Abstract
Basic leucine zipper (bZIP) transcription factors comprise one of the largest gene families in plants. They play a key role in almost every aspect of plant growth and development and also in biotic and abiotic stress tolerance. In this study, we report isolation and characterization of EcbZIP17, a group B bZIP transcription factor from a climate smart cereal, finger millet (Eleusine coracana L.). The genomic sequence of EcbZIP17 is 2662 bp long encompassing two exons and one intron with ORF of 1722 bp and peptide length of 573 aa. This gene is homologous to AtbZIP17 (Arabidopsis), ZmbZIP17 (maize) and OsbZIP60 (rice) which play a key role in endoplasmic reticulum (ER) stress pathway. In silico analysis confirmed the presence of basic leucine zipper (bZIP) and transmembrane (TM) domains in the EcbZIP17 protein. Allele mining of this gene in 16 different genotypes by Sanger sequencing revealed no variation in nucleotide sequence, including the 618 bp long intron. Expression analysis of EcbZIP17 under heat stress exhibited similar pattern of expression in all the genotypes across time intervals with highest upregulation after 4 h. The present study established the conserved nature of EcbZIP17 at nucleotide and expression level.
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-0984-2) contains supplementary material, which is available to authorized users.
Keywords: Finger millet, bZIP transcription factor, ER stress, Heat stress, Conserved gene
Introduction
Finger millet (Eleusine coracana L.) is an annual tetraploid cereal grass, mainly grown under semi-arid and subtropical rainfed conditions across the world. It is considered as an important staple food in drought prone areas and as an assured crop with respect to food security (Upadhyaya et al. 2007). Compared to other cereals and millets, finger millet can be considered as nutrient rice because of its high nutritional properties. Compared to white rice, it has higher amount of fibre, minerals (344 mg calcium, and 408 mg potassium per 100 g), vitamins and sulphur containing amino acids (Shobana et al. 2013; Chandra et al. 2016; Gull et al. 2016). It is a reservoir of many useful alleles with respect to biotic and abiotic stresses. The characteristic traits of drought tolerance, tolerance to high salinity, water logging and few diseases make it as a valuable resource crop for studying gene expression (Dida et al. 2007). Still it remains as an understudied crop, owing to lack of adequate genetic and genomic resources (Singh et al. 2014). However, in recent years, a few studies have been undertaken to characterize its genome and transcriptome (Sood et al. 2016; Hittalmani et al. 2017).
To enhance the genetic potential, the available germplasm resources need to be relooked for potential alleles. Allele mining, i.e. identification of allelic variation of a known gene with respect to a particular mechanism or trait in diverse genotypes is an important approach to mine potential novel alleles (Kumar et al. 2010). Among the transcription factor families, bZIP is one of the largest and diverse families (Ying et al. 2012). Transcription factors belonging to this group harbour a highly conserved bZIP domain and it constitutes two components, i.e. basic region of around 18 aa with an invariant motif (N-x7-R/K-x9) for nuclear localization and sequence specific DNA binding, and a less conserved leucine zipper region composed of several heptad repeats of leucine or hydrophobic amino acids for dimerization specificity (Liu et al. 2014). Plant bZIP TFs contain a relaxed binding specificity for ACGT motif and other motifs, such as A-box (TACGTA), G-box (CACGTG), and C-box (GACGTC). Regulation of binding specificity is mediated by flanking nucleotides (Wang et al. 2011). Plant bZIP TFs have been found to play an essential role in seed germination, floral induction, embryogenesis, vascular development, and photomorphogenesis (Nijhawan et al. 2008; Tang et al. 2012). They also have been found to play essential role in a variety of biotic and abiotic stress responses such as salinity, heat, drought, cold, pathogen infection, hormone signalling (ABA and ethylene) and endoplasmic reticulum (ER) stress (Liu et al. 2008, 2014; Xiang et al. 2008; Gao et al. 2011). ER stress results in accumulation of unfolded proteins in the ER, which activates the signalling pathway known as unfolded protein response (UPR) (Deng et al. 2013). A group of bZIP transcription factors, involved in UPR stress response, are known as membrane-tethered transcription factors (Seo 2008). In this study, we isolated genomic sequence of a bZIP transcription factor gene EcbZIP17 from different finger millet genotypes to examine the allelic variation. We also checked expression of this gene under heat stress condition in these different genotypes at different time points to understand its role in heat stress.
Materials and methods
Plant material and stress imposition
Finger millet genotypes were procured from ICAR-All India Coordinated Research Project on Small Millets, GKVK, Bangalore, India. Seeds were allowed to grow under 16 h light/8 h dark cycle at 25 °C in a culture room. Samples for DNA isolation were collected from 15-day-old seedlings. Same stage seedlings were also used for imposing stress. For heat stress treatment, seedlings were subjected to 42 °C in growth chamber and samples were collected after 2, 4 and 24 h along with seedlings grown at optimum temperature. The samples were immediately frozen in liquid nitrogen. All samples were stored at −80 °C till DNA or RNA isolation.
Genome walking
To isolate the complete gene sequence of EcbZIP17, we performed genome walking according to manufacturer’s instructions (Clontech Genome walker kit, USA) using genomic DNA of finger millet. For this, genomic DNA was isolated according to manufacturer’s instructions (GenElute Plant Genomic DNA Miniprep Kit, Sigma, USA) and purified twice to achieve high purity. Quality and quantity of isolated genomic DNA were analysed by 0.8% agarose gel electrophoresis and Nanodrop spectrophotometer (Thermo Scientific, USA). 3 µg of purified genomic DNA was subjected to restriction digestion with four different blunt enzymes, i.e. EcoRV, DraI, StuI and PvuII, respectively. Genome walking adapters were ligated to restricted genomic DNA fragments, and performed nested primary PCR using 1 µL as template from each library. Primary PCR product was diluted to 1:50 dilution and from this 1 µL was used as template to perform secondary PCR. Different amplicons obtained from genome walking nested PCRs were gel extracted using SureExtract PCR/Gel Extraction kit, and ligated with pGEMT easy vector (Promega) using manufacturer’s instructions. The ligation mixture was transformed into freshly prepared E. coli DH5α competent cells. The recombinant positive clones were screened by blue-white screening using X-gal (50 µg/mL), IPTG (0.5 mM), ampicillin (100 mg/L), and sequenced using SP6 and T7 primers. Clones were sequenced and aligned into contigs using Lasergene software to generate the consensus sequence (Clewley and Arnold 1997).
In silico analysis
Major domains in EcbZIP17 protein were confirmed using SMART online tool (http://smart.embl-heidelberg.de/). The secondary structure of EcbZIP17 protein was predicted by SOPMA online tool (Combet et al. 2000). Since EcbZIP17 protein is a transcription factor, its nucleotide (DNA) binding and protein binding regions were predicted with PROFisis Predict Protein server tool (https://www.predictprotein.org/). The amino acid composition of the protein was analysed through Bioedit software (Hall 1999). Various physiochemical properties of the EcbZIP17 protein were analysed through ProtParam package of the ExPASy web server (http://web.expasy.org/protparam/) (Gasteiger et al. 2005). To determine the exact mass of the EcbZIP17, Isotopident package of the ExPASy server was used (http://education.expasy.org/student_projects/isotopident/htdocs/). Hydrophilic index of this protein was analysed using ProtScale package of the ExPASy server (http://web.expasy.org/protscale/). Three-dimensional structure of the EcbZIP17 protein was predicted by submitting the aa sequence to I-TASSER server (Roy et al. 2010; Yang et al. 2015). The predicted 3D model was assessed by generating Ramachandran plot using PROCHECK server (Laskowski et al. 1996; Lovell et al. 2002). Sequences obtained from 16 genotypes were assembled through Laser gene software.
Allele mining
50 ng of genomic DNA from each of the 16 genotypes was used as template for PCR amplification. The total volume of PCR reaction was 25 µL containing 50 ng of DNA, 1X Taq buffer, 10 mM dNTP, 1 µM of each primer bZIP17FL-F and bZIP17FL-R (Table S1), and 1 U Taq DNA polymerase (Thermo Scientific). Amplification conditions were performed in a programmed thermal cycler (Eppendorf), with initial denaturation at 94 °C for 5 min, annealing at 52 °C for 1 min, and extension at 72 °C for 3 min, followed by final extension of 7 min. The amplicons were analysed in 1% agarose gel and PCR products were excised from the gel using a scalpel, and the amplicons were extracted using SureExtract PCR/Gel Extraction kit (Genetix) for clean fragment. These PCR products were further sequenced with amplicon specific and overlapping internal primers to get complete sequence of the gene.
RNA isolation, cDNA synthesis and qRT-PCR analysis
RNA was isolated from samples using Spectrum plant total RNA kit (Sigma), DNA contamination from RNA was eliminated during RNA isolation through On-Column DNase I Digestion Set, Sigma, USA. The quality and quantity of RNA samples were analysed through agarose gel electrophoresis and Nanodrop spectrophotometer (Thermo scientific, USA). From the total isolated RNA, 1 µg of RNA from every sample was used for synthesis of cDNA using Prime script 1st strand cDNA synthesis kit (Takara, Japan) as per manufacturer’s instructions. Expression analysis of EcbZIP17 in different genotypes of finger millet under heat stress condition was quantified by qRT-PCR. Each cDNA was diluted to 1:5 dilutions and 1 µL diluted cDNA was used as a template for qRT-PCR reaction which was carried out in a Roche Lightcycler 480 system using 2X Brilliant SYBR Green qPCR master mix (Agilent). Primers bZIPqPCR-F and bZIPqPCR-R (Table. S1) were used for qRT-PCR reaction. The reaction conditions for qRT-PCR were as follows: 95 °C for 5 min, followed by 38 cycles of 95 °C for 5 s, 58 °C for 10 s and 72 °C for 25 s. β-Tubulin was used as the internal reference gene to normalise the gene expression (Pudake et al. 2017). Changes in fold expression were calculated using delta CT method (Livak and Schmittgrn 2001; Kanakachari et al. 2016). For gene amplification and qRT-PCR expression analysis of EcbZIP17, primers were designed using Primer3 software (http://frodo.wi.mit.edu/primer3/). The designed primers were analysed by IDT-Oligo analyzer tool (https://eu.idtdna.com/calc/analyzer) for homo and heterodimer formation, melting temperature, and priming efficiency.
Results
Genome walking of EcbZIP17
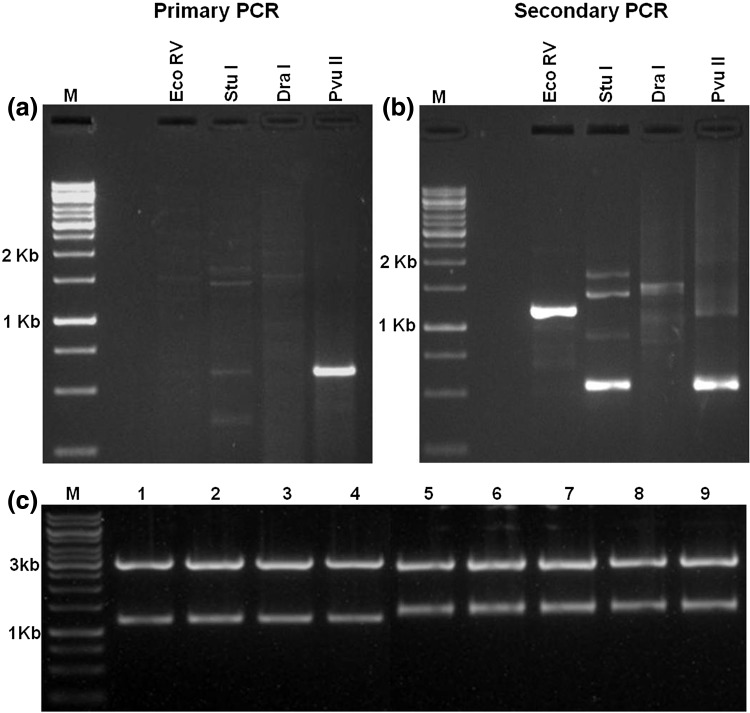
Earlier we have identified a heat responsive EST of bZIP gene from MR1 genotype of finger millet using Suppression subtractive hybridization (SSH) (Fig. S1a) and further complete cDNA is submitted to NCBI (Accession No. KF245640.1). To identify complete gene sequence, we followed genome walking strategy. Results of genome walking primary PCR showed that there was a single bright band of around 600 bp in PvuII library, whereas all other three libraries had low intensity faint bands ranging from 300 to 3 kb (Fig. 1a). The diluted primary PCR products were used as template for secondary PCR and the results shown that EcoRV, StuI, and PvuII libraries contained bright intensity bands ranging from 500 bp to 1.8 kb, whereas no such banding pattern was observed in DraI library (Fig. 1b). All these bands were extracted from the gel and cloned into pGEMT Easy vector, confirmed by restriction digestion (Fig. 1c) and sequenced. Alignment of sequenced clones with previous EST sequence resulted in a stretch of 2662 bp (Fig. S1b), having complete bZIP gene and encoding 573aa (Fig. S1c) which we further renamed as EcbZIP17.
Fig. 1.
Genome walking for EcbZIP17 gene. Four genomic DNA libraries, i.e. EcoRI, StuI, DraI and PvuII were prepared and performed primary PCR using each genomic library as template (a). The primary PCR product was diluted to 1:50 and used as template to perform secondary PCR (b). pGEMT cloning and restriction digestion confirmation analysis of genome walking secondary PCR products of EcoRV library (1–4) and StuI library (5–9) (c)
Comparison of EcbZIP17 gene structure with Arabidopsis and rice bZIP17
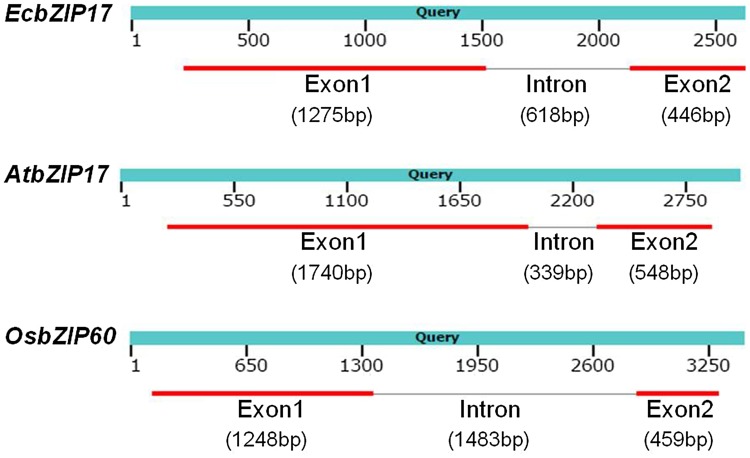
Comparison of EcbZIP17 gene structure with homologs from Arabidopsis (AtbZIP17; At2g40950) and rice (OsbZIP60; LOC_Os07g44950) revealed similarity in the gene structure. In all the three homologs, the two exons are separated by single intron having variable size. When the CDS regions of all three sequences were aligned with respective genomic regions (Fig. 2), the longest intron was observed in OsbZIP60. The length of gene including exons and intron is 2339 bp in EcbZIP17, 2627 bp in AtbZIP17 and 3237 bp in OsbZIP60, whereas the length of intron is 339 bp in AtbZIP17, 618 bp in EcbZIP17 and 1483 bp in OsbZIP60. Among all three sequences, Arabidopsis had the highest length of ORF. Although rice is closer to finger millet, intron of EcbZIP17 is smaller than its counterpart in rice.
Fig. 2.
Alignment of gene sequence with respective CDS regions of bZIP17 from finger millet, Arabidopsis, and rice. Length of introns and exons indicated, respectively
Structural analysis of EcbZIP17 protein
SMART analysis for major domains in EcbZIP17 protein revealed that BRLZ (basic region leucine zipper) domain extended from 120 to 184 aa, whereas TM (trans membrane) domain spanned from 249 to 271 aa (Fig. S2). The secondary structure analysis of EcbZIP17 protein revealed that 154 amino acid residues (26.88%) were involved in alpha helices, 78 residues (13.61%) in extended strands, 45 residues (7.85%) in beta turns and 296 residues (51.66%) in random coils (Fig. S3a, b). Protein binding regions of EcbZIP17 protein were identified between 1 and 62 aa, whereas polynucleotide binding region was identified between 124 and 155 aa (Fig. S3c). Aliphatic index of this protein was 71.05, whereas grand average of hydropathicity was −0.429, confirming the hydrophilic nature of the protein (Fig. S3d). Prediction of EcbZIP17 protein subcellular localization results in nucleus (Fig. S4). The EcbZIP17 protein contains 69 negatively charged (Asp + Glu) and 58 positively charged (Arg + Lys) aa. Atomic composition revealed that 8511 atoms (C2665 H4225 N743 O855 S23) were present in the EcbZIP17 protein, and the extinction coefficient was 33,725 M−1 cm−1. Mono-isotopic and exact mass of the protein calculated were 61,053.35 and 61,088.45 Da (Table 1). The EcbZIP17 protein was found to be rich in serine, alanine, proline, leucine and glycine aa (Table S2).
Table 1.
Primary structural properties of EcbZIP17 protein
| S. No | Primary structural properties | Theoretical prediction |
|---|---|---|
| 1 | Molecular weight (kDa) | 61.1 |
| 2 | Isoelectric point | 5.49 |
| 3 | Total no. of negatively charged residues (Asp + Glu) | 69 |
| 4 | Total no. of positively charged residues (Arg + Lys) | 58 |
| 5 | Extinction coefficient (M−1 cm−1, at 280 nm) | 33,725 |
| 6 | Aliphatic index | 71.05 |
| 7 | Grand average of hydropathicity (GRAVY) | −0.429 |
Molecular modelling of EcbZIP17
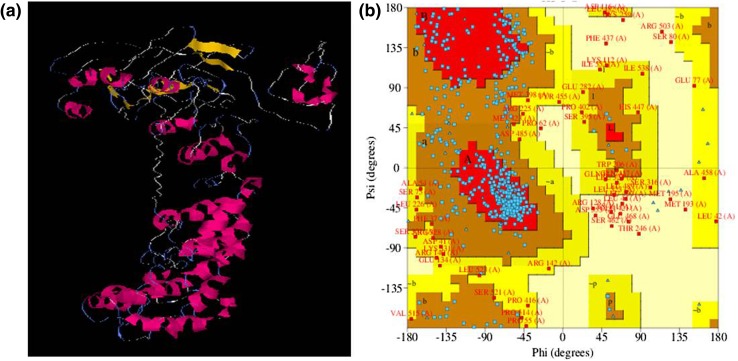
The 3D model of EcbZIP17 protein was generated by I-TASSER (Iterative Threading ASSEmbly Refinement) online tool (Fig. 3a). The major proteins used by the server are transcription factor MafB (MafB protein) and Runt-related transcription factor 1 (RUNX1 protein); both are bZIP transcription factors. This server generated five models using threading approach for the given amino acid sequence of the EcbZIP17 protein. Of these, model1 had the best C-Score (confidence score) of −1.29. C-score is a confidence score for estimating the quality of models generated by I-TASSER. It was calculated based on the convergence parameters of the structure assembly simulations and significance of threading template alignments. It usually lies in the range of −5 to 2. 3D protein structure of EcbZIP17 is having two major domains, i.e. BRLZ (basic region leucine zipper) domain required for sequence-specific DNA-binding transcription factor activity and TM (transmembrane) domain required for anchoring to the membrane and facilitates transfer of signals. The PROCHECK analysis of model1 from I-TASSER server generated Ramachandran plot (Fig. 3b). Statistical results of Ramachandran plot showed that 59.5% of amino acid residues were in the most favoured region while 29.5% of residues were in additionally allowed region, 7.6% residues in generously allowed region, and 3.4% residues in disallowed region. Structural analysis using Ramachandran plot results in the unusual category of G factor value. The overall average of G factors value for predicted model was −0.79. G factor is the important aspect of the Ramachandran plot and is a combination of different parameters such as dihedral angles (phi–psi distribution, chain1–chain2 distribution, chain1, chain3 & chain4, and Omega) and main-chain covalent forces (main-chain bond lengths, main-chain bond angles).
Fig. 3.
3D structure and Ramachandran plot prediction for EcbZIP17 protein. Predicted 3D model of EcbZIP17 protein through I-TASSER server, representing model 1 from the server (a). Ramachandran plot for predicted model 1 through PROCHECK server (b)
Amplification and sequencing of EcbZIP17 from different genotypes of finger millet
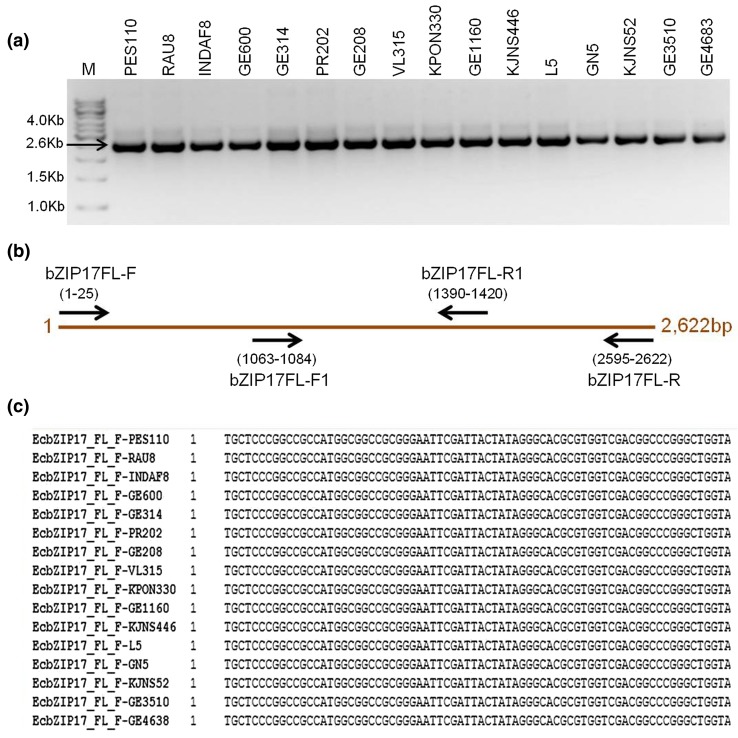
PCR products from the 16 finger millet genotypes were of identical size with no size variation (Fig. 4a). The first round of sequencing resulted in around 700 bp sequence information from both the ends of the PCR amplicon. To obtain the sequence of internal region, two internal primers were designed (bZIP17FL-F1 and bZIP17FL-R1) and the same amplicons were again sequenced to get complete sequence of entire amplicons (Fig. 4b). Assembly using all sequences was run using Lasergene resulted into single contig sequence with no change at SNP or indel level (Fig. 4c).
Fig. 4.
PCR amplification of EcbZIP17 gene in different genotypes of finger millet (a). Different primers and their respective positions used in sequencing to complete the sequencing of 2.6 kb amplicon (b). Snap shot of alignment representing nucleotide sequences obtained from sequencing results of 16 genotypes (c)
Expression analysis of EcbZIP17 in different genotypes of finger millet under heat stress
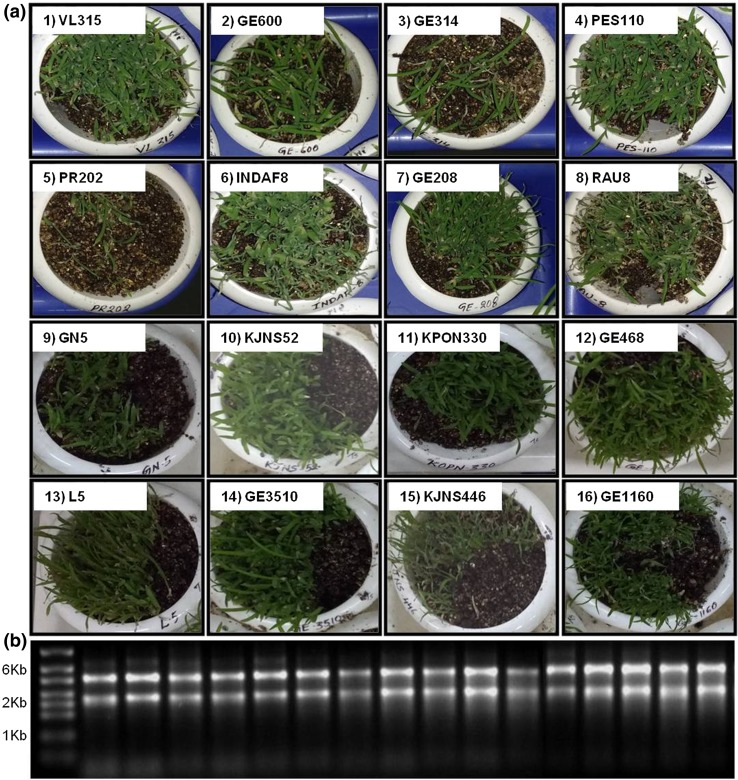
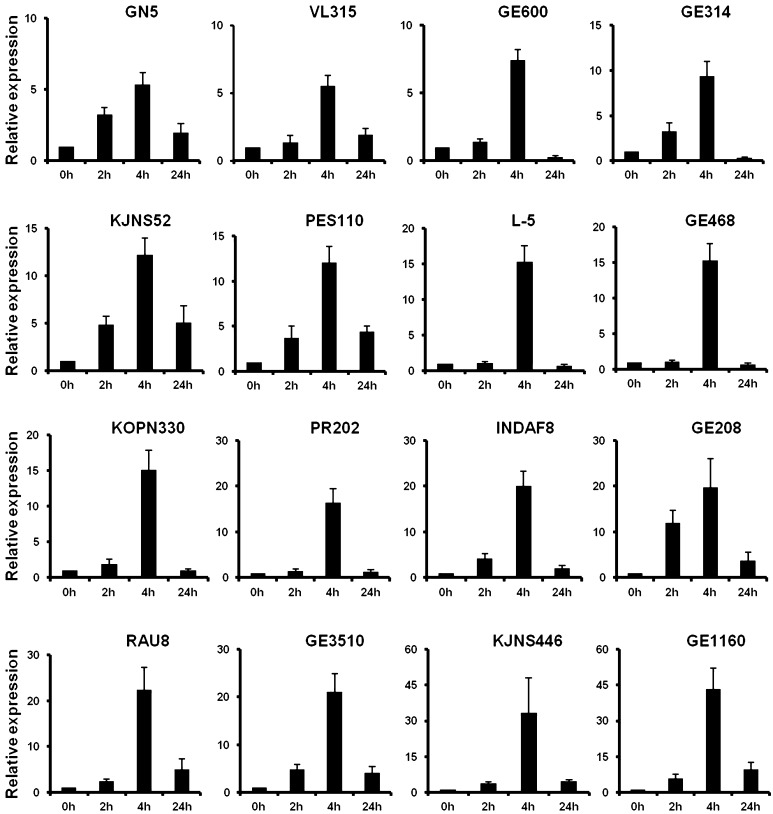
A total of 16 finger millet genotypes were subjected to heat stress at 42 °C and cDNA was prepared from isolated RNA (Fig. 5a, b). These cDNA were further used for expression analysis of EcbZIP17 by qRT-PCR. The results showed that the expression increases at 2 h but maximum expression was observed at 4 h stress in all the genotypes which was coming down at 24 h. In genotypes GN5, VL315, GE600, GE314, and KJNS52, the expression is ranging from 5.4- to 12-fold, whereas in genotypes L5, GE4683, KOPN330, PR202, INDAF8, GE208, RAU8, GE3510, KJNS446, and GE1160, the expression is ranging from 15- to 42-fold (Fig. 6). The highest expression was observed in the GE1160 genotype of 42-fold, whereas the lowest expression was observed at GN5 genotype of 5.4-fold with respect to 4 h stress.
Fig. 5.
Different genotypes of finger millet subjected to heat stress at 42 °C for 4 h (a). Gel image with isolated RNA from the heat stressed finger millet samples (b)
Fig. 6.
Expression analysis of EcbZIP17 in different finger millet genotypes subjected to heat stress at respective time periods
Discussion
Majority of transcription factors belonging to bZIP group regulates essential plant processes and protect plants from various biotic and abiotic stresses (Wang et al. 2011; He et al. 2012). In this study, we have isolated a heat stress responsive EcbZIP17 gene which belongs to group B of bZIP transcription factor family, members of which contains a bZIP domain, a transmembrane (TM) domain and protease sites (Jakoby et al. 2002). Literature shows that group B bZIPs are membrane-tethered transcription factors (MTTF) which remain in the membrane of ER under normal conditions. However, under stress conditions, proteolysis occurs resulting in activated ΔbZIP and the activated form moves to the nucleus to activate stress responsive genes. This plays a key role in multiple stress tolerance and the entire process is referred to as unfolded protein response (UPR) (Howell 2013). It was also observed that via endoplasmic reticulum MTTF bZIP activates brassinosteroid signalling pathway and promotes acclimation (Che et al. 2010). In rice, OsbZIP39 is a regulator of ER stress (Takahashi et al. 2012). Finger millet EcbZIP60 conferred abiotic stress tolerance through ER signalling pathway in tobacco (Babitha et al. 2015). ZmbZIP17 is a key regulator in ER quality control and ABA signalling in maize (Yang et al. 2013) and AtbZIP28 conferred heat stress response in Arabidopsis through ER pathway (Gao et al. 2008).
In this study, we isolated and characterized EcbZIP17 gene at sequencing and expression level in 16 finger millet genotypes. However, we did not observe any variation at nucleotide level across all the genotypes, not even in intron region of the gene. Expression analysis of EcbZIP17 in different genotypes of finger millet showed uniform heat responsive nature of this gene. Essential genes often code for basic cellular functions necessary for the viability of an organism and hence are conserved (Bergmiller et al. 2012). Since EcbZIP17 is highly conserved at nucleotide sequence, functional domains, and expression level under heat stress across the genotypes, we infer that it is a heat responsive gene in finger millet may playing an essential role in protein folding in ER and UPR under stress conditions like its homologs from Arabidopsis and maize.
Conclusion
Climate resilience and high nutritional value of finger millet makes it a selective crop for mining superior alleles. Here, we isolated a MTTF EcbZIP17 from finger millet and showed that the isolated EcbZIP17 gene is conserved at nucleotide as well as expression level among 16 finger millet genotypes. Expression analysis of this gene revealed its heat responsive nature.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We acknowledged All India Coordinated Research Project on Small Millets, ICAR, GKVK, Bangalore-560065 for providing seed material and ‘National Innovations in Climate Resilient Agriculture’ Project (NICRA), a flagship project of Indian Council of Agricultural Research (ICAR), New Delhi, India for providing financial support.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-0984-2) contains supplementary material, which is available to authorized users.
References
- Babitha KC, Ramu SV, Nataraja KN, Sheshshayee MS, Udayakumar M. EcbZIP60, a basic leucine zipper transcription factor from Eleusine coracana L. improves abiotic stress tolerance in tobacco by activating unfolded protein response pathway. Mol Breed. 2015;35:181. doi: 10.1007/s11032-015-0374-6. [DOI] [Google Scholar]
- Bergmiller T, Ackermann M, Silander OK. Patterns of evolutionary conservation of essential genes correlate with their compensability. PLoS Genet. 2012;8:e1002803. doi: 10.1371/journal.pgen.1002803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra D, Chandra S, Pallavi Sharma AK. Review of Finger millet (Eleusine coracana (L.) Gaertn): a power house of health benefiting nutrients. Food Sci Hum Wellness. 2016;5:149–155. doi: 10.1016/j.fshw.2016.05.004. [DOI] [Google Scholar]
- Che P, Bussell JD, Zhou W, Estavillo GM, Pogson BJ, Smith SM. Signaling from the endoplasmic reticulum activates brassinosteroid signaling and promotes acclimation to stress in Arabidopsis. Sci Signal. 2010;3:ra69. doi: 10.1126/scisignal.2001140. [DOI] [PubMed] [Google Scholar]
- Clewley JP, Arnold C. MEGALIGN. The multiple alignment module of LASERGENE. Methods Mol Biol. 1997;70:19–129. [PubMed] [Google Scholar]
- Combet C, Blanchet C, Geourjon C, Deleage G. NPS@: network protein sequence analysis. Trends Biochem Sci. 2000;25:147–150. doi: 10.1016/S0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- Deng Y, Srivastava R, Howell SH. Endoplasmic reticulum (ER) stress response and its physiological roles in plants. Int J Mol Sci. 2013;14:8188–8212. doi: 10.3390/ijms14048188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dida MM, Ramakrishnan S, Bennetzen JL, Gale MD, Devos KM. The genetic map of finger millet, Eleusine coracana. Theor Appl Genet. 2007;114:321–332. doi: 10.1007/s00122-006-0435-7. [DOI] [PubMed] [Google Scholar]
- Gao H, Brandizzi F, Benning C, Larkin RM. A membrane-tethered transcription factor defines a branch of the heat stress response in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105:16398–16403. doi: 10.1073/pnas.0808463105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao SQ, Chen M, Xu ZS, Zhao CP, Li L, Xu HJ, Tang YM, Zhao X, Ma YZ. The soybean GmbZIP1 transcription factor enhances multiple abiotic stress tolerances in transgenic plants. Plant Mol Biol. 2011;75:537–553. doi: 10.1007/s11103-011-9738-4. [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the Expasy server. In: Walker JM, editor. The proteomics protocols handbook. Totowa: Humana; 2005. pp. 571–607. [Google Scholar]
- Gull A, Ahmad NG, Prasad K, Kumar P. Technological, processing and nutritional approach of finger millet (Eleusine coracana)-a mini review. J Food Process Technol. 2016;7:8–11. doi: 10.4172/2157-7110.1000593. [DOI] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- He S, Shan W, Kuang J, Xie H, Xiao Y, Lu W, Chen J. Molecular characterization of a stress-response bZIP transcription factor in banana. Plant Cell Tissue Organ Cult. 2012;113:173–187. doi: 10.1007/s11240-012-0258-y. [DOI] [Google Scholar]
- Hittalmani S, Mahesh HB, Shirke MD, Biradar H, Uday G, Aruna YR, Lohithaswa HC, Mohanrao A. Genome and Transcriptome sequence of Finger millet (Eleusine coracana (L.) Gaertn.) provides insights into drought tolerance and nutraceutical properties. BMC Genom. 2017;18:465. doi: 10.1186/s12864-017-3850-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell SH. Endoplasmic reticulum stress responses in plants. Annu Rev Plant Biol. 2013;64:477–499. doi: 10.1146/annurev-arplant-050312-120053. [DOI] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Droge Laser W, Vicente Carbajosa J, Tiedemann J, Kroj T, Parcy F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7:106–111. doi: 10.1016/S1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- Kanakachari M, Solanke AU, Prabhakaran N, Ahmad I, Dhandapani G, Jayabalan N, Kumar PA. Evaluation of suitable reference genes for normalization of qPCR gene expression studies in Brinjal (Solanum melongena L.) during fruit developmental stages. Appl Biochem Biotechnol. 2016;178:433–450. doi: 10.1007/s12010-015-1884-8. [DOI] [PubMed] [Google Scholar]
- Kumar GR, Sakthivel K, Sundaram RM, Neeraja CN, Balachandran SM, Shobha Rani N, Viraktamath BC, Madhav MS. Allele mining in crops: prospects and potentials. Biotechnol Adv. 2010;28:451–461. doi: 10.1016/j.biotechadv.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Liu JX, Srivastava R, Howell SH. Stress-induced expression of an activated form of AtbZIP17 provides protection from salt stress in Arabidopsis. Plant Cell Environ. 2008;31:1735–1743. doi: 10.1111/j.1365-3040.2008.01873.x. [DOI] [PubMed] [Google Scholar]
- Liu J, Chen N, Chen F, Cai B, Santo SD. Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera) BMC Genom. 2014;15:281. doi: 10.1186/1471-2164-15-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lovell SC, Davis IW, Arendall WB, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by C alpha geometry: phi, psi and C beta deviation. Proteins: structure. Funct Genet. 2002;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- Nijhawan A, Jain M, Tyagi AK, Khurana JP. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008;146:333–350. doi: 10.1104/pp.107.112821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pudake RN, Mehta CM, Mohanta TK, Sharma S, Varma A, Sharma AK. Expression of four phosphate transporter genes from Finger millet (Eleusine coracana L.) in response to mycorrhizal colonization and Pi stress. 3 Biotech. 2017;7:17. doi: 10.1007/s13205-017-0609-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ. Recent advances in plant membrane—bound transcription factor research: emphasis on intracellular movement. J Integr Plant Biol. 2008;56:334–342. doi: 10.1111/jipb.12139. [DOI] [PubMed] [Google Scholar]
- Shobana S, Krishnaswamy K, Sudha V, Malleshi NG, Anjana RM, Palaniappan L, Mohan V. Finger Millet (Ragi, Eleusine coracana L.): a review of its nutritional properties, processing, and plausible health benefits. Adv Food Nutr Res. 2013;69:1–391. doi: 10.1016/B978-0-12-410540-9.00001-6. [DOI] [PubMed] [Google Scholar]
- Singh RK, Phanindra MLV, Singh VK, Sonam Raghavendrarao S, Solanke AU, Kumar PA. Isolation and characterization of drought responsive EcDehydrin7 gene from finger millet (Eleusine coracana (L.) Gaertn.) Indian J Genet. 2014;74:456–462. [Google Scholar]
- Sood S, Kumar A, Babu BK, Gaur VS, Pandey D, Kant L, Pattnayak A. Gene discovery and advances in finger millet [Eleusine coracana (L.) Gaertn.] genomics an important nutri-cereal of future. Front Plant Sci. 2016;7:1–17. doi: 10.3389/fpls.2016.01634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Kawakatsu T, Wakasa Y, Hayashi S, Takaiwa F. A rice transmembrane bZIP transcription factor, OsbZIP39, regulates the endoplasmic reticulum stress response. Plant Cell Physiol. 2012;53:144–153. doi: 10.1093/pcp/pcr157. [DOI] [PubMed] [Google Scholar]
- Tang N, Zhang H, Li X, Xiao J, Xiong L. Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiol. 2012;158:1755–1768. doi: 10.1104/pp.111.190389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyaya HD, Gowda CLL, Reddy VG. Morphological diversity in finger millet germplasm introduced from southern and eastern Africa. ICRISAT. 2007;3:1–3. [Google Scholar]
- Wang J, Zhou J, Zhang B, Vanitha J, Ramachandran S, Jiang SY. Genome-wide expansion and expression divergence of the basic leucine zipper transcription factors in higher plants with an emphasis on sorghum. J Integr Plant Biol. 2011;53:212–231. doi: 10.1111/j.1744-7909.2010.01017.x. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Tang N, Du H, Ye H, Xiong L. Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol. 2008;148:1938–1952. doi: 10.1104/pp.108.128199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YG, Lv WT, Li MJ, Wang B, Sun DM, Deng X. Maize membrane-bound transcription factor Zmbzip17 is a key regulator in the cross-talk of ER quality control and ABA signaling. Plant Cell Physiol. 2013;54:2020–2033. doi: 10.1093/pcp/pct142. [DOI] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER suite: protein structure and function prediction. Nat Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying S, Zhang DF, Fu J, Shi YS, Song YC, Wang TY, Li Y. Cloning and characterization of a maize bZIP transcription factor, ZmbZIP72, confers drought and salt tolerance in transgenic Arabidopsis. Planta. 2012;235:253–266. doi: 10.1007/s00425-011-1496-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.