 −/− CA1 neurons were specific to apical dendrites and were not observed in basal dendrites (Fig. 6). However, we restricted our spine analysis to apical oblique dendrites in the stratum radiatum and tertiary basal dendrites in the stratum oriens, so it is possible that additional changes in spine density or morphology may occur in other branches of CA1 apical and basal dendrites. As Bdnf Ex6 transcripts differentially impact dendrite complexity at various distances from the soma in defined synaptic regions, it will be important to determine whether loss of transcripts in specific dendritic compartments locally impacts structural and functional plasticity of dendritic spines. It is also likely that individual CA1 neurons show differential expression of Bdnf Ex6 transcripts; therefore, selectively analyzing neurons that rely heavily on BDNF derived from promoter VI, potentially for autocrine signaling in dendritic spines, may reveal more exaggerated differences in Bdnf-e6 mutants.
In conclusion, our results provide in vivo evidence that Bdnf 5′UTR mRNA splice variants play a key role in structural organization of key cell types in the hippocampus. These data support the hypothesis that differences in the expression and localization of individual Bdnf transcripts creates a spatial code that facilitates local structural and functional plasticity in specific synaptic connections between hippocampal neurons.
Supplementary Material
Supplementary Figure 1. Representative confocal images of GFP positive neurons in the hippocampus of a Thy1M transgenic mouse. a. Isolated CA1 and CA3 neurons were selected from the CA1 region (blue arrows) and CA3 region (red arrows). b. Illustrative example of an isolated CA1 neuron selected for reconstruction (b). Scale bar 100 um (a) and 50 um (b).
Supplementary Table 1. ANOVA table for data presented in Figures 1h–l and 3f–j. Mean ± SEM is presented for each group as well as ANOVA F and p values. The final 10 columns present the results of Tukey posthoc tests.
Supplementary Table 2. ANOVA table for data presented in Figure 2e–h. Mean ± SEM is presented for each group at each distance. The final 4 columns present the results of Bonferroni post-hoc tests (adjusted p-values). P values for ANOVA are provided below.
Supplementary Table 3. ANOVA table for data presented in Figure 2i–l. Mean ± SEM is presented for each group at each distance. The final 4 columns present the results of Bonferroni post-hoc tests (adjusted p-values). P values for ANOVA are provided below.
Supplementary Table 4. ANOVA table for data presented in Figure 4e–h. Mean ± SEM is presented for each group at each distance. The final 4 columns present the results of Bonferroni post-hoc tests (adjusted p-values). P values for ANOVA are provided below.
Supplementary Table 5. ANOVA table for data presented in Figure 5d. Mean ± SEM is presented for each genotype and bin. The final column presents the results of Bonferroni post-hoc tests. P values for ANOVA are provided below.
Acknowledgments
Funding
Funding for these studies was provided by the Lieber Institute for Brain Development and the National Institute for Mental Health (T32MH01533037 to KRM and RO1MH105592 to KM).
We thank Dr. Daniel Hoeppner for microscopy assistance and Dr. Daniel Weinberger for critical reading of the manuscript.
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
References
- Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T. Mouse and rat BDNF gene structure and expression revisited. Journal of neuroscience research. 2007;85:525–535. doi: 10.1002/jnr.21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso M, Medina JH, Pozzo-Miller L. ERK1/2 activation is necessary for BDNF to increase dendritic spine density in hippocampal CA1 pyramidal neurons. Learn Mem. 2004;11:172–178. doi: 10.1101/lm.67804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An JJ, et al. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134:175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baj G, et al. Physical exercise and antidepressants enhance BDNF targeting in hippocampal CA3 dendrites: further evidence of a spatial code for BDNF splice variants. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2012;37:1600–1611. doi: 10.1038/npp.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baj G, Leone E, Chao MV, Tongiorgi E. Spatial segregation of BDNF transcripts enables BDNF to differentially shape distinct dendritic compartments. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16813–16818. doi: 10.1073/pnas.1014168108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belichenko PV, Wright EE, Belichenko NP, Masliah E, Li HH, Mobley WC, Francke U. Widespread changes in dendritic and axonal morphology in Mecp2-mutant mouse models of Rett syndrome: evidence for disruption of neuronal networks. The Journal of comparative neurology. 2009;514:240–258. doi: 10.1002/cne.22009. [DOI] [PubMed] [Google Scholar]
- Chakravarthy S, et al. Postsynaptic TrkB signaling has distinct roles in spine maintenance in adult visual cortex and hippocampus. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1071–1076. doi: 10.1073/pnas.0506305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaruttini C, Sonego M, Baj G, Simonato M, Tongiorgi E. BDNF mRNA splice variants display activity-dependent targeting to distinct hippocampal laminae. Mol Cell Neurosci. 2008;37:11–19. doi: 10.1016/j.mcn.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Chowdhury TG, Barbarich-Marsteller NC, Chan TE, Aoki C. Activity-based anorexia has differential effects on apical dendritic branching in dorsal and ventral hippocampal CA1. Brain structure & function. 2014;219:1935–1945. doi: 10.1007/s00429-013-0612-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Cory S, Kidane AH, Shirkey NJ, Marshak S. Brain-derived neurotrophic factor and the development of structural neuronal connectivity. Dev Neurobiol. 2010;70:271–288. doi: 10.1002/dneu.20774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, et al. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Gao X, Smith GM, Chen J. Impaired dendritic development and synaptic formation of postnatal-born dentate gyrus granular neurons in the absence of brain-derived neurotrophic factor signaling. Exp Neurol. 2009;215:178–190. doi: 10.1016/j.expneurol.2008.10.009. [DOI] [PubMed] [Google Scholar]
- Gourley SL, Swanson AM, Koleske AJ. Corticosteroid-induced neural remodeling predicts behavioral vulnerability and resilience. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:3107–3112. doi: 10.1523/JNEUROSCI.2138-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harward SC, et al. Autocrine BDNF-TrkB signalling within a single dendritic spine. Nature. 2016;538:99–103. doi: 10.1038/nature19766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick NG, Harward SC, Hall CE, Murakoshi H, McNamara JO, Yasuda R. Rho GTPase complementation underlies BDNF-dependent homo- and heterosynaptic plasticity. Nature. 2016;538:104–108. doi: 10.1038/nature19784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JL, et al. Loss of promoter IV-driven BDNF expression impacts oscillatory activity during sleep, sensory information processing and fear regulation. Transl Psychiatry. 2016;6:e873. doi: 10.1038/tp.2016.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horch HW, Katz LC. BDNF release from single cells elicits local dendritic growth in nearby neurons. Nature neuroscience. 2002;5:1177–1184. doi: 10.1038/nn927. [DOI] [PubMed] [Google Scholar]
- Horch HW, Kruttgen A, Portbury SD, Katz LC. Destabilization of cortical dendrites and spines by BDNF. Neuron. 1999;23:353–364. doi: 10.1016/s0896-6273(00)80785-0. [DOI] [PubMed] [Google Scholar]
- Huang EJ, Reichardt LF. Neurotrophins: roles in neuronal development and function. Annu Rev Neurosci. 2001;24:677–736. doi: 10.1146/annurev.neuro.24.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Pang PT, Feng L, Lu B. Cyclic AMP controls BDNF-induced TrkB phosphorylation and dendritic spine formation in mature hippocampal neurons. Nature neuroscience. 2005;8:164–172. doi: 10.1038/nn1381. [DOI] [PubMed] [Google Scholar]
- Liu QR, Lu L, Zhu XG, Gong JP, Shaham Y, Uhl GR. Rodent BDNF genes, novel promoters, novel splice variants, and regulation by cocaine. Brain Res. 2006;1067:1–12. doi: 10.1016/j.brainres.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Lom B, Cohen-Cory S. Brain-derived neurotrophic factor differentially regulates retinal ganglion cell dendritic and axonal arborization in vivo. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1999;19:9928–9938. doi: 10.1523/JNEUROSCI.19-22-09928.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsutani S, Yamamoto N. Brain-derived neurotrophic factor induces rapid morphological changes in dendritic spines of olfactory bulb granule cells in cultured slices through the modulation of glutamatergic signaling. Neuroscience. 2004;123:695–702. doi: 10.1016/j.neuroscience.2003.10.030. [DOI] [PubMed] [Google Scholar]
- Maynard KR, et al. Functional Role of BDNF Production from Unique Promoters in Aggression and Serotonin Signaling. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2016;41:1943–1955. doi: 10.1038/npp.2015.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard KR, Stein E. DSCAM contributes to dendrite arborization and spine formation in the developing cerebral cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32:16637–16650. doi: 10.1523/JNEUROSCI.2811-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister AK, Katz LC, Lo DC. Opposing roles for endogenous BDNF and NT-3 in regulating cortical dendritic growth. Neuron. 1997;18:767–778. doi: 10.1016/s0896-6273(00)80316-5. [DOI] [PubMed] [Google Scholar]
- McAllister AK, Katz LC, Lo DC. Neurotrophins and synaptic plasticity. Annu Rev Neurosci. 1999;22:295–318. doi: 10.1146/annurev.neuro.22.1.295. [DOI] [PubMed] [Google Scholar]
- McAllister AK, Lo DC, Katz LC. Neurotrophins regulate dendritic growth in developing visual cortex. Neuron. 1995;15:791–803. doi: 10.1016/0896-6273(95)90171-x. [DOI] [PubMed] [Google Scholar]
- Murphy DD, Cole NB, Segal M. Brain-derived neurotrophic factor mediates estradiol-induced dendritic spine formation in hippocampal neurons. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:11412–11417. doi: 10.1073/pnas.95.19.11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattabiraman PP, Tropea D, Chiaruttini C, Tongiorgi E, Cattaneo A, Domenici L. Neuronal activity regulates the developmental expression and subcellular localization of cortical BDNF mRNA isoforms in vivo. Mol Cell Neurosci. 2005;28:556–570. doi: 10.1016/j.mcn.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Pruunsild P, Kazantseva A, Aid T, Palm K, Timmusk T. Dissecting the human BDNF locus: bidirectional transcription, complex splicing, and multiple promoters. Genomics. 2007;90:397–406. doi: 10.1016/j.ygeno.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauskolb S, et al. Global deprivation of brain-derived neurotrophic factor in the CNS reveals an area-specific requirement for dendritic growth. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:1739–1749. doi: 10.1523/JNEUROSCI.5100-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathanoori M, Dias BG, Nair AR, Banerjee SB, Tole S, Vaidya VA. Differential regulation of multiple brain-derived neurotrophic factor transcripts in the postnatal and adult rat hippocampus during development, and in response to kainate administration. Brain Res Mol Brain Res. 2004;130:170–177. doi: 10.1016/j.molbrainres.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Shimada A, Mason CA, Morrison ME. TrkB signaling modulates spine density and morphology independent of dendrite structure in cultured neonatal Purkinje cells. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1998;18:8559–8570. doi: 10.1523/JNEUROSCI.18-21-08559.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmusk T, Palm K, Metsis M, Reintam T, Paalme V, Saarma M, Persson H. Multiple promoters direct tissue-specific expression of the rat BDNF gene. Neuron. 1993;10:475–489. doi: 10.1016/0896-6273(93)90335-o. [DOI] [PubMed] [Google Scholar]
- Tolwani RJ, Buckmaster PS, Varma S, Cosgaya JM, Wu Y, Suri C, Shooter EM. BDNF overexpression increases dendrite complexity in hippocampal dentate gyrus. Neuroscience. 2002;114:795–805. doi: 10.1016/s0306-4522(02)00301-9. [DOI] [PubMed] [Google Scholar]
- Tongiorgi E. Activity-dependent expression of brain-derived neurotrophic factor in dendrites: facts and open questions. Neurosci Res. 2008;61:335–346. doi: 10.1016/j.neures.2008.04.013. [DOI] [PubMed] [Google Scholar]
- Tongiorgi E, et al. Brain-derived neurotrophic factor mRNA and protein are targeted to discrete dendritic laminas by events that trigger epileptogenesis. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2004;24:6842–6852. doi: 10.1523/JNEUROSCI.5471-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tongiorgi E, Righi M, Cattaneo A. Activity-dependent dendritic targeting of BDNF and TrkB mRNAs in hippocampal neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1997;17:9492–9505. doi: 10.1523/JNEUROSCI.17-24-09492.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigers AJ, Amin DS, Talley-Farnham T, Gorski JA, Xu B, Jones KR. Sustained expression of brain-derived neurotrophic factor is required for maintenance of dendritic spines and normal behavior. Neuroscience. 2012;212:1–18. doi: 10.1016/j.neuroscience.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth MJ, Brun A, Grabert J, Patz S, Wahle P. Accelerated dendritic development of rat cortical pyramidal cells and interneurons after biolistic transfection with BDNF and NT4/5. Development. 2003;130:5827–5838. doi: 10.1242/dev.00826. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Representative confocal images of GFP positive neurons in the hippocampus of a Thy1M transgenic mouse. a. Isolated CA1 and CA3 neurons were selected from the CA1 region (blue arrows) and CA3 region (red arrows). b. Illustrative example of an isolated CA1 neuron selected for reconstruction (b). Scale bar 100 um (a) and 50 um (b).
Supplementary Table 1. ANOVA table for data presented in Figures 1h–l and 3f–j. Mean ± SEM is presented for each group as well as ANOVA F and p values. The final 10 columns present the results of Tukey posthoc tests.
Supplementary Table 2. ANOVA table for data presented in Figure 2e–h. Mean ± SEM is presented for each group at each distance. The final 4 columns present the results of Bonferroni post-hoc tests (adjusted p-values). P values for ANOVA are provided below.
Supplementary Table 3. ANOVA table for data presented in Figure 2i–l. Mean ± SEM is presented for each group at each distance. The final 4 columns present the results of Bonferroni post-hoc tests (adjusted p-values). P values for ANOVA are provided below.
Supplementary Table 4. ANOVA table for data presented in Figure 4e–h. Mean ± SEM is presented for each group at each distance. The final 4 columns present the results of Bonferroni post-hoc tests (adjusted p-values). P values for ANOVA are provided below.
Supplementary Table 5. ANOVA table for data presented in Figure 5d. Mean ± SEM is presented for each genotype and bin. The final column presents the results of Bonferroni post-hoc tests. P values for ANOVA are provided below.