Abstract
The contribution of RNA processing to tumorigenesis is understudied. Here, we report that the human RNA debranching enzyme (hDBR1), when inappropriately regulated, induces oncogenesis by causing RNA processing defects, for example, splicing defects. We found that wild-type p53 and hypoxia-inducible factor 1 co-regulate hDBR1 expression, and insufficient hDBR1 leads to a higher rate of exon skipping. Transcriptomic sequencing confirmed the effect of hDBR1 on RNA splicing, and metabolite profiling supported the observation that neoplasm is triggered by a decrease in hDBR1 expression both in vitro and in vivo. Most importantly, when modulating the expression of hDBR1, which was found to be generally low in malignant human tissues, higher expression of hDBR1 only affected exon-skipping activity in malignant cells. Together, our findings demonstrate previously unrecognized regulation and functions of hDBR1, with immediate clinical implications regarding the regulation of hDBR1 as an effective strategy for combating human cancer.
Introduction
It is becoming increasingly clear that defective RNA processing events have roles in the development of diseases. During processing, both coding and non-coding RNAs in a given transcriptome associate with RNA processing machineries in a dynamic manner to influence cellular events and ultimately initiate cell fate decisions.1, 2 Alternative splicing (AS), a major RNA processing event, is a regulated process in gene expression, through which a single gene can encode multiple proteins. It allows different exons in a gene to be included in or excluded from a final messenger RNA.3, 4 These processes in AS are catalyzed by spliceosomes, which are large RNA–protein complexes mainly composed of five small nuclear ribonucleoproteins (snRNPs), the U1, U2, U4, U5 and U6 snRNPs.4 The cycling of these snRNPs is essential during RNA splicing and involves dynamic interactions between snRNPs and associated factors.5 How specific RNA process influences transcriptomes and, thus, cell fate decisions, is attracting an increasing amount of interest.
During RNA splicing, introns are removed from pre-mRNAs. The removed introns form lariat structures with 2',5'-phosphodiester bonds between their 5′ ends and branching sites. The major component of the lariat intron turnover pathway is the RNA debranching enzyme (DBR1),6, 7, 8, 9, 10 which is highly conserved in species as diverse as yeast and humans. DBR1 cleaves the 2′–5′ phosphodiester linkage at the branch point and converts a lariat intron into a linear molecule that is then rapidly processed in vivo,6, 11 possibly relevant to multiple other RNA biogenesis including intron turnover,9, 12, 13 mirtron production14 and etc. Studies in Schizosaccharomyces pombe and Arabidopsis have indicated that DBR1 activity is a rate-limiting step in intron turnover and that deficiency of DBR1 can result in aberrant growth.9, 15, 16 These observations suggest that DBR1 deficiency may promote disease in humans because rapid intron turnover is more important in higher eukaryotes, considering that their genes contain abundant introns. However, the regulation and function of DBR1 in humans remain unclear.
p53 is a pivotal surveillance protein that is essential for suppressing the development of human cancers.17 Many transcriptional targets of p53 have been identified and shown to mediate the functions of p53 in responses to cellular stressors, such as genotoxic agents. However, the mechanisms underlying the functions of p53 under hypoxic conditions are not well studied. Here, we report that the regulation of human DBR1 (hDBR1) expression is wild-type p53 (wtp53) dependent under hypoxic conditions and that its functions are at least partially mediated through AS. Together, our findings show that RNA processing can determine cellular fates and that there is a direct connection between RNA processing events and human cancer.
Results
hDBR1 expression is regulated by wtp53 in a hypoxic stress-dependent manner
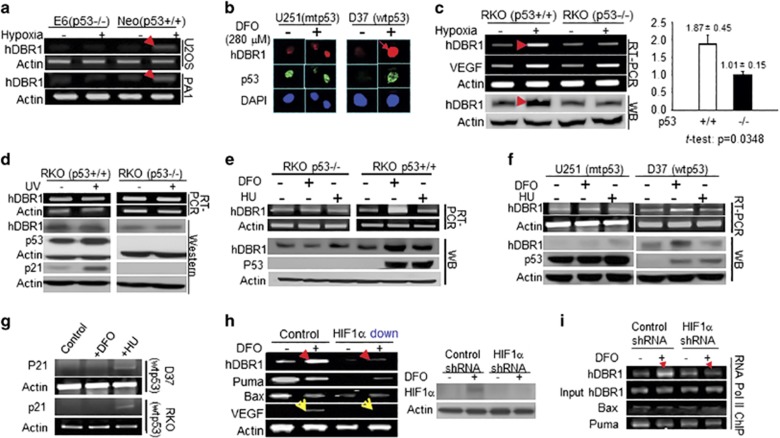
Our understanding of how p53 functions in cells under hypoxia, a major tumor-induced environmental stressor, is not as clear as for other stressors known to activate p53. In our previous observation by chance, we suspected that DBR1 may be a novel mediator of p53 under hypoxia. We confirmed that higher levels of hDBR1 expression were dependent on wtp53 in five different human cancer cell lines when treated with hypoxia or a hypoxia-mimicking drug, desferrioxamine (Figures 1a and c). In contrast, no substantial increase in hDBR1 gene expression (RNA or protein) was observed in any of the cell lines when treated with agents that induce DNA damage, such as ultraviolet light (Figure 1d) or hydroxyurea (Figures 1e and f). As expected for a DNA damage response, the expression of p21 was elevated in hydroxyurea- or ultraviolet-treated cells, but not in desferrioxamine-treated cells harboring wtp53 (Figures 1d and g). These data indicate that among the stressors tested, hDBR1 expression is substantially elevated in response to hypoxia in cells carrying wtp53.
Figure 1.
hDBR1 expression is regulated by p53 in a hypoxic stress-dependent manner. (a) A higher level of hDBR1 expression was identified through RT–PCR in U2OS-neo and PA1-neo cells treated with 0.1% O2 (neo cells harbor a functional p53) than in similarly treated E6 cells, in which p53 function is deficient. (b) In situ immunofluorescent staining showed that the protein expression of hDBR1 correlated with the level of the functional p53 protein in the wtp53 (D37) and mtp53 (U251)-carrying brain tumor cell lines. The intensity of red fluorescence (anti-hDBR1 antibody) was high in treated D37 cells, but not in treated U251 cells (green fluorescence represents the mt or wtp53 protein (anti-p53 antibody)). In U251 cells, mtp53 accumulation was observed as bright green fluorescence under either normal or hypoxic conditions, whereas wtp53 was only highly expressed in D37 cells under hypoxic conditions and not in the normal control. Images were taken at a magnification of × 600. Western blotting and RT–PCR showed similar results in RKO cells (p53+/+ or p53-/-) exposed to hypoxia (c), but not in cells exposed to ultraviolet (UV) (d). (e, f) Western blotting and RT–PCR showed a substantially higher level of hDBR1 expression, but not DNA damage, in wtp53-containing cells than in mtp53 or p53 null cells under hypoxia. (g) RT–PCR showed that p21 expression was increased in wtp53 cells following exposure to hydroxyurea (HU) but was not clearly increased in cells under hypoxic stress. (h) The mRNA expression level of hDBR1, but not Bax or Puma, was dependent on the level of HIF1α (HIF1α levels are shown in the right panel). VEGF (a target of HIF1) expression was used as the positive control (yellow arrowheads). (i) RNA pol II ChIP analysis showed that HIF1 enhanced the recruitment of RNA pol II to the promoter regions of the hDBR1 gene, but not to the corresponding regions of the Bax and Puma genes.
To determine the mechanisms by which p53 clearly upregulates hDBR1 expression under hypoxic conditions, but little in response to ultraviolet or hydroxyurea, we performed experiments to explore whether hDBR1 expression is affected by the level of hypoxia-inducible factor 1 (HIF1), which is a key mediator of hypoxic effects.18 We found that hDBR1, but not Puma or Bax (induced by wtp53 upon DNA damage),19 was affected by HIF1 expression levels (Figure 1h), suggesting that DBR1 may be a mediator of wtp53, relatively stress specific. We also performed RNA polymerase II-chromatin immunoprecipitation (ChIP) assays and found that much more pol II was recruited to the promoter region of DBR1 under hypoxia among the tested targets (Figure 1i), indicating that its induction is influenced by hypoxia (Figure 1h) and suggesting co-regulation of hDBR1 expression by wtp53 and HIF1.
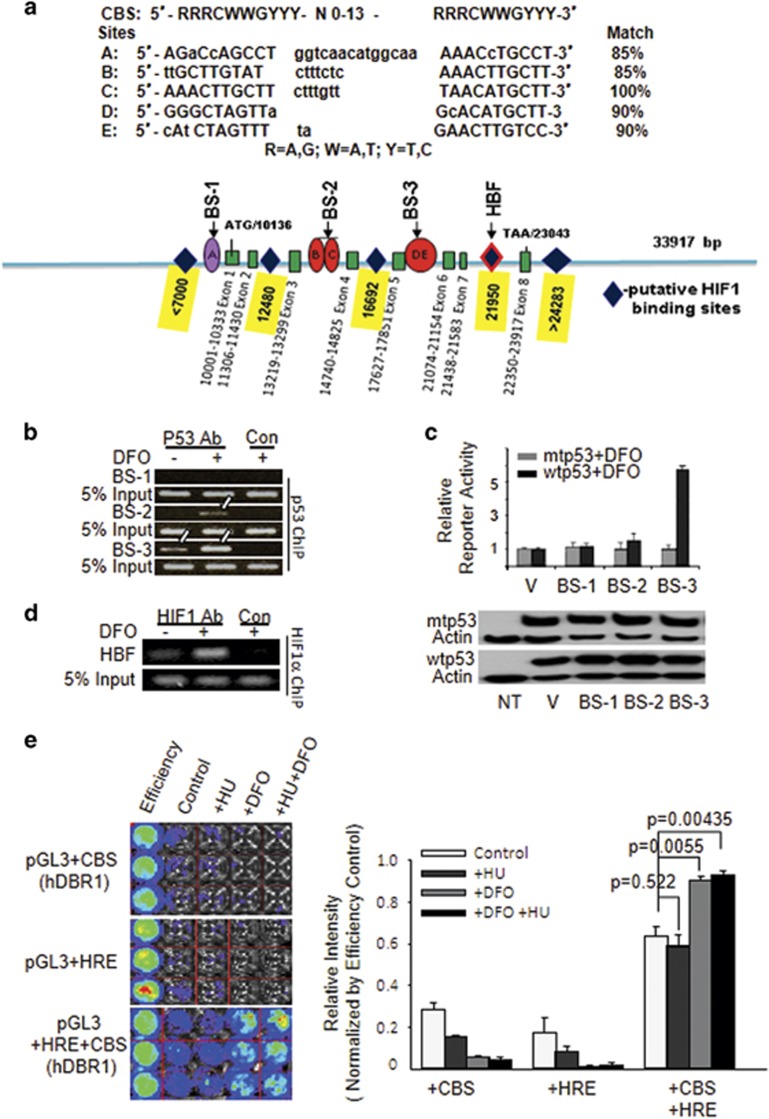
To explore the possibility of synergy between p53 and HIF1 at the cis-element level, we analyzed the hDBR1 genomic sequence and found that the hDBR1 gene contains five putative p53-binding sites with 85% or greater identity to the consensus p53 DNA-binding sequence (CBS) and several HIF1-binding motifs (G/ACGTG) dispersed within the hDBR1 locus (Figure 2a). To determine whether p53 can bind directly to these putative binding sites within the hDBR1 gene, we performed a p53 ChIP assay. Specific precipitation of DNA fragments corresponding to the BS-2 and BS-3 sites, but not the BS-1 site, was observed in p53 antibody pull-down pellets (Figure 2b). To confirm that the binding of DNA segments confers p53-dependent transcriptional activity, three DNA fragments (BS-1, -2 and -3) containing putative p53 CBS sites (Figure 2a) were individually cloned into the upstream region of the minimal SV40 promoter in the pGL3 luciferase reporter plasmid (because of physical distance, five putative p53 CBS sites were included within three DNA fragments). We found that, consistent with the p53 ChIP assay (Figure 2b), the BS-3 reporter was activated by exogenous wtp53, but not by exogenous mtp53 (Figure 2c). Via a HIF1α ChIP assay, the HIF1-binding motif near BS-3 (Figure 2a) was clearly pulled down using anti-HIF1α antibodies (Figure 2d). These results implied that HIF1 could work in concert with p53 directly at hDBR1 genomic loci to influence hDBR1 transcription. Next, both the p53-binding sequence 3 (BS-3) and HIF1 response elements (HREs) were cloned upstream of a luciferase reporter. As shown in Figure 2e, luciferase activity was substantially elevated only in the cells transfected with the reporter plasmid containing dual cis-elements (BS-3 and HRE). This finding shows that p53 and HIF1-binding regions work together in cis under hypoxic conditions to promote high transcriptional activity of the luciferase reporter, as high levels of hDBR1 expression were observed in wtp53-containing cells under conditions of hypoxia (Figure 1). Together, these results show a previously unknown feature of the regulation of hDBR1 expression under hypoxic conditions: wtp53 and HIF1 co-regulate hDBR1 expression.
Figure 2.
The DBR1 gene is a direct target of p53 and is also regulated by HIF1. (a) Five putative p53-binding sites (A, B, C, D and E) are illustrated with their corresponding sequences (nucleotides in uppercase indicate homology, whereas lowercase indicates a mismatch with the consensus). Diamonds represent putative HIF1-binding sites on the basis of the HIF1-binding motif. All symbols drawn are approximately proportional to the physical distance between them. (b) p53 ChIP assays were conducted using desferrioxamine (DFO)-treated cells, and fragments BS-3 and BS-2 were both immunoprecipitated by p53 antibodies. (c) The BS-3 reporter was activated by wtp53, but not mtp53. The level of p53 protein expression was equal between Calu-6 (p53-/-) cells transfected with wtp53 and mtp53 (lower panel). (d) In a HIF1 ChIP assay, the HIF1-binding fragment (HBF) was pulled down with anti-HIF1 antibodies, but not with control rabbit IgGs. (e) When the p53-binding sequence and HREs were placed upstream of the luciferase gene, the luciferase activity of pGL3 was substantially increased only when cells were exposed to hypoxic stress. No changes were observed in the cells under normal conditions (relative luciferase activity is shown to the right and is plotted according to photon counts).
Deficiency of hDBR1 changes AS
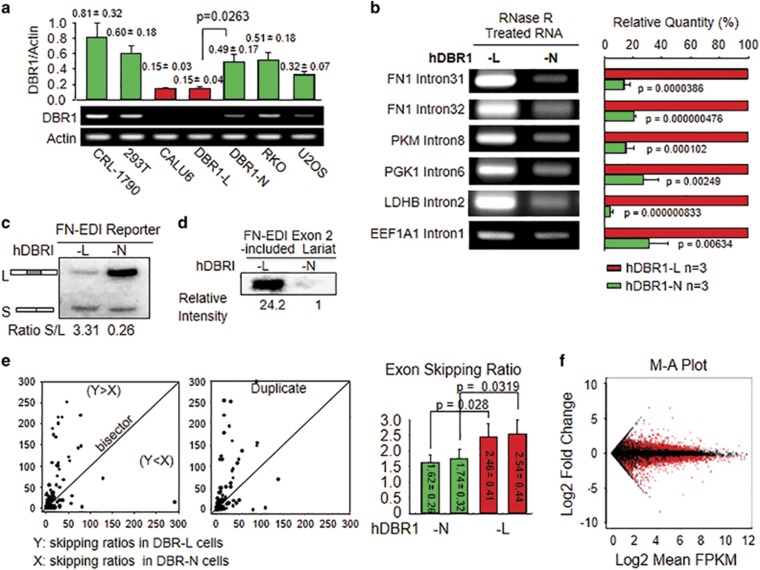
Based on the known function of DBR1 in processing lariat introns,6 as well as yeast studies,15, 16 a pathological change in DBR1 levels may lead to an abnormal level of lariat RNAs in a given transcriptome. We detected DBR1 expression in multiple human cell lines and found that DBR1 levels varied and that lung cancer Calu-6 (p53 null) cells expressed the lowest level of DBR1 among the tested cells (Figure 3a). We generated a pair of cells that were derived from Calu-6 and expressed either exogenous DBR1 at comparable normal levels (DBR1-N) or empty vector (DBR1-L) to more accurately assess the effects of DBR1 (Figure 3a and Supplementary Figure S1a). We found that the levels of specifically tested lariats were higher in DBR1-L cells than in DBR1-N cells (Figure 3b). This observation was further confirmed using a relatively direct measurement: a systematic approach using labeled reverse transcription (RT) with lariat-enriched RNA samples (Supplementary Figure S1c). These differences in lariat intron levels observed within human cells are consistent with the results of studies examining debranching in yeast in vitro and in vivo.19 Therefore, human cells expressing DBR1 at low levels contain elevated levels of lariat RNAs.
Figure 3.
Deficiency of hDBR1 expression changes AS activities. (a) Among all the tested cell lines, Calu-6 cells expressed the lowest levels of DBR1 (P<0.05. All results came from three separate experiments, and the data are presented as the mean±s.d.). (b) Lariat RNA levels were higher in DBR1-L cells than in DBR1-N cells. Lariat-enriched RNA samples (Supplementary Figure S1b) were used to perform reverse transcription (RT) and quantitative real-time polymerase chain reactions (qPCRs). Lariat intron-specific primers were used to examine the levels of the corresponding lariats. The relative levels of lariat introns corresponding to different genes are shown in the right panel. Three separate experiments were performed. (c) hDBR1 increased the inclusion of FN-EDI reporter exons in artificial minigene-derived FN mRNAs. (d) hDBR1 promoted a low level of lariat introns, including exon 2 (the controls for the input RNA and the transfection efficiency of the AS reporter are shown in Supplementary Figure S1g). (e) For each exon, the exon-skipping ratio was significantly higher in DBR1-L cells than in DBR1-N cells (shown when the value of the y coordinate is greater than the value of the x coordinate). The right bar graph shows the average exon-skipping ratio. In each group, n=2229; the results are presented as the mean±SE. The reproducibility is shown in Supplementary Figure S1h. (f) The plot shows the log fold change of the normalized FPKM (DBR1-L over DBR1-N) versus the average log expression.
End-product inhibition is a common phenomenon in biological systems. We hypothesized that an increased amount of lariat introns (reaction products) may interfere with the activities of enzymes responsible for RNA splicing. Therefore, compromised function of hDBR1 leading to an increase in lariat introns may have affected exon splicing. We then examined RNA splicing events in DBR1-N and DBR1-L cells into which an AS reporter system had been introduced.19, 20 We found that DBR1-N cells contained a dominant long form versus the short form of an artificial minigene transcript generated from the AS reporter21 (L: exons 1+2+3 and S: exons 1+3), whereas DBR1-L cells retained a relatively high level of the short form (Figure 3c). In addition, the expression levels of two functioning splicing variants (FAVL22, 23 and a caspase-9 variant)21 were higher in DBR1-L cells than in DBR1-N cells (Supplementary Figure S1e). We further specifically analyzed the AS reporter (three exons and two introns) (Supplementary Figure S1d) and found that the level of exon 2-containing lariat introns was significantly higher in DBR1-L cells than in DBR1-N cells (Figure 3d). These results indicate that DBR1 may influence exon skipping, in addition to directly processing lariat RNAs in mammalian cells, and agree with the observation of introns included in mRNAs in DBR1-deleted yeast.19
To validate the effect of hDBR1 on AS in depth, we sequenced transcriptomes prepared in duplicate from DBR1-N and DBR1-L cells. We used TopHat bioinformatics software (Johns Hopkins University, Baltimore, MD, USA) to count the exon junctions and compound ratios of abnormal exon fusions (exons that were not fused with the immediately following exon) with normal exon fusions (exons that were fused with the immediately following exon) for each exon (Supplementary Figure S1f). As shown in Figure 3e, each dot was plotted with X and Y coordinates, where X represented the skipping ratio of an exon in DBR1-N cells, and Y represented the skipping ratio of the same exon in DBR1-L cells. When Y>X, the dot was located in the area above the bisector, and when X>Y, the dot was located under the bisector (if X≈Y, the dots were located near the bisector, which indicated that the rate at which a given exon was skipped was similar between two types of cells). We found that the exon-skipping ratio was significantly higher in DBR1-L cells than in DBR1-N cells in terms of specific exon-exon junctions (overall Y>X with a P <0.05). Our transcriptome-wide analysis supported the elevated level of exon skipping indicated by the AS reporter assay in DBR1-L cells (Figures 3c and d). Through further analysis of RNA sequencing data using the Cuffdiff module of the Cufflinks software suite (http://cole-trapnell-lab.github.io/cufflinks/cuffdiff), we calculated the normalized FPKM for each transcript within each group and then tested for differential expression. The plot shows the log fold change of the normalized FPKM (comparing DBR1-L and DBR1-N cells) versus the average log expression. There were approximately 5000 differentially expressed transcripts between the DBR1-L and DBR1-N cell groups (shown in red, Figure 3f). Then, we input these results into the Ingenuity Pathway Analysis software (QIAGEN, Redwood City, CA, USA) for systemic analysis. The results showed that ‘cell growth and proliferation’, ‘cell death and survival’ and ‘cancer’ were the most significantly altered cell functions triggered by different levels of DBR1 expression in Calu-6 cells (Supplementary Figure S1i). These results show another previously unrecognized role of hDBR1 in regulating mammalian cellular RNA splicing, thereby contributing to maintaining appropriate RNA populations in the transcriptome (that is, ‘transcriptome integrity’) and, ultimately cell fate decisions. When hDBR1 is deregulated, the transcriptome fills with variant RNAs, and this alteration is at least partially attributed to the compromised function of hDBR1.
hDBR1 modulates the availability of active splicing complexes for introns that harbor a weak branch site
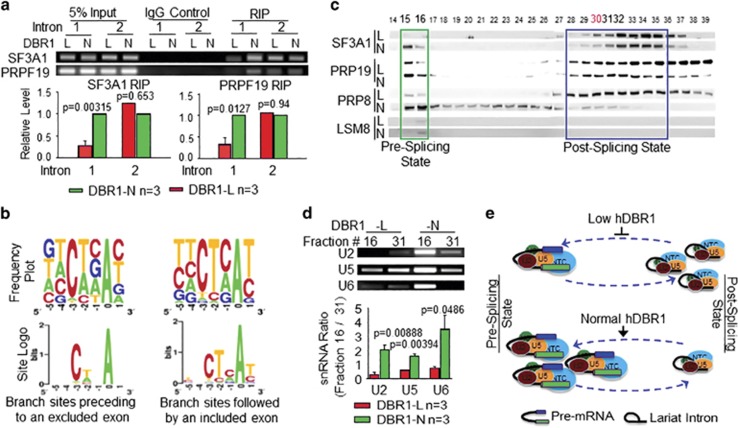
Whether an exon is included in or excluded from an mRNA molecule depends heavily on whether U2 snRNPs can properly bind to branch sites at the 3' end of an intron preceding a given exon.24 As a substantial proportion of exon 2 of the reporter minigene was excluded from the mRNAs in DBR1-L cells (Figures 3c and d), we suspected that in these cells, U2 snRNPs may bind to intron 1 in a manner that depends on the level of DBR1 expression. We performed RNA immunoprecipitation (RIP) assays for SF3A1 (a core component of the U2 snRNP) using DBR1-L and DBR1-N cells that were equally transfected with the AS reporter (Supplementary Figure S2a). As shown in Figure 4a (left), the amount of SF3A1 that interacted with the 3' end of intron 1 in pre-mRNAs was indeed lower in DBR1-L cells than in DBR1-N cells, but a lesser difference was observed in the interactions with intron 2. To determine whether active splicing complexes were affected by the level of DBR1 expression in addition to U2 snRNPs, we performed RIP for PRP19 (a core component of the NTC or NineTeen Complex, which joins U5 snRNPs to assemble the active splicing complex), and the results were the same (Figure 4a, right). These results showed that a smaller number of active splicing complexes were associated with the branch site of intron 1 in DBR1-L cells than in DBR1-N cells. We then compared the U2-binding sequences (branch sites) in both introns of the AS reporter and found that intron 2 carries a branch site that perfectly matches the U2-binding consensus sequence19, 25 (Supplementary Table S3), whereas intron 1 does not. We also analyzed randomly selected introns followed by an exon that was either included in or excluded from an mRNA molecule (using TopHat exon-skipping, Figure 3e), the latter of which is responsible for elevated levels of variants (for example, FAVL) in DBR1-L cells (Supplementary Table S3). We found that the introns preceding excluded exons carry branch sites that contain the following motif: N N C/A U/A N A N, sharing <50% homology with the authentic binding motif (Figure 4b, left column). In contrast, the introns followed by an included exon harbor the branch motif U/C N C/U U/C C/A A U/C, sharing higher homology with the consensus binding site.26, 27 U2 snRNPs bind to the latter site, regardless of the level of hDBR1 expression (Figure 4b, right column).
Figure 4.
hDBR1 modulates the availability of splicing complexes in introns harboring a weak branch site. (a) RIP assays were performed using antibodies targeting SF3A1 or PRP19 in DBR1-L and DBR1-N cells. The SF3A1 or PRP19 antibodies pulled down much greater amounts of pre-mRNA fragments composed of the 3' end of FN-EDI intron 1 and the beginning portion of exon 2 in DBR1-N cells than in DBR1-L cells. RT–qPCR was used to detect the relevant RNA fragments. Top panels: representative qPCR products; Bottom panels: qPCR results from three separate experiments. The DBR1 RNA levels in the input samples and the transfection efficiency of the AS reporter are shown in Supplementary Figure S2a. (b) The RNA sequences of the branch sites in the introns followed by excluded or included exons showed weak or strong homology, respectively, to the authentic branch site. (c) Gel filtration analyses of DBR1-L and DBR1-N cells for the dynamics of pre- or post-splicing complexes. (d) U2, U5 and U6 RNAs are accordingly altered in the two states of the splicing complexes. Top panels: representative qPCR products. Bottom panels: qPCR results from three separate experiments. (e) A schematic working model for a previously unknown function of DBR1: the level of DBR1 expression influences the dynamics of pre- or post-splicing complexes. When DBR1 expression levels are low, more snRNPs are retained in the post-splicing state, which affects the amount of snRNPs that are available to recycle back for the active splicing complex assembly.
Studies in yeast showed that snRNPs can be dynamically assembled into functional units in different states. A full-sized active splicing complex is approximately 4.8 MDa and interacts with introns in pre-mRNAs; whereas a post-catalytic splicing complex, without mRNAs but carrying lariat introns, is approximately 2 MDa in size.28, 29, 30, 31 Both of these complexes contain U2, U5 and U6 snRNPs. The amount of each form of the complexes is heavily dependent upon snRNP recycling,32, 33 and can be examined on the basis of their sizes through gel filtration analyses (Supplementary Figure S2b). In addition to the size difference, LSM8 and DHX35, which are specific to active splicing complexes, can be used to distinguish the two states of complexes.34, 35, 36 These proteins were abundant in earlier fractions (that is, the larger, or active complex) and were nearly undetectable in the later fractions (that is, the smaller complex related to the post-catalytic/splicing complex) (Figure 4c and Supplementary Figure S2c). Thus, the locations of these two proteins could be used as internal markers of the pre/post-splicing complexes for the subsequent filtration assays. We next explored the dynamics of the splicing complexes in cells containing different levels of DBR1 and found that the level of U2 snRNPs (indicated by the level of the complex core component, SF3A1) was lower in the pre-catalytic splicing state and higher in the post-catalytic splicing state in DBR1-L cells, whereas the opposite situation was observed in DBR1-N cells. The same differences were observed in the distribution of U5 snRNPs and NTC according to western blot analyses of their core components, PRP8 and PRP19, respectively (Figure 4c), although their expression levels were the same in DBR1-N and DBR1-L cells (Supplementary Figure S2d). These results were further validated via DBR1 RNA interference silencing (Supplementary Figure S2e). Therefore, a low level of DBR1 can lead to the retention of snRNPs in a post-catalytic splicing state and affect their availability to assemble into new, active splicing complexes. This conclusion was also fully supported by the relatively higher levels of the U2, U5 and U6 RNAs observed in pre-splicing complex-containing fraction #16 derived from DBR1-N cells compared with those in DBR1-L cells (Figure 4d). Collectively, these data indicate that a reduced level of hDBR1 increases the proportion of lariat introns and hinders snRNP recycling from the post-splicing state to the pre-splicing state (Figure 4e). The compromised availability of active RNA splicing complexes subsequently results in loose associations between splicing complexes and introns containing weak U2-binding sites, which ultimately results in the immediate exon being skipped and a decrease in splicing introns containing weak branch sites.
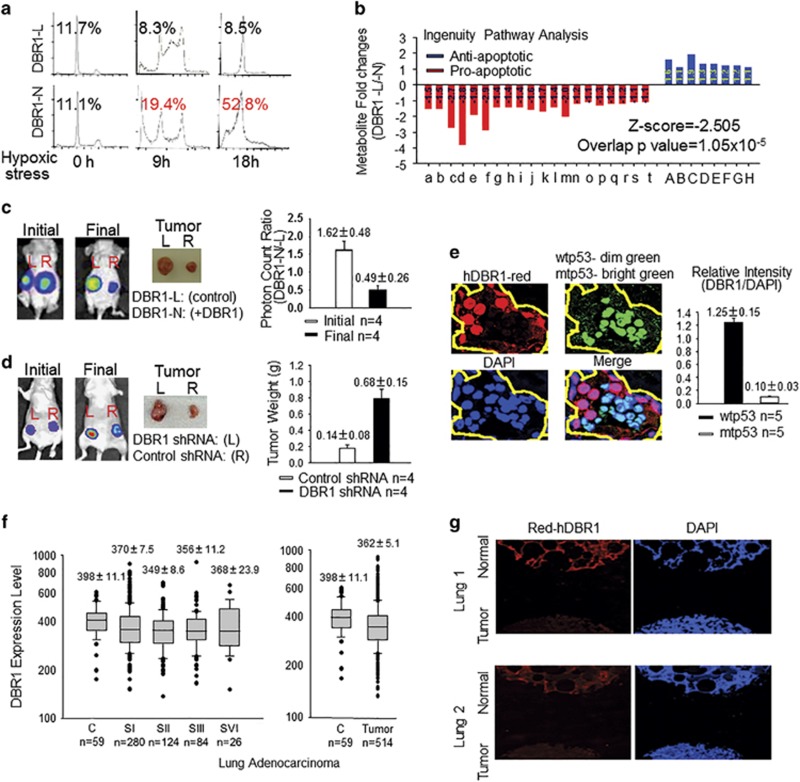
Reduced hDBR1 expression contributes to tumorigenicity
In DBR1-L cells, the elevated expression of FAVL, an oncogenic factor, and a caspase-9 variant that acts as an anti-apoptotic factor suggested that an appropriate level of hDBR1 may be essential for maintaining a normal growth potential in cells. As shown in Figure 5a, DBR1-L cells contained a substantially lower proportion of the sub-G1 population than the DBR1-N cells. This finding is strongly in agreement with the results of metabolite profiling (Figure 5b). The profile of metabolites in DBR1-L cells indicated that these cells possess less apoptotic activity than DBR1-N cells, according to ingenuity pathway analysis. This reduction of the overall apoptotic potential will naturally affect the rate of cell growth over time. We examined the corresponding cell growth rates by both overexpressing (Supplementary Figure S3b) and silencing hDBR1 (Supplementary Figures S3c, d and e) in vitro to demonstrate the suppressive effects of hDBR1, which were further shown in in vivo experiments (Figures 5c and d and Supplementary Figure S3f). To further verify the role of hDBR1 in p53-mediated tumor-suppressing activities during the development of human cancer, we analyzed their relationship in vivo. Using confocal immunofluorescence, we found that the expression level of hDBR1 was higher in adjacent tissue cells carrying wtp53 but was lower in p53-mutated lung or ovarian cancer cells (Figure 5e and Supplementary Figure S3g). Furthermore, we examined the general level of hDBR1 expression in human tissues and found that the low level of DBR1 expression was associated with neoplasm via analyzing publicly available data sets and performing immunofluorescence analyses (Figures 5f and g, and Supplementary Figures S3h and i). These analyses of hDBR1 expression in human cells or tissues demonstrated that hDBR1 has functionally significant roles in tumor suppression during the development of human tumors.
Figure 5.
hDBR1 contributes to p53 tumor-suppressor activity. (a) The sub-G1 percentage was clearly lower in DBR1-L cells than in DBR1-N cells. (b) The same set of cells (Supplementary Figure S3a) was subsequently prepared for metabolomics analysis. Nonparametric statistical analyses were used to screen the metabolites that were significantly different between the two groups (P<0.05). An ingenuity pathway analysis (IPA) showed that a lower level of apoptotic activity occurred in DBR1-L cells (Z-score=−2.505, overlapping P-value=1.05 × 10−5) (for the list of metabolites, see Supplementary Table S4). (c, d) DBR1-L and DBR1-N cells, or A549 control and DBR1-silenced cells (Supplementary Figure S3d) were used for xenograft analyses performed in groups of four nude mice. The xenografts generated from DBR1-N cells or A549 control cells grew much slower than the corresponding xenografts derived from DBR1-L cells or A549 DBR1-silenced cells, respectively. (P=0.00616 and 0.00481, respectively). (e) A higher level of hDBR1 expression was associated with wtp53 status in situ in human ovarian cancer. The fluorescence intensity was measured with ImageJ (NIH, Bethesda, MD, USA) (P=0.00000018). (The images were taken in the same microscopic field at a magnification of × 600). (f) Through the analysis of gene expression data sets for DBR1 expression in lung adenocarcinoma from the TCGA database (http://gdac.broadinstitute.org/), we found that tumor tissues generally have a lower level of DBR1 expression than para-carcinoma non-malignant tissues. However, there is no significant difference between different stages of malignant tumors. The results are presented as the mean±s.e., P=0.022. (g) The level of hDBR1 protein expression was detected using immunofluorescence in a lung tissue array that included a total of 24 cases. In 20 of these lung cancer cases, the level of hDBR1 expression was low compared with that in the four benign samples. (Images were acquired in the same microscopic field at a magnification of × 40).
Discussion
Regulation of hDBR1 expression by wtp53
We have shown that DBR1 expression is governed by both wtp53 and HIF1 through a synergetic mechanism (Figure 1). An increase of either wtp53 or HIF1 could affect DBR1 levels independently, as shown in Figures 1b and e, but DBR1 is increased to a maximum level when the levels of both wtp53 and HIF1 are increased. We have also demonstrated that both wtp53 and HIF1 can directly regulate hDBR1 expression under hypoxic conditions (Figure 2). This dual regulation of DBR1 expression influences the accessibility of RNA polymerase II to and its interaction with the promoter region of the hDBR1 gene (Figure 1i), which affects hDBR1 transcription. Importantly, experiments using a pGL3 reporter engineered to contain both the p53-binding site and the HIF1-binding motif supported essential roles for both wtp53 and HIF1 during hDBR1 transcription (Figure 2e). hDBR1 can therefore facilitate p53-mediated tumor-suppressor activity during the development of solid tumors, in which hypoxia is generally present. As hDBR1 is a housekeeping gene, the mechanism by which hDBR1 is regulated under normal conditions and what additional regulatory factors are required for its maintenance remain to be explored. Addressing this latter issue may help us understand why DBR1 is generally expressed at low levels in human cancers (Figures 5e and g), given that p53 signaling is not always defective in human tumors.
The roles of hDBR1 in modulating AS
hDBR1 linearizes lariat introns to promote their degradation by disrupting their 2′–5′ phosphodiester bonds.6, 11 How this process affects lariat introns and their neighboring exons has not been extensively studied in human cells. As illustrated in Figure 4e, a compromised level of hDBR1 expression leads to an elevated number of lariat introns and the retention of snRNPs in post-splicing complexes. An increased level of trapped snRNPs leads to a deficiency in the amount of active splicing complexes interacting with introns that carry a weak branch site. The active splicing complexes will therefore easily skip over an exon that immediately follows such an intron (Figures 3c and d). This conclusion was further supported by analyzing intron sequences in the AS reporter minigene and corresponding RIP assays, in addition to transcriptome sequencing analyses of DBR1-N and DBR1-L cells (Figures 3c and f and Figures 4a and b). Interestingly, an intron immediately followed by an excluded exon usually carries a weak U2-binding/branch site, whereas an intron located immediately before an included exon is not always required to contain a strong U2-binding/branch site (Figure 4b and Supplementary Table S3). Therefore, it is the strong interaction between the active splicing complex and a given intron, not a definite U2-binding site, which defines whether the following exon is included in an mRNA molecule. This finding is reasonable because enhancers, repressors and many other factors all may contribute to the interactions between introns and active splicing complexes.37 Nevertheless, U2-binding sequences that were randomly selected from the results of TopHat analysis (Figure 3e) indicated that an intron before an excluded exon would tend to exhibit lower homology (<50%) to the authentic binding site than an intron followed by an included exon (Figure 4b). This is because analysis of those cis-elements nearby 3’ ends of introns, possibly affecting the interaction of U2 with its binding site, for example, polypyrimidine tract38 in this case, seemed to not show significant differences (Supplementary Figure S1j). Our study demonstrates that hDBR1 modulates the availability of pre-splicing complexes by affecting snRNP recycling (Figures 4c and d), in combination with the cis effects in the preceding intron defining an excluded or included exon. During human tumor development, an insufficient amount of hDBR1 expression (Figures 5f and g, and Supplementary Figures S3h and i) will eventually promote rising levels of a variety of variants (Figures 3c and f) and thereby lead to tumorigenesis (Figure 5).
Functions of hDBR1 during the maintenance of transcriptome-wide RNA processing
Recent increases in our understanding of the important roles played by RNA in addition to simple coding have extended far beyond what we could have previously imagined.39, 40, 41 One of the NCI’s provocative questions relates to how changes in RNA processing contribute to human cancer. One answer to this question is provided by the results of this study. Although most of the known targets of p53 remain unaffected under conditions of hypoxia, we revealed that hDBR1 is a novel target of p53 more in a hypoxic stress-dependent manner (Figures 1 and 2). As abnormal growth is observed in yeast and Arabidopsis when DBR1 expression is compromised,9, 16 we hypothesized that hDBR1 also affects human cell behaviors and is associated with diseases when its expression is insufficient. Indeed, we found that reduced levels of hDBR1 promoted tumorigenesis in human cells (Figure 5). This cellular effect may result from an increase in exon skipping (Figures 3c and e). We believe that this study may explain a common phenomenon—increases in transcript variants—that has long been observed in human tumors.42 Nearly 85% of human cancers exhibit a defective p53 signaling pathway, which can in turn lead to insufficient levels of hDBR1 expression (Figures 1 and 5) and abnormal RNA processing activity (Figure 3). Together, our findings suggest a logical rationale for the existence of increased transcript variants in tumors. On the other hand, increases in variants, which are common in human tumors, may be an important and previously unrecognized factor that contributes to the tumorigenicity of a defective p53 pathway.
Immediate translational impact of this study
As shown in Figures 5a and b, a compromised level of hDBR1 expression can modulate the cellular apoptotic potential, which is generally low in human malignant tissues (Figures 5f and g and Supplementary Figures S3h and i). These results may provide insights to support the development of a novel approach by which hDBR1 levels could be boosted to ‘correct’ an abnormal transcriptome. In this case, tumor resistance may be avoidable because a balanced transcriptome delivers a net biological effect. Interestingly, we also found that when the endogenous level of DBR1 expression was not low (Supplementary Figure S4a), ectopic expression of hDBR1 (Supplementary Figure S4b) did not influence AS and the resulting variant populations in the examined host cells (Supplementary Figures S4c, d and e). This unexpected result provides a sound rationale for an effective strategy boosting the level of DBR1 expression because it would not affect ‘normal cells’. Furthermore, our metabolome analysis provides promising insights into the development of better biomarkers that can be used in such putative transcriptome-wide approaches, as well as many other approaches to determine the effectiveness of a given treatment. For example, stearic acid and other metabolites that we identified may serve as new markers to indicate cellular apoptotic activity (Figure 5b).
Materials and methods
The establishment of hDBR1-expressing stable cell lines and luciferase-expressing retrovirus and reporter plasmids, as well as transfection, gel filtration, reporter imaging assays and xenograft formation assays were all performed as previously described.22, 23, 43, 44, 45
Cell lines
All cell lines, with an exception of those specially engineered in house, were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). The PA1-Neo, PA1–E6, U2OS-Neo and U2OS-E6 cell lines have been previously described.44 The RKO p53–/– and RKO p53+/+ cells were generously provided by the laboratory of Dr Bert Vogelstein (Johns Hopkins University).
Antibodies
β-Actin peroxidase antibody (A3854) were purchased from Sigma (St Louis, MO, USA); p53 (sc-126), HIF1 alpha (sc-10790), Pol II (sc-899), LSm8 (sc-404816) antibodies, and goat anti-mouse IgG-HRP (sc-2005) were from Santa Cruz (Dallas, TX, USA); hDBR1 (H00051163-B01P), SF3A1 (NB100-79843), PRPF8 (NBP2-22274), PRP19 (H00027339-A01) and DHX35 (NBP1-57348) antibodies were from Novus Biologicals (Littleton, CO, USA). Rabbit IgG-HRP secondary antibody (A120-101 P) was from Bethyl Laboratories (Montgomery, TX, USA). Alexa Fluor 568 rabbit IgG (H+L) secondary antibody (A-11011) and Alexa Fluor 488 mouse IgG (H+L) secondary antibody (A-11001) were from Thermo Fisher Scientific (Waltham, MA, USA).
ChIP assay
A ChIP assay was performed as previously described.44 The following primer sequences were used:
BS-1F: 5′-TGGGTGCAGTAGCTCACAC-3′, BS-1B: 5′-CCAGGCTGAAGTGCAATGG-3′, BS-2F: 5′-CTCAGTACCTGTTAACCAGC-3′, BS-2F: 5′-TGCCAGGCAGTCACTGTG-3′, BS-2B: 5′-CGATGTGAATCCAAGCTATTCC-3′, BS-3F: 5′-GGAGTTCAGGTTTTAACTAGAGATGC-3′, BS-3B: 5′-GAAACACAAAGCAAGTTAGTGCTC-3′, HBF-F: 5′-GAAACAGGGTCTAGCTCTGTTA-3′, and HBF-B: 5′-CAAAGGTGGAGGCCAGATAC-3′.
Reverse transcription–PCR
The following PCR primer sequences were used for RT–PCR: hDBR1-F, 5′-GGAAACCATGAAGCCTCAAA-3′ and hDBR1-B, 5′-GGCAGCTGGACTTCCTAATG-3′ actin-F, 5′-CCGTGTGAACCATGTGACTT-3′ and actin-B, 5′-CTAAGTTGCCAGCCCTGCTA-3′ caspase-9-F, 5′-GCTCTTCCTTTGTTCATCTCC-3′ and caspase-9-B, 5′-CATCTGGCTCGGGGTTACTGC-3′ and EDI-F, 5′-GACCCGGTCAACTTCAAGCTCC-3′ and EDI-B: 5′-GCGGCCAGGGGTCACGAT-3′.46, 47
Xenograft formation
Nude mice (4–6 weeks old) were obtained from Charles River Laboratory (San Diego, CA, USA) and injected with 300 μl of Matrigel (Invitrogen Waltham, MA, USA) slurry (prepared at a 1:1 ratio with 1X phosphate-buffered saline) containing 10 million cells that constitutively expressed luciferase. Luciferase activity was measured by injecting d-luciferin (50 mg/kg mouse) into anesthetized mice abdominally. We then acquired live images using a Xenogen IVIS (PerkinElmer, New York, NY, USA) as previously described.44
AS/exon-skipping assay
Calu-6 control and high-hDBR1 cells were transfected with 500 ng of the FN-EDI reporter minigene.21 Total RNA was subsequently extracted from the cells. Minigene gene expression was analyzed using radioactive RT–PCR. The PCR products were run in a 5% non-denaturing polyacrylamide gel and then examined using autoradiography.
Housekeeping gene lariat intron RT–qPCR
Total RNA was extracted from cells overexpressing hDBR1 or EV control cells using a standard TRIzol method. Total RNA (20 μg) was isolated from each sample and then treated with RNase R (Epicentre, Madison, WI, USA). RNA extraction and RT–qPCR were performed as described elsewhere.23 β-Actin and untreated total RNA were used as the internal control and the transfection efficiency control, respectively. All primers are listed in Supplementary Table S1.
hDBR1 siRNA oligo transfection
siRNA oligo transfections were performed as previously described.44 Five siRNA oligonucleotides targeting human hDBR1 were designed and purchased from Dharmacon (Lafayette, CO, USA): 5′-GTATCGTCACATGCAAACCTT-3′, 5′-GCAGCCTATAGATATATTCTT-3′, 5′-CGAGGCGGATCTACGCTGCTT-3′, 5′-GGATCGGTGGAATCTCTGGTT-3′, 5′-GGCTCCAGTTCTCACGCTCTT-3′.
Gene knockdown using lentivirus shRNA
A set of five pLKO.1 plasmids containing shRNAs constructed to target HIF1α (purchased from Thermo Scientific, Waltham, MA, USA; Open BioSystems, Lafayette, CO, USA) and the empty pLKO.1 vector were used to generate the corresponding lentiviruses. U2OS cells were then infected with these viruses according to the manufacturer’s protocol. Infected cells were pool-selected using puromycin at 48 h post-infection, and the efficiency of HIF1α knockdown was verified using HIF1α antibodies (Santa Cruz).
RIP assay
RIP was performed according to the procedure in our previous publication.48 The primer sequences that were used in these experiments are listed in Table S2.
RT–PCR of U2, U5 and U6 snRNAs
PCR primers are as follows:
U2 snRNA: forward 5′-CGTCCTCTATCCGAGGACAATA-3′,
reverse 5′-GTACTGCAATACCAGGTCGATG-3′;
U5 snRNA: forward 5′-CTGGTTTCTCTTCAGATCGCATAA-3′,
reverse 5′-AGACTCAGAGTTGTTCCTCTCC-3′
U6 snRNA: forward 5′-GCTTCGGCAGCACATATACTA-3′,
reverse 5′-CGAATTTGCGTGTCATCCTTG-3′.
Analysis of AS
The RNA sequencing results were read using TopHat (http://ccb.jhu.edu/software/tophat/index.shtml) to identify splice junctions between exons and to determine the skipping ratio of each exon (Supplementary Figure S1f). A statistically significant change in the skipping ratio was used for all samples to subtract background noise (paired t-test, P<0.05). Two skipping ratios were determined for the same exon form DBR1-L and DBR1-N cells, and these two ratios are presented as a set of coordinates (x, y; with x representing the ratio derived from DBR1-N cells and y representing the same exon-skipping ratio from DBR1-L cells).
Metabolomics analysis
The metabolomics test and analysis were performed as previously described.49 The cell line samples were prepared and analyzed according to methods described elsewhere.50, 51, 52
Sample size estimation and statistics
Laboratory quantification experiments, including mouse xenograft studies, underwent a sample size estimation for the t-test by SigmaPlot (Systat Software, San Jose, CA, USA) when designing experiments. In addition, all data were examined by the two-tailed t-test for statistical significance.
Acknowledgments
This study was supported in part by NIH grants (R01CA136532 and R01CA188251) to PF and Chinese National Science Foundation (CNSF#81372171) to PF. We thank Dr Alberto R Kornblihtt (Universidad de Buenos Aires, Argentina) for providing the FN-EDI reporter construct.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Bischof O, Martinez-Zamudio RI. MicroRNAs and lncRNAs in senescence: a re-view. IUBMB Life 2015; 67: 255–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye M, Blanco S. Post-transcriptional modifications in development and stem cells. Development 2016; 143: 3871–3881. [DOI] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature 2010; 463: 457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Wang Z. A day in the life of the spliceosome. Nat Rev Mol Cell Biol 2014; 15: 108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstens RP. Networking in an alternative splicing world. Mol Cell 2014; 54: 903–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruskin B, Green MR. An RNA processing activity that debranches RNA lariats. Science 1985; 229: 135–140. [DOI] [PubMed] [Google Scholar]
- Clark NE, Katolik A, Roberts KM, Taylor AB, Holloway SP, Schuermann JP et al. Metal dependence and branched RNA cocrystal structures of the RNA lariat debranching enzyme Dbr1. Proc Natl Acad Sci USA 2016; 113: 14727–14732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N, Dobashi I, Hagiwara M, Ohno M. hDbr1 is a nucleocytoplasmic shuttling protein with a protein phosphatase-like motif essential for debranching activity. Sci Rep 2013; 3: 1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wang S, Cheng J, Su C, Zhong S, Liu Q et al. Intron Lariat RNA Inhibits MicroRNA Biogenesis by Sequestering the Dicing Complex in Arabidopsis. PLoS Genet 2016; 12: e1006422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Vuong BQ, Vaidyanathan B, Lin JY, Huang FT, Chaudhuri J. Non-coding RNA generated following lariat debranching mediates targeting of AID to DNA. Cell 2015; 161: 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PA, Eisenberg D. The evolution of catalytic function. Science 1987; 238: 729–730 807. [DOI] [PubMed] [Google Scholar]
- Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife 2015; 4: e07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hube F, Ulveling D, Sureau A, Forveille S, Francastel C. Short intron-derived ncRNAs. Nucleic Acids Res 2017; 45: 4768–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kock KH, Kong KW, Hoon S, Seow Y. Functional VEGFA knockdown with artificial 3'-tailed mirtrons defined by 5' splice site and branch point. Nucleic Acids Res 2015; 43: 6568–6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam K, Lee G, Trambley J, Devine SE, Boeke JD. Severe growth defect in a Schizosaccharomyces pombe mutant defective in intron lariat degradation. Mol Cell Biol 1997; 17: 809–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman KB, Boeke JD. Isolation and characterization of the gene encoding yeast debranching enzyme. Cell 1991; 65: 483–492. [DOI] [PubMed] [Google Scholar]
- Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene 2005; 24: 2899–2908. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene 2010; 29: 625–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitton DA, Rallis C, Jeffares DC, Smith GC, Chen YY, Codlin S et al. LaSSO, a strategy for genome-wide mapping of intronic lariats and branch points using RNA-seq. Genome Res 2014; 24: 1169–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt AR, Pesce CG, Alonso CR, Cramer P, Srebrow A, Werbajh S et al. The fibronectin gene as a model for splicing and transcription studies. FASEB J 1996; 10: 248–257. [DOI] [PubMed] [Google Scholar]
- Munoz MJ, Perez Santangelo MS, Paronetto MP, de la Mata M, Pelisch F, Boireau S et al. DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell 2009; 137: 708–720. [DOI] [PubMed] [Google Scholar]
- Park HK, Wang H, Zhang J, Datta S, Fei P. Convergence of Rad6/Rad18 and Fanconi anemia tumor suppressor pathways upon DNA damage. PLoS ONE 2010; 5: e13313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panneerselvam J, Park HK, Zhang J, Dudimah FD, Zhang P, Wang H et al. FAVL impairment of the Fanconi anemia pathway promotes the development of human bladder cancer. Cell Cycle 2012; 11: 2947–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Rio DC. Mechanisms and regulation of alternative pre-mRNA splicing. Annu Rev Biochem 2015; 84: 291–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kol G, Lev-Maor G, Ast G. Human-mouse comparative analysis reveals that branch-site plasticity contributes to splicing regulation. Hum Mol Genet 2005; 14: 1559–1568. [DOI] [PubMed] [Google Scholar]
- Pastuszak AW, Joachimiak MP, Blanchette M, Rio DC, Brenner SE, Frankel AD. An SF1 affinity model to identify branch point sequences in human introns. Nucleic Acids Res 2011; 39: 2344–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao K, Masuda A, Matsuura T, Ohno K. Human branch point consensus sequence is yUnAy. Nucleic Acids Res 2008; 36: 2257–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto R, Kataoka N, Okawa K, Ohno M. Isolation and characterization of post-splicing lariat-intron complexes. Nucleic Acids Res 2009; 37: 891–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N, Fujita M, Ohno M. Functional association of the microprocessor complex with the spliceosome. Mol Cell Biol 2009; 29: 3243–3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azubel M, Wolf SG, Sperling J, Sperling R. Three-dimensional structure of the native spliceosome by cryo-electron microscopy. Mol Cell 2004; 15: 833–839. [DOI] [PubMed] [Google Scholar]
- Ohi MD, Ren L, Wall JS, Gould KL, Walz T. Structural characterization of the fission yeast U5.U2/U6 spliceosome complex. Proc Natl Acad Sci USA 2007; 104: 3195–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourmann JB, Schmitzova J, Christian H, Urlaub H, Ficner R, Boon KL et al. Dissection of the factor requirements for spliceosome disassembly and the elucidation of its dissociation products using a purified splicing system. Genes Dev 2013; 27: 413–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Luhrmann R. SnapShot: spliceosome dynamics I. Cell 2015; 161: 1474–e1471. [DOI] [PubMed] [Google Scholar]
- Chan SP, Kao DI, Tsai WY, Cheng SC. The Prp19p-associated complex in spliceosome activation. Science 2003; 302: 279–282. [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell 2009; 136: 701–718. [DOI] [PubMed] [Google Scholar]
- Ilagan JO, Chalkley RJ, Burlingame AL, Jurica MS. Rearrangements within human spliceosomes captured after exon ligation. RNA 2013; 19: 400–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Wang Q, Kennedy CJ, Silver PA. An alternative splicing network links cell-cycle control to apoptosis. Cell 2010; 142: 625–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerte C, Hall KB. The domains of polypyrimidine tract binding protein have distinct RNA structural preferences. Biochemistry 2009; 48: 2063–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell 2016; 29: 452–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullenger BA, Nair S. From the RNA world to the clinic. Science 2016; 352: 1417–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoronos AA, Engelman DM, Slack FJ. OncomiR or tumor suppressor? The duplicity of microRNAs in cancer. Cancer Res 2016; 76: 3666–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward AJ, Cooper TA. The pathobiology of splicing. J Pathol 2010; 220: 152–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D, Dudimah FD, Zhang J, Pickering A, Paneerselvam J, Palrasu M et al. Recruitment of DNA polymerase eta by FANCD2 in the early response to DNA damage. Cell Cycle 2013; 12: 803–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei P, Wang W, Kim SH, Wang S, Burns TF, Sax JK et al. Bnip3L is induced by p53 under hypoxia, and its knockdown promotes tumor growth. Cancer Cell 2004; 6: 597–609. [DOI] [PubMed] [Google Scholar]
- Park HK, Panneerselvam J, Dudimah FD, Dong G, Sebastian S, Zhang J et al. Wip1 contributes to cell homeostasis maintained by the steady-state level of Wtp53. Cell Cycle 2011; 10: 2574–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panneerselvam J, Pickering A, Han B, Li L, Zheng J, Zhang J et al. Basal level of FANCD2 monoubiquitination is required for the maintenance of a sufficient number of licensed-replication origins to fire at a normal rate. Oncotarget 2014; 5: 1326–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panneerselvam J, Pickering A, Zhang J, Wang H, Tian H, Zheng J et al. A hidden role of the inactivated FANCD2: upregulating DeltaNp63. Oncotarget 2013; 4: 1416–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B, Shen Y, Zhang P, Jayabal P, Che R, Zhang J et al. Overlooked FANCD2 variant encodes a promising, portent tumor suppressor, and alternative polyadenylation contributes to its expression. Oncotarget 2017; 8: 22490–22500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panneerselvam J, Xie G, Che R, Su M, Zhang J, Jia W et al. Distinct metabolic signature of human bladder cancer cells carrying an impaired Fanconi anemia tumor-suppressor signaling pathway. J Proteome Res 2016; 15: 1333–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Cai G, Zhou B, Li D, Zhao A, Xie G et al. A distinct metabolic signature of human colorectal cancer with prognostic potential. Clin Cancer Res 2014; 20: 2136–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Zhou B, Su M, Baxter S, Zheng X, Zhao X et al. Mass spectrometry-based quantitative metabolomics revealed a distinct lipid profile in breast cancer patients. Int J Mol Sci 2013; 14: 8047–8061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A, Green J, Pollard J Jr, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 2014; 30: 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.