Abstract
Defensins as a prominent family of antimicrobial peptides (AMP) are major effectors of the innate immunity with a broad range of immune modulatory and antimicrobial activities. Particularly, defensins are the only recognized fast-response molecules that can neutralize a broad range of bacterial toxins, many of which are among the deadliest compounds on the planet. For a decade, the mystery of how a small and structurally conserved group of peptides can neutralize a heterogeneous group of toxins with little to no sequential and structural similarity remained unresolved. Recently, it was found that defensins recognize and target structural plasticity/thermodynamic instability, fundamental physicochemical properties that unite many bacterial toxins and distinguish them from the majority of host proteins. Binding of human defensins promotes local unfolding of the affected toxins, destabilizes their secondary and tertiary structures, increases susceptibility to proteolysis, and leads to their precipitation. While the details of toxin destabilization by defensins remain obscure, here we briefly review properties and activities of bacterial toxins known to be affected by or to be resilient to defensins and discuss how recognized features of defensins correlate with the observed inactivation.
Keywords: defensins, bacterial toxins, thermodynamic instability, unfolding
INTRODUCTION
Bacterial protein toxins are recognized as the deadliest compounds on our planet. Given the outstanding killing efficiency of toxins (lethal doses for some of which are as low as nanograms of pure protein), their effective neutralization by host organisms is essential for survival. The adaptive immune system of jawed vertebrates (Gnathostomata) does so by producing neutralizing antibodies, which are highly specific and therefore exceptionally potent effector molecules. However, their production takes several days – a too-long term to protect from immediate threats. In contrast, the innate immunity acts promptly by recognizing conserved and distinct molecular patterns of pathogens. But what strategy can be employed by the innate immunity to recognize bacterial toxins, most of which do not share common structures and are highly diverse and mutable? One possibility would be to produce effector molecules for each toxin class and target them based on their specific properties. However, in such case, an emergence of a new class of microbial toxins would leave eukaryotic hosts (whose evolution is much slower than that of microbes) unprotected and, therefore, destined to extinction. Is there a more universal defense mechanism that could target many various toxins?
Antimicrobial peptides (AMP) are major effectors of the innate immunity with a broad range of immune modulatory and direct antimicrobial activities (Wang, 2014; Zasloff, 2002). The AMP database (http://aps.unmc.edu/AP/) as of April 2017 contains 2846 records on AMPs from all kingdoms of life, among which 118 are human host defense peptides (Wang, 2015). Just over a decade ago, a group of AMPs called defensins was reported to efficiently neutralize anthrax toxin both in vitro and in an animal model (Kim et al., 2005). In the following years, it became increasingly clear that a small set of defensins are universal effector molecules capable of neutralizing numerous toxins, many of which are unrelated to each other (Giesemann et al., 2008; Hooven et al., 2012; Kim et al., 2006; Kudryashova et al., 2014b; Kudryashova et al., 2015; Lehrer et al., 2009; Wang et al., 2006; Welkos et al., 2011).
Term “defensins” was introduced in 1985, when three peptides with antibacterial, antifungal, and antiviral activities were isolated from human neutrophil granules and named human neutrophil peptides 1 through 3 (HNP1, 2, and 3 respectively; (Ganz et al., 1985)). In the following years, numerous defensin-like peptides with low sequence identity, but high degree of structural similarity were found in invertebrates, plants, and fungi. All of them share a triple-stranded antiparallel β-sheet stabilized by several intermolecular disulfide bonds and were originally thought to have common origin, but were recently recognized to belong to two major super-families whose similarity resulted from convergent evolution (Shafee et al., 2016). The first group (cis-defensins) comprises defensins from plants, fungi, and invertebrates, while another family (trans-defensins) – the invertebrate big defensins and vertebrate α-, β-, and θ-defensins (Figure 1A–D). Interaction of the latter three with bacterial proteinaceous toxins is the focus of this review.
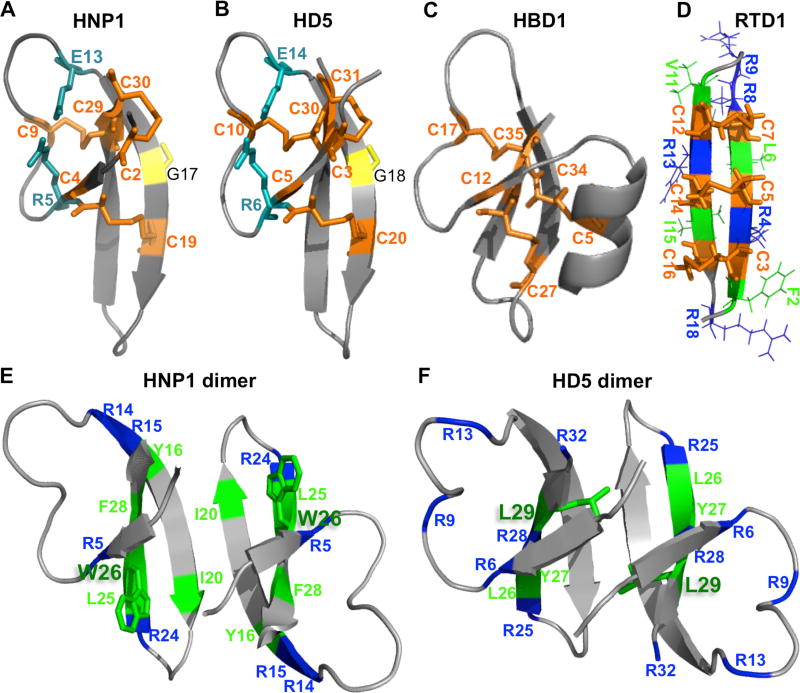
Figure 1. Defensin structures.
(A–D) Structures of human α-defensins HNP1 (PDB:3GNY) and HD5 (PDB:1ZMP), human β-defensin hBD1 (PDB:1IJU), and rhesus θ-defensin RTD1 (PDB:2LYF). Cysteines are shown for all structures in orange; for α-defensins, invariant Gly residue is colored in yellow and a salt bridge between oppositely charged R and E residues – in blue; for RTD1, arginines (dark blue) and hydrophobic residues (green) are shown. (E, F) Dimeric structures of α-defensins HNP1 and HD5. Arginines (dark blue) and several important hydrophobic residues (green) including the key residues W26 (sticks in HNP1) and L29 (sticks in HD5) are shown.
Six human α-defensins (Das et al., 2010) can be subdivided into myeloid and enteric peptides based on the place of production. Myeloid (or neutrophil) HNP1–4 peptides are produced by neutrophils, of which HNP1–3 are different only by a single N-terminal amino acid residue. Enteric peptides HD5 and HD6 are produced by Paneth cells in the intestinal epithelium. Although more than thirty β-defensin genes have been discovered in human (Scheetz et al., 2002), to date only about a dozen of β-defensin peptides have been recognized and partially characterized at the protein level (Jarczak et al., 2013). θ-defensins are cyclic peptides unique for Old World monkeys. In most primates, including humans, they are not translated and present in some tissues only as mRNA transcripts expressed from pseudogenes with a stop codon in the signal peptide sequence (Cole et al., 2002; Nguyen et al., 2003). Synthetic analogues of human θ-defensins, named retrocyclins (RC), demonstrate antimicrobial properties similar to their native counterparts found in non-human primates (Lehrer et al., 2012). Structurally, α-defensins are shorter than their β-counterparts as they are missing the N-terminal α-helix characteristic for β-defensins (Figure 1A–C); whereas θ-defensins are the shortest of the three (Figure 1D) as they are produced upon head to tail cyclization of two 9-a.a.-long fragments evolutionary related to α-defensins (Tang et al., 1999).
All three groups of defensins display rather amazing functional versatility. They demonstrate antimicrobial activity against Gram-negative and Gram-positive bacteria and against enveloped and non-enveloped viruses; next, all defensins are signaling molecules that modulate inflammatory response, wound healing, and angiogenesis in peptide- and tissue-specific manner (recently reviewed in (Mattar et al., 2016; Zhao and Lu, 2014)). Finally, defensins can selectively interfere with functional activity of different classes of pathogen effector proteins: various families of pore-forming, non-enzymatic, and enzymatic toxins (Giesemann et al., 2008; Hooven et al., 2012; Kim et al., 2005; Kim et al., 2006; Kudryashova et al., 2014b; Kudryashova et al., 2015; Lehrer et al., 2009; Wang et al., 2006; Welkos et al., 2011) and various viral proteins (Flatt et al., 2013; Gounder et al., 2012; Kudryashova et al., 2016b; Smith et al., 2010). The combination of the antitoxin, microbicidal, and immunomodulatory activities of α- and β-defensins is believed to play an important protective role during infection (Arnett and Seveau, 2011). Produced by barrier tissues and immune cells, these multifunctional peptides are ideally positioned to intercept toxins and compromise vitality of bacterial, fungal, and viral pathogens. Thus, the foodborne pathogen Listeria monocytogenes is targeted by α-defensins produced by small intestine Paneth cells and by neutrophils (latter of which migrate to various L. monocytogenes affected organs). Furthermore, neutrophil-produced HNPs not only affect L. monocytogenes directly, but also neutralize a major bacterial pathogenicity factor, a cholesterol-dependent cytolysin (CDC) listeriolysin O (LLO), as well as prevent its release by the bacteria (Arnett et al., 2011). Macrophages can either produce defensins on their own or acquire defensins released by neutrophils from the extracellular environment. Thus, rabbit macrophages produce MCP-1 and -2 α-defensins constitutively (Selsted et al., 1983) while human macrophages can produce β-defensins when challenged by hypoxia and/or bacteria (Hielpos et al., 2015; Nickel et al., 2012). Human macrophages can also acquire α-defensins released by neutrophils (Tan et al., 2006) and such cooperation between neutrophils and macrophages was shown to strongly boost the macrophages’ antilisterial activity, likely via neutralization of the LLO within the phagosome (Arnett et al., 2011). The above observations likely apply to the ability of defensins to mitigate the activity of CDC toxins produced by other Gram-positive pathogens.
To date, over a dozen of bacterial toxins and effector proteins beyond the CDC family has been recognized as potential targets of human defensins (Table 1), few toxins have been found to be unaffected, while many more have never been examined. This review will focus on known defensin-susceptible toxin targets (Table 1) and discuss the toxins’ and defensins’ structural determinants responsible for this targeting (Figure 1, Tables 2 and 3).
Table 1.
Bacterial toxins neutralized by defensins
| Class | Defensin | Toxin | Toxin Type | Effect of Defensins | References |
|---|---|---|---|---|---|
| α | HNP1–3 | B. anthracis lethal factor (LF) | Enzymatic | Inhibition of LF activity, protect mice in vivo | (Kim et al., 2005) |
| HNP1–3 | Diphtheria toxin (DT), Pseudomonas exotoxin A (ETA) | Enzymatic | Inhibition of enzymatic activity of DT and ETA | (Kim et al., 2006) | |
| HNP1, HNP3, HD5 | C. difficile toxin B (TcdB) | Enzymatic | Inhibition of enzymatic activity of TcdB | (Giesemann et al., 2008) | |
| HNP-1, HNP-3, HD-5 | Cholesterol-dependent cytolysins (CDCs) | Pore-forming | Inhibition of hemolysis mediated by CDCs | (Lehrer et al., 2009) | |
| HNP1, HD5 | V. cholerae and A. hydrophila MARTX | Enzymatic | Inhibition of enzymatic activity of actin crosslinking domain (ACD) and cysteine protease domain (CPD) | (Kudryashova et al., 2014b) | |
| HNP3 | S. aureus Panton-Valentine leukocidin (PVL) | Pore-forming | Partially decreases PVL pore formation and PMN lysis | (Cardot-Martin et al., 2015) | |
|
| |||||
| β | hBD1, hBD2, hBD4 | Neisseria gonorrhoeae gNarE | Enzymatic | Inhibition of auto-ADP-ribosyltransferase activity | (Rodas et al., 2016) |
| hBD3 | B. anthracis LF | Enzymatic | Inhibition of LF | (Wei et al., 2009) | |
|
| |||||
| θ | RTD1–3, Retrocyclins | B. anthracis LF | Enzymatic | Inhibition of enzymatic activity of LF, protect mice in vivo | (Wang et al., 2006; Welkos et al., 2011) |
| RC1, RC101 | V. cholerae and A. hydrophila MARTX | Enzymatic | Inhibition of enzymatic activity of ACD | (Kudryashova et al., 2015) | |
| RC101 | Gardnerella vaginalis vaginolysin (VLY) | Pore-forming | Inhibition of cytolysis mediated by VLY | (Hooven et al., 2012) | |
Table 2.
Correlation between structural elements of human α-defensins and their antibacterial and antitoxin activities
| Structural elements of α- defensins* |
Direct antibacterial Activity |
Toxin inhibition | |
|---|---|---|---|
| Gram-(−) | Gram-(+) | ||
| Disulfide bonds | −** | +++ | +++ |
| Hydrophobicity | +++ | +++ | +++ |
| Dimerization | − | ++ | ++ |
| Cationicity | +++ | +++ | ++ |
| High Arg content | ++ | +++ | +++ |
| Stereo-specificity | − | +++ | + |
| Conserved salt bridge | − | − | − |
The structure-function correlations presented in the table are i) a cumulative imprint from numerous, occasionally contradictory studies, ii) often representative of a complex sum of different, only partially understood mechanisms, and iii) not easily quantifiable, and therefore, are likely to need adjustments as more data and clearer understanding of the involved mechanisms emerge.
Relative roles of structural features on functional properties of defensins are expressed from being unimportant (−), i.e. demonstrating no influence upon removal, to mild (+), moderate (++) and essential (+++).
Table 3.
Properties of several defensins and other antimicrobial peptides (AMPs)
| AMP name | AMP sequence | Number of residues |
R/10a | K/10b | “+”/10c | “−”/10d | pI | GRAVYe | Monomer MW, kDa |
Neutralizes toxins |
|---|---|---|---|---|---|---|---|---|---|---|
| HNP1 |
|
30 | 1.33 | 0 | 1.33 | 0.33 | 8.7 | 0.300 | 3.45 | Yes |
| HNP2 |
|
29 | 1.38 | 0 | 1.38 | 0.34 | 8.7 | 0.248 | 3.38 | Yes |
| HNP3 |
|
30 | 1.33 | 0 | 1.33 | 0.67 | 8.3 | 0.123 | 3.49 | Yes |
| HNP4 |
|
33 | 1.52 | 0 | 1.52 | 0.30 | 9.0 | 0.661 | 3.72 | Nog |
| HD5 |
|
32 | 1.88 | 0 | 1.88 | 0.63 | 9.0 | −0.113 | 3.59 | Yes |
| HD6 |
|
32 | 0.94 | 0 | 1.56 | 0.31 | 8.4 | 0 | 3.71 | Nog |
| hBD1 |
|
36 | 0.28 | 1.11 | 1.39 | 0.28 | 8.9 | −0.272 | 3.93 | Noh |
| hBD2 |
|
41 | 0.49 | 1.21 | 1.71 | 0.24 | 9.3 | −0.102 | 4.33 | Noh |
| hBD3 |
|
45 | 1.56 | 1.33 | 2.89 | 0.44 | 10.1 | −0.700 | 5.16 | Yes/Nog |
| hBD4 |
|
50 | 1.60 | 0.80 | 2.40 | 1.20 | 9.3 | −1.008 | 5.89 | N/d |
| hBD5 |
|
51 | 0.78 | 0.78 | 1.57 | 0.98 | 8.3 | −0.610 | 5.78 | N/d |
| hBD6 |
|
44 | 0.23 | 1.59 | 1.81 | 1.14 | 8.6 | −0.473 | 4.89 | N/d |
| hBD14 |
|
43 | 1.16 | 1.16 | 2.32 | 2.32 | 6.4 | −1.012 | 5.22 | N/d |
| RC1 |
|
18 | 1.67 | 0.56 | 2.22 | 0 | 9.0 | 0.778 | 1.91 | Yes |
| RC101 |
|
18 | 1.11 | 1.11 | 2.22 | 0 | 9.0 | 0.811 | 1.89 | Yes |
| Cathelicidin LL-37 |
|
39 | 1.28 | 1.54 | 2.82 | 1.28 | 10.6 | −0.569 | 4.71 | No |
| Pig protegrin1 |
|
19 | 3.16 | 0 | 3.16 | 0 | 10.7 | −0.258 | 2.22 | N/d |
| Mouse cryptdin-1 |
|
35 | 2.00 | 0.57 | 2.86 | 0.57 | 9.6 | −0.551 | 4.12 | N/d |
| Plant cyO2 |
|
30 | 0.33 | 0.67 | 1.00 | 0.33 | 8.3 | 0.443 | 3.16 | No |
| Plant cyO19 |
|
31 | 0 | 0.97 | 0.97 | 0.65 | 7.8 | 0.471 | 3.25 | No |
| Plant kB1 |
|
29 | 0.34 | 0 | 0.34 | 0.34 | 6.0 | 0.152 | 2.92 | No |
| Plant kB7 |
|
29 | 0.34 | 0.34 | 0.68 | 0.34 | 7.8 | 0.038 | 3.10 | No |
Acidic residues are shown in red, basic residues are in blue, hydrophobic uncharged residues are in green, cysteines are in bold.
Parameters calculated using ExPASy ProtParam tool:
number of arginines per 10 residues;
number of lysines per 10 residues;
number of positively charged residues per 10 residues;
number of negatively charged residues per 10 residues;
grand average of hydropathicity (GRAVY).
Cyclic peptide.
Did not show protective activity against CDCs in a single study (Lehrer et al., 2009).
Weak destabilization was demonstrated only under low salt conditions (Kudryashova et al., 2014b).
N/d, Not determined.
Defensin-susceptible bacterial toxins/effector proteins
Enzymatic toxins
Bacillus anthracis lethal factor
Anthrax toxin is a three-component toxin comprised of pore-forming protective antigen (PA), lethal factor (LF), and edema factor (EF). EF is an adenylate cyclase that increases cellular cAMP level (Leppla, 1982), while LF is a highly toxic metalloprotease that proteolytically inactivates MAP kinase kinases leading to detrimental inhibition of the MAPK signal transduction pathway (Collier and Young, 2003; Duesbery et al., 1998). Seminal study by Kim and co-authors (Kim et al., 2005) demonstrated that HNP1 reversibly and noncompetitively inhibits anthrax LF, protecting MAPK signaling in LeTx-treated macrophages. Furthermore, HNP1–3 delivered immediately after injection of the anthrax holotoxin protected mice against Bacillus anthracis-induced intoxication and death in striking contrast with other cationic AMPs, magainin I and LL-37, which were ineffective. Preservation of the three intramolecular disulfide bridges of HNP1 was essential for the LF neutralization. Furthermore, in the extensive study by Wang and co-authors (Wang et al., 2006) rhesus θ-defensins (RTD1–3) and retrocyclins (RCs) also noncompetitively inhibited LF. Chirality, polarity, conserved tri-disulfide ladder, cationicity (arginine residues), and cyclic backbone were important for the RC ability to neutralize LF. Surface plasmon resonance experiments showed that RCs bind to LF with high affinity and at multiple sites, whereas preincubation of LF with RC further enhances the inhibitory activity, suggesting that post-binding steps (e.g. oligomerization or precipitation) contribute to the LF inhibition. RCs were also effective against murine anthrax infection in vivo (Welkos et al., 2011).
Mono-ADP-ribosyltransferase toxins
The next two toxins recognized as targets of α-defensins were diphtheria toxin (DT) and Pseudomonas aeruginosa exotoxin A (ETA) (Kim et al., 2006). These are two deadly toxins of a mono-ADP-ribosyltransferase (ART) family, which inactivate eukaryotic elongation factor 2 (eEF2) thus shutting down protein production and causing cell death (Collier, 2001; Michalska and Wolf, 2015). Similarly to the effects on anthrax toxin, HNP1–3, but not LL-37, protected cells from DT- and ETA-induced cell death. The mechanism of inactivation was not straightforward as defensins behaved as competitive inhibitors in respect to the eEF2 substrate but as noncompetitive in respect to the NAD+ substrate (Kim et al., 2006). Surprisingly, in contrast to inactivation of most other toxins, where preservation of the disulfide bonds of defensins is crucial for their activity, a DTT-reduced form of HNP1 appeared to be active against these two toxins.
The list of bacterial ARTs inhibited by defensins extends to NarE, ADP-ribosylating toxins from Neisseria meningitidis (mNarE toxin; (Castagnini et al., 2012)) and N. gonorrhoeae (gNarE toxin; (Rodas et al., 2016)), while cholera toxin (CT) and pertussis toxin (PT) were not affected by the peptides, implying that properties unrelated to the specific catalytic activity of the toxins were targeted by the defensins.
Interestingly, human arginine-specific mono-ADP-ribosyltransferase ART1 present in airway epithelial cells can ADP-ribosylate HNP1 in vitro and ADP-ribosylated HNP1 was detectable in vivo in patients with asthma and idiopathic pulmonary fibrosis (Paone et al., 2006). Such modification attenuated the defensin’s antimicrobial and cytotoxic activities, but preserved its immunomodulatory abilities (Paone et al., 2002). To assess whether bacterial ARTs, might employ similar mechanisms of defensin inactivation, P. aeruginosa exoenzyme S (ExoS), as well as CT, PT, DT, and ETA were examined but found to be unable to ADP-ribosylate HNP1 (Kim et al., 2006; Paone et al., 2002). In partial contradiction, more sensitive detection techniques revealed that CT and another bacterial ART toxin, Escherichia coli heat labile enterotoxin (LT) can ADP-ribosylate HNP1 and hBD1 in vitro (Castagnini et al., 2012). Given that only a small fraction of defensins was modified under ideal experimental conditions in vitro (Castagnini et al., 2012), the in vivo relevance of this modification by bacterial ART toxins is uncertain.
Clostridium difficile large toxins: TcdA and TcdB
C. difficile toxins A (TcdA) and B (TcdB) are main virulence factors that glucosylate and thus inactivate small GTP-binding proteins (Rho, Rac, and Cdc42) eventually leading to disruption of the actin cytoskeleton and cell death (Just et al., 1995a; Just et al., 1995b). It was discovered that α-defensins strongly inhibited the cytotoxicity implemented by TcdB, but not by TcdA (Giesemann et al., 2008). Accordingly, enzymatic activity of TcdB, but not TcdA, was inhibited by α-defensins HNP1, HNP3, and less potently by HD5. Breaking the disulfide bonds of the peptides with DTT abolished their activity. Neither β-defensin hBD1 nor cathelicidin LL-37 showed inhibition of either of the toxins. The inhibition of TcdB by α-defensins was associated with aggregation of the toxin and its co-precipitation with HNP1 and HD5. A molar ratio for HNP1 to TcdB in the aggregates was estimated to be as high as 14:1 suggesting binding at multiple sites. Dilution of the enzyme/defensin mixture partially reversed the inhibition, pointing on equilibrium-based mechanisms. Interestingly, TcdA also was pulled by defensins to the aggregates, albeit to a lesser extent than TcdB (Giesemann et al., 2008). In a latter study, it was confirmed that TcdB is more prone to the defensin-promoted unfolding, while TcdA was affected to a notably lesser degree (Kudryashova et al., 2014b). Given the structural similarity between the toxins, they represent a natural tool for evaluating parameters of toxin susceptibility and deciphering the antitoxin mechanisms of defensins.
Effector domains of Vibrio cholerae and Aeromonas hydrophila MARTX toxins
MARTX is a multifunctional autoprocessing repeat-in-toxin toxin produced by Vibrio and related species of Gram-negative bacteria (Satchell, 2015). The toxin is comprised of several well-defined components: i) a variable number (from one to five) of effector domains with distinct toxicities found in different combinations; ii) a cysteine protease domain (CPD) always positioned C-terminally to the effector domains and responsible for domain liberation upon delivery into a host cell (Prochazkova et al., 2009; Shen et al., 2009); and iii) the N- and C-terminal glycine-rich Ca2+-binding repeats that serve for membrane integration and translocation across the membrane (Kim et al., 2015). The specific enzymatic activity of one of the MARTX effector domains, actin cross-linking domain (ACD; (Heisler et al., 2015; Kudryashova et al., 2016a)) produced by V. cholerae and A. hydrophila, was tested in vitro and found to be potently inhibited by α- (HNP1, HD5; (Kudryashova et al., 2014b)) and θ-defensins (RC1 and RC101; (Kudryashova et al., 2015)). Cytotoxicity of both ACD and Rho inhibitory effector domain (RID; (Ahrens et al., 2013)) was prevented by HNP1 in cell culture experiments (Kudryashova et al., 2014b). Additionally, MARTX autoprocessing was impaired by defensins implying that the catalytic activity of CPD was also inhibited (Kudryashova et al., 2014b). Essentially, defensins induced unfolding and precipitation of all tested MARTX toxin effector domains of V. cholerae (all three domains) and A. hydrophila (two domains tested out of four total) as confirmed by characteristic shifts of circular dichroism (CD) and differential scanning fluorimetry (DSF) melting curves, higher exposure of toxins’ tryptophan residues to solution quenchers, exposure of otherwise protected sites for limited proteolysis, and disproportionally increased protein cross-section detected by surface-induced dissociation (SID) mass spectrometry (Kudryashova et al., 2014b; Kudryashova et al., 2015). These data strongly favored the hypothesis that the defensin-induced unfolding of the toxins is the key element of their inactivation. Similar to the situation with most other toxins, preservation of the defensins’ tertiary structure was essential as preincubation of the peptides with a reducing agent TCEP abolished their antitoxin activity. The effects of the only tested in these studies β-defensin hBD2 on toxin unfolding were weak and manifested only in the absence of salt.
Pore-forming toxins
Cholesterol-dependent cytolysins (CDCs)
The cholesterol-dependent cytolysin (CDC) family is the largest family of bacterial pore-forming toxins and belongs to the MACPF (Membrane Attack Complex and Perforin)/CDC superfamily, which also comprises eukaryotic pore-forming proteins (Anderluh et al., 2014). CDCs are major virulence factors that generally recognize membrane cholesterol as their binding receptor and are used by pathogenic bacteria to exert multiple functions during infection. CDCs can perforate the plasma membrane exerting a lethal effect on the target cell, while at sub-lethal concentrations, CDCs can facilitate host cell invasion, affect gene expression, and modulate inflammation (Hamon et al., 2007; Rabes et al., 2016; Seveau, 2014; Vadia et al., 2011). α-Defensins (HNP1–3 and, to a lesser extent, HD5) prevented lysis of red blood cells by listeriolysin O (LLO), anthrolysin O (ALO), and pneumolysin (PLY) (presented in the order of decreasing susceptibility to defensin inhibition) via binding of multiple copies of HNPs to a single CDC molecule (Lehrer et al., 2009). Intact HNP structure stabilized by three disulfide bonds is required for the inhibitory activity, since substitution of a single pair of cysteines (9 and 29) in HNP1 for α-aminobutyric acid (Abu) caused a 10-fold reduction of the activity, while a fully linearized HNP1 analog with all six cysteines replaced by Abu was ineffective and did not bind to CDCs. Surface plasmon resonance experiments with immobilized defensins showed reversibility of HNP/CDC binding upon addition of fetuin (a highly glycosylated serum protein). HNP4, HD6, and β-defensins hBD1–3 had no inhibitory potential on the same toxins at least at the two tested concentrations of 5 and 10 µg/ml (Lehrer et al., 2009). However, CDCs can also be inhibited by retrocyclins RC1, as was shown for LLO (Arnett et al., 2011), and RC101, as demonstrated for vaginolysin (VLY), a human-specific cholesterol-dependent cytolysin from Gardnerella vaginalis (Hooven et al., 2012). Most recently, it was found that both α- and θ-defensins neutralize CDCs by potentiating their unfolding (Kudryashova et al., 2014b; Kudryashova et al., 2015).
Staphylococcus aureus Panton-Valentine leukocidin (PVL)
S. aureus-produced Panton-Valentine leukocidin (PVL) is a cytolytic two-component toxin with hemolytic activity against immune cells (Kaneko and Kamio, 2004; Vandenesch et al., 2012). Its water-soluble monomeric components (LukF-PV and LukS-PV) bind to the cell surface and oligomerize into a pre-pore. In a somewhat similar to CDC fashion, the formation of transmembrane β-barrel pores from the mature PVL hetero-octamers undergoing major conformational rearrangements leads to host cell lysis (Pedelacq et al., 1999; Pedelacq et al., 2000). HNP3, but not HNP1 or 2, was reported to partially (~30%) protect neutrophils from PVL-induced lysis (Cardot-Martin et al., 2015). This is a rather intriguing result given that the three defensins differ from each other by a single N-terminal amino acid and share the majority of activities. Impairment of PVL conformational transitions by the defensin during pore formation has been suggested as a mechanism, but remains speculative in the lack of experimental evidence.
Non-enzymatic defensin-neutralizing virulence factors
Given that many bacterial toxins can be intercepted by defensins, it is reasonable to expect that microorganisms would engage mechanisms of neutralization. Indeed, it has been proposed that defensins can be neutralized upon their sequestering association with pathogen-produced molecules and molecular assemblies, e.g. Bacteroides fragilis-produced outer membrane vesicles (Nakayama-Imaohji et al., 2016), human adenovirus serotype 3 subviral penton-dodecahedral particles (Vragniau et al., 2017), and streptococcal effector proteins (Fernie-King et al., 2007). Although such inhibition should be a common phenomenon, surprisingly few bacterial proteins are recognized as effective decoy inhibitors of defensins likely reflecting insufficient attention to this topic rather than small number of such inhibitors. Strictly speaking, any affected toxin can be considered as an inhibitor of defensins as defensins are consumed upon interaction with toxins. However, given that most of the toxins are produced in small quantities and therefore are unlikely to cause a notable level of peptide neutralization, only abundant effector proteins of non-enzymatic nature will be discussed here as possible physiological defensin inhibitors.
Staphylococcus aureus staphylokinase
Historically, staphylokinase (SAK) was the first bacterial effector protein identified as a binding partner for defensins (Bokarewa and Tarkowski, 2004). Stoichiometric binding of SAK to plasminogen induces conformational changes that facilitate its conversion to plasmin, a broad-spectrum proteolytic enzyme whose activation facilitates bacterial invasion. SAK is a non-enzymatic exoprotein produced by a majority of S. aureus strains. High-affinity multivalent interaction of HNP1 and HNP2 with SAK (Jin et al., 2004) mutually reduces SAK’s thrombolytic potency and defensin bactericidal activity, which allowed to propose the role of SAK in mechanisms of resistance to host immune peptides (Bokarewa et al., 2006). Up to 6-fold molar excess of HNP1 can be complexed with SAK suggesting multiple binding sites for HNP, which is typical for defensin-toxin interactions in general. SAK also binds other AMPs such as melittin (a component of honey bee venom), tritrpticin (cathelicidin family AMP from porcine neutrophils), and bovine lactoferricin. This further supports its potential role as a sink for bioactive peptides, although the functional consequences of these interactions for SAK or AMP activities have not been analyzed (Nguyen and Vogel, 2016).
Streptococcal secreted virulence factors
Another inhibitor of the antibacterial activity of α- and β-defensins is a 31-kDa protein streptococcal inhibitor of complement (SIC), which is abundantly secreted by highly virulent M1 strains of Streptococcus pyogenes (Fernie-King et al., 2004, 2006; Frick et al., 2003). In addition to defensins, SIC inhibits such immune factors as LL-37, lysozyme, leukocyte proteinase inhibitor (SLPI), and the membrane attack complex of complement (MAC) (Fernie-King et al., 2002; Fernie-King et al., 2001; Frick et al., 2003). Such versatility can be partially explained by a highly acidic nature of SIC (pI ~4), the capacity to hydrophobic interactions, and partially by a rapid selection and highly polymorphic character of the sic gene and its product (Hoe et al., 2001; Stockbauer et al., 1998).
Other Streptococcus strains secrete virulence factors CRS (closely related to SIC) and DRS (distantly related to SIC) that share different degrees of structural and functional homology to SIC. Thus, DRS was found to bind and inhibit antibacterial activity of β-defensins hBD2 and hBD3 (Fernie-King et al., 2007). Binding of DRS to both defensins showed positive enthalpy, suggesting its hydrophobic nature. Interestingly, defensins and other immune peptides are able to inhibit the secretion of various virulence factors via ExPortal (unique microdomain of the S. pyogenes membrane specialized for protein secretion and processing), but not the secretion of SIC (Vega and Caparon, 2012).
Common features of toxins and inactivation mechanisms by defensins
As can be concluded from the first part of this review (summarized in Table 1), toxins inactivated by defensins belong to various enzymatic and non-enzymatic (pore-forming) families, and most of them share little to no sequence similarity. In contrast, most tested mammalian proteins are not affected by defensins. Such enigmatic selective susceptibility can be explained by some shared common properties that are both essential for toxins’ functionality and vulnerable for host defense factors. Obvious similarities in inactivation mechanisms (multivalent interaction, a partially reversible nature of inhibition, a requirement for preservation of disulfide bonds, and, finally, the potentiation of unfolding) further support this idea.
Such susceptibility-granting properties were proposed to be structural/conformational plasticity reflected in marginal thermodynamic stability of the toxins (Kudryashova et al., 2014b). This marginal stability is dictated by the necessity to make pores or cross the host membranes under unfavorable conditions, i.e. in the absence of folding/unfolding assisting chaperones and ATP. To exemplify, pore-forming CDC toxins (Marriott et al., 2008; Seveau, 2014) and anthrax PA (Jiang et al., 2015) undergo dramatic conformational reshaping upon transition from soluble to membrane-bound states in response to ligand binding or environmental changes. Thus, CDCs are released into the extracellular environment as water-soluble monomers (or dimers) and oligomerize upon binding to cholesterol-containing membranes into a pre-pore complex of 35 to 50 CDC subunits. This oligomerization is accompanied by dramatic conformational changes induced by a combination of protein-protein and protein-lipid interactions resulting in the conversion of α-helices into β-strands for the formation of a large transmembrane β-barrel pore (Hotze and Tweten, 2012; Lukoyanova et al., 2016).
Similarly, but for a different reason, many cell-penetrating exotoxins (e.g. effector domains of anthrax, diphtheria, and MARTX toxins) also demonstrate metastability essential for translocation across a narrow (~12–16 Å) pore in their partially or fully unfolded states (Kudryashova et al., 2014a; Montagner et al., 2007; Thoren and Krantz, 2011). The mechanism of anthrax effector domains translocation across the host membrane is one of the most thoroughly studied and best comprehended. LF and EF bind to PA ring-shaped oligomers associated with host cell membrane proteins. Following the endocytosis of the toxin complexes and acidification of the endosomes, PA forms a membrane-spanning channel and facilitates a proton gradient-driven passage of LF and EF through this channel in their unfolded states (Feld et al., 2010). Equilibrium chemical denaturation studies using fluorescence and CD spectroscopy demonstrated that LF and EF unfolding proceeds though a molten globule state with 60% increase in buried surface area exposure (Krantz et al., 2004). Hydrophobic sites inside the PA channel, and particularly a phenylalanine clamp, support LF and EF destabilization by providing favorable hydrophobic environment for interaction with the unfolded LF and EF (Krantz et al., 2005; Thoren et al., 2009). Although low-pH conditions and interaction with pore-forming subunits facilitate unfolding (Feld et al., 2012), toxins’ inherent plasticity per se appears to be crucial, given that toxin cross-linking and/or stabilization can block its passage through the membrane (Wesche et al., 1998).
Defensin-resistant bacterial toxins
Among a couple of dozens of tested bacterial toxins, only a few enzymatic or pore-forming toxins were found to be resistant to defensins, namely C. difficile TcdA, S. pneumoniae PLY, and V. cholerae CT. Assuming that marginal thermodynamic stability is the key element targeted by defensins, it would be reasonable to expect that the resistant toxins are more stable than their susceptible counterparts. This is indeed so for the CT holotoxin, comprised of a single CTA subunit bound to five CTB subunits. CT holotoxin is highly stable with Tm~74°C (Goins and Freire, 1988) and protein plasticity/instability required for translocation to the cytoplasm is achieved upon a proteolytic cleavage of CTA to disulfide-linked CTA1 and CTA2 subunits followed by a reduction of the disulfide bond in the ER. On the other hand, the catalytic domain of TcdA has similar stability to that of TcdB while demonstrating substantially higher resistance to destabilization by defensin and the similar situation is observed for a less susceptible PLY as compared to other CDC toxins (Kudryashova et al., 2014b). The latter examples suggest that low thermodynamic stability per se is insufficient to render toxins susceptible to defensins and additional properties play important roles in these interactions.
Human protein partners of defensins
As compared to bacterial toxins and effector proteins, mammalian proteins, in general, are less prone to destabilization by defensins, raising the question of the difference between the two groups. On the other hand, several human proteins were reported to interact with defensins. Reviewing these interactions is beyond the scopes of this work, but several general comments would be appropriate to consider due to physiological importance of such interactions.
First, defensins are either contained within intracellular granules or released to the extracellular space, which strictly limits the incidence of their interaction with cytoplasmic proteins. Cell membrane penetration by defensins has been reported, but its scale remains controversial. Regardless, high cytoplasmic concentrations of defensins are unlikely under any circumstances; therefore, their major physiologically relevant partners should be sought among extracellular proteins and membrane receptors. Indeed, several physiological partners of defensins were proposed among serum proteins: 2-macroglobulin, serpins, complement components, fibrin, plasminogen, and von Willebrand factor (Higazi et al., 1996; Panyutich and Ganz, 1991; Panyutich et al., 1995; Panyutich et al., 1994; Pillai et al., 2016).
Second, binding of defensins does not necessarily lead to destabilization and/or loss of functional activities of the targets as it is observed for bacterial toxins. Among about a dozen of structural and enzymatic mammalian cytoplasmic proteins tested by our group, none showed a decreased stability in the presence of defensins; while two proteins got stabilized upon such interaction (Kudryashova et al., 2014b). This is not entirely unexpected as interaction of a ligand with a native state of a target protein should in general result in stabilization of the native state (Waldron and Murphy, 2003). Why defensins destabilize toxin proteins? The observed destabilization of toxins can be explained by stronger interaction of defensins with the unfolded (partially or deeply) states of the targets, which is dictated by a particular combination and cumulative effect of all forces contributing to folding of a partner protein.
Inactivation Mechanism
Defensins’ ability to affect various proteins is non-specific, yet broadly selective towards pathogen effector proteins. Toxin determinants of susceptibility to defensin actions are poorly understood, but certainly include a marginal thermodynamic stability as discussed above. Structural features of selected defensins and their correlation with functional properties (particularly the bactericidal activities, but also interaction with selected bacterial toxins and viral proteins) have been meticulously addressed by Wuyuan Lu and collaborators (specific references will be given in the following text). Among all human defensins, the correlation between structural features and functionalities was the most comprehensively characterized for HNP1, followed by HD5 and synthetic retrocyclins. Since the ability of defensins to neutralize bacterial toxins was mostly tested on a single model toxin (LF of anthrax toxin) and only for very few defensins, the current level of knowledge does not allow to formulate a unifying hypothesis on the molecular mechanisms engaged by defensins and awaits a more thorough investigation and generalization.
Correlation between determinants essential for toxin inactivation and other activities of defensins (Table 2)
Disulfide bonds
Stabilization of defensin structure by three conserved disulfide bonds (Figure 1A–D) is crucial for toxin inactivation activity. Inhibition of LF (Kim et al., 2005; Wang et al., 2006; Wei et al., 2009), TcdB (Giesemann et al., 2008), MARTX effector domain ACD (Kudryashova et al., 2014b; Kudryashova et al., 2015), CDCs (Lehrer et al., 2009) required preserved α-defensin tertiary structure. Linearization of α-defensins HNP1 and HD5 (via reducing disulfide bonds or replacing Cys with Ala or α-aminobutiric acid) abolished their antitoxin activity (Lehrer et al., 2009; Rajabi et al., 2012; Wei et al., 2009). Only inhibition of DT and ETA by HNP1 was reported to be insensitive to the linearization of the defensin structure by a reducing agent (Kim et al., 2006). This is an interesting observation that either implies that DT and ETA are inhibited via different mechanisms than all other toxins, or possibly, that the reduction was not complete in these cases as defensins’ cores show substantial resistance to reducing agents. Of note, linearization of hBD3 had much milder effect on its anti-LF activity than that of α-defensins (Wei et al., 2009) possibly due to a positive role of the N-terminal α-helix (missing in α-defensins) in stabilization of the peptides’ tertiary structure.
It is important to understand that despite high structural similarity, defensins are highly diverse in their mechanisms and various functional activities of the same defensin rely on different structural elements. Thus, structure stabilization by disulfide bonds is essential for some, but not other activities of the toxin-reactive defensins (e.g. HNP1–3, HD5), while antibacterial activity of other defensins (e.g. hBD1 and HD6) is activated only upon reduction of disulfide bonds (Schroeder et al., 2015; Schroeder et al., 2011). The ability of HNP1 and HD5 to inhibit growth of Gram-negative, but not Gram–positive, bacteria appear to not require intact tertiary structure and may occur in the presence of reducing agents (de Leeuw et al., 2007; Wei et al., 2009). Killing Gram-negative bacteria is likely the result of the membrane disintegrating activity of defensins, which is arguably the least specific activity as it is shared by numerous other cationic antimicrobial peptides; whereas the ability to inactivate bacterial effector proteins is so far reserved strictly to selected defensins (Kudryashova et al., 2014b; Kudryashova et al., 2015). Therefore, while certain functions of certain defensins may require a high degree of disorder (as proposed in a recent review (Mattar et al., 2016)), this requirement should not be generalized, as other functions require structural stability. In fact, defensins the most active against bacterial toxins (HNP1–3) are also among the most ordered in the defensin’s family (Mattar et al., 2016).
Other structural elements of α-defensins: β-bulge, salt bridge, and chirality
In addition to disulfide connectivity, an invariant Gly residue is preserved in mammalian α-defensins (Xie et al., 2005) as a structural constraint for a β-bulge, which can only be met by achiral Gly or D-amino acids (Figure 1A,B). L-Ala can be tolerated in the defensin β-bulge structure, but it introduces quaternary structure distortion, which reduces the ability of defensins to form dimers (see below) and attenuates both antitoxin and antimicrobial activities of defensins, although the effect is rather mild (2-fold difference; (Zhao et al., 2012)).
Another conserved element of mammalian α-defensins, a salt bridge between oppositely charged R5 and E13 residues (HNP1 numbering; Figure 1A,B), renders defensins resistant to proteolysis by neutrophil elastase found in azurophilic granules together with the mature HNP peptides (Wu et al., 2005). However, the salt bridge was dispensable for antimicrobial activity against both Gram-negative and -positive bacteria. Similarly, a salt bridge-deficient E15D mutant of mouse Paneth cell α-defensin Criptdin-4 was functionally comparable to WT and, although insensitive to degradation by MMP-7 (Rosengren et al., 2006), was prone for trypsinization (Andersson et al., 2012). Although the role of the salt bridge in antitoxin activities of defensins was not investigated and remains unknown, it is conceivable that the salt bridge conservation is dictated by its role in folding and resistance to proteolysis rather than by specific defensin activities.
Since stereo-specificity is a key component of most of the complex, high-affinity interactions between chiral partners, a high-affinity interaction of natural proteins assembled of L-amino acids with L- and D-defensin enantiomers should vary dramatically, unless it is represented by multiple simple non-stereospecific interactions. Thus, L- and D-enantiomers are equally bactericidal when acting upon membrane lipids of E. coli, but D-enantiomers are less active against S. aureus, suggesting a more complex and more specific interaction of native L-defensins with some cellular components (e.g. lipid II; (de Leeuw et al., 2010)) in the latter case. Unnatural α-D-defensins and D-retrocyclins were moderately less potent but did not lose their ability to inhibit the activity of LF toxin (Wang et al., 2006; Wei et al., 2009) implying that the interactions are largely non-stereospecific. This interesting observation may suggest that the specificity of toxin targeting by defensins is primarily dictated by structural peculiarities of the affected toxins (see below) and by non-stereo-specific properties of defensins (e.g. charge, hydrophobicity, dimerization). We would like to speculate that such mechanism, which certainly requires a deeper analysis, tentatively enables broad selectivity against proteins allotted with certain properties (e.g. toxins) without a need for a particular spatial arrangement of the partners’ chemical entities.
Hydrophobicity
Comprehensive alanine scanning mutagenesis on HNP1 (Wei et al., 2010) revealed that both cationic and hydrophobic residues contribute to the abilities of HNP1 to kill Gram-positive bacteria S. aureus, inhibit bacterial toxins (e.g. anthrax LF), and bind to glycosylated viral proteins (e.g. HIV gp120). Most of the mutations were found to be silent or affected the activities of the peptide only moderately: mostly negatively, but occasionally also positively. Thus, the replacement of the only acidic residue E13 with alanine enhanced the ability of HNP1 to kill S. aureus, while having no effect on inhibition of LF. On the other hand, G23A mutant was a more potent inhibitor of LF, but had no notable effect on inhibition of S. aureus growth. This suggests that the peptide has evolved to be active against a broad range of targets rather than to be highly potent against any particular molecule. Interestingly, the most detrimental mutations were all in hydrophobic residues and their severity correlated well with an overall hydrophobicity of the affected residues in the following order: W26>F28>Y16>I20>L25. Particularly, by far the strongest detrimental effects were observed upon alanine substitution for the conserved Trp (W26A mutation), likely owing to one or both of the following: i) its role in stabilization of the HNP1 dimer’s hydrophobic interface (see below), and ii) overall contribution of the residue to hydrophobicity of the peptide (Figure 1E). Indeed, introduction of straight aliphatic unnatural amino acids at this position partially rescued the peptide activities and the rescue positively correlated with the length (and therefore, hydrophobicity) of the side chain (Wei et al., 2010). Higher hydrophobicity of a residue at this position could positively contribute to improved antimicrobial activities of the HNP1 mutants by enhancing their dimerization potential (see below), yet artificial dimerization of the W26A mutant via an additional intermolecular disulfide bond was not sufficient to improve the peptide’s activity against S. aureus, suggesting that hydrophobicity of this residue also plays a role in antimicrobial mechanisms.
Quadruple Ala mutant of HNP1 with substituted hydrophobic residues important for monomer interaction in a dimer (Y16 and F28) or dimer-dimer interaction (I20 and L25) has greatly reduced inhibitory activity towards LF (95 fold), comparable to that of the W26A mutant (Zhao et al., 2013). Since the quadruple mutant still can form dimers and tetramers (albeit likely less efficiently), self-association is probably less important than hydrophobicity for toxin binding and inactivation. Along the trend, the I20A mutant, which failed to form dimers upon crystallization, had a much milder activity reduction (less than 2-fold reduction of anti-LF activity). Interestingly, if dimer formation is disrupted by N-methylation of I20 (Pazgier et al., 2012), the effects are more dramatic (11-fold reduction of LF inhibitory activity, see below). This difference points on methodological limitations precluding accurate assessment of the defensin individual properties, our inability to fully separate those properties (e.g. hydrophobicity and dimerization) and/or our overall poor understanding of the inactivation mechanisms.
On intestinal α-defensin HD5, Ala scanning mutagenesis revealed L29 (Figure 1F) as a crucial residue for its antitoxin and antibacterial activity (Rajabi et al., 2012). Further substitution of this residue with a bulky hydrophobic F or lengthening the aliphatic side chain restored the activity. Notably, mutations of other hydrophobic residues in the C-terminal region (L26 and Y27) also led to moderately reduced abilities to bind and neutralize LF; and the same residues were critical for antiviral activity of the peptide (Tenge et al., 2014). The latter suggests that similar fundamental mechanisms might be involved in destabilization of bacterial toxins and viruses (Kudryashova et al., 2016b). Similarly to W26 in HNP1, L29 strongly contributes to a hydrophobic interface within the HD5 dimer (Figure 1E,F), making it difficult to separate the effects of hydrophobicity from those of impaired dimerization.
Among other tested defensins, reducing hydrophobicity in a conserved patch surrounding the key tryptophan (corresponding to W26 of HNP1) in mouse α-defensin Cryptdin-4 and rhesus myeloid α-defensin RMAD4 attenuated bactericidal activity (Tai et al., 2014), but effects on bacterial toxins were not explored. From all tested β-defensins, only arginine-rich hBD3 demonstrated prominent ability to neutralize toxins (Wei et al., 2009).
Dimerization
α-Defensins dimerize in a conserved manner via their central (2nd out of three) β-strands in antiparallel orientation to form symmetrical dimers (Figure 1E,F; (Wei et al., 2009)). Dimers are stabilized by four hydrogen bonds and entropic hydrophobic interactions. In human HNP1, the H-bonds are formed between backbone amide and carboxyl groups of T18 and I20, while hydrophobic interactions are between Y21 of one subunit with Y16, F21, and C2-C30 disulfide bond on the opposing subunit. W26 indirectly and substantially contributes to the hydrophobic interactions by affecting orientation of other hydrophobic residues. To address a recognized correlation between the dimerization ability of defensins with their functionality, Pazgier and colleagues (Pazgier et al., 2012) created constructs of HNP1 with impaired and enhanced abilities to form dimers. Breaking two of the four H-bonds and introducing steric interference at the dimer interface by N-methylation of I20 strongly impaired the ability of HNP1 to form dimers and resulted in a substantially weakened ability to inhibit the catalytic activity of LF (11-fold increase in IC50 from 148 nM to 1.6 µM). Similarly, weakening the HNP1 dimer interface by the W26A mutation resulted in a 20-fold higher IC50 of LF inhibition, while both mutations led synergistically to 300-fold higher IC50, as compared to wild type HNP1. On the other hand, stabilization of the dimer by an intermolecular disulfide bond connecting the N-termini of the subunits improved slightly the inhibitory activity of HNP1 (2-fold lower IC50) and even the W26A mutation in such cross-linked dimer had only very limited negative effect (IC50 is identical to that of uncross-linked wild type HNP1). Binding of HNP1 mutants to HIV gp120 protein reproduced well the above trend, suggesting that the same or highly similar mechanisms are behind binding and inactivation of the pathogen effector proteins in both cases. Interestingly, native disulfide-stabilized conformation and dimerization of HNP1 was essential for killing Gram-positive S. aureus, but not Gram-negative E. coli strains of bacteria.
The requirement for dimerization might not be universal. Thus, N-methylation of E21 in HD5 (similarly to N-methylation of I20 in HNP1) also weakened LF binding but to a much lesser degree than for HNP1 (2-fold versus 11-fold reduction; (Rajabi et al., 2012)), suggesting that disruption of dimerization is less deleterious for HD5 than for HNP1. Forming a non-canonical L29Nle-HD5 dimer with monomers arranged in a parallel fashion upon replacing L29 with norleucine (Nle) did not affect the LF-binding ability of HD5.
Cationicity
The majority of antimicrobial peptides, including defensins, are polycationic. Ala scanning mutagenesis revealed that for HNP1 (Wei et al., 2010) and HD5 (de Leeuw et al., 2009; Rajabi et al., 2012) the decrease in hydrophobicity is functionally more deleterious than decrease in net cationicity, yet the latter certainly is the essential component of the defensins’ functionality. Higher cationicity in general, and higher arginine content in particular, appear to correlate with higher salt resistance and higher antimicrobial potential for both α- and β-defensins (Llenado et al., 2009; Mathew and Nagaraj, 2015; Olli et al., 2013; Wang et al., 2015; Zou et al., 2007). Yet, the net cationicity does not appear to correlate well with the antitoxin activities of defensins as moderately basic α-defensins HNP1–3 (pI 8.3–8.7) are substantially more potent against most tested toxins than β-defensins hBD1–3 (pI 8.9–10.1) and therefore additional parameters must be considered. The net cationicity, density of charged residues, balance between arginine and lysine residues, and the presence of acidic residues vary substantially between defensins (Table 3). Among all the parameters, only high density of arginine residues (over 11% of total number of residues) appear to reasonably correlate with the high antitoxin potency of defensins. Indeed, human α-defensins demonstrate overall higher antitoxin activity and stronger selection for R over K (arginine content ranges from 9.4% of the total number of residues in HD6 to 18.8% in HD5). This also holds true for θ-defensins/retrocyclins (16.7% in RC1), while β-defensins vary significantly in this regard and contain members with as little as 2.8% (hBD1) to as much as 16% arginines (hBD3 and hBD4; Table 2). Accordingly, while most tested α-defensins (i.e. HNP1–3, HD5) and retrocyclins (RC1 and RC101) are potent inhibitors of bacterial toxins, only β-defensins with high R-content (i.e. hBD3) have a detectable neutralizing activity against selected toxins (Wei et al., 2009), whereas β-defensins with low R-content do not have this ability or express it very moderately (Kudryashova et al., 2014b). On the negative side, this correlation at least partially fails in case of hBD3 and HNP4 defensins, both of which have high arginine density (~15%) and overall high cationicity (pI 9.0 and 10.1, respectively) but demonstrate no ability to inhibit hemolytic activity of several CDC toxins (Lehrer et al., 2009). Interestingly, the ability of hBD3 to bind and neutralize LF toxin was moderately less efficient as compared to α-defensins, but also it was substantially less sensitive to linearization (Wei et al., 2009), possibly reflecting its overall more stable structure owing to its larger size and the N-terminal α-helix. In general, neither high arginine content, nor disulfide bond stabilized structure and high hydrophobicity are sufficient on their own. Indeed, human cathelicidin LL-37 (whose R-content is comparable to that of HNP1–3) and plant cyclotides (whose hydrophobic cores are stabilized by three disulfide bonds) demonstrate no antitoxin activity (Giesemann et al., 2008; Kim et al., 2005; Kim et al., 2006; Kudryashova et al., 2016b), similarly to the linearized forms of α-defensins. This suggests that a right combination of all the above factors (e.g. properly stabilized structure, ability to form dimers, hydrophobicity and cationicity/high R-content) contribute to the destabilization/inactivation mechanisms of defensins in complex mechanisms that require further investigation.
CONCLUSION
Human defensins act as natural “anti-chaperones” inactivating bacterial toxins by destabilizing their tertiary structure and promoting untimely or unnatural conformational transitions. Of note, a similar mechanism based on unfolding of viral proteins may play a role in the defensin-promoted inactivation of viruses. However, the exact molecular details of these inactivating mechanisms and the nature of selectivity towards toxins and viral effector proteins remain poorly understood. Given that only few defensins were tested for their ability to inactivate toxins and that only a small number (often only one) of toxins were used in most of the studies, the common features defining toxins’ susceptibility to the defensin inactivation are yet to be fully established and their roles in the inactivation mechanisms to be understood. Examining other defensins and their artificial derivatives and utilizing various bacterial toxins in experimental and computational studies should enable the transition from the current stage of data accumulation to formulating a unifying mechanism behind bacterial toxin inactivation by defensins.
Acknowledgments
This work was supported by The OSU Public Health Preparedness for Infectious Diseases (PHPID) program (New Investigator Grant to D.S.K.), the National Institute of General Medical Sciences of the NIH under award number R01GM114666 (to D.S.K.), and the National Institute of Health under award R01AI107250 (to S.M.S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Ahrens S, Geissler B, Satchell KJ. Identification of a His-Asp-Cys catalytic triad essential for function of the Rho inactivation domain (RID) of Vibrio cholerae MARTX toxin. J Biol Chem. 2013;288:1397–1408. doi: 10.1074/jbc.M112.396309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderluh G, Kisovec M, Krasevec N, Gilbert RJ. Distribution of MACPF/CDC proteins. Subcell Biochem. 2014;80:7–30. doi: 10.1007/978-94-017-8881-6_2. [DOI] [PubMed] [Google Scholar]
- Andersson HS, Figueredo SM, Haugaard-Kedstrom LM, Bengtsson E, Daly NL, Qu X, Craik DJ, Ouellette AJ, Rosengren KJ. The alpha-defensin salt-bridge induces backbone stability to facilitate folding and confer proteolytic resistance. Amino Acids. 2012;43:1471–1483. doi: 10.1007/s00726-012-1220-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnett E, Lehrer RI, Pratikhya P, Lu W, Seveau S. Defensins enable macrophages to inhibit the intracellular proliferation of Listeria monocytogenes. Cell Microbiol. 2011;13:635–651. doi: 10.1111/j.1462-5822.2010.01563.x. [DOI] [PubMed] [Google Scholar]
- Arnett E, Seveau S. The multifaceted activities of mammalian defensins. Curr Pharm Des. 2011;17:4254–4269. doi: 10.2174/138161211798999348. [DOI] [PubMed] [Google Scholar]
- Bokarewa M, Tarkowski A. Human alpha -defensins neutralize fibrinolytic activity exerted by staphylokinase. Thromb Haemost. 2004;91:991–999. doi: 10.1160/TH03-11-0696. [DOI] [PubMed] [Google Scholar]
- Bokarewa MI, Jin T, Tarkowski A. Staphylococcus aureus: Staphylokinase. Int J Biochem Cell Biol. 2006;38:504–509. doi: 10.1016/j.biocel.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Cardot-Martin E, Casalegno JS, Badiou C, Dauwalder O, Keller D, Prevost G, Rieg S, Kern WV, Cuerq C, Etienne J, et al. alpha-Defensins partially protect human neutrophils against Panton-Valentine leukocidin produced by Staphylococcus aureus. Lett Appl Microbiol. 2015;61:158–164. doi: 10.1111/lam.12438. [DOI] [PubMed] [Google Scholar]
- Castagnini M, Picchianti M, Talluri E, Biagini M, Del Vecchio M, Di Procolo P, Norais N, Nardi-Dei V, Balducci E. Arginine-specific mono ADP-ribosylation in vitro of antimicrobial peptides by ADP-ribosylating toxins. PLoS One. 2012;7:e41417. doi: 10.1371/journal.pone.0041417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole AM, Hong T, Boo LM, Nguyen T, Zhao C, Bristol G, Zack JA, Waring AJ, Yang OO, Lehrer RI. Retrocyclin: a primate peptide that protects cells from infection by T- and M-tropic strains of HIV-1. Proc Natl Acad Sci U S A. 2002;99:1813–1818. doi: 10.1073/pnas.052706399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier RJ. Understanding the mode of action of diphtheria toxin: a perspective on progress during the 20th century. Toxicon. 2001;39:1793–1803. doi: 10.1016/s0041-0101(01)00165-9. [DOI] [PubMed] [Google Scholar]
- Collier RJ, Young JA. Anthrax toxin. Annu Rev Cell Dev Biol. 2003;19:45–70. doi: 10.1146/annurev.cellbio.19.111301.140655. [DOI] [PubMed] [Google Scholar]
- Das S, Nikolaidis N, Goto H, McCallister C, Li J, Hirano M, Cooper MD. Comparative genomics and evolution of the alpha-defensin multigene family in primates. Mol Biol Evol. 2010;27:2333–2343. doi: 10.1093/molbev/msq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw E, Burks SR, Li X, Kao JP, Lu W. Structure-dependent functional properties of human defensin 5. FEBS Lett. 2007;581:515–520. doi: 10.1016/j.febslet.2006.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw E, Li C, Zeng P, Li C, Diepeveen-de Buin M, Lu WY, Breukink E, Lu W. Functional interaction of human neutrophil peptide-1 with the cell wall precursor lipid II. FEBS Lett. 2010;584:1543–1548. doi: 10.1016/j.febslet.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw E, Rajabi M, Zou G, Pazgier M, Lu W. Selective arginines are important for the antibacterial activity and host cell interaction of human alpha-defensin 5. FEBS Lett. 2009;583:2507–2512. doi: 10.1016/j.febslet.2009.06.051. [DOI] [PubMed] [Google Scholar]
- Duesbery NS, Webb CP, Leppla SH, Gordon VM, Klimpel KR, Copeland TD, Ahn NG, Oskarsson MK, Fukasawa K, Paull KD, Vande Woude GF. Proteolytic inactivation of MAP-kinase-kinase by anthrax lethal factor. Science. 1998;280:734–737. doi: 10.1126/science.280.5364.734. [DOI] [PubMed] [Google Scholar]
- Feld GK, Brown MJ, Krantz BA. Ratcheting up protein translocation with anthrax toxin. Protein Sci. 2012;21:606–624. doi: 10.1002/pro.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feld GK, Thoren KL, Kintzer AF, Sterling HJ, Tang II, Greenberg SG, Williams ER, Krantz BA. Structural basis for the unfolding of anthrax lethal factor by protective antigen oligomers. Nat Struct Mol Biol. 2010;17:1383–1390. doi: 10.1038/nsmb.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie-King BA, Seilly DJ, Binks MJ, Sriprakash KS, Lachmann PJ. Streptococcal DRS (distantly related to SIC) and SIC inhibit antimicrobial peptides, components of mucosal innate immunity: a comparison of their activities. Microbes Infect. 2007;9:300–307. doi: 10.1016/j.micinf.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Fernie-King BA, Seilly DJ, Davies A, Lachmann PJ. Streptococcal inhibitor of complement inhibits two additional components of the mucosal innate immune system: secretory leukocyte proteinase inhibitor and lysozyme. Infect Immun. 2002;70:4908–4916. doi: 10.1128/IAI.70.9.4908-4916.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie-King BA, Seilly DJ, Lachmann PJ. The interaction of streptococcal inhibitor of complement (SIC) and its proteolytic fragments with the human beta defensins. Immunology. 2004;111:444–452. doi: 10.1111/j.0019-2805.2004.01837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie-King BA, Seilly DJ, Lachmann PJ. Inhibition of antimicrobial peptides by group A streptococci: SIC and DRS. Biochem Soc Trans. 2006;34:273–275. doi: 10.1042/BST20060273. [DOI] [PubMed] [Google Scholar]
- Fernie-King BA, Seilly DJ, Willers C, Wurzner R, Davies A, Lachmann PJ. Streptococcal inhibitor of complement (SIC) inhibits the membrane attack complex by preventing uptake of C567 onto cell membranes. Immunology. 2001;103:390–398. doi: 10.1046/j.1365-2567.2001.01249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt JW, Kim R, Smith JG, Nemerow GR, Stewart PL. An intrinsically disordered region of the adenovirus capsid is implicated in neutralization by human alpha defensin 5. PLoS One. 2013;8:e61571. doi: 10.1371/journal.pone.0061571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick IM, Akesson P, Rasmussen M, Schmidtchen A, Bjorck L. SIC, a secreted protein of Streptococcus pyogenes that inactivates antibacterial peptides. J Biol Chem. 2003;278:16561–16566. doi: 10.1074/jbc.M301995200. [DOI] [PubMed] [Google Scholar]
- Ganz T, Selsted ME, Szklarek D, Harwig SS, Daher K, Bainton DF, Lehrer RI. Defensins. Natural peptide antibiotics of human neutrophils. J Clin Invest. 1985;76:1427–1435. doi: 10.1172/JCI112120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesemann T, Guttenberg G, Aktories K. Human alpha-defensins inhibit Clostridium difficile toxin B. Gastroenterology. 2008;134:2049–2058. doi: 10.1053/j.gastro.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Goins B, Freire E. Thermal stability and intersubunit interactions of cholera toxin in solution and in association with its cell-surface receptor ganglioside GM1. Biochemistry. 1988;27:2046–2052. doi: 10.1021/bi00406a035. [DOI] [PubMed] [Google Scholar]
- Gounder AP, Wiens ME, Wilson SS, Lu W, Smith JG. Critical determinants of human alpha-defensin 5 activity against non-enveloped viruses. J Biol Chem. 2012;287:24554–24562. doi: 10.1074/jbc.M112.354068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamon MA, Batsche E, Regnault B, Tham TN, Seveau S, Muchardt C, Cossart P. Histone modifications induced by a family of bacterial toxins. Proc Natl Acad Sci U S A. 2007;104:13467–13472. doi: 10.1073/pnas.0702729104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler DB, Kudryashova E, Grinevich DO, Suarez C, Winkelman JD, Birukov KG, Kotha SR, Parinandi NL, Vavylonis D, Kovar DR, Kudryashov DS. ACTIN-DIRECTED TOXIN. ACD toxin-produced actin oligomers poison formin-controlled actin polymerization. Science. 2015;349:535–539. doi: 10.1126/science.aab4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hielpos MS, Ferrero MC, Fernandez AG, Bonetto J, Giambartolomei GH, Fossati CA, Baldi PC. CCL20 and Beta-Defensin 2 Production by Human Lung Epithelial Cells and Macrophages in Response to Brucella abortus Infection. PLoS One. 2015;10:e0140408. doi: 10.1371/journal.pone.0140408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higazi AA, Ganz T, Kariko K, Cines DB. Defensin modulates tissue-type plasminogen activator and plasminogen binding to fibrin and endothelial cells. J Biol Chem. 1996;271:17650–17655. doi: 10.1074/jbc.271.30.17650. [DOI] [PubMed] [Google Scholar]
- Hoe NP, Vuopio-Varkila J, Vaara M, Grigsby D, De Lorenzo D, Fu YX, Dou SJ, Pan X, Nakashima K, Musser JM. Distribution of streptococcal inhibitor of complement variants in pharyngitis and invasive isolates in an epidemic of serotype M1 group A Streptococcus infection. J Infect Dis. 2001;183:633–639. doi: 10.1086/318543. [DOI] [PubMed] [Google Scholar]
- Hooven TA, Randis TM, Hymes SR, Rampersaud R, Ratner AJ. Retrocyclin inhibits Gardnerella vaginalis biofilm formation and toxin activity. J Antimicrob Chemother. 2012;67:2870–2872. doi: 10.1093/jac/dks305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotze EM, Tweten RK. Membrane assembly of the cholesterol-dependent cytolysin pore complex. Biochim Biophys Acta. 2012;1818:1028–1038. doi: 10.1016/j.bbamem.2011.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarczak J, Kosciuczuk EM, Lisowski P, Strzalkowska N, Jozwik A, Horbanczuk J, Krzyzewski J, Zwierzchowski L, Bagnicka E. Defensins: natural component of human innate immunity. Hum Immunol. 2013;74:1069–1079. doi: 10.1016/j.humimm.2013.05.008. [DOI] [PubMed] [Google Scholar]
- Jiang J, Pentelute BL, Collier RJ, Zhou ZH. Atomic structure of anthrax protective antigen pore elucidates toxin translocation. Nature. 2015;521:545–549. doi: 10.1038/nature14247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T, Bokarewa M, Foster T, Mitchell J, Higgins J, Tarkowski A. Staphylococcus aureus resists human defensins by production of staphylokinase, a novel bacterial evasion mechanism. J Immunol. 2004;172:1169–1176. doi: 10.4049/jimmunol.172.2.1169. [DOI] [PubMed] [Google Scholar]
- Just I, Selzer J, Wilm M, von Eichel-Streiber C, Mann M, Aktories K. Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature. 1995a;375:500–503. doi: 10.1038/375500a0. [DOI] [PubMed] [Google Scholar]
- Just I, Wilm M, Selzer J, Rex G, von Eichel-Streiber C, Mann M, Aktories K. The enterotoxin from Clostridium difficile (ToxA) monoglucosylates the Rho proteins. J Biol Chem. 1995b;270:13932–13936. doi: 10.1074/jbc.270.23.13932. [DOI] [PubMed] [Google Scholar]
- Kaneko J, Kamio Y. Bacterial two-component and hetero-heptameric pore-forming cytolytic toxins: structures, pore-forming mechanism, and organization of the genes. Biosci Biotechnol Biochem. 2004;68:981–1003. doi: 10.1271/bbb.68.981. [DOI] [PubMed] [Google Scholar]
- Kim BS, Gavin HE, Satchell KJ. Distinct roles of the repeat-containing regions and effector domains of the Vibrio vulnificus multifunctional-autoprocessing repeats-in-toxin (MARTX) toxin. MBio. 2015:6. doi: 10.1128/mBio.00324-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Gajendran N, Mittrucker HW, Weiwad M, Song YH, Hurwitz R, Wilmanns M, Fischer G, Kaufmann SH. Human alpha-defensins neutralize anthrax lethal toxin and protect against its fatal consequences. Proc Natl Acad Sci U S A. 2005;102:4830–4835. doi: 10.1073/pnas.0500508102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Slavinskaya Z, Merrill AR, Kaufmann SH. Human alpha-defensins neutralize toxins of the mono-ADP-ribosyltransferase family. Biochem J. 2006;399:225–229. doi: 10.1042/BJ20060425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krantz BA, Melnyk RA, Zhang S, Juris SJ, Lacy DB, Wu Z, Finkelstein A, Collier RJ. A phenylalanine clamp catalyzes protein translocation through the anthrax toxin pore. Science. 2005;309:777–781. doi: 10.1126/science.1113380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krantz BA, Trivedi AD, Cunningham K, Christensen KA, Collier RJ. Acid-induced unfolding of the amino-terminal domains of the lethal and edema factors of anthrax toxin. J Mol Biol. 2004;344:739–756. doi: 10.1016/j.jmb.2004.09.067. [DOI] [PubMed] [Google Scholar]
- Kudryashova E, Heisler D, Zywiec A, Kudryashov DS. Thermodynamic properties of the effector domains of MARTX toxins suggest their unfolding for translocation across the host membrane. Mol Microbiol. 2014a;92:1056–1071. doi: 10.1111/mmi.12615. [DOI] [PubMed] [Google Scholar]
- Kudryashova E, Heisler DB, Kudryashov DS. Pathogenic Mechanisms of Actin Cross-Linking Toxins: Peeling Away the Layers. Curr Top Microbiol Immunol. 2016a doi: 10.1007/82_2016_22. [DOI] [PubMed] [Google Scholar]
- Kudryashova E, Koneru PC, Kvaratskhelia M, Stromstedt AA, Lu W, Kudryashov DS. Thermodynamic instability of viral proteins is a pathogen-associated molecular pattern targeted by human defensins. Sci Rep. 2016b;6:32499. doi: 10.1038/srep32499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudryashova E, Quintyn R, Seveau S, Lu W, Wysocki VH, Kudryashov DS. Human defensins facilitate local unfolding of thermodynamically unstable regions of bacterial protein toxins. Immunity. 2014b;41:709–721. doi: 10.1016/j.immuni.2014.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudryashova E, Seveau S, Lu W, Kudryashov DS. Retrocyclins neutralize bacterial toxins by potentiating their unfolding. Biochem J. 2015;467:311–320. doi: 10.1042/BJ20150049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrer RI, Cole AM, Selsted ME. theta-Defensins: cyclic peptides with endless potential. J Biol Chem. 2012;287:27014–27019. doi: 10.1074/jbc.R112.346098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrer RI, Jung G, Ruchala P, Wang W, Micewicz ED, Waring AJ, Gillespie EJ, Bradley KA, Ratner AJ, Rest RF, Lu W. Human alpha-defensins inhibit hemolysis mediated by cholesterol-dependent cytolysins. Infect Immun. 2009;77:4028–4040. doi: 10.1128/IAI.00232-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppla SH. Anthrax toxin edema factor: a bacterial adenylate cyclase that increases cyclic AMP concentrations of eukaryotic cells. Proc Natl Acad Sci U S A. 1982;79:3162–3166. doi: 10.1073/pnas.79.10.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llenado RA, Weeks CS, Cocco MJ, Ouellette AJ. Electropositive charge in alpha-defensin bactericidal activity: functional effects of Lys-for-Arg substitutions vary with the peptide primary structure. Infect Immun. 2009;77:5035–5043. doi: 10.1128/IAI.00695-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukoyanova N, Hoogenboom BW, Saibil HR. The membrane attack complex, perforin and cholesterol-dependent cytolysin superfamily of pore-forming proteins. J Cell Sci. 2016;129:2125–2133. doi: 10.1242/jcs.182741. [DOI] [PubMed] [Google Scholar]
- Marriott HM, Mitchell TJ, Dockrell DH. Pneumolysin: a double-edged sword during the host-pathogen interaction. Curr Mol Med. 2008;8:497–509. doi: 10.2174/156652408785747924. [DOI] [PubMed] [Google Scholar]
- Mathew B, Nagaraj R. Antimicrobial activity of human alpha-defensin 6 analogs: insights into the physico-chemical reasons behind weak bactericidal activity of HD6 in vitro. J Pept Sci. 2015;21:811–818. doi: 10.1002/psc.2821. [DOI] [PubMed] [Google Scholar]
- Mattar EH, Almehdar HA, Yacoub HA, Uversky VN, Redwan EM. Antimicrobial potentials and structural disorder of human and animal defensins. Cytokine Growth Factor Rev. 2016;28:95–111. doi: 10.1016/j.cytogfr.2015.11.002. [DOI] [PubMed] [Google Scholar]
- Michalska M, Wolf P. Pseudomonas Exotoxin A: optimized by evolution for effective killing. Front Microbiol. 2015;6:963. doi: 10.3389/fmicb.2015.00963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagner C, Perier A, Pichard S, Vernier G, Menez A, Gillet D, Forge V, Chenal A. Behavior of the N-terminal helices of the diphtheria toxin T domain during the successive steps of membrane interaction. Biochemistry. 2007;46:1878–1887. doi: 10.1021/bi602381z. [DOI] [PubMed] [Google Scholar]
- Nakayama-Imaohji H, Hirota K, Yamasaki H, Yoneda S, Nariya H, Suzuki M, Secher T, Miyake Y, Oswald E, Hayashi T, Kuwahara T. DNA Inversion Regulates Outer Membrane Vesicle Production in Bacteroides fragilis. PLoS One. 2016;11:e0148887. doi: 10.1371/journal.pone.0148887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Vogel HJ. Staphylokinase has distinct modes of interaction with antimicrobial peptides, modulating its plasminogen-activation properties. Sci Rep. 2016;6:31817. doi: 10.1038/srep31817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TX, Cole AM, Lehrer RI. Evolution of primate theta-defensins: a serpentine path to a sweet tooth. Peptides. 2003;24:1647–1654. doi: 10.1016/j.peptides.2003.07.023. [DOI] [PubMed] [Google Scholar]
- Nickel D, Busch M, Mayer D, Hagemann B, Knoll V, Stenger S. Hypoxia triggers the expression of human beta defensin 2 and antimicrobial activity against Mycobacterium tuberculosis in human macrophages. J Immunol. 2012;188:4001–4007. doi: 10.4049/jimmunol.1100976. [DOI] [PubMed] [Google Scholar]
- Olli S, Rangaraj N, Nagaraj R. Effect of selectively introducing arginine and D-amino acids on the antimicrobial activity and salt sensitivity in analogs of human beta-defensins. PLoS One. 2013;8:e77031. doi: 10.1371/journal.pone.0077031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panyutich A, Ganz T. Activated alpha 2-macroglobulin is a principal defensin-binding protein. Am J Respir Cell Mol Biol. 1991;5:101–106. doi: 10.1165/ajrcmb/5.2.101. [DOI] [PubMed] [Google Scholar]
- Panyutich AV, Hiemstra PS, van Wetering S, Ganz T. Human neutrophil defensin and serpins form complexes and inactivate each other. Am J Respir Cell Mol Biol. 1995;12:351–357. doi: 10.1165/ajrcmb.12.3.7873202. [DOI] [PubMed] [Google Scholar]
- Panyutich AV, Szold O, Poon PH, Tseng Y, Ganz T. Identification of defensin binding to C1 complement. FEBS Lett. 1994;356:169–173. doi: 10.1016/0014-5793(94)01261-x. [DOI] [PubMed] [Google Scholar]
- Paone G, Stevens LA, Levine RL, Bourgeois C, Steagall WK, Gochuico BR, Moss J. ADP-ribosyltransferase-specific modification of human neutrophil peptide-1. J Biol Chem. 2006;281:17054–17060. doi: 10.1074/jbc.M603042200. [DOI] [PubMed] [Google Scholar]
- Paone G, Wada A, Stevens LA, Matin A, Hirayama T, Levine RL, Moss J. ADP ribosylation of human neutrophil peptide-1 regulates its biological properties. Proc Natl Acad Sci U S A. 2002;99:8231–8235. doi: 10.1073/pnas.122238899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazgier M, Wei G, Ericksen B, Jung G, Wu Z, de Leeuw E, Yuan W, Szmacinski H, Lu WY, Lubkowski J, et al. Sometimes it takes two to tango: contributions of dimerization to functions of human alpha-defensin HNP1 peptide. J Biol Chem. 2012;287:8944–8953. doi: 10.1074/jbc.M111.332205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedelacq JD, Maveyraud L, Prevost G, Baba-Moussa L, Gonzalez A, Courcelle E, Shepard W, Monteil H, Samama JP, Mourey L. The structure of a Staphylococcus aureus leucocidin component (LukF-PV) reveals the fold of the water-soluble species of a family of transmembrane pore-forming toxins. Structure. 1999;7:277–287. doi: 10.1016/s0969-2126(99)80038-0. [DOI] [PubMed] [Google Scholar]
- Pedelacq JD, Prevost G, Monteil H, Mourey L, Samama JP. Crystal structure of the F component of the Panton-Valentine leucocidin. Int J Med Microbiol. 2000;290:395–401. doi: 10.1016/S1438-4221(00)80050-8. [DOI] [PubMed] [Google Scholar]
- Pillai VG, Bao J, Zander CB, McDaniel JK, Chetty PS, Seeholzer SH, Bdeir K, Cines DB, Zheng XL. Human neutrophil peptides inhibit cleavage of von Willebrand factor by ADAMTS13: a potential link of inflammation to TTP. Blood. 2016;128:110–119. doi: 10.1182/blood-2015-12-688747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochazkova K, Shuvalova LA, Minasov G, Voburka Z, Anderson WF, Satchell KJ. Structural and molecular mechanism for autoprocessing of MARTX toxin of Vibrio cholerae at multiple sites. J Biol Chem. 2009;284:26557–26568. doi: 10.1074/jbc.M109.025510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabes A, Suttorp N, Opitz B. Inflammasomes in Pneumococcal Infection: Innate Immune Sensing and Bacterial Evasion Strategies. Curr Top Microbiol Immunol. 2016;397:215–227. doi: 10.1007/978-3-319-41171-2_11. [DOI] [PubMed] [Google Scholar]
- Rajabi M, Ericksen B, Wu X, de Leeuw E, Zhao L, Pazgier M, Lu W. Functional determinants of human enteric alpha-defensin HD5: crucial role for hydrophobicity at dimer interface. J Biol Chem. 2012;287:21615–21627. doi: 10.1074/jbc.M112.367995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodas PI, Alamos-Musre AS, Alvarez FP, Escobar A, Tapia CV, Osorio E, Otero C, Calderon IL, Fuentes JA, Gil F, et al. The NarE protein of Neisseria gonorrhoeae catalyzes ADP-ribosylation of several ADP-ribose acceptors despite an N-terminal deletion. FEMS Microbiol Lett. 2016:363. doi: 10.1093/femsle/fnw181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosengren KJ, Daly NL, Fornander LM, Jonsson LM, Shirafuji Y, Qu X, Vogel HJ, Ouellette AJ, Craik DJ. Structural and functional characterization of the conserved salt bridge in mammalian paneth cell alpha-defensins: solution structures of mouse CRYPTDIN-4 and (E15D)-CRYPTDIN-4. J Biol Chem. 2006;281:28068–28078. doi: 10.1074/jbc.M604992200. [DOI] [PubMed] [Google Scholar]
- Satchell KJ. Multifunctional-autoprocessing repeats-in-toxin (MARTX) Toxins of Vibrios. Microbiol Spectr. 2015:3. doi: 10.1128/microbiolspec.VE-0002-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheetz T, Bartlett JA, Walters JD, Schutte BC, Casavant TL, McCray PB., Jr Genomics-based approaches to gene discovery in innate immunity. Immunol Rev. 2002;190:137–145. doi: 10.1034/j.1600-065x.2002.19010.x. [DOI] [PubMed] [Google Scholar]
- Schroeder BO, Ehmann D, Precht JC, Castillo PA, Kuchler R, Berger J, Schaller M, Stange EF, Wehkamp J. Paneth cell alpha-defensin 6 (HD-6) is an antimicrobial peptide. Mucosal Immunol. 2015;8:661–671. doi: 10.1038/mi.2014.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder BO, Wu Z, Nuding S, Groscurth S, Marcinowski M, Beisner J, Buchner J, Schaller M, Stange EF, Wehkamp J. Reduction of disulphide bonds unmasks potent antimicrobial activity of human beta-defensin 1. Nature. 2011;469:419–423. doi: 10.1038/nature09674. [DOI] [PubMed] [Google Scholar]
- Selsted ME, Brown DM, DeLange RJ, Lehrer RI. Primary structures of MCP-1 and MCP-2, natural peptide antibiotics of rabbit lung macrophages. J Biol Chem. 1983;258:14485–14489. [PubMed] [Google Scholar]
- Seveau S. Multifaceted activity of listeriolysin O, the cholesterol-dependent cytolysin of Listeria monocytogenes. Subcell Biochem. 2014;80:161–195. doi: 10.1007/978-94-017-8881-6_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafee TM, Lay FT, Hulett MD, Anderson MA. The Defensins Consist of Two Independent, Convergent Protein Superfamilies. Mol Biol Evol. 2016;33:2345–2356. doi: 10.1093/molbev/msw106. [DOI] [PubMed] [Google Scholar]
- Shen A, Lupardus PJ, Albrow VE, Guzzetta A, Powers JC, Garcia KC, Bogyo M. Mechanistic and structural insights into the proteolytic activation of Vibrio cholerae MARTX toxin. Nat Chem Biol. 2009;5:469–478. doi: 10.1038/nchembio.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JG, Silvestry M, Lindert S, Lu W, Nemerow GR, Stewart PL. Insight into the mechanisms of adenovirus capsid disassembly from studies of defensin neutralization. PLoS Pathog. 2010;6:e1000959. doi: 10.1371/journal.ppat.1000959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockbauer KE, Grigsby D, Pan X, Fu YX, Mejia LM, Cravioto A, Musser JM. Hypervariability generated by natural selection in an extracellular complement-inhibiting protein of serotype M1 strains of group A Streptococcus. Proc Natl Acad Sci U S A. 1998;95:3128–3133. doi: 10.1073/pnas.95.6.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai KP, Le VV, Selsted ME, Ouellette AJ. Hydrophobic determinants of alpha-defensin bactericidal activity. Infect Immun. 2014;82:2195–2202. doi: 10.1128/IAI.01414-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan BH, Meinken C, Bastian M, Bruns H, Legaspi A, Ochoa MT, Krutzik SR, Bloom BR, Ganz T, Modlin RL, Stenger S. Macrophages acquire neutrophil granules for antimicrobial activity against intracellular pathogens. J Immunol. 2006;177:1864–1871. doi: 10.4049/jimmunol.177.3.1864. [DOI] [PubMed] [Google Scholar]
- Tang YQ, Yuan J, Osapay G, Osapay K, Tran D, Miller CJ, Ouellette AJ, Selsted ME. A cyclic antimicrobial peptide produced in primate leukocytes by the ligation of two truncated alpha-defensins. Science. 1999;286:498–502. doi: 10.1126/science.286.5439.498. [DOI] [PubMed] [Google Scholar]
- Tenge VR, Gounder AP, Wiens ME, Lu W, Smith JG. Delineation of interfaces on human alpha-defensins critical for human adenovirus and human papillomavirus inhibition. PLoS Pathog. 2014;10:e1004360. doi: 10.1371/journal.ppat.1004360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoren KL, Krantz BA. The unfolding story of anthrax toxin translocation. Mol Microbiol. 2011;80:588–595. doi: 10.1111/j.1365-2958.2011.07614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoren KL, Worden EJ, Yassif JM, Krantz BA. Lethal factor unfolding is the most force-dependent step of anthrax toxin translocation. Proc Natl Acad Sci U S A. 2009;106:21555–21560. doi: 10.1073/pnas.0905880106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadia S, Arnett E, Haghighat AC, Wilson-Kubalek EM, Tweten RK, Seveau S. The pore-forming toxin listeriolysin O mediates a novel entry pathway of L. monocytogenes into human hepatocytes. PLoS Pathog. 2011;7:e1002356. doi: 10.1371/journal.ppat.1002356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenesch F, Lina G, Henry T. Staphylococcus aureus hemolysins, bi-component leukocidins, and cytolytic peptides: a redundant arsenal of membrane-damaging virulence factors? Front Cell Infect Microbiol. 2012;2:12. doi: 10.3389/fcimb.2012.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega LA, Caparon MG. Cationic antimicrobial peptides disrupt the Streptococcus pyogenes ExPortal. Mol Microbiol. 2012;85:1119–1132. doi: 10.1111/j.1365-2958.2012.08163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vragniau C, Hubner JM, Beidler P, Gil S, Saydaminova K, Lu ZZ, Yumul R, Wang H, Richter M, Sova P, et al. Studies on the interaction of tumor-derived HD5 alpha defensins with adenoviruses and implications for oncolytic adenovirus therapy. J Virol. 2017 doi: 10.1128/JVI.02030-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron TT, Murphy KP. Stabilization of proteins by ligand binding: application to drug screening and determination of unfolding energetics. Biochemistry. 2003;42:5058–5064. doi: 10.1021/bi034212v. [DOI] [PubMed] [Google Scholar]
- Wang C, Shen M, Gohain N, Tolbert WD, Chen F, Zhang N, Yang K, Wang A, Su Y, Cheng T, et al. Design of a potent antibiotic peptide based on the active region of human defensin 5. J Med Chem. 2015;58:3083–3093. doi: 10.1021/jm501824a. [DOI] [PubMed] [Google Scholar]
- Wang G. Human antimicrobial peptides and proteins. Pharmaceuticals (Basel) 2014;7:545–594. doi: 10.3390/ph7050545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. Improved methods for classification, prediction, and design of antimicrobial peptides. Methods Mol Biol. 2015;1268:43–66. doi: 10.1007/978-1-4939-2285-7_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Mulakala C, Ward SC, Jung G, Luong H, Pham D, Waring AJ, Kaznessis Y, Lu W, Bradley KA, Lehrer RI. Retrocyclins kill bacilli and germinating spores of Bacillus anthracis and inactivate anthrax lethal toxin. J Biol Chem. 2006;281:32755–32764. doi: 10.1074/jbc.M603614200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G, de Leeuw E, Pazgier M, Yuan W, Zou G, Wang J, Ericksen B, Lu WY, Lehrer RI, Lu W. Through the looking glass, mechanistic insights from enantiomeric human defensins. J Biol Chem. 2009;284:29180–29192. doi: 10.1074/jbc.M109.018085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G, Pazgier M, de Leeuw E, Rajabi M, Li J, Zou G, Jung G, Yuan W, Lu WY, Lehrer RI, Lu W. Trp-26 imparts functional versatility to human alpha-defensin HNP1. J Biol Chem. 2010;285:16275–16285. doi: 10.1074/jbc.M110.102749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welkos S, Cote CK, Hahn U, Shastak O, Jedermann J, Bozue J, Jung G, Ruchala P, Pratikhya P, Tang T, et al. Humanized theta-defensins (retrocyclins) enhance macrophage performance and protect mice from experimental anthrax infections. Antimicrob Agents Chemother. 2011;55:4238–4250. doi: 10.1128/AAC.00267-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesche J, Elliott JL, Falnes PO, Olsnes S, Collier RJ. Characterization of membrane translocation by anthrax protective antigen. Biochemistry. 1998;37:15737–15746. doi: 10.1021/bi981436i. [DOI] [PubMed] [Google Scholar]
- Wu Z, Li X, de Leeuw E, Ericksen B, Lu W. Why is the Arg5-Glu13 salt bridge conserved in mammalian alpha-defensins? J Biol Chem. 2005;280:43039–43047. doi: 10.1074/jbc.M510562200. [DOI] [PubMed] [Google Scholar]
- Xie C, Prahl A, Ericksen B, Wu Z, Zeng P, Li X, Lu WY, Lubkowski J, Lu W. Reconstruction of the conserved beta-bulge in mammalian defensins using D-amino acids. J Biol Chem. 2005;280:32921–32929. doi: 10.1074/jbc.M503084200. [DOI] [PubMed] [Google Scholar]
- Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- Zhao L, Ericksen B, Wu X, Zhan C, Yuan W, Li X, Pazgier M, Lu W. Invariant gly residue is important for alpha-defensin folding, dimerization, and function: a case study of the human neutrophil alpha-defensin HNP1. J Biol Chem. 2012;287:18900–18912. doi: 10.1074/jbc.M112.355255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Lu W. Defensins in innate immunity. Curr Opin Hematol. 2014;21:37–42. doi: 10.1097/MOH.0000000000000005. [DOI] [PubMed] [Google Scholar]
- Zhao L, Tolbert WD, Ericksen B, Zhan C, Wu X, Yuan W, Li X, Pazgier M, Lu W. Single, double and quadruple alanine substitutions at oligomeric interfaces identify hydrophobicity as the key determinant of human neutrophil alpha defensin HNP1 function. PLoS One. 2013;8:e78937. doi: 10.1371/journal.pone.0078937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou G, de Leeuw E, Li C, Pazgier M, Li C, Zeng P, Lu WY, Lubkowski J, Lu W. Toward understanding the cationicity of defensins. Arg and Lys versus their noncoded analogs. J Biol Chem. 2007;282:19653–19665. doi: 10.1074/jbc.M611003200. [DOI] [PubMed] [Google Scholar]