SUMMARY
Outbreaks of multidrug-resistant bacteria present a frequent threat to vulnerable patient populations in hospitals around the world. Intensive care unit (ICU) patients are particularly susceptible to nosocomial infections due to indwelling devices such as intravascular catheters, drains, and intratracheal tubes for mechanical ventilation. The increased vulnerability of infected ICU patients demonstrates the importance of effective outbreak management protocols to be in place. Understanding the transmission of pathogens via genotyping methods is an important tool for outbreak management. Recently, whole-genome sequencing (WGS) of pathogens has become more accessible and affordable as a tool for genotyping. Analysis of the entire pathogen genome via WGS could provide unprecedented resolution in discriminating even highly related lineages of bacteria and revolutionize outbreak analysis in hospitals. Nevertheless, clinicians have long been hesitant to implement WGS in outbreak analyses due to the expensive and cumbersome nature of early sequencing platforms. Recent improvements in sequencing technologies and analysis tools have rapidly increased the output and analysis speed as well as reduced the overall costs of WGS. In this review, we assess the feasibility of WGS technologies and bioinformatics analysis tools for nosocomial outbreak analyses and provide a comparison to conventional outbreak analysis workflows. Moreover, we review advantages and limitations of sequencing technologies and analysis tools and present a real-world example of the implementation of WGS for antimicrobial resistance analysis. We aimed to provide health care professionals with a guide to WGS outbreak analysis that highlights its benefits for hospitals and assists in the transition from conventional to WGS-based outbreak analysis.
KEYWORDS: bioinformatics, intensive care units, next-generation sequencing, nosocomial infections, outbreak analysis, outbreak management, pathogen surveillance, point of care, whole-genome sequencing
INTRODUCTION
While several improvements have been made to limit the burden of health care-associated infections, outbreaks of especially-multidrug-resistant (MDR) bacteria still present a frequent threat to vulnerable patient populations in hospitals around the world (1). The EPIC II study, which assessed outcomes and prevalences of infections in 13,796 intensive care unit (ICU) patients worldwide, reported that 36% of ICU patients were infected with MDR bacteria, eventually leading to a doubling of their mortality rate compared to uninfected ICU patients (2). ICU patients are the patient group that is most vulnerable to bacterial infections due to their immune systems being compromised by, for instance, indwelling devices and severe underlying illness. In addition to the vulnerable nature of ICU patients, the prolonged overuse of broad-spectrum antibiotics during and after surgical procedures, inadequate nurse-to-patient ratios, and overcrowding lead to the unintended promotion of MDR bacteria and an eventual increase in the number of bacterial outbreaks in hospitals (3, 4). The increased vulnerability and consequent high mortality rates of infected ICU patients demonstrate the need for effective and standardized outbreak management protocols to be in place (5).
As part of most outbreak management protocols, several phenotypic and molecular methods for pathogen characterization are conventionally used to monitor and curb the spread of resistant bacterial pathogens in hospitals worldwide (6). However, conventional outbreak control approaches often fail to distinguish closely related outbreak strains or detect virulence/resistance features. This is due largely to the limited genomic resolution of conventional molecular methods and the target-specific nature of outbreak analysis approaches; e.g., during infections by antimicrobial-resistant organisms, genotypic tests are employed, which detect only antimicrobial resistance (AMR) genes but not virulence genes, which, if detected concurrently, can provide additional phylogenetic information and improve outbreak analysis (7). To overcome these caveats of conventional outbreak management, novel technologies that provide higher genomic resolution and full genetic information on the entire bacterial genome are needed. Whole-genome sequencing (WGS) can cover all these relevant genomic characteristics, but clinicians have long been hesitant to implement WGS in standard outbreak analysis protocols due to high costs and the cumbersome nature of early next-generation sequencing (NGS) technologies (8–10). Recent advances in sequencing technologies and analysis tools have rapidly increased the output and analysis speed as well as reduced the costs of WGS (11, 12). There is now an ever-increasing body of evidence showing that WGS can provide a fast and affordable outbreak analysis method with a markedly higher resolution than those of conventional methods (13–15). In several countries, such as the United States, Denmark, the United Kingdom, Germany, and The Netherlands, WGS-based pathogen typing is already in the trial phase for implementation as a routine tool for the monitoring and detection of MDR pathogens (16–19) as well as for the early detection of outbreaks (20–22). Still, one has to bear in mind that PCR-based techniques offer relatively cheap and fast typing of isolates and screening for gene functions using dedicated primer sets at a lower resolution.
A number of excellent reviews have covered next-generation sequencing technologies and analysis tools in great detail (23–27). Several important sequencing technologies are not discussed in our review, such as the 454 genome sequencer (Roche) (8), the Ion Torrent personal genome machine (Life Technologies) (9), and the Sequencing by Oligonucleotide Ligation and Detection (SOLiD) system (Applied Biosystems) (10), as they have been superseded by other sequencing technologies. Instead, we assessed the performances of today's most frequently used sequencing technologies as well as the latest developments in sequencing technologies. Furthermore, the performances of selected bioinformatics tools for assembly, genome characterization, comparative genomics, and phylogeny were reviewed. In an attempt to provide a representative overview of the vast number of bioinformatics tools to a broad audience, our analysis included both well-established and recently developed algorithms, which span over three different user interface types and require various levels of bioinformatics skills. Finally, we discuss the benefits and drawbacks of using the selected sequencing technologies and analysis tools and provide a future outlook for the real-world implementation of WGS-based outbreak analyses.
OUTBREAK DEFINITION
According to the Centers for Disease Control and Prevention (CDC), an outbreak is defined as “the occurrence of more cases of disease than expected in a given area or among a specific group of people over a particular period of time” (https://www.cdc.gov/). Instead of disease, one may also consider the state of carrying a specific pathogen, such as a multidrug-resistant Pseudomonas strain. An outbreak alert might be triggered by a cluster of patients colonized with the same drug-resistant Gram-negative bacterium (GNB) in an ICU ward (3). According to a study by Gastmeier et al., which reviewed the 2005 worldwide database of health care-associated outbreaks (https://www.outbreak-database.com/), outbreaks in neonatal ICUs are due mainly to Klebsiella spp. (20.3%) and Staphylococcus spp. (15.9%), with the majority of infections being bloodstream infections (62.7%) and gastrointestinal infections (20.7%) (28). In other ICUs, the majority of infections are due largely to Staphylococcus spp. (20.1%) and Acinetobacter spp. (15.9%), with the majority of infections being bloodstream infections (46.8%) and pneumonia (20.7%) (28). The majority of infection sources are reportedly unknown, followed by infections originating from patients, the environment, medical equipment, and health care personnel (28, 29).
CONVENTIONAL MOLECULAR CHARACTERIZATION METHODS
For many years, the large majority of clinical microbiology laboratories used several methods for characterizing bacterial strains, including serotyping (30, 31), antimicrobial susceptibility testing (32, 33), and mass spectrometry (MS)-based (34) methods that are still considered the gold standard of phenotypic characterization of pathogenic bacteria. In an extensive review, van Belkum et al. provide a detailed description of conventional phenotypic and molecular characterization methods (6). While conventional phenotypic characterization methods have proven to be successful in identifying and controlling outbreaks in ICUs, they all have the common disadvantage of being time-consuming and providing low taxonomic resolution (35, 36). In recent years, pathogen characterization has therefore moved to more sensitive genomic analysis techniques. The early beginnings of genomic analysis were made by the use of several genetic analysis tools that focus on small parts of the bacterial genome (6). In the focus of our review, the most frequently used non-amplification- and amplification-based genomic methods are described briefly.
Non-Amplification-Based Typing Technologies
Restriction fragment length polymorphism methods.
In restriction endonuclease analysis (REA), one of the first restriction fragment length polymorphism (RFLP) methods, a bacterial chromosome is subjected to a digestion step, where restriction enzymes cut the chromosome into smaller fragments, which are then separated by size via gel electrophoresis (37). Under a standardized protocol, this method is relatively fast, discriminatory, and easy to reproduce, yet the complex nature of the produced patterns makes interpretation of the results difficult and hampers data exchange between different research groups (6). To improve the interpretation of results, a combination of RFLP and ribotyping can be used, where, in addition to genome digestion, a second step is added, which hybridizes an rRNA gene-complementary probe to the genome fragments. Certain hybridization probes that are species specific can be used, such as during IS6110 typing, in which standardized typing of Mycobacterium tuberculosis can be achieved (38). However, despite these improvements, studies have shown that RFLP clusters lack discriminatory power and can be further subdivided by newer WGS-based typing methods (39, 40). The higher resolution of such WGS methods could enable clinicians to better distinguish outbreak strains from nonoutbreak strains.
Several other non-amplification-based methods are commonly used, such as DNA-DNA reassociation, which assesses the hybridization of DNA fragment pools to infer genetic distances between organisms (41), and plasmid typing, which distinguishes bacteria based on their unique profiles of plasmids (42).
Matrix-assisted laser desorption ionization–time of flight mass spectrometry.
Matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) MS (43) is a molecular typing technique that identifies bacterial isolates based on unique protein profiles. For detection, a protein spectrum is obtained and compared to a reference database of bacterial protein spectra to identify the isolate. MALDI-TOF MS has been established as a frequently used method for the identification of bacterial pathogens during routine screenings (44, 45) and for the distinction of bacterial strains during nosocomial outbreaks in intensive care units (46–48). For an extensive description of further applications of MALDI-TOF MS in microbiological diagnostics, the reader is referred to a review by Wieser et al. (49).
Recently, Schlebusch et al. described the complementary use of MALDI-TOF MS and WGS for the investigation of a vancomycin-resistant Enterococcus faecium (VRE) outbreak (50). That study highlighted the inconsistency of MALDI-TOF MS results based on potential biases in phenotypic typing data from various protein expression levels. Even though MALDI-TOF MS was able to distinguish outbreak strains with shorter turnaround times (TATs), WGS analysis provided far-higher discriminatory power, which ultimately allowed an improved understanding of transmission events. That study hence argued that in an outbreak scenario, MALDI-TOF MS could be used to complement WGS as a rapid initial analysis tool until WGS data are generated.
Pulsed-field gel electrophoresis.
Pulsed-field gel electrophoresis (PFGE) is a typing technique that differentiates bacterial isolates at the strain level. During PFGE, a fingerprint (pulsotype) of DNA fragments is generated on a gel and compared to a database, the extent of which can vary largely depending on the bacterial species, to identify the bacterial isolate (51).
A major disadvantage of this method is the inconsistency in results caused by the use of multiple standardized protocols and variations of restriction enzymes from the same or between different manufacturers (52). However, PFGE networks such as PulseNET present examples where the coordinated implementation of standardized workflows can result in the successful implementation of this technique at the national level (53).
Despite its widely accepted use as a highly sensitive typing method, PFGE is a laborious method due to its finicky sample preparation, long run time, and dependence on bacterial culture (51, 54). Even though the costs of PFGE are still approximately half of those associated with newer WGS-based typing methods (55), the superiority of WGS over PFGE in bacterial typing has been successfully demonstrated in analyses of bacterial transmission events. Several studies have shown the higher discriminatory power of WGS than of PFGE in identifying transmission events during outbreaks of methicillin-resistant Staphylococcus aureus (56) and Escherichia coli O157:H7 (57) infections.
Amplification-Based Typing Technologies
Multiple-locus variable-number tandem-repeat analysis.
The limitations of PFGE have led to the development of cheaper, faster, and more detailed PCR-based typing methods. Multiple-locus variable-number tandem-repeat (VNTR) analysis (MLVA) is a typing method that discriminates closely related bacterial strains based on their numbers of VNTRs. PCR primers are chosen to be outside the VNTR region, producing DNA fragments of various lengths depending on the number of repeats. PCR products are then analyzed through capillary electrophoresis to determine their size via the use of specific software. Results are usually reported as a string of numbers representing the VNTRs at each tested locus (58), allowing universal interpretation. One example of MLVA is spa typing, where strains of S. aureus are discriminated based on the staphylococcal protein A (spa) gene (59).
PCR-based MLVA was demonstrated to be a faster and more available alternative to PFGE, as it is able to discriminate between highly related bacterial strains. However, Bertrand et al. demonstrated that for clinical isolates of Salmonella enterica serovar Enteritidis, it was possible with other typing methods to further discriminate the most common MLVA profile identified into five phage subtypes (58). Hence, when investigations are performed on bacterial isolates with a highly common MLVA profile, the technique should be accompanied by complementary typing methods, such as WGS-based approaches, to achieve unique subtyping results and increased resolution. In fact, WGS-based typing has been shown to be less expensive, less labor-intensive, and of higher resolution for strain distinction than MLVA (60).
Multilocus sequence typing.
Multilocus sequence typing (MLST) is a typing technique that identifies bacteria based on sequence differences in housekeeping genes. MLST can be performed through either a single-gene sequencing or a WGS approach; a detailed description of the latter follows later in this review. For usually at least seven housekeeping genes, the sequence differences for a bacterial isolate are assigned a distinct allele. The alleles at each of the loci (genes) are described as the allelic profile or sequence type (ST). This ST can then be used as a barcode to differentiate isolates and establish evolutionary relationships via designated analysis tools (61).
MLST has been shown to be effective in identifying pathogenic bacterial strains with high resolution (62, 63); however, the high level of variation of housekeeping genes among different bacteria makes it possible to create MLST schemes only for bacterial pathogens that are highly related at the genus-to-species levels (61). MLST furthermore does not provide discrimination between variants of a single clone, which is relevant for asexual pathogens such as Bacillus anthracis and Yersinia pestis, which can make this method insufficient as an outbreak analysis tool for such pathogens (64). In organisms with considerable levels of recombination, the same MLST type may hide considerable biological diversity, which may result in inappropriate conclusions on the clonal nature of strains (65–67).
Virulence gene typing.
In addition to typing, PCR can be used to identify bacterial pathogens based on specific virulence factors such as toxins, adhesins, or capsules. As in PCR-based genotyping, species-specific virulence genes are assessed as PCR primer targets and amplified for the characterization of a pathogen in a sample (68–70). Traditional PCR detection of virulence genes has the disadvantage of being able to identify only one gene or species per reaction, which limits its use in high-throughput outbreak analyses. Multiplex PCR methods have hence been established to detect multiple species and genes in one sample with the use of multiple target-specific primers. The multiplex method is a well-established method for the fast and reliable detection of virulence genes and has been shown by several studies to be successful in detecting virulence, antibiotic resistance, and toxin (VAT) genes in Campylobacter species and virulence-associated genes in Arcobacter species, to name only a few examples (69, 71). However, limitations in resolution and the superiority of WGS over PCR-based detection of virulence genes at comparable TATs have been demonstrated (20). Therefore, WGS-based detection of virulence genes might be more suitable than PCR-based methods in outbreak situations where high-resolution detection of virulence determinants could lead to improved pathogenicity characterization and, consequently, outbreak control.
In addition to the methods described above, several other amplification-based methods are used for pathogen characterization, such as amplified rRNA restriction analysis, a modified RFLP method that analyzes the 16S rRNA gene (72); random amplified polymorphic DNA (RAPD) analysis, where PCR using arbitrary primers amplifies random DNA sequences to create a semiunique DNA fragment profile for isolate identification (73); and amplified fragment length polymorphism (AFLP), a PCR method that amplifies restriction fragments from genomic DNA digests to create DNA fingerprints for the identification of bacterial isolates (74).
NEED FOR WGS FOR OUTBREAK ANALYSIS
The above-described amplification-based and non-amplification-based methods are used to investigate only small fragments of the bacterial genome, which limits these approaches to species-dependent protocols. WGS-based typing of bacterial pathogens includes mobile genetic elements and could provide unprecedented resolution in discriminating even highly related lineages, thereby obviating the use of species-dependent protocols. By sequencing the entire genome (chromosome and mobile genetic elements), WGS immediately provides information on pathogen detection and identification, epidemiological typing, and drug susceptibility, which is crucially important information that in conventional outbreak management is achievable only through the use of multiple methods.
Of additional importance is the fact that resistance/virulence genes detected via WGS might not be expressed under conditions of phenotypic testing in vitro or, for that matter, in vivo. In particular, there have been reports of the “in vivo-only” expression of virulence gene promoters in S. aureus and Salmonella enterica serovar Typhimurium (75, 76). The detection of such pathogenicity features via WGS could help clinicians identify potential nosocomial transmission events earlier and manage bacterial outbreaks before conventional phenotypic tests can detect them.
Despite the concerns of high operational costs associated with WGS, which are frequently voiced by health care professionals (77–79), WGS pipelines could potentially reduce overall costs for hospital practices through savings of indirect costs. Of note is a recent study by Mellmann et al., which assessed the performance of a novel WGS typing pipeline for monitoring bacterial transmission in a multibed-room, tertiary hospital in Germany (55). That study successfully demonstrated that WGS typing was more precise in excluding the majority of bacterial isolates from nosocomial transmission clusters than conventional typing methods such as PFGE. These results prompted a reduction in the number of patient isolation procedures over a 6-month period, which in turn enabled cost savings of more than $230,000, largely due to reduced workloads and indirect savings from the avoidance of blocked beds.
METHODS
For this review, sequencing technologies were assessed based on sequence coverage, output quantity, consumables and instrument costs, read length, number of reads per run, cost per gigabase, run time, and error rates. Sequencing coverage describes the average number of aligned read fragments that cover a specific nucleotide in the reconstructed sequence and is calculated by dividing the total output by the target genome size and dividing this result by the number of samples per run. To provide examples of coverage for each sequencing technology, this review calculated coverage based on the genome size of S. aureus strain MRSA252. Presented coverages can then be compared to reference values of 35-fold to 50-fold for small genomes, as previously recommended (80). Output describes the amount of sequence information produced per sequencing run. Error rates were analyzed from reported benchmarks of “raw” sequence data after a sequencing run was completed. As possible improvements in error values through data cleaning can vary highly depending on data sets, sequencing technology, and sample preparation, etc., we decided not to mention error values after additional improvement of the data. By doing so, this review aims to present the reader with an unbiased picture of the machine performance of each technology described.
Tools for the analysis of WGS data were divided into five groups: assembly, genome characterization, comparative genomics, phylogeny, and complete outbreak analysis software suites. Assembly tools were assessed based on sequencing technology, computational requirements, speed, and assembly quality. Computational requirements were based on the reported random-access memory (RAM) usage for various benchmarking data sets, speed was based on the reported run time for various benchmarking data sets, and assembly quality was based on reported N50 values and percentages of identity for various benchmarking data sets. In a given set of assembled contigs, the N50 value describes the base pair length of the shortest contig in an assembly, such that the sum of all contigs of longer or identical lengths results in a minimum of half the total base pair length of all contigs of the original assembly. Genome characterization tools were assessed mainly based on input/output types. Tools for comparative genomics and phylogeny estimations were assessed based on input/output type, run time, and topology score/accuracy. The complete outbreak analysis software suites were assessed based on RAM compatibility, the number of schemes, price, and run time.
SEQUENCING TECHNOLOGIES
Ever since the first report of a complete bacterial genome sequence in 1995 (81), sequencing technologies have rapidly improved. As presented in Table 1, second-generation sequencing platforms allow whole bacterial genomes to be sequenced within hours, while third-generation sequencing platforms, that provide longer reads and additional information, such as methylation sites, with even higher speed have been developed (82). This review assesses the performance of popular sequencing platforms as well as emerging state-of-the art technologies that were available at the time of writing of this review. The results of the performance assessment are shown in Table 1.
TABLE 1.
Performance analysis of sequencing platformsa
| Platform | Read length (bp) | Output (Gb) | Coverageb | Run time (h) | No. of reads | Cost per Gb ($) | Consumables cost ($) | Instrument cost ($) | Error rate | Dimensions (width × depth × ht) (cm) | Source(s) (reference[s]) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sequencing by synthesis | |||||||||||
| Illumina MiniSeq Mid Output | 2 × 150c | 2.1–2.4c | 8.6 | 17c | 14 million–16 millionc | 2,584–2,953d | 6,201c | 55,411c | 0.1% in >80% of base callsc | 45.6 × 48 × 51.8c | Illumina |
| Illumina MiniSeq High Output | 1 × 75c | 1.7–1.9c | 6.8 | 7c | 22 million–25 millionc | 3,264–3,648d | 6,201c | 55,411c | 0.1% in >85% of base callsc | 45.6 × 48 × 51.8c | Illumina |
| 2 × 75c | 3.3–3.8c | 13.6 | 13c | 44 million–50 millionc | 1,632–1,879d | 6,201c | 55,411c | 0.1% in >85% of base callsc | 45.6 × 48 × 51.8c | Illumina | |
| 2 × 150c | 6.6–7.5c | 26.9 | 24c | 44 million–50 millionc | 827–940d | 6,201c | 55,411c | 0.1% in >80% of base callsc | 45.6 × 48 × 51.8c | Illumina | |
| Illumina MiSeq Reagent kit v2 | 1 × 36c | 0.54–0.61c | 2.2 | 4c | 12 million–15 millionc | 7,946–8,976d | 4,847c | 108,244c | 0.1% in >90% of base callsc | 68.6 × 56.5 × 52.3c | Illumina |
| 2 × 25c | 0.75–0.85c | 3.1 | 5.5c | 24 million–30 millionc | 5,702–6,463d | 4,847c | 108,244c | 0.1% in >90% of base callsc | 68.6 × 56.5 × 52.3c | Illumina | |
| 2 × 150c | 4.5–5.1c | 18.3 | 24c | 24 million–30 millionc | 950–1,077d | 4,847c | 108,244c | 0.1% in >80% of base callsc | 68.6 × 56.5 × 52.3c | Illumina | |
| 2 × 250c | 7.5–8.5c | 30.5 | 39c | 24 million–30 millionc | 570–646d | 4,847c | 108,244c | 0.1% in >75% of base callsc | 68.6 × 56.5 × 52.3c | Illumina | |
| Illumina MiSeq Reagent kit v3 | 2 × 75c | 3.3–3.8c | 13.6 | 21c | 44 million–50 millionc | 1,362–1,568d | 5,174c | 108,244c | 0.1% in >85% of base callsc | 68.6 × 56.5 × 52.3c | Illumina |
| 2 × 300c | 13.2–15c | 53.8 | 56c | 44 million–50 millionc | 345–392d | 5,174c | 108,244c | 0.1% in >70% of base callsc | 68.6 × 56.5 × 52.3c | Illumina | |
| Illumina NextSeq 500 Mid Output | 2 × 75c | 16.3–20c | 71.8 | 15c | <260 millionc | 318–391d | 6,369c | 266,835c | 0.1% in >75% of base callsc | 53.3 × 63.5 × 58.4c | Illumina |
| 2 × 150c | 32.5–39c | 140 | 26c | <260 millionc | 163–196d | 6,369c | 266,835c | 0.1% in >80% of base callsc | 53.3 × 63.5 × 58.4c | Illumina | |
| Illumina NextSeq 500 High Output | 1 × 75c | 25–30c | 107.7 | 11c | <400 millionc | 312–374d | 9,347c | 266,835c | 0.1% in >80% of base callsc | 53.3 × 63.5 × 58.4c | Illumina |
| 2 × 75c | 50–60c | 215.3 | 18c | <800 millionc | 156–187d | 9,347c | 266,835c | 0.1% in >80% of base callsc | 53.3 × 63.5 × 58.4c | Illumina | |
| 2 × 150c | 100–120c | 430.6 | 29c | <800 millionc | 78–93d | 9,347c | 266,835c | 0.1% in >75% of base callsc | 53.3 × 63.5 × 58.4c | Illumina | |
| Single-molecule real-time sequencing | |||||||||||
| Pacific Biosciences RS II P6-C4 chemistry | >20,000c | 8–16e | 57.4 | 0.5–4c | 55,000c | 250–500d | 4,000e | 695,000 | 14% errors per base | 203.0 × 90.0 × 160.0c | PacBio, AllSeqf (89) |
| Pacific Biosciences Sequel system | >20,000c | 80–160e | 574.2 | 0.5–6c | 370,000c | 70–140d | 11,200e | 350,000 | 14% errors per base | 92.7 × 86.4 × 167.6c | PacBio, AllSeqf (89) |
| Oxford Nanopore MinION Mk1 (1D) | >882,000 | 10–20c | 71.8 | 1.67–72c | 138,000 | 49.95–99.9d | 999c | 1,000c | 12% errors per base | 10.5 × 3.3 × 2.3c | Oxford Nanopore Technologies, Loman Labsg (231) |
| Oxford Nanopore MinION Mk1 (2D) | >882,000 | 10–20c | 71.8 | 1.67–72c | 138,000 | 49.95–99.9d | 999c | 1,000c | 15% errors per base | 10.5 × 3.3 × 2.3c | Oxford Nanopore Technologies, Loman Labsg (231, 232) |
| Oxford Nanopore PromethION single flow cell | <300,000c | 233c | 836.2 | 1.67–>72c | 26 millionc | NA | NA | 135,000 (PEAP)c | NA | 44.0 × 24.0 × 40.0c | Oxford Nanopore Technologies |
| Oxford Nanopore PromethION 48 flow cells | <300,000c | 11,000c | 3,9475.8 | 1.67–>72c | 1.25 billionc | NA | NA | 135,000 (PEAP)c | NA | 44.0 × 24.0 × 40.0c | Oxford Nanopore Technologies |
All quantitative performance measures were taken from previously reported data, as indicated. Consumables costs were calculated as follows: Illumina costs included PhiX Control kit v3, the Nextera XT DNA sample preparation kit (96 samples)/Nextera DNA library preparation kit (96 samples), and Nextera XT Index kit v2 (96 indexes and 384 samples), the highest-output reagent kit. PEAP, PromethION Early-Access Program; NA, no data available.
Calculated for 96 samples and the genome size of S. aureus strain MRSA252 (2,902,619 bp).
Manufacturer's data.
Estimated calculation for consumables.
For 16 SMRT cells.
Illumina
Principle of technology.
The Illumina sequencing platforms use fluorescently labeled nucleotides (deoxynucleoside triphosphates [dNTPs]) to determine the genetic sequence of DNA fragments. Here we focus on three Illumina model series: MiniSeq, the smallest, most affordable Illumina sequencer; MiSeq, a simple system for rapid sequencing with relatively low outputs; and NextSeq, a midsized, flexible system with options for high- and mid-range outputs.
The Illumina sequencing-by-synthesis (SBS) technology begins with several library preparation steps (83). Initially, purified sample DNA is fragmented by either mechanical shearing, e.g., via sonication, or enzymatic shearing, e.g., via transposases. Unique adaptor sequences (and, optionally, barcodes) are then ligated to either end of the DNA fragments and loaded onto a reagent cartridge that is inserted into the sequencer. The sequencer then loads the mix of reagents and DNA fragments into a solid-surface flow cell that is coated with primers complementary to the adaptor sequences. The ligated fragment ends then bind to the cell surface, and a DNA polymerase amplifies the fragments to produce several copies of the initial DNA fragment, called clusters. Next, four different fluorescently labeled nucleotides (A, C, G, and T) are added to the flow cell and incorporated by a polymerase into a new DNA strand one base at a time. The MiniSeq and NextSeq systems use a two-fluorophore system, instead of the four-fluorophore system used by the MiSeq system (23). After a wash step, the fluorescence of incorporated nucleotides is imaged by using one of four different imaging channels. Next, the fluorescent dyes are cleaved off and washed away, and the process is repeated. The sequencer documents the color changes after nucleotide addition to construct the genetic sequence of the DNA clusters. Either results can be analyzed as single-end reads or a second strand can be synthesized, and the process is repeated for paired-end reads. Paired-end reads provide more sequencing information but increase the sequencing cost and time needed for sequencing.
Specifications.
Whereas enzymatic reactions take very little time, the major contributor to run time is the imaging of the flow cell. Illumina has reduced the run time of previous models considerably by reducing the imaged surface area on the flow cell. As shown in Table 1, total run times, including cluster generation, sequencing, and base calling, can hence be reduced on the Illumina MiSeq system to 4 h and 56 h at the lowest-output (reagent kit v2) and highest-output (reagent kit v3) settings, respectively. However, with a decrease in the imaged surface area, the total number of generated data points per run decreases, which in turn increases the sequencing cost per nucleotide considerably (24).
On the fastest setting, the MiSeq system (reagent kit v2) can produce a minimum of 0.54 to 0.61 Gb of data with a single-end read length of 36 bp. On the more powerful NextSeq 500 system, a data output of 100 to 120 Gb can be achieved in the highest-output mode with a paired-end read length of 150 bp.
The average sequencing cost presented here is either taken from the literature or estimated based on the listed prices for consumables and output by the manufacturer, as indicated in Table 1. Most Illumina sequencing machines require a PhiX DNA control kit, a DNA library preparation kit, an indexing primer kit to allow the sequencing of up to 96 pooled samples, and a reagent kit. The sequencing costs per gigabase decrease with higher total outputs and hence start from $7,946 to $8,976/Gb with the MiSeq system (reagent kit v2, 1- by 36-bp read length) and can be decreased to around $78 to $93/Gb with the NextSeq 500 system (high output, 2- by 150-bp read length), the latter of which is the lowest range of sequencing costs per gigabase of the sequencers described in this study. Here it must be noted that multiple bacterial genomes can be run on the Illumina sequencers at a time, which reduces the costs per genome accordingly. As shown in Table 1, Illumina sequencers are offered at competitive instrument prices compared to those of other technologies, such as those of Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (ONT). With prices ranging from $55,411 to $266,835 for the Illumina MiniSeq and the Illumina NextSeq 500 systems, respectively, instrument costs are lower than those of the PacBio system but well above those of the cheapest ONT sequencers. The relation between instrument cost and other parameters, such as instrument footprint, is an important aspect to consider when evaluating the costs of WGS infrastructures for specific hospital needs.
On Illumina systems, error rates in base calling are predicted by a quality score. A quality score of 30 (Q30) predicts an error rate of 0.1 or an error of 1 in 1,000 base callings. The MiSeq system (reagent kit v2) achieves the highest quality score, 0.1% for >90% of base callings, and the MiSeq system (reagent kit v3) produces the lowest score, 0.1% for >70% of base callings.
The Illumina platforms have already been used for pathogen detection during outbreaks, and several studies have demonstrated their applicability and superiority over conventional methods in terms of outbreak control in clinical settings. A study by McGann et al. used WGS to study an outbreak of VRE that occurred among three ICU patients at a tertiary care hospital in Honolulu, HI (84). TATs for the Illumina MiSeq sequencer were determined to assess its applicability in a clinical setting during outbreaks. The initial epidemiological assessment was based on the timeline of the outbreak and suggested linear nosocomial transmission of the outbreak pathogen from a source patient (patient A) to a second patient (patient B) and, consequently, to a third patient (patient C). However, in contrast to the initial assessment, sequence data generated on the Illumina MiSeq system revealed that isolates of patient A differed from the isolates of the two other patients (patients B and C) by one single nucleotide polymorphism (SNP). This indicated that instead of the initially suspected linear transmission route, two separate events of transmission from patient A to both patients B and C most likely occurred. WGS therefore improved the understanding of the outbreak transmission network, which, in retrospect, could have potentially enhanced the outbreak control response at that time. The sequencer provided superior resolution with a TAT, including overnight culturing, of 48.5 h, which would allow a faster and more comprehensive response by infection control teams than with conventional detection methods with TATs of several weeks (15).
Another evaluation of the use of WGS for outbreak surveillance was recently conducted by Kwong et al. in the context of Listeria monocytogenes surveillance in Australia (60). That study compared the performance of WGS via the Illumina NextSeq or MiSeq system to those of conventional typing methods, including binary typing, PCR serotyping, MLST, MLVA, and PFGE. Besides being highly concordant (>99%) with results of binary typing, MLST, and serotyping, WGS enabled the identification of separate nested clusters among isolate groups that were undetectable with conventional methods. During additional routine epidemiological surveillance over a 12-month period, WGS allowed higher resolution in linking point source outbreaks than conventional typing. Based on these results, Kwong and colleagues were able to develop a nationwide risk-based alert system for WGS data to inform epidemiologists of sequence similarities and possible events of transmission of bacterial pathogens at discriminatory powers far superior to those with conventional typing-based surveillance.
Pacific Biosciences
Principle of technology.
While Illumina sequencers have proven their accurate performance, there are limitations in their short reads, creating problems with the determination and assembly of complex genomic regions. PacBio's third-generation sequencing platforms, the Sequel system and RSII, aim to solve this issue by implementing single-molecule real-time (SMRT) sequencing (85). The SMRT technology achieves this in two main steps. First, a so-called SMRT bell is generated by ligating both ends of a double-stranded target DNA with hairpin adaptors. The SMRT bell is then loaded onto a SMRT cell that contains a number of microscopic chambers, called zero-mode wave guides (ZMWs), that act as a detection space during sequencing. As the SMRT bell is loaded onto the cell, its hairpin adaptor binds to an immobilized DNA polymerase at the bottom of the ZMW. Next, fluorescently labeled nucleotides (A, C, G, and T) are added to the cell. As the polymerase begins to incorporate labeled nucleotides into a new DNA strand, the fluorescent labels are cleaved off and produce light pulses of emission spectra unique to each base. The light pulses are detected by a laser beam and recorded in real time to determine the nucleotide sequence as a continuous long read (CLR) (86). With this technology, it is possible to simultaneously detect thousands of single-molecule sequencing reactions at high speeds. Whereas the individual light signals are recorded in real time, the data cannot be observed in real time unless the run is stopped for observation.
Specifications.
Due to the lack of amplification, SMRT sequencing makes the PacBio sequencers some of the fastest on the market, with total run times of 0.5 to 4 h on the RSII (P6-C4) system. This makes the technology extremely valuable for outbreak analyses, where quick identification leads to faster treatment and, eventually, reductions in costs and loss of life.
As shown in Table 1, the output of PacBio systems is one of the lowest available on the market, with only 500 Mb to 1 Gb per SMRT cell on the RSII (P6-C4) system and 5 to 10 Gb per SMRT cell on the Sequel system. However, as indicated in Table 1, both the RSII and Sequel systems allow the running of up to 16 SMRT cells at once, which increases total outputs. The low output is due mainly to the focus on long reads for genome assembly, making it possible to achieve read lengths of >20 kb.
The sequencing costs per gigabase for PacBio sequencers are comparatively cheap, at $250 to $500 for the RSII (P6-C4) system and $70 to $140 with the Sequel system. However, the sequencers are expensive, at $695,000 for the RSII (P6-C4) system and $350,000 for the Sequel system, making PacBio technology one of the costlier options for clinical outbreak analysis.
One error specific to this technique is that during DNA replication in the ZMW, detection of nucleotides that are dwelling long enough at the active site of the polymerase can occur without these nucleotides actually being incorporated into the new DNA strand. These errors accumulate during the sequencing run and increase the overall error rate of the final read (87). Whereas the SMRT sequencing technique allows some of the longest reads available today, the small number of reads per run and technique-specific errors increase the error rate to around 14% of all bases read during a sequencing run to be falsely identified.
As the PacBio sequencers are designed to provide exceptionally long reads, they are especially well suited for the de novo assembly of reference genomes from outbreak strains. In a comparative analysis of MDR Acinetobacter baumannii (MDRAB) outbreaks, Kanamori et al. used NGS to investigate a 3-year outbreak between 2007 and 2010 at a large academic burn center at a hospital in North Carolina (88). That study used a PacBio RSII system to sequence the genome of an isolate from the first detected case and assemble a draft genome in order to compare isolates against an outbreak-specific reference genome. By utilizing the high sequencing speed and long reads, that group was able to quickly provide a case-specific reference genome to analyze detailed phylogeny and transmission events. With this approach, the use of generic reference genomes was avoided, as they may have masked small evolutionary differences between outbreak isolates.
In October 2015, PacBio introduced the Sequel system, its newest sequencing platform (89). With 1 million ZMWs per SMRT cell, PacBio claims that the Sequel system will deliver up to seven times more reads at less than half the instrument cost and with a considerably smaller instrument size than its predecessor, which is the size of a laboratory bench (90). At the time of this review, no studies on the use of the Sequel system in a clinical setup have been reported.
Oxford Nanopore Technologies
Principle of technology.
Another sequencing technology that enables single-molecule sequencing is ONT technology. It is sometimes referred to as fourth-generation sequencing, as it is capable of single-molecule sequencing but does not rely on sequence replication (91). At the heart of the technology is a protein nanopore that is inserted into an electrically resistant polymer membrane. The membrane is connected to an electrical current, which flows through only the aperture of the nanopore. For sequencing, complexes of DNA strands and processive enzymes are added to the membrane and bind to the nanopore. As single DNA molecules pass through the nanopore, they cause characteristic disruptions in the electrical current. By measuring variations in the current flowing through the pore, individual nucleotides can be identified based on these specific disruptions. If DNA strands are prepared with a hairpin structure at the opposite end, the nanopore can read both DNA ends in one continuous read, which enables higher-quality reads and reduces overall error rates. The nanopore will proceed to read more DNA molecules until the pore life span is exhausted or until a desired sequence coverage or mutation is detected in real time and the run is terminated by the user. The ability to analyze data in real time presents a major advantage of the ONT system in clinical scenarios, where fast detection of specific mutations can provide epidemiological information, such as the relatedness of outbreak strains or AMR and virulence genes, that directly impacts initial management decisions during hospital outbreaks (92).
ONT currently offers two sequencers for commercial use. The MinION Mk1 system is the first pocket-sized, real-time sequencer and enables DNA, RNA, and protein analyses. It can be connected to a laptop via a USB 3 connection and enables sequencing in virtually any working environment. The PromethION system is a benchtop sequencer that utilizes the same technology with a higher output. It provides docking stations for 48 individual flow cells, allowing the parallel sequencing of 144,000 nanopores at once. At the time of this review, the PromethION was available only with a subscription for early access.
As this review focuses on the newest flow cell technologies, R9 and one early-access-only platform, only a limited number of studies on machine performance was found. Most performance specifications advertised by ONT were therefore used.
Specifications.
ONT advertises run times until sufficient coverage is achieved. However, the flow cell lifetime limits possible run times from 1 min to 72 h. Nevertheless, the flexibility to choose the end of each run in real time presents an advantage over other technologies, which enables the optimal use of run time and sequencing capabilities.
As shown in Table 1, at the default run time of 48 h and in the fastest mode, the maximum outputs are 20 Gb on the MinION system and up to a theoretical maximum of 11 Tb on the PromethION system, which would make them the sequencing platforms with the highest outputs currently on the market. The high output is due mainly to the ability to read DNA fragments at a near-original input length and the long run time per chip. Nevertheless, output data for the PromethION system are presented as advertised by ONT and have not been confirmed by any independent experimental benchmark study to date.
Due to the quick preparation and low reagent costs, the MinION system presents one of the cheapest options for WGS to date, with sequencing costs of $49.95/Gb and an instrument cost of $1,000. At the time of this review, no data on reagents or sequencing costs for the PromethION system were available.
Despite its many advantages in cost, run time, and output, the ONT system is still a technology under development. System-specific errors and a lack of standard protocols produce an inconsistent quality of reads and lead to high error rates of up to 15% per base. Another disadvantage is the inherent sensitivity of biological nanopores to changes in experimental conditions, such as the salt concentration, pH, and temperature. Nevertheless, with the introduction of the PromethION system and further improvements in chip technology, such as solid-state synthetic nanopores, error rates are advertised to be close to 1% per base.
With its short run times and long real-time reads, the ONT system is best suited for rapidly identifying and distinguishing outbreak strains. In a study on foodborne outbreaks of Salmonella, Quick et al. assessed the performance of the MinION platform in sequencing an outbreak strain and a nonoutbreak strain of Salmonella enterica (92). During an initial 3-week outbreak in a UK hospital, that study first sequenced initial outbreak isolates on the Illumina MiSeq and HiSeq 2500 systems to assemble de novo draft genomes for reference use and the general detection of transmission events. An outbreak strain and a nonoutbreak strain, previously identified on the MiSeq instrument, were then chosen for the assessment of the MinION system. The results showed that the MinION system allowed confident species-level assignment within 20 min and serotype-level assignment within 40 min. In <2 h, the real-time sequencing system achieved differentiation between the outbreak-causing and nonoutbreak strains (92). That study demonstrated that in combination with other sequencing technologies for de novo assembly, the ONT system is able to rapidly offer reliable clinical information during outbreaks while providing real-time sequencing insight. The potential of the MinION platform was also illustrated by a recent study in which this sequencer was used to directly identify pathogens, and the resistance genes that they acquired, from clinical urine samples (93). In addition, the MinION system was also used to rapidly map the reservoir of antibiotic resistance genes in the gut microbiota of a critically ill patient (94). At the time of this review, no studies on the use of PromethION for outbreak analysis have been reported.
Read Length, Read Depth, and Error Rate in Perspective
Illumina sequencers are very popular and, as mentioned above, deliver high-quality bases and very high sequencing capacities although with shorter reads. The upside of these sequence data is that assembly is straightforward. However, the generated contigs tend to end at either paralogous genes (genes copied in the same genome) or repetitive elements. From an outbreak analysis perspective, this might not be a big problem, as phylogenetic trees are not reconstructed based on repetitive elements and the probability of pathogenic functionality not being represented in contigs is low. Nonetheless, from a comparative genomics perspective, one might be interested in genomic rearrangements and operons, which might be affected by these smaller contigs.
Single-molecule sequencers deliver longer reads but still suffer from lower read quality and lower throughput. Longer reads result in longer contigs, allowing more straightforward comparative genomics. However, contigs might still contain erroneous bases, potentially influencing the phylogenetic signal, which is based on SNPs present in coding regions.
WGS OUTBREAK ANALYSIS TOOLS
In the first step of WGS outbreak analysis, a completed sequencing run produces fragmented DNA sequence reads for multiple outbreak samples. Sequencing data need to be separated to obtain one file per sample or, for paired-end sequencing, two files per sample. In order to extract essential genomic information from these sequence reads, several analysis steps can be applied. Assembly tools are first used to assemble fragmented reads into larger contigs that can be constructed into near-complete genomes. From this assembled DNA sequence, tools can be applied for genome characterization. This characterization is achieved by determining the bacterial identity of the sample, annotating genes, and identifying genes of clinical importance, such as AMR and virulence genes. To determine the relatedness between outbreak strains and pinpoint the source of the outbreak, comparative genomics tools can be applied, which determine genomic differences and similarities between strains. By utilizing the analysis data up to this point, phylogeny tools are implemented to establish detailed networks of transmission between different patients and ultimately inform appropriate patient isolation protocols that could aid in the control of an outbreak.
Web-Based Tools
Web-based tools can be accessed through the Internet, and it is possible to use them within a clinical setting. With access to sequencing data and a stable Internet connection, these tools can be included in daily practice. The drawback of Web-based tools is that in the case of either server failure at the host side or large, undocumented changes made to the server, the utilization of these tools becomes impossible. If, in this case, the clinician is relying on these tools, the outcome will be delayed, which could eventually lead to increased costs for the hospital and may affect patient outcomes. Additional drawbacks of Web-based tools may lie in the unwillingness of hospital laboratories to share patient data with other groups, a prerequisite for updated databases, and the fact that performing analyses via Web-based tools often requires more time than local analyses. Finally, the use of Web-based tools bears a constant risk of compromised data on unsecure servers. Hence, the potential loss of confidential patient information might prompt hospitals to opt for a local user interface instead of Web-based tools.
Command Line Tools
Nearly all outbreak analysis tools are available as so-called command line tools that can be used free of charge. To use this type of tool, bioinformatics expertise and access to Unix-based computers are needed. As not every clinical microbiology laboratory would have access to these kinds of computers, Unix-based tools might be of variable relevance in a given setting. Nevertheless, if access to such expertise is available, the fast development and accessibility of such tools would prove to be of high value to the clinic. The presence of an experienced bioinformatician would therefore provide a great advantage, especially in a more research-driven clinical laboratory. For the optimal use of command line-based tools, installation on a Linux or Mac machine is preferable. Alternatively, tools could also be installed on a Windows 10 machine using the Windows Subsystem for Linux or by installing a virtual machine. One example of a virtual machine is Bio-Linux, which contains a suite of various bioinformatics tools and can be run either as a stand-alone operating system or “live” from a DVD or USB stick (http://www.environmentalomics.org/bio-linux/). The requirements for certain analysis steps can require considerable amounts of computer resources, and therefore, computers with multiple cores and hundreds of gigabytes of RAM or access to a computing cluster is highly advisable.
Complete Analysis Software Suites
Complete analysis software suites have the benefit of operating on a very user-friendly graphical user interface (GUI) and therefore seem ideal for clinicians to use in combination with practical routines. The use of these suites often needs little to no bioinformatics knowledge (95). Some packages are able to perform only a small fraction of all WGS outbreak analysis steps, whereas others are able to perform all steps in a single suite. For ease of use, a single suite that includes all needed tools and methods would fit best into daily routines, yet these all-in-one solutions come with a large price tag. A computer with multiple cores and a large amount of RAM is needed for the optimal performance of these tools, and the developers should be consulted for individual system requirements. One inconvenience of these suites is the fact that clinicians will be trained in how to use the packages while not knowing how the underlying algorithms and methods work. This could lead to misinterpretations of results or unreliable outcomes due to a lack of competencies in troubleshooting and system maintenance. To avoid such problems, it is important that staff or collaborators who have a deeper understanding and more knowledge of the underlying algorithms and methods are present. Nearly all algorithms and methods used in commercial suites are also available as free-to-use, command line versions.
What follows are detailed descriptions of Web-based tools, command line tools, and complete analysis software suites for the various steps of WGS outbreak analysis (assembly, genome characterization, comparative genomics, and phylogeny).
Assembly
Once DNA fragments are sequenced as reads in FASTQ or BAM format (for PacBio sequencers), an assembly algorithm is implemented to compile reads into larger sequences (contigs) that eventually represent a genome. Whereas it is desirable to assemble reads into contigs that are identical to the original genome sequence, this is close to impossible during short-read sequencing due to the presence of long repeat regions. Repeat regions in a target genome can be significantly longer than sequence reads and hence limit the correct assembly of these regions to the maximum read lengths produced by a given sequencing technology. The use of paired-end or long reads partially or completely overcomes this limitation.
By comparing contigs to a reference sequence, differences in the contig sequence can be found, which originate from either assembly errors or biological differences. An indel is a group term for an insertion or a deletion in a contig, where a short nucleotide sequence is either added or deleted at a specific position, respectively, compared to the reference sequence. Another error can occur where a contig aligns with the reference sequence at all but one nucleotide position, where a mismatch has occurred.
Based on these errors, an assembly problem (AP) was defined by Boisvert et al. as a criterion to assess the quality of assemblies (96). With a given group of reads, the AP arises from assembling contigs in such a way that (i) the number of contigs is minimal, (ii) the extent of genome coverage is maximal, and (iii) the number of assembly errors is minimal.
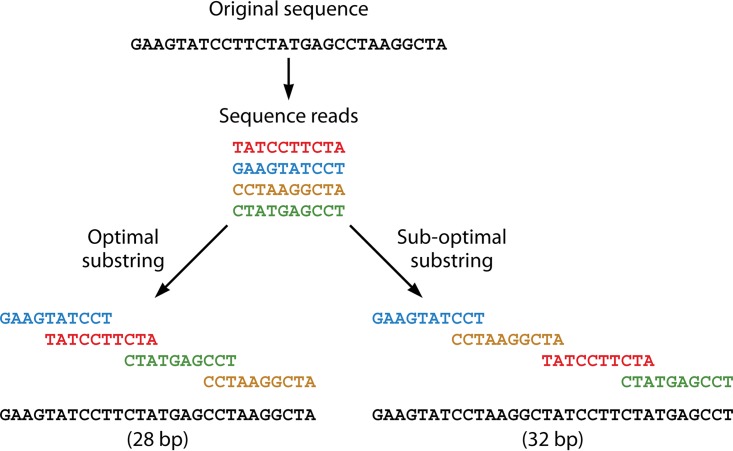
To address the first aspect of the assembly problem, the shortest common substring (SCS), or, in other words, the shortest path through a string of contigs with the largest overlap, is identified. Figure 1 illustrates the SCS construction with a simplified example. If the original sequence is repeat rich, it becomes crucial to identify the right minimal length of reads. For example, given the sequence ACGGGGGTATGCTTA, a read length of 3 would not be efficient, as there is a repetitive element of 5 bases (GGGGG). The read length must be longer than the repetitive sequence in order to cover the repeat during assembly. Some algorithms tackle this problem by providing scripts that automate the determination of optimal fragment or k-mer lengths. Furthermore, sequencing technologies are attempting to resolve this by providing paired-end reads and more recently are aiming to increase read lengths to cover repetitive regions in the genome.
FIG 1.
Simplified SCS construction.
Assembly algorithms can either assemble reads by using a single sequencing technology or use reads from multiple sequencing platforms in a hybrid approach. The single-sequencing-technology method will be most frequently applied for outbreak analyses. As this review focuses on widely applied sequencing technologies, only a short description of hybrid assemblers and a detailed description of specific assemblers are given. In a common WGS outbreak workflow, the assembly step is the most resource-demanding step and requires dedicated hardware. The tools are all available as command line tools, but some of them can be accessed via Web-based interfaces or software suites.
Identifying the best assembly tool can be challenging due to the many variabilities during sequencing that range from DNA isolation methods (97–100) to library preparation protocols, sequencing technologies, performance per sequencer, and sequence kit batches. All of these variabilities can affect the composition of the sequencing data set in size, quality, error rates, and sequencing characteristics, consequently influencing the quality of the assembly. Nevertheless, the following overall advice can be given: it is highly recommended that one use a single workflow for all samples included in the outbreak analysis, to reduce errors and variability introduced along the analysis step. In some cases, if external samples need to be included in the outbreak analysis, it is preferred that the sequencing data be reassembled by using the same assembly tool applied to all other samples. A helpful tool to assess assembly quality by using a well-defined reference genome is QUAST (101).
Table 2 shows a performance comparison of technology-specific and hybrid assembly tools for assembly.
TABLE 2.
Performance analysis of assembly toolsa
| Analysis tool (reference[s]) | Concept | Computational requirement | Speed | Assembly quality | Preferred sequencing technology(ies) | Web address(es) | Input format | Output format(s) |
|---|---|---|---|---|---|---|---|---|
| Web based | ||||||||
| Velvet (103, 126) | de Bruijn graph-based assembly that resolves repeat-rich regions; can be used for de novo or reference-guided assembly; requires paired reads with 20- to 25-fold coverage | Mid* | Medium* | Low* | Illumina | https://cge.cbs.dtu.dk/services/Assembler/ | FASTA, FASTQ, SAM, or BAM | AMOS, modified FASTA |
| SPAdes/hybridSPAdes (112) | de Bruijn graph-based assembler for de novo assembly of short and long reads | Low** | Low** | Mid*/** | Mixed input (Illumina, Ion Torrent, PacBio CLR, Oxford Nanopore) | https://cge.cbs.dtu.dk/services/SPAdes/ | FASTA, FASTQ, or BAM | FASTA, FASTQ, FASTG |
| Command line | ||||||||
| IDBA-UD (108) | de Bruijn graph-based assembly designed for assembly of repeat-rich reads of various sequencing depths | Low* | Medium* | Mid* | Illumina | http://i.cs.hku.hk/~alse/hkubrg/projects/idba_ud/ | FASTA | FASTA |
| RAY (96) | de Bruijn graph-based assembly that uses seeds instead of Eulerian walks; used for de novo assembly; designed for short reads | Low*** | Fast*** | Low*** | Mixed input (454, Illumina, Ion Torrent) | http://denovoassembler.sourceforge.net/ | FASTA, FASTQ, or SFF | FASTA, TXT |
| Minimap/miniasm (116) | OLC framework that computes overlaps and performs read trims and unitig construction; can be used for de novo or reference-guided assembly | Low** | High** | High*/** | PacBio, Oxford Nanopore | https://github.com/lh3/minimap, https://github.com/lh3/miniasm | FASTA | GFA, PAF |
| Canu (118) | OLC framework that computes overlaps and performs read correction, read trims, and unitig construction; used for de novo assembly | Mid** | Low** | High*/** | PacBio, Oxford Nanopore | https://github.com/marbl/canu | FASTA or FASTQ | FASTA |
All quantitative performance measures were taken from data reported previously, as indicated. CLR, continuous long reads; GFA, graphical fragment assembly; PAF, pairwise mapping format; SFF, standard flowgram format (454 data format); *, E. coli K-12 MG1655 data set (110); **, Enterobacter kobei data set (233); ***, Illumina data from E. coli (SRA accession number SRX000429) (234). Note that for SPAdes, only the nonhybrid tool is accessible as a Web-based tool.
Technology-specific short reads.
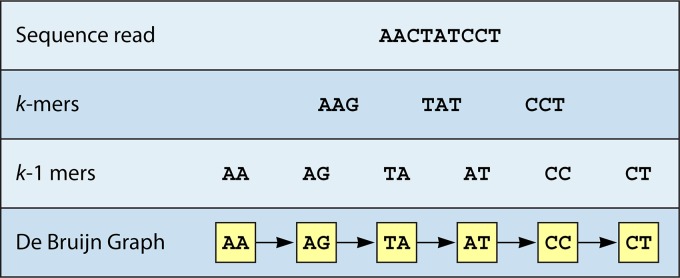
(i) de Bruijn graph-based assemblers.
One common assembly issue is that algorithms have difficulties in distinguishing read errors from sequence repeats when short reads overlap (96). This can lead to the exclusion of sequences between repeat sections during assembly. To solve this problem, a de Bruijn graph (DBG) breaks down original short reads into smaller sequences called k-mers, which are further reduced into k-1-mers. An algorithm then identifies a Eulerian walk, which describes the shortest possible path through these k-1-mers. In this way, the DBG reduces the chance of an incorrect assembly of repeat regions. Figure 2 illustrates k-mer construction with a simplified example.
FIG 2.
Simplified k-mer construction during de Bruijn graph assembly.
DBG-based assemblers are dependent on high-quality reads and could become less suitable in clinical settings that use long-read sequencers with intrinsically higher error rates (26, 102).
(a) Velvet.
One such DBG-based algorithm is Velvet, an assembler that generates multiple contigs from raw sequencing data (103). The algorithm is used for de novo genome assembly to reconstruct novel strains. Such de novo assemblies are especially relevant during outbreak scenarios, where the source strain might be unclassified and undetectable with conventional characterization methods. The Velvet assembler comprises velveth and velvetg, which are tools used for k-mer construction (hashing) and graph building from error-corrected k-mer alignments, respectively. A Perl script called VelvetOptimizer was developed by Simon Gladman and Torsten Seemann to automate the optimization of parameters such as k-mer length (http://www.vicbioinformatics.com/software.velvetoptimiser.shtml). By creating k-mers and identifying sequencing errors in the DBG, Velvet increases the probability of a correct assembly of strains with repeat-rich regions.
Velvet is the most frequently used assembler for Illumina sequencing data and has been applied to a variety of species, e.g., E. faecium (104), S. aureus (19, 105, 106), Clostridium difficile (105), E. coli (80), and Brachybacterium faecium (106). This tool is most frequently executed as a command line version; however, the Center for Genomic Epidemiology (CGE) provides a server that allows the user to assemble raw reads using a Web-based user interface (https://cge.cbs.dtu.dk/services/Assembler/). Furthermore, Velvet is incorporated as an assembler in multiple-tool workflows, e.g., the CGE Bacterial Analysis Pipeline (BAP) (107) and Ridom SeqSphere+ (19).
(b) IDBA-UD.
IDBA-UD is another DBG-based assembler for short reads of various sequencing depths (108). If a set of short reads with uneven coverage is to be analyzed, three major problems can arise with the determination of optimal k-mer lengths (109): (i) sequencing errors can produce incorrect or erroneous k-mers; (ii) if k is too small, repeat-rich regions or erroneous reads can introduce gaps into the DBG; and (iii) if k is too large, k-mers of low sequence coverage can be missing in the DBG.
To resolve erroneous reads in the first scenario and created gaps in the second scenario, the IDBA-UD assembler uses progressive relative sequencing depth thresholds determined by sequencing depths of neighboring contigs (108). To resolve gaps from low-depth repeat regions (third major problem), IDBA-UD performs a local assembly of paired-end reads. By following this approach, longer k-mers that are missing in the short reads can be assembled from the information contained in paired-end reads. The final corrected DBG is then used to extract contigs for scaffold construction. For a short-read E. coli data set, the IDBA-UD assembler is able to achieve an identity of 99.93% in 31 min with 2 GB RAM using an 8-core central processing unit (CPU) (110).
IDBA-UD is not frequently used, is available only as a command line tool, and therefore is less user friendly for the clinic, as it requires bioinformatics or informatics knowledge. Nonetheless, studies have shown that the assembly performance is comparable to or even improved compared to that of Velvet (80, 101, 106) and more or less equal to that of SPAdes (101, 106). Assemblies for a number of species, including E. coli (101), S. aureus (106), and B. faecium (106), showcase the tool's application range. IDBA-UD presents an advantage in cases where the coverage depths differ because of sequencing bias or the presence of a plasmid(s).
(c) RAY.
RAY is the third assembler that is based on DBGs. Instead of relying on Eulerian walks, this algorithm defines specific sequence subsets called seeds, which are extended into contigs (96). The extension process is controlled by heuristics or commands in such a way that the process stops as soon as a family of reads does not have overlaps that clearly identify a specific direction in the graph. The contig length is therefore limited, but overall assembly errors are minimized. To account for the various coverages of different sequencing technologies, a coverage distribution is calculated by RAY.
RAY has been used to assemble various bacterial genomes, e.g., Streptococcus pneumoniae (96), E. coli (96), S. aureus (106), B. faecium (106), and a Francisella tularensis genome, by using 454 Roche GS Junior and Illumina MiSeq data in a hybrid assembly (111). However, overall, this tool is not widely used for outbreak analyses. To apply RAY to raw sequencing data, command lines are needed. This assembler could be of value if sequencing data sets from multiple platforms per outbreak strain are available, but at present, this still remains a rare scenario.
(d) SPAdes.
SPAdes is the fourth DBG-based de novo assembler for short reads from multiple sequencing platforms (112). This algorithm follows four steps, where (i) an assembly graph is constructed as a multisized graph with modified error correction algorithms, (ii) estimations are made for the distance between k-mers in the DBG, (iii) a paired-assembly graph is constructed, and, finally, (iv) contigs are constructed, and initial are reads mapped against them, to determine the final contig sequences.
SPAdes is primarily a command line tool but can also be accessed via a Web-based interface (https://cge.cbs.dtu.dk/services/SPAdes/). It is the second most applied assembler for Illumina sequencing data and is distributed with the BioNumerics software suite (95). The applicability of SPAdes has been shown for several bacterial species such as E. coli (95, 101), S. aureus (106), and B. faecium (106). In multiple studies, SPAdes showed an improved performance compared to that of Velvet and showed results comparable to those of IDBA-UD (80, 101, 106). Compared to Velvet, IDBA-UD, and RAY, SPAdes is the only assembler that is still under development, resulting in continued improvements in performance and outcome.
Technology-specific long reads.
(i) Overlap layout consensus.
The overlap layout consensus (OLC) is a framework in which overlaps between reads are identified as contigs (96). Assemblies that follow this framework include Arachne (113), Celera (114), as well as short read algorithms such as EDENA (115). Long-read assembly algorithms usually follow the same four-step approach, where (i) all-versus-all raw read mapping is first conducted, followed by (ii) raw read error correction, where the directed graph is trimmed; (iii) the assembly of error-corrected reads; and, finally, (iv) contig consensus polish, where final contigs are compared to original reads to identify the final matching sequence.
(a) Minimap/miniasm.
The minimap/miniasm toolbox is used for the assembly of long reads, such as those of obtained from PacBio and ONT sequencers. The toolbox consists of two algorithms that implement the overlap and layout approaches of the OLC framework without using the consensus stage (116). Minimap overlaps raw reads, and miniasm assembles the overlaps de novo. It is also possible to use outputs of other assembly and overlap programs by converting them into GFA and PAF formats.
Due to the lack of error correction during the consensus stage, the final assembly with minimap/miniasm produces unpolished contigs with the same error rates as those of the initial input reads. Despite some improvements in performance that can be achieved when ONT sequence data are improved with Nanopolish, the minimap/miniasm assembler is outperformed by other tools (117). Whereas this presents a major disadvantage in assembly quality, it also significantly reduces the run time by skipping the time-intensive computation for error correction, a crucial benefit during outbreak analysis, where fast assemblies are needed to quickly determine the identity of the outbreak strain.
(b) Canu.
Canu is an algorithm designed to assemble high-noise long reads from single-molecule sequencing platforms such as the PacBio and ONT platforms (118). The assembly pipeline consists of overlap computing followed by read correction, read trimming, and unitig construction. A unitig is a subset of overlapping sequence read fragments. An advantage of Canu is the high alignment accuracy of over 99% for ONT reads (117). However, memory usage of up to 8 GB of RAM for two consecutive hours is common for an Enterobacter kobei genome assembly (119). The developers of Canu supply a detailed online tutorial for all possible uses of the program with detailed explanations, which improves user-friendliness (http://canu.readthedocs.io/en/stable/tutorial.html). The program can be run with several operating systems and theoretically any hardware; however, a minimum of 32 GB RAM is recommended for larger assemblies.
Compared to other assemblers such as miniasm and SPAdes, Canu performs best for assemblies from PacBio and ONT sequence data (118). With mixed data sets that contain ONT and Illumina reads, other assemblers can provide boosts in performance for some bacterial genomes. It must be noted that there are only a few studies to date that provide comparable benchmark values for the performance of Canu in comparison to those of other tools.
Hybrid assemblers.
Hybrid assemblers process sequence reads from multiple sequencing technologies and thus decrease the number of correlated read errors. The simultaneous assembly of hybrid reads allows improved de novo assemblies but results in higher sequencing costs. RAY can be used as a hybrid assembler, using several kinds of input reads; however, the only documented use was for read mixtures from short-read technologies such as Roche 454, Illumina, and Ion Torrent; e.g., for a mixed data sat comprising Illumina and Roche 454 data, a final identity of up to 98.31% was reported (96, 102). If long reads from other technologies are available, the hybridSPAdes algorithm can also be used to increase repeat resolution and fill gaps in the assembly graph (120).
Genome Characterization
Once sequence reads are assembled into a set of contigs, clinicians would next be interested in further classifying the sequenced bacterial isolate and infer an epidemiological profile from genes contained in the bacterial genome. Here several questions could be addressed. (i) What is the species of the sequenced isolate? (ii) Which genes are contained in the genome, and do they infer virulence or AMR? Genome characterization tools aim to address these questions by comparing several reference databases of known genes and reference genomes to contigs. The results of a comparison of genome characterization tools are presented in Table 3.
TABLE 3.
Overview of genome characterization toolsa
| Analysis tool (reference[s]) | Concept(s) | Input type(s) | Input format(s) | Output format(s) | Web address |
|---|---|---|---|---|---|
| Identification | |||||
| Web based | |||||
| KmerFinder (121, 122) | Uses k-mers to identify strain using WGS data | Raw sequences, contigs | FASTQ, FASTA | Tab delimited, online | https://cge.cbs.dtu.dk/services/KmerFinder/ |
| NCBI BLASTb (123) | NCBI Web-based interface for performing BLAST searches; searches hits in the database that match the given sequence | Contigs | FASTA | Online, tab delimited | https://blast.ncbi.nlm.nih.gov/Blast.cgi |
| MLST Web server (125) | Web-based database that identifies STs from short sequencing reads or draft genomes | Raw sequences, contigs | FASTQ, FASTA | Online | https://cge.cbs.dtu.dk/services/MLST/ |
| Command line | |||||
| PathoScope 2.0 (127) | Complete framework based on Bayesian missing-data approach, for direct strain identification | Raw sequences | FASTQ, FASTA | Tab delimited | https://sourceforge.net/p/pathoscope/wiki/Home/ |
| Annotation | |||||
| Web based | |||||
| RAST (129) | Web-based server for localization and identification of tRNA, rRNA, and coding sequences; includes a browser for screening the output | Contigs | FASTA | GenBank, EMBL, GFF3, GTF, Excel, and tab delimited | http://rast.nmpdr.org/ |
| Command line | |||||
| PROKKA (132) | Rapid annotation tool for localization and identification of rRNA, tRNA, tmRNA, signal peptides, noncoding RNA, and coding sequences | Contigs | FASTA | FASTA, tab delimited, SQN, GenBank file, GFF3 | http://www.vicbioinformatics.com/software.prokka.shtml |
| Virulence | |||||
| Web based | |||||
| VirulenceFinder | Detect virulence genes in WGS data using the BLAST algorithm | Raw sequences, contigs | FASTQ, FASTA | Tab-delimited summary, FASTA | https://cge.cbs.dtu.dk/services/VirulenceFinder/ |
| VFDB (138) | Source of virulence information, including Web-based service to perform BLAST to detect virulence genes | Contigs | FASTA | Online, tab delimited | http://www.mgc.ac.cn/VFs/ |
| Antimicrobial resistance | |||||
| Web based | |||||
| ResFinder | Detects resistance genes in WGS data | Raw sequences, contigs | FASTQ, FASTA | Tab-delimited summary, FASTA | https://cge.cbs.dtu.dk/services/ResFinder/ |
| RGI/CARD (144–146) | Web-based as well as command line versions available to perform resistance gene detection using the CARD database | Contigs, GenBank accession no. | FASTA, GenBank accession no. (nucleotide or protein) | JSON, tab-delimited summary, FASTA, heat map PDF | https://card.mcmaster.ca/analyze/rgi |
| PlasmidFinder | Tool to detect plasmids in WGS data | Raw sequences, contigs | FASTQ, FASTA | Tab-delimited summary, FASTA | https://cge.cbs.dtu.dk/services/PlasmidFinder/ |
| CGE BAP (107) | Web-based suite for automated genomic characterization; if raw sequence reads are provided, performs assembly; set of tools is applied to the contigs, ResFinder, VirulenceFinder, and PlasmidFinder | Raw sequences, contigs | FASTQ, FASTA | Tab-delimited summaries, FASTA | https://cge.cbs.dtu.dk/services/cge/ |
ND, no data; NA, not applicable; EMBL, sequence file format; JSON, JavaScript Object Notation; SQN, GenBank submission file; GFF3, General Feature Format 3.
Also available as a command line tool and as GUI via prfectBLAST (124).
Identification.
To address the first question (what is the species of the sequenced isolate?), identification tools that are able to identify species from either raw sequence reads or contigs are needed. Some useful and user-friendly tools that can be applied in the clinic are discussed below.
(i) Web-based tools.
(a) KmerFinder.
To identify species from raw sequencing data or contigs, KmerFinder (121, 122) is a relatively fast solution. The command line version is able to identify isolates to the species level using contigs, based on benchmarks, in an average of 9 s. When applied to raw reads, computational times, depending on the amount of data, of an average of 3 min 10 s have been reported (121). The tool is accessible as a Web-based tool (https://cge.cbs.dtu.dk/services/KmerFinder/), where two different scoring methods can be applied. The “standard” method will give an overview of all k-mers matching all template species, and a ranking will be based on the amount of k-mers matching each template. The other method, “winner takes it all,” will count k-mers only once and is therefore ideal to determine if the data originate from a single strain. The default setting for KmerFinder is the winner-takes-it-all method.
(b) NCBI BLAST.
The National Center for Biotechnology Information (NCBI) has a Basic Local Alignment Search Tool (BLAST) service available (https://blast.ncbi.nlm.nih.gov/Blast.cgi) (123). Multiple BLAST variants are available via a Web-based interface. To identify the species origin from single or multiple contigs, megaBLAST is the most advisable tool, which is also the default when standard nucleotide BLAST is performed. The default database is the Nucleotide Collection (nr/nt) which is a large database containing all sequences present in the NCBI database. Another workaround is selecting the RefSeq Representative Genome Database, which is faster due to the smaller database size. If no desired hit is identified with this smaller database, the nr/nt database should be used. In cases where no proper result is obtained by using either database, the contigs are assembled incorrectly, or the query is from a novel strain. Interpretation of the results is critical when using BLAST; hence, it is advised that one should always compare the query length with the length of the hits in combination with query coverage and identity percentages before drawing conclusions. BLAST also has a stand-alone application, which requires minor command line skills (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/). An easier and elegant way would be to use a commercial software package that includes a GUI or to use prfectBLAST (124). The main advantage of using a stand-alone version of BLAST is that there is full control in constructing one's own BLAST database and parameters. This could be relevant for the clinic in some dedicated cases, i.e., for the detection of specific markers.
(c) MLST Web server.
The MLST Web server (https://cge.cbs.dtu.dk/services/MLST/) is an online database of MLST alleles of 66 bacterial species (125). It uses short sequencing reads or draft genomes as the input and identifies the best-matching MLST alleles by using a BLAST-based ranking method. The identified MLST alleles are then combined to determine the STs of the samples. If short reads are used as the input, the MLST Web server uses assembly algorithms, such as Velvet version 1.1.04 for Illumina reads (126), to create contigs and scaffolds prior to ST determination. Most MLST schemes include at least 7 housekeeping genes, and for six bacterial species (Acinetobacter baumannii, Clostridium difficile, Pasteurella multocida, E. coli, Streptococcus thermophilus, and Leptospira species), there are two or three different schemes available. With short sequencing reads from 3 different platforms, this Web server was able to correctly identify 83.3% of known STs (125). For outbreak analysis, MLST derived from WGS data can be the first step in analysis after assembly and is a good alternative to conventional MLST, yet this will be applicable only if WGS is well adopted in routine diagnostics. However, this will not make use of the full potential of WGS; MLST derived from WGS has a lower resolution than core genome MLST (cgMLST), whole-genome MLST (wgMLST), or core single nucleotide polymorphism (coreSNP) methods, which are described later in this review.
(ii) Command line tools.
(a) PathoScope 2.0.
PathoScope 2.0 is a comprehensive framework for direct strain identification from raw sequence reads without the need for assembly (127). The framework comes with an additional program called Clinical PathoScope, which is adapted for clinical samples. The framework consists of four core modules that allow (i) the generation of custom reference genome libraries (PathoLib), (ii) read alignment and filtering against host and filter references (PathoMap), (iii) reassignment of ambiguous reads for strain identification (PathoID), and, finally, (iv) the generation of a detailed results report (PathoReport). The framework additionally offers the two optional modules, PathoDB, a database that provides gene, taxonomy, and protein product annotation information to complement the NCBI nucleotide database input, and PathoQC, which improves raw sequencing reads for identification by filtering low-complexity reads and trimming low-quality bases and adaptors. With Clinical PathoScope, pathogen detection from multispecies samples can be achieved in 25 min with 94.7% accuracy (128). Due to the default priors of the Bayesian framework used, it is possible that PathoScope fails to distinguish between closely related outbreak substrains when assigning best fits to only complete reference genomes. However, this can be changed by adjusting informative priors and ensuring a minimum coverage of at least 20-fold to distinguish closely related strains or substrains.
Annotation.
Annotation tools aim to answer the second question (which genes are contained in the genome, and do they infer virulence or AMR?) by retrieving the gene content from assembled contigs. This has a number of applications ranging from the detection of novel antibiotic or virulence genes to the detection of efflux pumps involved in AMR or simply to obtain more insight into gene content, all of which are pathogen characteristics highly relevant to the clinician during outbreak analysis.
(i) Web-based tool (RAST).
Rapid Annotation Using Subsystem Technology (RAST) is a fully automated Web-based tool that can be used to annotate contigs. After submission, the annotated genome is available within 12 to 24 h (129). Results are presented in multiple file formats, as shown in Table 3. It uses a hierarchical structure to annotate tRNA, rRNA, and coding sequences and is built upon the SEED framework (130). The algorithm uses GLIMMER3, BLASTP, BLASTX, BLAST (123), and a SEED k-mer-based annotation algorithm (131) to obtain the best annotation possible. Manual curation of annotations is possible throughout the Web-based analysis process. Because RAST algorithms rely on closely related isolates, RAST is not able to operate on mixed or contaminated cultures. myRAST is the stand-alone version of RAST, which can be installed on a local computer and has the same functionalities as the RAST server, which would be useful for the clinic if full operational independence is desired. To install myRAST, informatics or bioinformatics expertise is needed. Running the Web-based RAST tool is the most advisable method for user-friendly use in the clinic.
(ii) Command line tool (PROKKA).
PROKKA is a rapid software tool that annotates bacterial contigs (132). This software orchestrates several feature prediction tools and identifies gene locations and function in two essential steps.
First, preassembled contigs are presented as complete sequences or a set of scaffolds in FASTA format. Second, several tools are utilized to identify possible genes within contigs: RNAmmer (133) is a computational predictor that uses hidden Markov models to predict rRNA genes, Aragorn (134) uses heuristic algorithms and a modified version of the BRUCE program to predict tRNA and transfer-messenger RNA (tmRNA) genes from assembled contigs, SignalP (135) uses a hidden Markov model to predict signal peptides, and, finally, Infernal (136) builds covariance models (CMs) to predict noncoding RNA.
In order to identify which predicted genes are transcribed, PROKKA relies on a number of prediction tools. In the first step, identifying the product of a candidate gene, the Prodigal tool is used to identify gene coordinates (137). Following this, PROKKA employs several other tools that compare the gene sequence to sequences in databases in a hierarchical manner. The most valuable output of PROKKA is a General Feature Format 3 (GFF3) table of annotated genomic features that can be used to identify the sampled bacterial species. With an E. coli K-12 reference genome, PROKKA is able to annotate genes with 99.63% accuracy in about 6 min using a quad-core CPU (132). A benchmark test showed that PROKKA outperforms RAST in terms of the number of predicted elements (132).
Virulence.
Tools for the detection of virulence genes can also be used to answer the second question (which genes are contained in the genome, and do they infer virulence or AMR?). The conventional detection of certain genes/markers via quantitative PCR (qPCR) would imply the need for a large investment in primers and probe design. The rapid evolution of bacteria leads to mutations in targets and results in possible negative qPCR results. Redesign of primers and probes will often delay diagnostics during outbreak analyses. WGS in combination with gene/marker detection tools using curated databases would partly overcome these qPCR issues, as most tools are competent to detect genes/markers harboring mutations.
(i) Web-based tools.
(a) VirulenceFinder.
The VirulenceFinder tool uses BLASTn (123) and can be accessed online (https://cge.cbs.dtu.dk/services/VirulenceFinder/). It has a GUI that is very similar to that of ResFinder. To use VirulenceFinder, one of the following four organisms have to be selected: Listeria, S. aureus, E. coli, or Enterococcus. The latest tool update is from 18 February 2017 and contains 503 virulence markers in total. This tool is adequate to detect genes that are mutated compared to the genes in the database. In a study that performed an outbreak analysis of verotoxigenic E. coli (VTEC), VirulenceFinder was used to automatically characterize virulence genes (20).
(b) VFDB.
The Virulence Factor Database (VFDB) (138) is a Web-based analysis tool (available at http://www.mgc.ac.cn/VFs/). This database contains data from 74 distinct genera, 926 bacterial strains, and a total of 1,796 identified virulence factors. The virulence factors are stratified into groups based on characteristics that are, for example, being used in offensive or defensive actions. Both nucleotide and protein sequences are contained in the database. A query can be entered to find matches to the virulence factors in the database using the full range of BLAST algorithms (123). Compared to VirulenceFinder, this database contains many more markers associated with virulence, and several genes involved in housekeeping functions are included in the database. The database is not as user-friendly and the content is not as well validated as VirulenceFinder. To use the full potential of the database, programming skills or minor bioinformatics expertise is needed. The main advantage of VFDB over VirulenceFinder is the extended background material present, including schematics and references to literature.
Antimicrobial resistance.
When attempting to answer the second question (which genes are contained in the genome, and do they influence the risks to patients?), the same applies to AMR gene analysis as discussed above for the annotation and virulence tools. McArthur and Tsang provided a historical overview of applications and databases that could be used for AMR detection from WGS data (139). Below is selection of tools that fit best to AMR detection in combination with WGS outbreak analyses. The criteria for the selection of tools in this review were that tools should be maintained, curated, applicable to multiple species, and easy to use.
(i) Web-based tools.
(a) ResFinder.
ResFinder is a tool that uses BLASTn (123) to detect acquired resistance genes in WGS data. It is accessible online (https://cge.cbs.dtu.dk/services/ResFinder/) and has a user-friendly GUI. The database has regular updates; the version from 17 February 2017 contained 2,166 markers for 14 different antibiotics, including the latest mcr gene, which was linked to transferable colistin resistance (140–143). Inputs for ResFinder can be either raw sequence reads from Roche 454, Illumina, Ion Torrent, or SOLiD sequencing or contigs.
(b) RGI/CARD.
The Resistance Gene Identifier (RGI) uses the CARD database, which contains curated AMR genes and mutated sequences (https://card.mcmaster.ca/analyze/rgi) (144–146). It is based on the older ARDB database (147). RGI allows the identification of AMR genes but also specific AMR-associated mutations. A GenBank accession number of either a nucleotide or a protein sequence can be used as the input, or in cases where the detection of AMR and AMR-associated mutations from WGS data is applicable, contigs can be uploaded by using the upload sequence(s) method. In contrast to ResFinder, RGI uses protein sequences to detect matches derived from Prodigal open reading frame detection (137). For the detection of AMR genes, it uses so-called “protein homolog models,” where BLAST sequence similarity is determined to detect functional homologs (123). For the detection of mutations associated with AMR, so-called “protein variant models” are applied. Searches can be applied by using 2 criteria: the default (perfect and strict hits only) or discovery (perfect, strict, and loose hits) setting. For outbreak analyses, the default setting is endorsed. The developers of this tool placed a disclaimer stating that constant curation changes to the database and cutoff values could potentially affect results. Hence, caution should be exercised when interpreting results.
(c) PlasmidFinder.
PlasmidFinder is a tool for the detection of plasmids that could harbor potential AMR or virulence genes (available at https://cge.cbs.dtu.dk/services/PlasmidFinder/). As plasmids harbor AMR genes, such as beta-lactamase genes (148), the simultaneous detection of plasmids and pathogen genomes could be of great value to outbreak analyses, as shown for the detection of plasmid pC15-1a, which was associated with an outbreak of extended-spectrum-beta-lactamase (ESBL)-producing E. coli (149). The detection of certain plasmids and the determination of plasmid frequencies could result in more dedicated screening methods when developing routine PCRs targeting specific plasmids. Most DNA isolation and library preparation protocols are suitable for obtaining WGS data for both genomes and plasmids. The Web-based interface is user-friendly, and a detailed user manual is present. Prior to plasmid detection, the user has to define the database type to be either Enterobacteriaceae, Enterococcus, Streptococcus, or Staphylococcus containing the Gram-positive plasmids. The latest version from 20 February 2017 contained 128 markers specific to Enterobacteriaceae plasmids and 141 specific to Gram-positive-organism-associated plasmids (150). Results give an indication of which plasmids are present, but further investigation is needed for confirmation.
(d) CGE Bacterial Analysis Pipeline.
The Bacterial Analysis Pipeline (BAP) (107) is a pipeline that requires either FASTA/FASTQ reads or contigs as the input and applies a series of tools to the data. If more samples need to be processed, a batch import option is available. Before submission, a metadata file has to be added and attached to the GUI, requesting extra information from the user. The analysis then starts in the following order: (i) de novo assembly to construct contigs and the simultaneous use of KmerFinder (121, 122) for species identification, (ii) MLST for identifying STs, (iii) gene detection using ResFinder (151) and VirulenceFinder (20), and, finally, (iv) detection and typing of plasmids using PlasmidFinder (150) and plasmid MLST (pMLST) (150). Results are presented in a report showing the outcomes of all tools in the pipeline. The use of BAG will minimize the overall workload in a clinical microbiology laboratory compared to that with running all tools separately. On average, a single sample will be analyzed in 19 to 28 min. For most organisms, a minimum of 50-fold genome coverage is advised, but, e.g., Salmonella isolates will benefit from having >100-fold genome coverage (107). BAP can be a valuable tool for the clinic to quickly produce an overview of the characteristics of the strain and would require minimal effort to incorporate into daily routines if WGS is already applied.
Comparative Genomics
Once a clinician has obtained contigs and optionally performed genome characterization of the WGS data, it would next be of interest to perform comparative genomics to detect relatedness between strains. Preferably, the species is known before applying comparative genomic tools, as the tools tend to perform best by using closely related strains of the same species. The questions that could be addressed with comparative genomics tools are as follows. (i) Which strains could be clonal? (ii) What is the source of the outbreak?
There are multiple tools available for comparative genomics, which can use different methodologies. For the clinician, it is of interest to be informed on the differences between methods, mainly because these methods differ in discriminatory power and ease of data sharing. What follows are descriptions of these different methods, before different tools for comparative genomics are described in detail. Table 4 shows a detailed comparison of comparative genomics tools.
TABLE 4.
Performance analysis of comparative genomics toolsa
| Analysis tool (reference[s]) | Concept | Method | Run time (h) | Topology score (%) | Web address(es) | Input type(s) | Input format(s) | Output format(s) |
|---|---|---|---|---|---|---|---|---|
| Web based | ||||||||
| PubMLST (158) | Web-accessible database where it is possible to run cgMLST and wgMLST analyses | cgMLST/wgMLST | NA | NA | https://pubmlst.org/ | Contigs | FASTA | cgMLST/wgMLST profile |
| CSI Phylogeny 1.4 (161) | High-quality SNP method using reference mapping of reads and mapping and SNP calling assessments | Reference-based SNP | ND | ND | https://cge.cbs.dtu.dk/services/CSIPhylogeny/ | Raw sequences, contigs | FASTA, FASTQ | ND |
| NDtree 1.2 (161) | Creates k-mers of reads and maps them to a reference; performs simple model to determine no. of SNPs | Statistical method | 3–3.5b | ND | https://cge.cbs.dtu.dk/services/NDtree/ | Raw sequences | FASTQ | Newick |
| Command line | ||||||||
| kSNP3 (154, 155) | Uses k-mer analyses to detect SNPs between strains without using either multiple-sequence alignment or a reference genome | Non-reference-based SNP | 0.5c | 91.80–95.80c,e | https://sourceforge.net/projects/ksnp/ | Raw sequences, contigs | FASTA | Newick, MSA |
| Roary (169) | Tool for constructing pangenomes from contigs | Pangenome | 4.30d | 100d | https://sanger-pathogens.github.io/Roary/ | Contigs | GFF3 | FASTA, TXT, CSV, Rtab |
| Pan-Seqf (175) | Pangenome assembler with additional locus finder for core/accessory gene allele profiles (a Web-based version is also available) | Pangenome | ND | ND | https://github.com/chadlaing/Panseq, https://lfz.corefacility.ca/panseq/ | Contigs | FASTA | TXT, FASTA |
| Lyve-SET (179) | High-quality SNP method using reference mapping of reads and mapping and SNP calling assessments | Reference-based SNP | 6.25c | 85c | https://github.com/lskatz/lyve-SET | Raw sequences, contigsg | FASTA, FASTQ | Matrix, FASTA, Newick, VCF |
| SPANDx (182) | Complete workflow for creating SNP/indel matrixes as well as locus presence/absence matrixes from raw sequencing reads from a range of NGS technologies | Reference-based SNP | 3.1c | 100c | https://sourceforge.net/projects/spandx/ | Raw sequences | FASTA, FASTQ | NEXUS |
All quantitative performance measures were taken from previously reported data, as indicated. ND, no data; NA, not applicable; MSA, multiple-sequence alignment; GFF3, General Feature Format 3; VCF, variant call format.
Based on 46 VTEC genomes (20).
Based on 21 E. coli genomes (167).
Wall time for 1,000 S. enterica serovar Typhi genomes (169).
Using core.
A Web-based version is also available.
Contigs are simulated to reads.
Non-reference-based SNP analysis.
Where MLST methods focus exclusively on genes and/or loci, SNP-based methods have the advantage of including intergenic regions. Studies show that intergenic regions harbor SNPs that are host specific and could help in studies of the evolution of E. coli (152, 153). SNP-based methods have the highest discriminatory power of all comparative genomics approaches. No SNP databases or nomenclature is available because of the diversity of the algorithms used to detect SNPs, making it useful for local outbreak detection and unsuitable for the global use of WGS data that would be shared and reanalyzed. An example of a tool using non-reference-based SNP analyses is kSNP, which is described in detail below (154, 155).
Reference-based SNP analysis.
Reference-based SNP methods use a single reference genome to detect SNPs, making it possible to detect SNPs in genes, loci, and intergenic regions present in the query genome (56, 57, 105, 156). Raw sequence reads can be used as the input, and these reads are then mapped onto a reference genome. The algorithm for mapping the reads allows for some variation between reads and the reference. The drawback of this method is that lineage-specific regions could be absent in the reference and therefore would be excluded. Reference-based SNP methods are therefore recommended only for genomes for which a high-quality reference is present.
Pangenome-based analysis.
Pangenome-based analysis compares both core and accessory genes between strains. The workflow for most pangenome tools is to identify genes via nucleotide or protein comparison to identify orthologs. The genes are often grouped by using a clustering method, with the main goal of grouping genes into families as accurately as possible while reducing computational complexity (157). Genes are stored to the pangenome if they differ from all other genes in the pangenome under specified parameters. The pangenome consists of the full gene pool of all strains used to build it. Often, genes are classified in two categories: core genes, which are present in all strains, and accessory genes, which are present in single or multiple strains but not all strains. Applying pangenome approaches in outbreak settings is possible, but the number of strains that they can be applied to is limited by computational resources. This method is therefore not widely applied in hospitals but can still be of value to study orthologs of genes between strains.
Core genome MLST.
cgMLST is an extension of conventional MLST that increases the number of core genome-related genes/loci. One example for S. aureus cgMLST uses 1,861 genes/loci, in comparison to just 7 with MLST (7). This increases the resolution drastically and makes it possible to detect isolate-specific genotypes, which, when used for outbreak management, enables the identification of novel transmission events (7). The use of cgMLST allows the user to create a species-specific nomenclature; i.e., strains with identical cgMLST results are grouped into a cluster type (CT). These CTs can be easily stored and shared by using central databases, enabling consistent outbreak management protocols among different hospitals. This has an advantage over, e.g., whole-genome MLST- or SNP-based methods that do not use such a nomenclature. At the time of this review, the Ridom SeqSphere+ cgMLST nomenclature server (http://www.cgmlst.org/ncs) was the only database applying the concept of CT. Other publically available databases that use cgMLST are PubMLST (https://pubmlst.org/), EnteroBase (https://enterobase.warwick.ac.uk/), and the Bacterial Isolate Genome Sequence Database (BIGSdb) (158).
Whole-genome MLST.
wgMLST is often used as an extension of cgMLST and uses core genome genes/loci and all accessory genes/loci to detect lineage-specific genes/loci. BioNumerics 7.6.2 (Applied Maths) uses wgMLST schemes for analyses (159).
For use in routine outbreak detection, it would be best to use a standardized method for comparative genomics that would be capable of using species-specific nomenclature. Out of the above-described methods, cgMLST would be best suited for this application. It must be noted, however, that wgMLST might offer higher resolution for closely linked clusters of outbreak strains. However, outbreak analyses must not be limited to a single method for comparative genomics, as other methods show higher discriminatory power and could be of high value in cases where more resolution is needed. What follows is a detailed description of analysis tools for comparative genomics.
Web-based tools.
(i) PubMLST.
PubMLST is a public database that can be used to perform typing of WGS data using both cgMLST and wgMLST schemes. The tool is hosted by the Department of Zoology, University of Oxford, United Kingdom (available at https://pubmlst.org/). The software used to set up this Web-based version is BIGSdb (158). Because it harbors a database containing data from multiple strains on which WGS was performed, data can be uploaded and retrieved at all times, which would allow a growing strain knowledge base within or across hospitals. By installing BIGSdb locally, all functionality would be retained, yet intermediate knowledge of informatics or bioinformatics is needed in order to install the software. With a locally installed version, it is possible to construct custom schemes by selecting multiple loci or alleles, e.g., an ampC scheme for A. baumannii (160). The numbers of schemes present at PubMLST are still increasing and, at the time of this review, included 129 MLST schemes, a bacteriophage MLST scheme, a pMLST scheme, and a ribosomal MLST (rMLST) scheme.
(ii) CSI Phylogeny 1.4.
As a replacement for snpTree, which was the method previously provided by the CGE and was not able to infer phylogeny when a too-distant reference was provided, two new methods were developed, one of which is CSI Phylogeny 1.4 (161). CSI Phylogeny 1.4 can be accessed via a Web-based interface (https://cge.cbs.dtu.dk/services/CSIPhylogeny/). This method is a reference-based high-quality SNP (hqSNP) method, which is characterized by defining SNPs using additional quality criteria and hence is more conservative but of higher certainty. To run CSI Phylogeny, a reference genome needs to be provided in FASTA format. For the strains of interest, WGS data in FASTA/FASTQ format or contigs in FASTA format can be uploaded as well. This method uses the Burrows-Wheeler Aligner (BWA) to align reads to the reference genome (162). SNPs are called and filtered according to user-adjustable parameters. This tool allows the user to control minimal sequence coverage depth at the SNP location, filter SNPs based on both the quality of mapping and SNP quality, set SNP density restrictions, and select a minimal Z-score. Finally, the method checks across all input genomes if an SNP is detected in other input genomes; if not, SNPs are neglected. To infer phylogeny, a maximum likelihood (ML) tree is built by using a modified, more accurate version of FastTree (163).
CSI Phylogeny was able to detect the outbreak source of hospital-acquired Legionella pneumophila infection among a total of 25 strains (164). In another study, CSI Phylogeny was able to distinguish different Streptococcus groups among 80 clinical Streptococcus strains (165).
(iii) NDtree 1.2.
The other method made available by the CGE is NDtree (161). This method creates k-mers of the reads and maps them to a reference. The number of bases per position is measured. A formula is used to count the number of nucleotide differences between strains. A threshold of a 10-fold-higher abundance than the next most abundant nucleotide is applied to accommodate low-quality positions (20). The number of nucleotide differences is placed into a matrix, and phylogenetic relatedness is calculated by using Phylip (http://evolution.genetics.washington.edu/phylip.html), which implements the unweighted pair group method with arithmetic mean (UPGMA) algorithm (166). At the Web-based interface, users upload sequence FASTQ files and need to select if input files are in a single-end or paired-end format. The template file is essentially the reference genome in FASTA format and can be uploaded by the user. A feature that has great value to the clinic is the ability to automatically predict a close reference by using KmerFinder (121, 122), if no reference is provided. Of note is that NDtree is conservative and sensitive to parameter settings, which could lead to inaccurate results (21).
NDtree was used to detect an outbreak of S. enterica, showing agreement with data from PFGE outbreak analyses and an SNP-based method after applying parameter optimization (21).
Command line tools.
(i) kSNP3.
kSNP3 is a tool that is able to detect SNPs between strains without using any reference genome and without performing multiple-sequence alignments (MSAs) (154, 155). It uses k-mer analyses to infer SNPs. Inferring SNPs is completely dependent on the input set of strains and can therefore be applied to any species. By default, kSNP3 uses no annotation and produces results in 0.5 h when applied to 21 E. coli genomes (167). To calculate the optimum k-mer size for a particular data set, a program called Kchooser is included. Kchooser tries multiple k-mer sizes to estimate the optimum k-mer size and calculates the fraction of core k-mers (FCK). The FCK is used to estimate the accuracy of kSNP3 parsimony trees and the reliability of phylogenetic trees; as sequence variation increases, the FCK decreases. An FCK of ≥0.1 is recommended. kSNP3 has an option to perform analyses on core SNPs, identifying SNPs and locations that are present in all strains, and is the recommended method for outbreak analyses, as it best reflects the evolutionary signal. kSNP3 is able to produce many output files along an MSA FASTA file containing all concatenated core SNPs per strain. This file can be used as the input for phylogenetic analyses. Furthermore, a summary file is produced, which includes how many core and noncore SNPs were found as well as parsimony trees, neighbor-joining (NJ) trees, and ML trees in Newick format. Additionally, kSNP3 has an option to add strains to a previously executed analysis, where it uses the existing detected SNPs and adds the new strains to the analysis. In a VRE outbreak, kSNP was compared to MALDI-TOF MS, showing that kSNP had a higher discriminatory power (50). A retrospective L. pneumophila outbreak analysis showed that by using kSNP, evolutionary relatedness between strains could be successfully studied (168).
(ii) Roary.
Roary is a command line tool for the rapid generation of pangenomes and performing outbreak analyses. The pangenome describes a set of genes that are present in one or more strains, thereby representing the complete gene content. Genes present in most strains are called core genes, and all other genes are called accessory genes. This software uses one annotated assembly per sample, given that all samples have to be from one species (169). Coding regions on the assembly are converted into protein sequences that are filtered and preclustered with the CD-HIT tool, which defines genes present in all isolates as core genes (170). Following this step, an all-against-all search is performed with BLASTP to identify protein sequences. The so-identified sequences are then grouped into families with the Markov cluster algorithm (MCL) (157) and finally merged with precluster results from CD-HIT. Isolates are grouped together based on the similarity of genes and the presence of genes in the accessory genome. Pangenome accuracy was shown to be 100%, and a data set of 1,000 annotated S. Typhimurium assemblies was analyzed in 4.3 h using a single CPU (169).
A number of outbreaks were analyzed by using Roary, such as an outbreak of carbapenemase-producing Citrobacter freundii in Miami, FL, which identified 3 clonal strains and 2 unrelated strains (171); an outbreak of 495 vanA VRE strains in Copenhagen, Denmark, where the tool detected the spread of a vanA-carrying plasmid as a possible outbreak cause (104); and emerging Serratia marcescens clones in the United Kingdom and Ireland (172). Roary can also be used to identify clade-specific gene markers (173, 174), which, compared to SNP-based methods, is an advantage because it enables to use the gene markers for creating target-specific PCR primers.
(iii) Pan-Seq.
Pan-Seq is another pangenome sequence analysis program to compare contigs (175). This program comprises a novel region finder (NRF), a core and accessory genome finder (CAGF), and a locus selector (LS). The NRF tool uses MUMmer (176) to identify novel sequences that cannot be aligned to a contig database and extract these sequences to a separate file. The CAGF uses the MUMmer alignment to identify sequences that are present in multiple contigs and adds these sequences to the initial pangenome. The pangenome is then divided into fragments, which are checked, once again, for sequence identity with the initial contig sequences. A sequence identity cutoff is determined with the BLASTn algorithm (123), and fragments above or below the cutoff are assigned to the core or accessory genome, respectively. From this set of genes, the LS can then identify gene variations between input sequences and distinct alleles present for each gene. At the time of this review, no studies that reported the accuracy or computing time of Pan-Seq were found. There have been studies of the applicability of Pan-Seq: the construction of a Brucella species pangenome, including calculating phylogenetic relatedness (177), and a study on S. enterica strains comparing phylogenetic outcomes using Pan-Seq versus MLST schemes, which showed that Pan-Seq has higher discriminatory power (178).
(iv) Lyve-SET.
A recently reported tool called Lyve-SET performs reference-based SNP analysis but could be characterized as an hqSNP pipeline. Lyve-SET identifies SNPs and performs a series of filtering steps, including applying minimal and maximum numbers of coverage and base call consistency and discarding clustered SNPs and SNPs not covered by both forward and reverse reads, to retain only reliable SNPs. There are additional options to exclude regions from analyses, e.g., phage-specific regions and repeat regions (179).
Lyve-SET was used to perform comparative analyses of ESBL CTX-M-65-producing Salmonella enterica serovar Infantis strains isolated from multiple sources, including human (180), and an S. enterica evolutionary analysis of samples originating from bovine and poultry sources (181). This application is focused on foodborne pathogens, although the tool is applicable to hospital-acquired strains as well. A study that compared multiple SNP tools with Lyve-SET showed that all methods identified outbreak isolates with >99.5% concordance to Lyve-SET results (179).
(v) SPANDx.
The Synergized Pipeline for Analysis of NGS Data in Linux (SPANDx) is a comparative genomics tool that integrates several well-validated tools into a single workflow for comparative analysis of raw sequence reads (182). It performs read mapping alignment with BWA-mem (162, 183). The reads are filtered and parsed by using SAMtools (184). Determining the core/accessory genome through the presence or absence of a genetic locus is performed by BEDTools. Data filtering is done by Picard, and base quality score recalibration, improved insertion-deletion (indel) calling, data filtering, and variant determination are done with the Genome Analysis Tool Kit (GATK). Indel matrix construction and the detection of SNPs are done with VCFtools, and variant annotation is done with SnpEff. The final output of the workflow is a matrix to identify core and accessory fragments and a filtered SNP matrix.
SPANDx was designed to minimize the workload and complexity of WGS analysis for inexperienced users. One example of this is the optimized variant calling with GATK. Through preoptimized variant calling, the usually subjective and time-consuming task of specifying call settings can be improved. If required, the program still enables the user to customize settings. On an experimental data set of 21 E. coli genomes, SPANDx was able to reach a topological score of 100%, whereas scores of only 87.20% were reached for simulated data (167).
Phylogeny
Genomic characteristics that are obtained by genome characterization and comparison tools can be used to estimate the phylogeny of pathogenic isolates. Estimated phylogenies allow clinicians to establish detailed networks of transmission of outbreak strains between different patients and inform appropriate patient isolation protocols. Here clinicians might want to address the following questions. (i) Are bacterial isolates from different patients nearly identical or only distantly related? (ii) Are different pathogenic isolates from the same outbreak cluster or from separate transmission events? (iii) Which patient harbors the initial outbreak source strain?
Several phylogeny algorithms that address these questions through computing phylogeny estimates via either Bayesian methods or ML methods are available (185). These phylogeny algorithms are able to model the evolutionary signal better than neighbor-joining and parsimony methods (186) but are less suitable for large numbers of strains because of the computational costs. A number of nucleotide substitution models need to be applied to infer phylogeny as accurately as possible, but the user should accept that they are all a simplification of the actual evolutionary signal (187). The general time-reversible (GTR) model is the model most frequently used to infer phylogeny from nucleotide and SNP data. To visualize phylogenetic trees, with the most common file formats being NEXUS and Newick, GUI tools such as FigTree (http://tree.bio.ed.ac.uk/software/figtree/), MEGA (188), and Archaeopteryx (https://sites.google.com/site/cmzmasek/home/software/archaeopteryx) (189) or Web-based applications such as iTOL (http://itol.embl.de/) (190) can be used. Table 5 shows a detailed performance analysis of phylogeny tools.
TABLE 5.
Performance analysis of phylogeny toolsa
| Command line analysis tool (reference) | Concept | Run time (h) | Accuracy (%) | Input format | Output format |
|---|---|---|---|---|---|
| RAxML (191) | Maximum likelihood phylogenetic tree estimator tool; slow but very accurate | 612b | 84.47c | PHYLIP or FASTA | Newick |
| FastTree (163) | Approximately maximum likelihood phylogenetic tree estimator; fast but slightly less accurate | 2.63b | 83.6c | PHYLIP or FASTA | Newick |
| MrBayes (198) | Bayesian-based phylogenetic tree; complex to define models and not user-friendly | ND | ND | NEXUS | NEXUS |
All quantitative performance measures were taken from previously reported data, as indicated. The input type for all of these tools is aligned reads/SNPs. ND, no data.
Averages for 3 large biological data sets aligned via 3 different methods (TrueAln, PartTree, and Quicktree) (197).
Accuracy = 100% − missing branch rates (%) for 3 large biological data sets aligned via 3 different methods (TrueAln, PartTree, and Quicktree) (197).
Command line tools.
(i) RAxML.
Randomized Axelerated Maximum Likelihood (RAxML) is an ML based large-scale statistical phylogeny estimator (191). This algorithm creates an initial phylogenetic tree that presents the shortest possible tree that describes the input sequence data. Several tree optimization steps are then implemented, where the tree is rearranged. If one of these rearrangements increases the likelihood of the tree representing the evolutionary relatedness of the sequences, the tree is updated, and the process is repeated until no better solutions can be found.
The latest RAxML version, version 8, offers four different methods to assess the reliability of branches, including bootstopping, rapid bootstrapping, Shimodaira-Hasegawa (SH) test-like support values, and standard nonparametric bootstrapping, all of which have been explained in detail in previous studies (192–194). Furthermore, several posttree analyses are available for more detailed and accurate tree construction (195).
In order to apply RAxML to nucleotide and SNP data, the user must use the GTRCAT model by adding “−m GTRCAT” when running on the command line to have a correct inference of phylogeny. However, this model is not recommended for sample sizes below 50 genomes.
(ii) FastTree.
FastTree is another ML-based phylogeny estimator (163) that operates in four stages: (i) FastTree creates a starting tree and stores profiles of the internal nodes, (ii) the length of the initial tree is reduced by swapping neighboring nodes and rearranging subtrees (196), (iii) the tree likelihood is maximized via a mathematical model (CAT) that estimates variations in evolution rates across sites, and (iv) the reliability of the tree splits is evaluated by comparing tree splits to alternative topologies with the SH test (196). FastTree runs result in an average tree accuracy of 83.6% for large data sets of around 27,000 sequences. Due to modified heuristics during ML estimations, fast average run times of 2.63 h can be achieved with either 8 or 16 core processors, depending on the size of the data set (197).
FastTree needs to be executed by using the GTR+CAT model when applied to nucleotide and SNP data for inferring accurate phylogeny. The user must add “−gtr” while in the command line in order to enable the correct inference.
(iii) MrBayes.
MrBayes is a Bayesian (198) method to infer phylogeny. It uses the Markov chain Monte Carlo (MCMC) method (199). It is a command line program that, due to its many options, is not very user-friendly to execute. It is therefore advised for less-experienced users that either RAxML or FastTree be applied. Windows-, Linux-, and Mac-compatible executables are available. Compared to ML methods, MrBayes takes significantly more computing time (197), but its outcome is comparable.
Complete Outbreak Analysis Software Suites
Complete software suites can provide an “all-in-one” solution for outbreak analyses. Such suites perform analyses starting from raw sequence data to phylogeny and are able to determine genomic characteristics. For outbreak control, most commercial suites, such as BioNumerics and Ridom SeqSphere+, use a minimal spanning tree (MST), which can be proficient in many outbreak scenarios (200–202). MSTs are widely applied to display results in a convenient manner (19, 48, 105, 203). What follows are detailed descriptions of the most widely used complete outbreak analysis software suites (Table 6).
TABLE 6.
Overview of complete analysis software suitesa
| Software suite | Concept | RAM compatibility (Gb) | Run time (h) | No. of schemes | Price ($) | Source or Web address | Input format | Output format(s) |
|---|---|---|---|---|---|---|---|---|
| Commercial | ||||||||
| BioNumerics 7.6.2 | Suite containing multiple modules, thereby having many functionalities; able to perform wgMLST | ND | ND | 14 | Request quoteb | Applied Maths | FASTQ | Depends on module |
| Ridom SeqSphere+ | Suite dedicated to outbreak analyses; customizable automation flows for processing raw reads to phylogeny using either cgMLST or wgMLST; for cgMLST, it includes CT definitions | 16–32c | ND | 7 | 2,500d | Ridom Bioinformatics | FASTQ | CT, phylogeny |
| Free | ||||||||
| NCBI Pathogen Detection (beta) | NCBI-provided Web service with main focus on detection of foodborne pathogens; automated flow from raw sequences to phylogeny inference | NA | NA | 19e | Free | https://www.ncbi.nlm.nih.gov/pathogens/ | FASTQ | Web-accessible SNP tree, AMR data |
ND, no data; NA, not applicable.
Cost needs to be requested and is dependent on the number of modules.
According to the manufacturer.
One-year, 2-user accounts (academic/governmental).
Numbers of species and groups of species, as no schemes apply.
Commercial software.
(i) BioNumerics 7.6.2.
BioNumerics is a commercial software package that consists of different modules holding unique sets of software-specific tools. Users should purchase these modules separately as needed, and we recommend a minimum of two data modules for outbreak analyses: the “character data module” and the “sequence data module.” For improved and deeper data analysis, the additional “tree and network inference model” and “genome analysis tools module” are recommended. With these modules, whole-genome SNP (wgSNP) analysis and wgMLST can be performed, where 14 schemes are present for wgMLST. However, with wgMLST analyses, it is not possible to obtain a species-specific CT. It is possible to modify the scheme to a cgMLST or rMLST scheme, if desired. This package includes the possibility of creating custom schemes that make it possible to analyze more species than those of the 14 accessible schemes.
This suite provides an all-in-one solution, where raw sequence reads can be used as the input and processed all the way until phylogeny is calculated. For the detection of resistance and virulence genes, additional tools need to be applied.
BioNumerics wgMLST was previously used to identify the source of an L. monocytogenes outbreak as contaminated laboratory culture media (204) and to determine the relatedness of 13 Y. pestis isolates (205).
(ii) Ridom SeqSphere+.
Ridom SeqSphere+ is a commercial software package that has a comprehensive range of tools, such as an automated workflow from raw sequencing data to contigs, WGS marker detection, and phylogeny inference. It is user-friendly and has a complete set of work-arounds. It is possible to predefine protocols based on a series of steps functioning as a pipeline to automatically process multiple samples. For cgMLST, publicly available schemes are available (see http://www.cgmlst.org/ncs) and comprise 7 schemes. For species with no available scheme, ad hoc schemes can be created. To create these schemes, reference genomes of the corresponding species are downloaded, and a predefined workflow is then run to identify the cgMLST markers. The user should be aware that these ad hoc schemes are not stable, which in essence means that it is not possible to define a species-specific CT. SeqSphere+ also has the ability to perform wgMLST, including inferring phylogeny.
A ring trial was performed to show the continuity of SeqSphere+ in combination with standardized protocols to be able to use this method at multiple hospitals. In that study, 5 hospitals were sent 20 strains for WGS to determine the reproducibility of this method. That study showed that this method is able to identify identical clusters corresponding to the correct strains among different hospitals (19). In another study, prospective isolates from infected patients were subjected to WGS and analyzed by using SeqSphere+. Isolates were sequenced with a TAT of 4.4 days and with a success rate of 87% for the first try, identifying 14 methicillin-resistant S. aureus (MRSA) and 2 E. coli clusters with probable transmission events (55).
Free software.
(i) NCBI Pathogen Detection (beta).
NCBI Pathogen Detection is a platform for sharing data on outbreak strains, with a strong emphasis on foodborne pathogen detection. As of 8 June 2017, a total of 142,574 isolates out of 19 groups of genera/species were present. Examples of groups available and interesting for analyses of outbreaks in hospitals are E. coli, Shigella, Klebsiella pneumoniae, Enterobacter, C. freundii, and M. tuberculosis. The NCBI Pathogen Detection Web interface gives access to a database of isolates, AMR genotypes, and SNP trees. Per genus/species group, multiple SNP trees can be viewed in a Web-based interactive browser. Here strains can be selected, and the number of SNP differences can be observed. To be able to contribute to this platform, input raw sequence reads can be submitted. For submission of data, authorization is needed, which can be acquired by contacting the developers. After submission, all data become publicly available, which could potentially violate confidentiality agreements between patients and hospitals.
This platform uses raw sequence reads to determine the most closely related reference, and contigs are constructed by using a de novo assembly combined with a reference-assisted assembly. The NCBI AMR Finder process identifies AMR genes. The SNP trees are calculated by using a maximum compatibility algorithm. This algorithm shows similar results when maximum parsimony trees are applied, but the author claims that it is less sensitive to WGS artifacts and works well for closely related strains (206). This platform has high value for sharing and exploring outbreak strains but is not suitable for real-time hospital-acquired outbreaks. Combined with the public availability of the submitted data, clinicians should exercise caution when applying this platform.
DISCUSSION
Whole-genome sequencing presents a promising tool for outbreak analysis and has been predicted to replace conventional methods in the near future (82). Nevertheless, the application specificity of current sequencing technologies and the wealth of analysis tools make it nearly impossible to identify a single WGS workflow that suits the needs and capabilities of every hospital and every situation (Fig. 3). To implement the switch from conventional outbreak analysis methods to WGS, hospitals should hence carefully identify their specific needs and capacities for outbreak analysis and choose sequencing platforms and bioinformatics tools accordingly, based on their benefits and drawbacks.
FIG 3.
WGS outbreak analysis tools. Different steps in the analysis of WGS data are shown in orange (assembly, genomic characterization, comparative genomics, and phylogeny). Analysis tools are grouped by the analysis step that they perform and are separated by user interface in shades of blue (complete analysis software suites, Web based, and command line).
Advantages and Limitations of WGS Technologies: a Clinical Perspective
As shown in Table 7, the Illumina sequencing-by-synthesis technology is still the most widely used WGS solution to date and appears to be best suited for outbreak protocols where high accuracy and reliability are prioritized. Due to its well-established position on the sequencing market, Illumina has developed a range of instruments that seek to fit a variety of sequencing demands and capabilities. Disadvantages of this technology are the relatively short reads, which prohibit the resolution of large-repeat/low-complexity regions, and the possibility of incomplete base extensions (phasing and prephasing), which together increase final error rates. The high output of most Illumina instruments furthermore increases total run times, a disadvantage that is particularly important for pathogen analysis, where the rapid acquisition of results directly affects successful outbreak control. Finally, the relatively high per-run costs of Illumina sequencers imply that if a sequencing run fails, it can cost several thousands of dollars and potentially jeopardize project budgets.
TABLE 7.
Pros and cons of sequencing platforms
| Platform | Pros | Cons |
|---|---|---|
| Sequencing by synthesis | ||
| Illumina | Technology used widely by the WGS industry; lowest per-Gb sequencing cost range; highest confirmed output; wide range of Illumina machines suited for a wealth of applications and demands; lowest error rates | Rehybridization of template strands and low-copy-no. yields during bridge amplification; use of potentially biased DNA polymerases during bridge amplification; incomplete base extension (phasing, prephasing); shortest read lengths; long sequence runs; high instrument costs; no real-time data access |
| Single-molecule real-time sequencing | ||
| Pacific Biosciences | Fast sequence runs; long reads suitable for assembly of draft genomes and completion of genome assemblies; possibility of obtaining epigenetic sequence information; real-time measurement of base incorporation | Possibility of false detection of unincorporated nucleotides during sequencing; largest instrument footprint; low output per run; high error rates |
| Oxford Nanopore Technologies | Fast sequencing; longest confirmed reads; smallest instrument footprint; lowest instrument and consumables costs; real-time measurement of base incorporation; real-time data output | Sensitivity of biological nanopores to changes in exptl environment; highest error rate of all platforms; the performance of the PromethION machine is not experimentally validated |
Pacific Biosciences provides a WGS solution with high sequencing speed and is best suited for when rapid confirmation of the identity of a pathogen is given priority. The long reads produced by PacBio sequencers provide sufficient coverage for bacterial genomes and are ideal for obtaining complete genomes, a feature that can be particularly useful when the production of a high-confidence genome of the first identified outbreak strain is required as a reference genome. Its low accuracy causes the PacBio system to be more suitable for identity confirmation through draft genome closure rather than a stand-alone pathogen analysis tool.
For hospitals with limited financial and spatial capabilities or simply in situations where rapid sequencing is desired to be close to the patient bed, Oxford Nanopore Technologies (ONT) presents a promising alternative. ONT currently offers the only truly mobile sequencing device that enables high flexibility and accessibility. With the lowest instrument and consumables costs, the MinION instrument is a cost-efficient solution for hospital environments with low output demands. With the longest confirmed reads, the ONT system is best suited for identity confirmation through draft genome closure and provides sufficient coverage of bacterial genomes (207). Nanopore sequencing is in continuous development, and many advances have been made since its introduction. A comprehensive review by Magi et al. about all progress made to date and all available tools can serve as guidance for implementing nanopore sequencing in the clinic (208).
Importance of Introducing WGS Analysis Tools of Various Interface Types
The bioinformatics tools described in this review present a wide range of applications for various steps of the WGS outbreak analysis workflow, and the advantages and limitations of each tool are described in Table 8. For each step, we describe Web-based and command-line-based tools as well as a separate section for commercial software suites. While these three interface types vary in their user-friendly applications, costs, and complexities, we are convinced of the need to describe all three for the following reasons:
The price and application range of most commercial software suites would likely exceed the need for and affordability of these suites for smaller, more specified hospital practices. In these particular clinical settings, open-source, application-specific analysis tools might be a good alternative to commercial software suites.
A key factor in the management of global outbreaks is the early and successful coordination of hospital practices and government agencies around the world. Global outbreak management can succeed only if WGS data can be acquired, stored, and, most importantly, shared consistently in a variety of clinical settings. Given that many global outbreaks originate in parts of the world where health care resources are limited, as seen with the recent Ebola virus outbreak, the argument of consistent global WGS workflows becomes particularly important. Hence, in geographically isolated parts of the world, where health care resources and Internet connectivity are limited, hospitals might find command line-based analysis tools more convenient and reliable for outbreak analysis.
Command line-based tools often allow more freedom in applying specific types of analysis than with Web-based tools and software suites, which might be more or less suitable for specific hospital setups. As the switch from conventional methods to WGS-based outbreak analysis is only just starting, we believe that it is important to present the reader a wide range of user-friendly software suites and Web-based tools as well as more demanding but specified command line-based tools to allow hospital practices to identify a WGS pipeline best suited for them.
TABLE 8.
Pros and cons of analysis tools
| Algorithm | Interface type(s) | Pro(s) | Con(s) |
|---|---|---|---|
| Assembly | |||
| Velvet | Web based | Designed for repeat-rich reads; automated parameter tuning for quality control; detailed tutorial; Web-based accessibility | Small N50 contig size; technology specific; coverage cutoff excludes potentially correct low-coverage vertices; high memory usage; suitable for short reads only |
| IDBA-UD | Command line | Designed for repeat-rich short reads with various sequencing depths; among the lowest memory usages; error correction after each iteration for quality control | Technology specific; no tutorial; suitable for short reads only |
| RAY | Command line | Hybrid assembly of multiple sequencing platform reads; heuristics for contig length determination that increase quality of sequence accuracy; automated parameter calculation; detailed tutorial | Small N50 contig size; poor performance with lower-quality reads; suitable for short reads only |
| SPAdes/hybridSPAdes | Web based | Hybrid assembly of multiple sequencing platform reads; suited for short and long reads; among the lowest memory usages; largest N50 contig size; closing of gaps and resolution of repeats in assembly graph for quality control; option to merge contigs from other assemblers; detailed tutorial; Web-based accessibility | Longest computing time |
| Minimap/miniasm | Command line | Shortest computing time; compatibility with other overlapping workflows when converted to PAF format; detailed tutorial | Technology specific; no sequencing error correction; missing overlaps and misassemblies during graph cleaning; suitable for long reads only |
| Canu | Command line | Large N50 contig size; detailed tutorial; initial read correction to remove noise for quality control | Long computing time; high memory usage; suitable for long reads only |
| Genome characterization | |||
| Identification | |||
| KmerFinder | Web based | No bioinformatics skills required; easy to use; easy to interpret output; raw sequence or contig input; possible to detect contamination | Method should be set properly; no assembly is performed |
| NCBI BLAST | Web based | Largest database; multiple databases; multiple tools available | Interpretation of results can be difficult; some BLAST knowledge is advised |
| MLST Web server | Web based | Simple online workflow; no bioinformatics skills required | Suitable for samples of single species only; accepts short reads only from Illumina, Roche 454, Ion Torrent, and SOLiD |
| PathoScope 2.0 | Command line | Able to detect contamination; quality control of raw sequencing reads; complete workflow that minimizes the need for intense computational background; detailed and understandable tutorial | When testing samples with multiple strains of one species, parsimony can lead to missing of strains due to reassignment; for nearly identical strains, a coverage of >20% is necessary to distinguish between them; long computing time |
| Annotation | |||
| RAST | Web based | Web accessible; KEGG connection; graph presentation | Long waiting times; must send data to server |
| PROKKA | Command line | Short computing time; parallel annotation with 5 tools in a single workflow; detailed tutorial | Decreased annotation performance with understudied or draft genomes; suitable only for samples of single species |
| Virulence | |||
| VirulenceFinder | Web based | Easy to use; fast results; parameter control; raw sequence or contig input | Not able to detect SNP-related virulence; available for only limited groups of species/genera |
| VFDB | Web based | Extended wealth of information; more markers associated with virulence than in VirulenceFinder | Function to detect virulence markers is not easy to use; not able to detect SNP-related virulence |
| AMR | |||
| ResFinder | Web based | Fast results; parameter control; raw sequence or contig input | Not able to detect SNP-related resistance; not able to detect ampC |
| RGI/CARD | Web based | Able to detect SNP-related resistance; accession no. input possible; raw sequence or contig input; access to antibiotic resistance ontology; BLAST present; graphical views | Limited contig upload size (<20 Mb); no raw sequence data input possible |
| PlasmidFinder | Web based | Raw sequence or contig input | Limited database; detects only plasmids and does not include the presence of AMR |
| CGE BAP | Web based | Complete suite for genome characterization; easy to use | Need for subscription for access; long computing times; no annotation performed |
| Comparative genomics | |||
| PubMLST | Web based | Creates source for both MLST and cgMLST as other sets of genes used for typing; built on BIGSdb, which makes it locally installable; all databases can be downloaded; user is able to contribute to the database | Finding correct data can be difficult; built to share data publically |
| CSI Phylogeny 1.4 | Web based | Raw read and contig input possible; hqSNPs by selecting SNPs based on strict criteria; many parameters can be set | Only reference-based comparison; need to provide reference sequence; amt of parameters could be confusing for clinician without bioinformatics knowledge |
| NDtree 1.2 | Web based | Raw read input, which makes it able to skip assembly; easy to use; automatic selection of best reference using KmerFinder | Method is not comparable to others; fixed parameters; lack of documentation; only reference-based comparison |
| kSNP3 | Command line | Very fast method; automatically skips regions with high mutation frequency; easily scalable; all-to-all comparison possible; works with raw sequence data and/or contigs as input | Compared to other comparative genomics tools, overall accuracy is slightly low; no hqSNP method; bioinformatics knowledge needed |
| Roary | Command line | Protein misprediction control; detailed manual; construction of pangenome | Input has to be contigs; slow computation with larger sample sizes; relies fully on annotation accuracy |
| Pan-Seq | Command line and Web based | Minimal user interaction needed; construction of pangenome | Input has to be contigs; no exptl data on computing speed and accuracy |
| Lyve-SET | Command line | Extensive SNP filtering (hqSNP); implementation for running on a computing cluster is present | Can be too conservative in SNP calling; only reference-based comparison; bioinformatics knowledge needed |
| SPANDx | Command line | Extensive error checking, filtering, and variant identification steps during quality control (hqSNP); complete workflow from raw reads to comparative analysis; quick variant visualization through automatically generated presence/absence matrixes and error-corrected SNP and indel matrixes; works with raw sequence data as input | Only reference-based comparison; bioinformatics knowhow needed |
| Phylogeny | |||
| RAxML | Command line | Enables standard nonparametric bootstrapping, rapid bootstrapping, bootstopping, and calculation of SH-like support values for quality control; CAT and Shimodaira-Hasegawa test for quality control; comprehensive workflow; detailed manual; GTR model available | Longest computing time; highest accuracy; computationally expensive |
| FastTree | Command line | Shortest computing time; CAT and Shimodaira-Hasegawa test for quality control; GTR model available; detailed manual | Lowest accuracy due to limited initial tree improvement |
| MrBayes | Command line | Possible to optimize a model; most models available for all phylogeny methods; detailed manual; GTR model available | Input and output formats in NEXUS; complex to use |
| Complete outbreak analysis software suites | |||
| BioNumerics 7.6.2 | Local suite | Easy to use; custom schemes possible; scheme modification; wgMLST; cgMLST; rMLST; most schemes present | Separate modules needed; no cluster types |
| Ridom SeqSphere+ | Local suite | Easy to use; use of cluster types; ad hoc schemes possible; cgMLST; wgMLST | Database can be slow with many samples; fewer schemes available than with BioNumerics |
| NCBI Pathogen Detection (beta) | Web-based suite | Free to use; direct link to foodborne pathogen outbreaks; data sharing; uses collection of strains | Registration needed; focus on foodborne pathogens; data are publically available; time-consuming to register new samples; not suitable for real-time hospital-acquired outbreaks |
Real-World Implementation of WGS Outbreak Analysis: Detection of Antimicrobial Resistance
Antibiotic susceptibility testing is traditionally performed by disk diffusion or broth microdilution tests, while in clinical laboratories, automated systems for phenotypic testing are widely used. The main drawback of phenotypic susceptibility testing is the relatively long time that it can take to determine a resistance profile for an isolate. This information is clinically important, as it may guide antibiotic therapy, and several innovative methods have been developed to replace current methods for susceptibility testing (209).
WGS holds considerable promise for antimicrobial susceptibility testing (27). Sequence data can be queried to identify the presence of both acquired antibiotic resistance genes and chromosomal mutations that contribute to antibiotic resistance. WGS-based inference of antibiotic resistance phenotypes can be >95% concordant with the outcome of phenotypic testing for Enterobacteriaceae (17, 210–212) and S. aureus (213, 214). However, WGS cannot entirely replace phenotypic susceptibility testing for a number of reasons, including the high costs of WGS and the potential emergence of novel antibiotic resistance genes or mutations. In addition, sequencing runs on the Illumina platform can take several days, lagging behind traditional methods of susceptibility testing (215). The relatively long time to a result for antibiotic resistance predictions by WGS is of less concern for slow-growing bacteria such as mycobacteria, for which identification by culture and susceptibility testing can take 3 to 6 weeks (216).
In particular, important progress has been made in the implementation of WGS for the detection of antibiotic resistance in M. tuberculosis. Antibiotic-resistant M. tuberculosis strains are increasingly common and greatly complicate antibiotic therapy. In 2015, an estimated 480,000 new cases of multidrug-resistant tuberculosis and an additional 100,000 cases of rifampin-resistant tuberculosis were reported (217). Horizontal transfer of antibiotic resistance genes does not occur in M. tuberculosis, and therefore, antibiotic resistance in M. tuberculosis is due exclusively to chromosomal point mutations (218). WGS-based diagnosis and in silico susceptibility testing of M. tuberculosis are 7% cheaper and 21 days faster than traditional phenotypic testing (219). However, PCR-based typing of strains with a monophyletic distribution of phenotypes using dedicated primer designs could prove to be even faster and cheaper. A recent study showed that by sequencing DNA that was previously isolated from respiratory samples, identification of M. tuberculosis could be achieved in 44 h. In 62% of the sequenced samples, sufficient data were collected for the inference of antibiotic susceptibilities, with all results being concordant with results of phenotypic laboratory testing (220). Notably, current databases of resistance mutations in M. tuberculosis may be skewed toward strains that have been isolated in high-income countries, suggesting that current databases of resistance-conferring mutations may need to be populated by additional M. tuberculosis sequence data from low- and middle-income countries (221).
The vast majority of microbial genome sequencing is still performed on Illumina sequencers (27). However, this platform does not allow the analysis of sequence data before the completion of a run, which contributes to the relatively long time to a result for most WGS studies on the detection of antibiotic resistance determinants in bacterial pathogens. Initially, sequence data that were generated by the first models of the Oxford Nanopore Technologies MinION sequencer and its flow cells could be used to detect the presence of antibiotic resistance genes, but resistance mutations in chromosomal genes could not be identified reliably due to the relatively high error rate of reads generated at that time (222). Recent technological advances have led to an increased sequence output by the MinION system, while the error rate has decreased (223). Due to the high sequence output and generally stochastic nature of MinION sequence errors, the current state of the technology allows the sequencing of bacterial genomes with high coverage and a limited number of errors in the consensus sequence. These technological advances allowed the rapid sequencing of an M. tuberculosis genome and the reliable identification of antibiotic resistance mutations in this genome in only 12.5 h (220).
Emerging Issues and Future Directions of WGS Outbreak Analysis
Standardization.
The use of different criteria for WGS analysis by leading institutes advances to quality issues and a consequent lack of standardization. Used protocols and applications vary extensively, which makes it challenging for multiple laboratories to come to similar results. Eventually, the outcome of a bacterial whole-genome outbreak analysis should be laboratory independent. To date, only a single study applied the same workflow in different laboratories (19). Hence, there is a clear need for harmonization and a reached consensus on desired standards. There are multiple strategies that can be used to obtain these standards, with one being assay validation. Most of the validation and regulations to date apply to the use of NGS for human testing in clinical laboratories. However, some of the criteria could easily be applied to WGS outbreak analyses. Examples of such criteria are analytical performance characteristics for NGS that assess precision, accuracy, analytic sensitivity, and specificity and the assay validation framework to perform analytical validation. Hence, the most challenging part of the implementation of WGS analysis would be selecting a bioinformatics strategy or pipeline that would work consistently for all hospital-associated pathogens. This selection is critical, as validated workflows are fixed according to regulations (224).
Proficiency testing, as commonly used in routine diagnostic laboratories, is the logical next step (224, 225). The Global Microbial Identifier (GMI) is an interlaboratory proficiency test (PT) for WGS in clinical settings (http://www.globalmicrobialidentifier.org/workgroups/about-the-gmi-proficiency-tests). The GMI project is a useful start for understanding and validating variations between laboratories that perform WGS outbreak analyses. Nonetheless, the GMI PT is currently limited to three species, and therefore, more such tests have to be conducted in order to cope with all hospital-acquired pathogens (224).
An alternative PT for bioinformatics analysis would be an in silico PT, which presents a simple, inexpensive, and flexible method to evaluate bioinformatics workflows (226). Multiple studies of non-outbreak-related strains have already shown the applicability of in silico PTs for WGS (227, 228). Nonetheless, no studies that involved in silico PTs for the evaluation of WGS outbreak analysis have been conducted, which is a missed opportunity and therefore should be explored in the near future.
It needs to be ensured that standardization of WGS outbreak analyses acquires a level of flexibility to be tailored to the needs of specific situations and health care practices with various resources and capabilities.
Current state.
The drawbacks mentioned above should not withhold laboratories from starting to implement WGS in their daily routines and outbreak management, as this technology still shows increased resolution and provides a wealth of additional information, which no other method to date is able to accomplish at the same scale (50, 56, 57). During the current era of sequencing technologies, data sharing is technically feasible but runs into issues of ownership and patient data privacy. With the sharing of WGS data, there is the potential to track specific pathogens across cities, regions, and even farther. Agreement on data-sharing practices between institutes is very much possible if patient privacy is adequately protected, which in turn enables the identification of larger regional outbreaks and thus presents an immense advancement compared to previous approaches (229, 230).
The success of patient treatment and outbreak management depends largely on how fast usable sequencing data can be produced and analyzed. Short TATs could enable quick preliminary screening of resistance/virulence markers in a patient sample and determine relatedness to isolates from other patients. The vast majority of the literature states that the TAT for WGS is within 5 days, which is comparable or even shorter than those of a number of conventional methods, and WGS provides more information (84, 95, 220). With new advances in sample preparation and sequencing technologies, the TAT is predicted to decrease in the coming years. The use of long-read sequencing technologies will even further improve WGS outbreak analysis in terms of TAT, applicability, and quality.
Future perspectives.
Future developments in real-time, long-read sequencing are predicted to overcome most of the remaining technology hurdles faced today. Additionally, solving computational challenges will lead to high-quality genome sequences. In order to scale up WGS to a routine technology, quality issues need to be tackled through harmonization, quality assurance, and proficiency testing, for which initiatives are under way (224–226). Besides producing high-quality sequence reads, the analysis tools also need to be rigorously assessed and benchmarked regarding their quality. The numbers of commercial analysis software platforms are increasing, and strict standards also need to be in place for these platforms. If the above-discussed hurdles of WGS technology and analysis tools will be overcome, one might imagine the following scenario for future outbreak analysis in hospitals.
Multiple patients in an ICU ward exhibit highly similar symptoms that are indicative of a bacterial infection of the gastrointestinal tract. A doctor collects rectal swabs from each patient and inserts the samples into a point-of-care device that performs automated sample preparation within 30 min. The prepared sample is loaded into a handheld sequencing device that is connected to a laptop, and DNA sequences from the freshly prepared samples start to appear in real time. After an hour, a number of long sequence reads are produced and automatically analyzed by a bioinformatics pipeline that assembles reads, identifies strains, identifies genomic features, and creates phylogenetic relationships between sampled strains. The genomic information is compared against a database of antibiotic resistance and virulence genes and plasmids to determine if such elements are present in the sample. Based on these features, an antibiogram and phenotypic characteristics are given. If potentially pathogenic strains with high relatedness are detected, an automated alert is triggered. The alert displays, on the laptop screen, a summary of the strain classification and clinically relevant genomic features, such as AMR and virulence genes. Based on the phylogenetic information obtained, the pipeline finally visualizes the estimated route of transmission of pathogenic isolates to identify the outbreak source strain. The doctor then initiates an outbreak management protocol and treats the patients accordingly.
CLOSING REMARKS
This review aims to educate health care professionals on the use of state-of-the-art WGS technology and bioinformatics tools for nosocomial outbreak analysis. We believe that an improved understanding of bioinformatics principles by health care professionals will greatly enhance the successful transition toward WGS outbreak analysis. Future analysis tools with a stronger emphasis on clinical utility could be developed by a new generation of clinical bioinformaticians.
ACKNOWLEDGMENTS
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
J.P.M.C. and W.V.S. participated in the MinION Access Programme of Oxford Nanopore Technologies.
Biographies

Scott Quainoo obtained his bachelor's of Water Management in 2015 at the HZ University of Applied Sciences, Vlissingen, The Netherlands. In June 2017, he graduated with an M.Sc. in Microbiology from Radboud University, Nijmegen, The Netherlands. In 2015, he worked as a Graduate Research Intern at the Laboratory of Pediatric Infectious Diseases at the Radboud University Medical Centre (UMC), Nijmegen, The Netherlands. In 2016, he worked as a Graduate Research Intern at the Department of Veterinary Medicine at the University of Cambridge, United Kingdom. In August 2017, he started his Ph.D. studies at The Novo Nordisk Foundation Center for Biosustainability (DTU Biosustain), Technical University of Denmark (DTU), where he works on novel treatment strategies against MDR bacterial pathogens. His research activities focus on WGS outbreak analysis as well as the nature and frequency of antibiotic resistance transfer in human and animal microbiomes. He has actively conducted infectious disease research for over 1 year.

Jordy P. M. Coolen obtained his bachelor's of applied science in 2009 at the Fontys Hogeschool, Eindhoven, The Netherlands. Since 2009, he has worked in molecular biology, specializing in bioinformatics and next-generation sequencing at the TNO Microbiology and Systems Biology Department, Zeist, The Netherlands. He finished his master's of science in the field of bioinformatics in 2016 at Wageningen University, The Netherlands. He conducted his final thesis on improving the assembly of a plant pathogen genome using sequence data from Oxford Nanopore Technologies. Currently, he is working as a bioinformatician at the Radboud University Medical Centre (UMC), Nijmegen, The Netherlands. His work is focused on implementing whole-genome sequencing in the clinic to innovate bacterial pathogen surveillance and typing. As well, he is doing research on a variety of topics, including plasmid detection/reconstruction and resistance development in Aspergillus. He has been working in the field of bioinformatics for a total of 5 years.

Sacha A. F. T. van Hijum is a molecular biologist by training and obtained his Ph.D. in 2004 (University of Groningen, The Netherlands). He held bioinformatics postdoctoral positions at the University of Groningen and the University of Greifswald (Germany). Since 2008, he has been working at NIZO (Ede, The Netherlands) as a principal scientist in microbiomics, and in his role as associate professor, he leads the bacterial (meta)genomics group (CMBI, Radboudumc, Nijmegen, The Netherlands). His group focuses on researching microbes in relation to human health through (meta)genomics approaches. He has been working in the field for over 10 years.

Martijn A. Huynen obtained his Ph.D. in 1993 at Utrecht University, The Netherlands. After his Ph.D., he did a postdoc with Alan Perelson at the Los Alamos National Laboratory and the Sante Fe Institute in New Mexico (1993 to 1996) and a postdoc with Peer Bork at the EMBL (1996 to 2001), Heidelberg, Germany. Since 2002, he has been a professor in comparative genomics at the Radboud University Medical Centre, Nijmegen, The Netherlands. The leading theme in his research is predicting protein functions and protein interactions from various types of genomics data. Where early in his career, he was instrumental in the development of general tools, e.g., STRING, in the last 10 years, besides his continued involvement in tool development, he has specifically focused on applying these tools to biological systems like mitochondria and on validating the predictions obtained.

Willem J. G. Melchers obtained his Ph.D. in 1989 at the Free University of Amsterdam, The Netherlands. Directly after his Ph.D., he started working at the Radboud University Medical Centre (UMC), Nijmegen, The Netherlands, as Head of the Molecular Microbiology and Molecular Diagnostics section, and since 2013, he has been the Head of the Clinical Laboratory of the Department of Medical Microbiology. He is an Associate Professor in Molecular Microbiology at the Department of Medical Microbiology, Radboudumc. His research interests are in molecular microbiology. His research activities are in the molecular diagnosis and pathogenesis of infectious diseases. Part of his work involves technology development and clinical applications for the benefit of public health. He investigates the pathogenesis of infectious diseases, the impact of molecular diagnostics, the oncogenesis of human papillomavirus (HPV) infections, and resistance development in Aspergillus. He has been working in the field for a total of 30 years.

Willem van Schaik is Professor in Microbiology and Infection at the Institute of Microbiology and Infection (IMI) of the University of Birmingham (United Kingdom). He recently moved to the United Kingdom from his position as Associate Professor in the Department of Medical Microbiology of the University Medical Centre Utrecht in The Netherlands. Dr. van Schaik obtained his Ph.D. from Wageningen University in The Netherlands in 2005 and performed postdoctoral research at the Institut Pasteur (Paris, France). Over the last decade, Professor van Schaik's main research interests have centered on the characterization of the mechanisms by which commensal bacteria evolve to become multidrug-resistant opportunistic pathogens and metagenomic analysis of the reservoir of antibiotic resistance genes (the “resistome”) in complex microbiomes.

Heiman F. L. Wertheim, M.D., Ph.D., is a professor in clinical microbiology and is head of the clinical microbiology department at Radboudumc (http://www.radboudumc.nl/). Until recently, he was director of the Oxford University Clinical Research Unit (OUCRU) (http://www.oucru.org/) in Hanoi, Vietnam, which is based at the National Hospital for Tropical Diseases. He is an Associate Professor at Oxford University, United Kingdom. His main research topics are respiratory infections and antibiotic resistance, and he has used molecular techniques, including whole-genome sequencing, to tackle these issues. He recently set up a whole-genome sequencing pipeline at Radboud University. He is the author of numerous scientific papers, an editorial board member of three medical journals, and chief editor of the Atlas of Human Infectious Diseases.
REFERENCES
- 1.Wong H, Eso K, Ip A, Jones J, Kwon Y, Powelson S, de Grood J, Geransar R, Santana M, Joffe AM, Taylor G, Missaghi B, Pearce C, Ghali WA, Conly J. 2015. Use of ward closure to control outbreaks among hospitalized patients in acute care settings: a systematic review. Syst Rev 4:152. doi: 10.1186/s13643-015-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vincent J, Rello J, Marshall J, Silva E, Anzueto A, Martin C, Moreno R, Lipman J, Gomersall C, Sakr Y, Reinhart K. 2009. International study of the prevalence and outcomes of infection in intensive care units. JAMA 302:2323–2329. doi: 10.1001/jama.2009.1754. [DOI] [PubMed] [Google Scholar]
- 3.Anthony M, Bedford-Russell A, Cooper T, Fry C, Heath PT, Kennea N, McCartney M, Patel B, Pollard T, Sharland M, Wilson P. 2013. Managing and preventing outbreaks of Gram-negative infections in UK neonatal units. Arch Dis Child Fetal Neonatal Ed 98:F549–F553. doi: 10.1136/archdischild-2012-303540. [DOI] [PubMed] [Google Scholar]
- 4.Ridge KW, Hand K, Sharland M, Abubakar I, Livermore DM. 2011. Antimicrobial resistance, p 73–86. In Walker D, Fowler T (ed), Annual report of the Chief Medical Officer, vol 2 Infections and the rise of antimicrobial resistance Department of Health, London, United Kingdom. [Google Scholar]
- 5.Birt J, Le Doarea K, Kortsalioudaki C, Lawn J, Heath PT, Sharland M. 2016. Lack of evidence for the efficacy of enhanced surveillance compared to other specific interventions to control neonatal healthcare-associated infection outbreaks. Trans R Soc Trop Med Hyg 110:98–106. doi: 10.1093/trstmh/trv116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Belkum A, Tassios PT, Dijksoon L, Haeggman S, Cookson B, Fry NK, Fussing V, Green J, Feil E, Gerner-Smidt P, Brisse S, Struelens M. 2007. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin Microbiol Infect 13(Suppl 3):1–46. doi: 10.1111/j.1469-0691.2007.01786.x. [DOI] [PubMed] [Google Scholar]
- 7.Leopold SR, Goering RV, Witten A, Harmsen D, Mellmann A. 2014. Bacterial whole-genome sequencing revisited: portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J Clin Microbiol 52:2365–2370. doi: 10.1128/JCM.00262-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen Y-J, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer MLI, Jarvie TP, Jirage KB, Kim J-B, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, et al. . 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rothberg JM, Hinz W, Rearick TM, Schultz J, Mileski W, Davey M, Leamon JH, Johnson K, Milgrew MJ, Edwards M, Hoon J, Simons JF, Marran D, Myers JW, Davidson JF, Branting A, Nobile JR, Puc BP, Light D, Clark TA, Huber M, Branciforte JT, Stoner IB, Cawley SE, Lyons M, Fu Y, Homer N, Sedova M, Miao X, Reed B, Sabina J, Feierstein E, Schorn M, Alanjary M, Dimalanta E, Dressman D, Kasinskas R, Sokolsky T, Fidanza JA, Namsaraev E, McKernan KJ, Williams A, Roth GT, Bustillo J. 2011. An integrated semiconductor device enabling non-optical genome sequencing. Nature 475:348–352. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- 10.Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, Peckham H, Zeng K, Malek JA, Costa G, McKernan K, Sidow A, Fire A, Johnson SM. 2008. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res 18:1051–1063. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris SR, Cartwright EJ, Török ME, Holden MT, Brown NM, Ogilvy-Stuart AL, Ellington MJ, Quail MA, Bentley SD, Parkhill J, Peacock SJ. 2013. Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infect Dis 13:130–136. doi: 10.1016/S1473-3099(12)70268-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Köser CU, Ellington MJ, Cartwright EJP, Gillespie SH, Brown NM, Farrington M, Holden MTG, Dougan G, Bentley SD, Parkhill J, Peacock SJ. 2012. Routine use of microbial whole genome sequencing in diagnostic and public health microbiology. PLoS Pathog 8:e1002824. doi: 10.1371/journal.ppat.1002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walker TM, Ip CLC, Harrell RH, Evans JT, Kapatai G, Dedicoat MJ, Eyre DW, Wilson DJ, Hawkey PM, Crook DW, Parkhill J, Harris D, Walker AS, Bowden R, Monk P, Smith EG, Peto TEA. 2013. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis 13:137–146. doi: 10.1016/S1473-3099(12)70277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.den Bakker HC, Allard MW, Bopp D, Brown EW, Fontana J, Iqbal Z, Kinney A, Limberger R, Musser KA, Shudt M, Strain E, Wiedmann M, Wolfgang WJ. 2014. Rapid whole-genome sequencing for surveillance of Salmonella enterica serovar Enteritidis. Emerg Infect Dis 20:1306–1314. doi: 10.3201/eid2008.131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price J, Gordon NC, Crook D, Llewelyn M, Paul J. 2013. The usefulness of whole genome sequencing in the management of Staphylococcus aureus infections. Clin Microbiol Infect 19:784–789. doi: 10.1111/1469-0691.12109. [DOI] [PubMed] [Google Scholar]
- 16.Pecora ND, Li N, Allard M, Li C, Albano E, Delaney M, Dubois A, Onderdonk AB, Bry L. 2015. Genomically informed surveillance for carbapenem-resistant enterobacteriaceae in a health care system. mBio 6:e01030-15. doi: 10.1128/mBio.01030-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zankari E, Hasman H, Kaas RS, Seyfarth AM, Agersø Y, Lund O, Larsen MV, Aarestrup FM, Agerso Y, Lund O, Larsen MV, Aarestrup FM. 2013. Genotyping using whole-genome sequencing is a realistic alternative to surveillance based on phenotypic antimicrobial susceptibility testing. J Antimicrob Chemother 68:771–777. doi: 10.1093/jac/dks496. [DOI] [PubMed] [Google Scholar]
- 18.Köser CU, Ellington MJ, Peacock SJ. 2014. Whole-genome sequencing to control antimicrobial resistance. Trends Genet 30:401–407. doi: 10.1016/j.tig.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mellmann A, Andersen PS, Bletz S, Friedrich AW, Kohl TA, Lilje B, Niemann S, Prior K, Rossen JW, Harmsen D. 2017. High interlaboratory reproducibility and accuracy of next-generation-sequencing-based bacterial genotyping in a ring trial. J Clin Microbiol 55:908–913. doi: 10.1128/JCM.02242-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, Nielsen EM, Aarestrup FM. 2014. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol 52:1501–1510. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leekitcharoenphon P, Nielsen EM, Kaas RS, Lund O, Aarestrup FM. 2014. Evaluation of whole genome sequencing for outbreak detection of Salmonella enterica. PLoS One 9:e87991. doi: 10.1371/journal.pone.0087991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eyre DW, Tracey L, Elliott B, Slimings C, Huntington PG, Stuart RL, Korman TM, Kotsiou G, McCann R, Griffiths D, Fawley WN, Armstrong P, Dingle KE, Walker AS, Peto TE, Crook DW, Wilcox MH, Riley TV. 2015. Emergence and spread of predominantly communityonset Clostridium difficile PCR ribotype 244 infection in Australia, 2010 to 2012. Euro Surveill 20:21059. doi: 10.2807/1560-7917.ES2015.20.10.21059. [DOI] [PubMed] [Google Scholar]
- 23.Goodwin S, McPherson JD, McCombie WR. 2016. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet 17:333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buermans HPJ, den Dunnen JT. 2014. Next generation sequencing technology: advances and applications. Biochim Biophys Acta 1842:1932–1941. doi: 10.1016/j.bbadis.2014.06.015. [DOI] [PubMed] [Google Scholar]
- 25.Quail M, Smith ME, Coupland P, Otto TD, Harris SR, Connor TR, Bertoni A, Swerdlow HP, Gu Y. 2012. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 13:341. doi: 10.1186/1471-2164-13-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagarajan N, Pop M. 2013. Sequence assembly demystified. Nat Rev Genet 14:157–167. doi: 10.1038/nrg3367. [DOI] [PubMed] [Google Scholar]
- 27.Schürch AC, van Schaik W. 2017. Challenges and opportunities for whole-genome sequencing-based surveillance of antibiotic resistance. Ann N Y Acad Sci 1388:108–120. doi: 10.1111/nyas.13310. [DOI] [PubMed] [Google Scholar]
- 28.Gastmeier P, Loui A, Stamm-Balderjahn S, Hansen S, Zuschneid I, Sohr D, Behnke M, Obladen M, Vonberg R-P, Rüden H. 2007. Outbreaks in neonatal intensive care units—they are not like others. Am J Infect Control 35:172–176. doi: 10.1016/j.ajic.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 29.Vonberg R-P, Weitzel-Kage D, Behnke M, Gastmeier P. 2011. Worldwide Outbreak Database: the largest collection of nosocomial outbreaks. Infection 39:29–34. doi: 10.1007/s15010-010-0064-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schrader KN, Fernandez-Castro A, Cheung WKW, Crandall CM, Abbott SL. 2008. Evaluation of commercial antisera for Salmonella serotyping. J Clin Microbiol 46:685–688. doi: 10.1128/JCM.01808-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prager R, Strutz U, Fruth A, Tschäpe H. 2003. Subtyping of pathogenic Escherichia coli strains using flagellar (H)-antigens: serotyping versus fliC polymorphisms. Int J Med Microbiol 292:477–486. doi: 10.1078/1438-4221-00226. [DOI] [PubMed] [Google Scholar]
- 32.Meyer C, Stolle A, Fredriksson-Ahomaa M. 2011. Comparison of broth microdilution and disk diffusion test for antimicrobial resistance testing in Yersinia enterocolitica 4/O:3 strains. Microb Drug Resist 17:479–484. doi: 10.1089/mdr.2011.0012. [DOI] [PubMed] [Google Scholar]
- 33.Lee M, Chung HS. 2015. Different antimicrobial susceptibility testing methods to detect ertapenem resistance in Enterobacteriaceae: VITEK2, MicroScan, Etest, disk diffusion, and broth microdilution. J Microbiol Methods 112:87–91. doi: 10.1016/j.mimet.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 34.Griffin PM, Price GR, Schooneveldt JM, Schlebusch S, Tilse MH, Urbanski T, Hamilton B, Ventera D. 2012. Use of matrix-assisted laser desorption ionization–time of flight mass spectrometry to identify vancomycin-resistant enterococci and investigate the epidemiology of an outbreak. J Clin Microbiol 50:2918–2931. doi: 10.1128/JCM.01000-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Outhred AC, Jelfs P, Suliman B, Hill-Cawthorne GA, Crawford ABH, Marais BJ, Sintchenko V. 2015. Added value of whole-genome sequencing for management of highly drug-resistant TB. J Antimicrob Chemother 70:1198–1202. doi: 10.1093/jac/dku508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dallman TJ, Byrne L, Launders N, Glen K, Grant KA, Jenkins C. 2015. The utility and public health implications of PCR and whole genome sequencing for the detection and investigation of an outbreak of Shiga toxin-producing Escherichia coli serogroup O26:H11. Epidemiol Infect 143:1672–1680. doi: 10.1017/S0950268814002696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Owen RJ. 1989. Chromosomal DNA fingerprinting—a new method of species and strain identification applicable to microbial pathogens. J Med Microbiol 30:89–99. doi: 10.1099/00222615-30-2-89. [DOI] [PubMed] [Google Scholar]
- 38.Radhakrishnan I, Manju YK, Kumar RA, Mundayoor S. 2001. Implications of low frequency of IS6110 in fingerprinting field isolates of Mycobacterium tuberculosis from Kerala, India. J Clin Microbiol 39:1683. doi: 10.1128/JCM.39.4.1683.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schürch AC, Kremer K, Daviena O, Kiers A, Boeree MJ, Siezen RJ, Van Soolingen D. 2010. High-resolution typing by integration of genome sequencing data in a large tuberculosis cluster. J Clin Microbiol 48:3403–3406. doi: 10.1128/JCM.00370-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gardy JL, Johnston JC, Sui SJH, Cook VJ, Shah L, Brodkin E, Rempel S, Moore R, Zhao Y, Holt R, Varhol R, Birol I, Lem M, Sharma MK, Elwood K, Jones SJM, Brinkman FSL, Brunham RC, Tang P. 2011. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N Engl J Med 364:730–739. doi: 10.1056/NEJMoa1003176. [DOI] [PubMed] [Google Scholar]
- 41.Stackebrandt E, Goebel BM. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Evol Microbiol 44:846–849. doi: 10.1099/00207713-44-4-846. [DOI] [Google Scholar]
- 42.Mayer LW. 1988. Use of plasmid profiles in epidemiologic surveillance of disease outbreaks and in tracing the transmission of antibiotic resistance. Clin Microbiol Rev 1:228–243. doi: 10.1128/CMR.1.2.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caprioli RM, Farmer TB, Gile J. 1997. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem 69:4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 44.Winkler MA, Uher J, Cepa S. 1999. Direct analysis and identification of Helicobacter and Campylobacter species by MALDI-TOF mass spectrometry. Anal Chem 71:3416–3419. doi: 10.1021/ac990135r. [DOI] [PubMed] [Google Scholar]
- 45.Dieckmann R, Malorny B. 2011. Rapid screening of epidemiologically important Salmonella enterica subsp. enterica serovars by whole-cell matrix-assisted laser desorption ionization–time of flight mass spectrometry. Appl Environ Microbiol 77:4136–4146. doi: 10.1128/AEM.02418-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ilahi A, Hadrich I, Goudjil S, Kongolo G, Chazal C, Léké A, Ayadi A, Chouaki T, Ranque S. 2017. Molecular epidemiology of a Malassezia pachydermatis neonatal unit outbreak. Med Mycol 2017:myx022. doi: 10.1093/mmy/myx022. [DOI] [PubMed] [Google Scholar]
- 47.Pulcrano G, Roscetto E, Iula VD, Panellis D, Rossano F, Catania MR. 2012. MALDI-TOF mass spectrometry and microsatellite markers to evaluate Candida parapsilosis transmission in neonatal intensive care units. Eur J Clin Microbiol Infect Dis 31:2919–2928. doi: 10.1007/s10096-012-1642-6. [DOI] [PubMed] [Google Scholar]
- 48.Berrazeg M, Diene SM, Drissi M, Kempf M, Richet H, Landraud L, Rolain J-M. 2013. Biotyping of multidrug-resistant Klebsiella pneumoniae clinical isolates from France and Algeria using MALDI-TOF MS. PLoS One 8:e61428. doi: 10.1371/journal.pone.0061428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wieser A, Schneider L, Jung J, Schubert S. 2012. MALDI-TOF MS in microbiological diagnostics—identification of microorganisms and beyond (mini review). Appl Microbiol Biotechnol 93:965–974. doi: 10.1007/s00253-011-3783-4. [DOI] [PubMed] [Google Scholar]
- 50.Schlebusch S, Price GR, Gallagher RL, Horton-Szar V, Elbourne LDH, Griffin P, Venter DJ, Jensen SO, Van Hal SJ. 2017. MALDI-TOF MS meets WGS in a VRE outbreak investigation. Eur J Clin Microbiol Infect Dis 36:495–499. doi: 10.1007/s10096-016-2824-4. [DOI] [PubMed] [Google Scholar]
- 51.Parizad EG, Parizad EG, Valizadeh A. 2016. The application of pulsed field gel electrophoresis in clinical studies. J Clin Diagn Res 10:DE01–DE04. doi: 10.7860/JCDR/2016/15718.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kušar D, Kavalič M, Ocepek M, Zdovc I. 2013. Report on overcoming the poor quality of ApaI pulsotypes with a short review on PFGE for Listeria monocytogenes. Pol J Microbiol 62:307–309. [PubMed] [Google Scholar]
- 53.Swaminathan B, Barrett TJ, Hunter SB, Tauxe RV, CDC PulseNet Task Force. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg Infect Dis 7:382–389. doi: 10.3201/eid0703.017303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goering RV. 2010. Pulsed field gel electrophoresis: a review of application and interpretation in the molecular epidemiology of infectious disease. Infect Genet Evol 10:866–875. doi: 10.1016/j.meegid.2010.07.023. [DOI] [PubMed] [Google Scholar]
- 55.Mellmann A, Bletz S, Böking T, Kipp F, Becker K, Schultes A, Prior K, Harmsen D. 2016. Real-time genome sequencing of resistant bacteria provides precision infection control in an institutional setting. J Clin Microbiol 54:2874–2881. doi: 10.1128/JCM.00790-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harris SR, Feil EJ, Holden MTG, Quail MA, Nickerson EK, Chantratita N, Gardete S, Tavares A, Day N, Lindsay JA, Edgeworth JD, de Lencastre H, Parkhill J, Peacock SJ, Bentley SD. 2010. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Turabelidze G, Lawrence SJ, Gao H, Sodergren E, Weinstock GM, Abubucker S, Wylie T, Mitreva M, Shaikh N, Gautom R, Tarr PI. 2013. Precise dissection of an Escherichia coli O157:H7 outbreak by single nucleotide polymorphism analysis. J Clin Microbiol 51:3950–3954. doi: 10.1128/JCM.01930-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bertrand S, De Lamine de Bex G, Wildemauwe C, Lunguya O, Phoba MF, Ley B, Jacobs J, Vanhoof R, Mattheus W. 2015. Multi locus variable-number tandem repeat (MLVA) typing tools improved the surveillance of Salmonella enteritidis: a 6 years retrospective study. PLoS One 10:e0117950. doi: 10.1371/journal.pone.0117950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strommenger B, Braulke C, Heuck D, Schmidt C, Pasemann B, Nübel U, Witte W. 2008. spa typing of Staphylococcus aureus as a frontline tool in epidemiological typing. J Clin Microbiol 46:574–581. doi: 10.1128/JCM.01599-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kwong JC, Mercoulia K, Tomita T, Easton M, Li HY, Bulach DM, Stinear TP, Seemann T, Howden BP. 2016. Prospective whole-genome sequencing enhances national surveillance of Listeria monocytogenes. J Clin Microbiol 54:333–342. doi: 10.1128/JCM.02344-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maiden MCJ, Jansen van Rensburg MJ, Bray JE, Earle SG, Ford SA, Jolley KA, McCarthy ND. 2013. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol 11:728–736. doi: 10.1038/nrmicro3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pinholt M, Larner-Svensson H, Littauer P, Moser CE, Pedersen M, Lemming LE, Ejlertsen T, Søndergaard TS, Holzknecht BJ, Justesen US, Dzajic E, Olsen SS, Nielsen JB, Worning P, Hammerum AM, Westh H, Jakobsen L. 2015. Multiple hospital outbreaks of vanA Enterococcus faecium in Denmark, 2012-13, investigated by WGS, MLST and PFGE. J Antimicrob Chemother 70:2474–2482. doi: 10.1093/jac/dkv142. [DOI] [PubMed] [Google Scholar]
- 63.Hammerum AM, Hansen F, Skov MN, Stegger M, Andersen PS, Holm A, Jakobsen L, Justesen US. 2015. Investigation of a possible outbreak of carbapenem-resistant Acinetobacter baumannii in Odense, Denmark using PFGE, MLST and whole-genome-based SNPs. J Antimicrob Chemother 70:1965–1968. doi: 10.1093/jac/dkv072. [DOI] [PubMed] [Google Scholar]
- 64.Achtman M, Morelli G, Zhu P, Wirth T, Diehl I, Kusecek B, Vogler AJ, Wagner DM, Allender CJ, Easterday WR, Chenal-Francisque V, Worsham P, Thomson NR, Parkhill J, Lindler LE, Carniel E, Keim P. 2004. Microevolution and history of the plague bacillus, Yersinia pestis. Proc Natl Acad Sci U S A 101:17837–17842. doi: 10.1073/pnas.0408026101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brodrick HJ, Raven KE, Harrison EM, Blane B, Reuter S, Török ME, Parkhill J, Peacock SJ. 2016. Whole-genome sequencing reveals transmission of vancomycin-resistant Enterococcus faecium in a healthcare network. Genome Med 8:4. doi: 10.1186/s13073-015-0259-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Raven KE, Reuter S, Reynolds R, Brodrick HJ, Russell JE, Török ME, Parkhill J, Peacock SJ. 2016. A decade of genomic history for healthcare-associated Enterococcus faecium in the United Kingdom and Ireland. Genome Res 26:1388–1396. doi: 10.1101/gr.204024.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Been M, Lanza VF, de Toro M, Scharringa J, Dohmen W, Du Y, Hu J, Lei Y, Li N, Tooming-Klunderud A, Heederik DJJ, Fluit AC, Bonten MJM, Willems RJL, de la Cruz F, van Schaik W. 2014. Dissemination of cephalosporin resistance genes between Escherichia coli strains from farm animals and humans by specific plasmid lineages. PLoS Genet 10:e1004776. doi: 10.1371/journal.pgen.1004776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bhatta DR, Cavaco LM, Nath G, Kumar K, Gaur A, Gokhale S, Bhatta DR. 2016. Association of Panton Valentine leukocidin (PVL) genes with methicillin resistant Staphylococcus aureus (MRSA) in Western Nepal: a matter of concern for community infections (a hospital based prospective study). BMC Infect Dis 16:199. doi: 10.1186/s12879-016-1531-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Laprade N, Cloutier M, Lapen DR, Topp E, Wilkes G, Villemur R, Khan IUH. 2016. Detection of virulence, antibiotic resistance and toxin (VAT) genes in Campylobacter species using newly developed multiplex PCR assays. J Microbiol Methods 124:41–47. doi: 10.1016/j.mimet.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 70.Nelson MU, Bizzarro MJ, Baltimore RS, Dembry LM, Gallagher PG. 2015. Clinical and molecular epidemiology of methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit in the decade following implementation of an active detection and isolation program. J Clin Microbiol 53:2492–2501. doi: 10.1128/JCM.00470-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Whiteduck-Léveillée J, Cloutier M, Topp E, Lapen DR, Talbot G, Villemur R, Khan IUH. 2016. Development and evaluation of multiplex PCR assays for rapid detection of virulence-associated genes in Arcobacter species. J Microbiol Methods 121:59–65. doi: 10.1016/j.mimet.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 72.Milan A, Furlanis L, Cian F, Bressan R, Luzzati R, Lagatolla C, Deiana ML, Knezevich A, Tonin E, Dolzani L. 2016. Epidemic dissemination of a carbapenem-resistant Acinetobacter baumannii clone carrying armA two years after its first isolation in an Italian hospital. Microb Drug Resist 22:668–674. doi: 10.1089/mdr.2015.0167. [DOI] [PubMed] [Google Scholar]
- 73.Odinot PT, Meis JF, Van den Hurk PJ, Hoogkamp-Korstanje J, Melchers WJ. 1995. PCR-based characterization of Yersinia enterocolitica: comparison with biotyping and serotyping. Epidemiol Infect 115:269–277. doi: 10.1017/S0950268800058398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wolf DG, Falk R, Hacham M, Theelen B, Boekhout T, Scorzetti G, Shapiro M, Block C, Salkin IF, Polacheck I. 2001. Multidrug-resistant Trichosporon asahii infection of nongranulocytopenic patients in three intensive care units. J Clin Microbiol 39:4420–4425. doi: 10.1128/JCM.39.12.4420-4425.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lowe AM, Beattie DT, Deresiewicz RL. 1998. Identification of novel staphylococcal virulence genes by in vivo expression technology. Mol Microbiol 27:967–976. doi: 10.1046/j.1365-2958.1998.00741.x. [DOI] [PubMed] [Google Scholar]
- 76.Cheung AL, Bayer AS, Zhang G, Gresham H, Xiong Y-Q. 2004. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol Med Microbiol 40:1–9. doi: 10.1016/S0928-8244(03)00309-2. [DOI] [PubMed] [Google Scholar]
- 77.Hasnain SE, O'Toole RF, Grover S, Ehtesham NZ. 2015. Whole genome sequencing: a new paradigm in the surveillance and control of human tuberculosis. Tuberculosis 95:91–94. doi: 10.1016/j.tube.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 78.Olive DM, Bean P. 1999. Principles and applications of methods for DNA-based typing of microbial organisms. J Clin Microbiol 37:1661–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lecuit M, Eloit M. 2014. The diagnosis of infectious diseases by whole genome next generation sequencing: a new era is opening. Front Cell Infect Microbiol 4:25. doi: 10.3389/fcimb.2014.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Desai A, Marwah VS, Yadav A, Jha V, Dhaygude K, Bangar U, Kulkarni V, Jere A. 2013. Identification of optimum sequencing depth especially for de novo genome assembly of small genomes using next generation sequencing data. PLoS One 8:e60204. doi: 10.1371/journal.pone.0060204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fleischmann R, Adams M, White O, Clayton R, Kirkness E, Kerlavage A, Bult C, Tomb J, Dougherty B, Merrick J, Al E. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 82.Land M, Hauser L, Jun S-R, Nookaew I, Leuze MR, Ahn T-H, Karpinets T, Lund O, Kora G, Wassenaar T, Poudel S, Ussery DW. 2015. Insights from 20 years of bacterial genome sequencing. Funct Integr Genomics 15:141–161. doi: 10.1007/s10142-015-0433-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Cheetham RK, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, Rasolonjatovo IMJ, Reed MT, Rigatti R, Rodighiero C, Ross MT, Sabot A, Sankar SV, Scally A, Schroth GP, Smith ME, Smith VP, Spiridou A, Torrance PE, Tzonev SS, Vermaas EH, Walter K, Wu X, Zhang L, Alam MD, Anastasi C, et al. . 2008. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McGann P, Bunin JL, Snesrud E, Singh S, Maybank R, Ong AC, Kwak YI, Seronello S, Clifford RJ, Hinkle M, Yamada S, Barnhill J, Lesho E. 2016. Real time application of whole genome sequencing for outbreak investigation—what is an achievable turnaround time? Diagn Microbiol Infect Dis 85:277–282. doi: 10.1016/j.diagmicrobio.2016.04.020. [DOI] [PubMed] [Google Scholar]
- 85.Rhoads A, Au KF. 2015. PacBio sequencing and its applications. Genomics Proteomics Bioinformatics 13:278–289. doi: 10.1016/j.gpb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, Bibillo A, Bjornson K, Chaudhuri B, Christians F, Cicero R, Clark S, Dalal R, DeWinter A, Dixon J, Foquet M, Gaertner A, Hardenbol P, Heiner C, Hester K, Holden D, Kearns G, Kong X, Kuse R, Lacroix Y, Lin S, Lundquist P, Ma C, Marks P, Maxham M, Murphy D, Park I, Pham T, Phillips M, Roy J, Sebra R, Shen G, Sorenson J, Tomaney A, Travers K, Trulson M, Vieceli J, Wegener J, Wu D, Yang A, Zaccarin D, et al. . 2009. Real-time DNA sequencing from single polymerase molecules. Science 323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 87.Mardis ER. 2013. Next-generation sequencing platforms. Annu Rev Anal Chem 6:287–303. doi: 10.1146/annurev-anchem-062012-092628. [DOI] [PubMed] [Google Scholar]
- 88.Kanamori H, Parobek CM, Weber DJ, van Duin D, Rutala WA, Cairns BA, Juliano JJ. 2016. Next-generation sequencing and comparative analysis of sequential outbreaks caused by multidrug-resistant Acinetobacter baumannii at a large academic burn center. Antimicrob Agents Chemother 60:1249–1257. doi: 10.1128/AAC.02014-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Heger M. 1 October 2015. PacBio launches higher-throughput, lower-cost single-molecule sequencing system. GenomeWeb, New York, NY: https://www.genomeweb.com/business-news/pacbio-launches-higher-throughput-lower-cost-single-molecule-sequencing-system. [Google Scholar]
- 90.Krol A. 1 October 2015. A worthy sequel: PacBio's new sequencing system. Bio-IT World, Needham, MA: http://www.bio-itworld.com/2015/10/1/a-worthy-sequel.aspx. [Google Scholar]
- 91.Ku C-S, Roukos DH. 2013. From next-generation sequencing to nanopore sequencing technology: paving the way to personalized genomic medicine. Expert Rev Med Devices 10:1–6. doi: 10.1586/erd.12.63. [DOI] [PubMed] [Google Scholar]
- 92.Quick J, Ashton P, Calus S, Chatt C, Gossain S, Hawker J, Nair S, Neal K, Nye K, Peters T, De Pinna E, Robinson E, Struthers K, Webber M, Catto A, Dallman TJ, Hawkey P, Loman NJ. 2015. Rapid draft sequencing and real-time nanopore sequencing in a hospital outbreak of Salmonella. Genome Biol 16:114. doi: 10.1186/s13059-015-0677-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schmidt K, Mwaigwisya S, Crossman LC, Doumith M, Munroe D, Pires C, Khan MA, Woodford N, Saunders NJ, Wain J, O'Grady J, Livermore DM. 2017. Identification of bacterial pathogens and antimicrobial resistance directly from clinical urines by nanopore-based metagenomic sequencing. J Antimicrob Chemother 72:104–114. doi: 10.1093/jac/dkw397. [DOI] [PubMed] [Google Scholar]
- 94.van der Helm E, Imamovic L, Hashim Ellabaan MM, van Schaik W, Koza A, Sommer MOA. 2017. Rapid resistome mapping using nanopore sequencing. Nucleic Acids Res 45:e61. doi: 10.1093/nar/gkw1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lindsey RL, Pouseele H, Chen JC, Strockbine NA, Carleton HA. 2016. Implementation of whole genome sequencing (WGS) for identification and characterization of Shiga toxin-producing Escherichia coli (STEC) in the United States. Front Microbiol 7:766. doi: 10.3389/fmicb.2016.00766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Boisvert S, Laviolette F, Corbeil J. 2010. Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comput Biol 17:1519–1533. doi: 10.1089/cmb.2009.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pitcher DG, Saunders NA, Owen RJ. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol 8:151–156. doi: 10.1111/j.1472-765X.1989.tb00262.x. [DOI] [Google Scholar]
- 98.Neumann B, Pospiech A, Schairer HU. 1992. Rapid isolation of genomic DNA from gram-negative bacteria. Trends Genet 8:332–333. doi: 10.1016/0168-9525(92)90269-A. [DOI] [PubMed] [Google Scholar]
- 99.Smith K, Diggle MA, Clarke SC. 2003. Comparison of commercial DNA extraction kits for extraction of bacterial genomic DNA from whole-blood samples. J Clin Microbiol 41:2440–2443. doi: 10.1128/JCM.41.6.2440-2443.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cheng H-R, Jiang N. 2006. Extremely rapid extraction of DNA from bacteria and yeasts. Biotechnol Lett 28:55–59. doi: 10.1007/s10529-005-4688-z. [DOI] [PubMed] [Google Scholar]
- 101.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Powers JG, Weigman VJ, Shu J, Pufky JM, Cox D, Hurban P. 2013. Efficient and accurate whole genome assembly and methylome profiling of E. coli. BMC Genomics 14:675. doi: 10.1186/1471-2164-14-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zerbino DR. 2010. Using the Velvet de novo assembler for short-read sequencing technologies. Curr Protoc Bioinformatics Chapter 11:Unit 11.5. doi: 10.1002/0471250953.bi1105s31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pinholt M, Gumpert H, Bayliss S, Nielsen JB, Vorobieva V, Pedersen M, Feil E, Worning P, Westh H. 2017. Genomic analysis of 495 vancomycin-resistant Enterococcus faecium reveals broad dissemination of a vanA plasmid in more than 19 clones from Copenhagen, Denmark. J Antimicrob Chemother 72:40–47. doi: 10.1093/jac/dkw360. [DOI] [PubMed] [Google Scholar]
- 105.Eyre DW, Golubchik T, Gordon NC, Bowden R, Piazza P, Batty EM, Ip CLC, Wilson DJ, Didelot X, O'Connor L, Lay R, Buck D, Kearns AM, Shaw A, Paul J, Wilcox MH, Donnelly PJ, Peto TEA, Walker AS, Crook DW. 2012. A pilot study of rapid benchtop sequencing of Staphylococcus aureus and Clostridium difficile for outbreak detection and surveillance. BMJ Open 2:e001124. doi: 10.1136/bmjopen-2012-001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Prjibelski AD, Vasilinetc I, Bankevich A, Gurevich A, Krivosheeva T, Nurk S, Pham S, Korobeynikov A, Lapidus A, Pevzner PA. 2014. ExSPAnder: a universal repeat resolver for DNA fragment assembly. Bioinformatics 30:i293–i301. doi: 10.1093/bioinformatics/btu266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Thomsen MCF, Ahrenfeldt J, Cisneros JLB, Jurtz V, Larsen MV, Hasman H, Aarestrup FM, Lund O. 2016. A bacterial analysis platform: an integrated system for analysing bacterial whole genome sequencing data for clinical diagnostics and surveillance. PLoS One 11:e0157718. doi: 10.1371/journal.pone.0157718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Peng Y, Leung HCM, Yiu SM, Chin FYL. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 109.Peng Y, Leung HCM, Yiu SM, Chin FYL. 2010. IDBA—a practical iterative de Bruijn graph de novo assembler, p 426–440. In Berger B. (ed), research in computational molecular biology. Springer, Berlin, Germany. [Google Scholar]
- 110.Zhu X, Leung HCM, Chin FYL, Yiu SM, Quan G, Liu B, Wang Y. 2014. PERGA: a paired-end read guided de novo assembler for extending contigs using SVM and look ahead approach. PLoS One 9:e114253. doi: 10.1371/journal.pone.0114253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Coolen JPM, Sjödin A, Maraha B, Hajer GF, Forsman M, Verspui E, Frenay HME, Notermans DW, de Vries MC, Reubsaet FAG, Paauw A, Roeselers G. 2013. Draft genome sequence of Francisella tularensis subsp. holarctica BD11-00177. Stand Genomic Sci 8:539–547. doi: 10.4056/sigs.4217923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Batzoglou S. 2002. ARACHNE: a whole-genome shotgun assembler. Genome Res 12:177–189. doi: 10.1101/gr.208902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, Flanigan MJ, Kravitz SA, Mobarry CM, Reinert KH, Remington KA, Anson EL, Bolanos RA, Chou HH, Jordan CM, Halpern AL, Lonardi S, Beasley EM, Brandon RC, Chen L, Dunn PJ, Lai Z, LIang Y, Nusskern DR, Zhan M, Zhang Q, Zheng X, Rubin GM, Adams MD, Venter JC. 2000. A whole-genome assembly of Drosophila. Science 287:2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- 115.Hernandez D, François P, Farinelli L, Østerås M, Schrenzel J. 2008. De novo bacterial genome sequencing: millions of very short reads assembled on a desktop computer. Genome Res 18:802–809. doi: 10.1101/gr.072033.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li H. 2016. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics 32:2103–2110. doi: 10.1093/bioinformatics/btw152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sović I, Križanović K, Skala K, Šikić M. 2016. Evaluation of hybrid and non-hybrid methods for de novo assembly of nanopore reads. Bioinformatics 32:2582–2589. doi: 10.1093/bioinformatics/btw237. [DOI] [PubMed] [Google Scholar]
- 118.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Judge K, Hunt M, Reuter S, Tracey A, Quail MA, Parkhill J, Peacock SJ. 2016. Comparison of bacterial genome assembly software for MinION data and their applicability to medical microbiology. Microb Genom 2:e000085. doi: 10.1099/mgen.0.000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Antipov D, Korobeynikov A, McLean JS, Pevzner PA. 2016. hybridSPAdes: an algorithm for hybrid assembly of short and long reads. Bioinformatics 32:1009–1015. doi: 10.1093/bioinformatics/btv688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Larsen MV, Cosentino S, Lukjancenko O, Saputra D, Rasmussen S, Hasman H, Sicheritz-Ponten T, Aarestrup FM, Ussery DW, Lund O. 2014. Benchmarking of methods for genomic taxonomy. J Clin Microbiol 52:1529–1539. doi: 10.1128/JCM.02981-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hasman H, Saputra D, Sicheritz-Ponten T, Lund O, Svendsen CA, Frimodt-Moller N, Aarestrup FM. 2014. Rapid whole-genome sequencing for detection and characterization of microorganisms directly from clinical samples. J Clin Microbiol 52:139–146. doi: 10.1128/JCM.02452-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 124.Santiago-Sotelo P, Ramirez-Prado JH. 2012. prfectBLAST: a platform-independent portable front end for the command terminal BLAST+ stand-alone suite. Biotechniques 53:299–300. doi: 10.2144/000113953. [DOI] [PubMed] [Google Scholar]
- 125.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Ponten T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hong C, Manimaran S, Shen Y, Perez-Rogers JF, Byrd AL, Castro-Nallar E, Crandall KA, Johnson W. 2014. PathoScope 2.0: a complete computational framework for strain identification in environmental or clinical sequencing samples. Microbiome 2:33. doi: 10.1186/2049-2618-2-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Byrd AL, Perez-Rogers JF, Manimaran S, Castro-Nallar E, Toma I, McCaffrey T, Siegel M, Benson G, Crandall KA, Johnson WE. 2014. Clinical PathoScope: rapid alignment and filtration for accurate pathogen identification in clinical samples using unassembled sequencing data. BMC Bioinformatics 15:262. doi: 10.1186/1471-2105-15-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the Rapid Annotation of Microbial Genomes Using Subsystems Technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Edwards RA, Olson R, Disz T, Pusch GD, Vonstein V, Stevens R, Overbeek R. 2012. Real time metagenomics: using k-mers to annotate metagenomes. Bioinformatics 28:3316–3317. doi: 10.1093/bioinformatics/bts599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 133.Lagesen K, Hallin P, Rødland EA, Stærfeldt HH, Rognes T, Ussery DW. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 136.Nawrocki EP, Eddy SR. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chen L, Yang J, Yu J, Yao Z, Sun L, Shen Y, Jin Q. 2005. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res 33:D325–D328. doi: 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.McArthur AG, Tsang KK. 2017. Antimicrobial resistance surveillance in the genomic age. Ann N Y Acad Sci 1388:78–91. doi: 10.1111/nyas.13289. [DOI] [PubMed] [Google Scholar]
- 140.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 141.Xavier BB, Lammens C, Ruhal R, Malhotra-Kumar S, Butaye P, Goossens H, Malhotra-Kumar S. 2016. Identification of a novel plasmid-mediated colistin resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill 21:30280. doi: 10.2807/1560-7917.ES.2016.21.27.30280. [DOI] [PubMed] [Google Scholar]
- 142.Di Pilato V, Arena F, Tascini C, Cannatelli A, Henrici De Angelis L, Fortunato S, Giani T, Menichetti F, Rossolini GM. 2016. mcr-1.2, a new mcr variant carried on a transferable plasmid from a colistin-resistant KPC carbapenemase-producing Klebsiella pneumoniae strain of sequence type 512. Antimicrob Agents Chemother 60:5612–5615. doi: 10.1128/AAC.01075-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yang Y-Q, Li Y-X, Song T, Yang Y-X, Jiang W, Zhang A-Y, Guo X-Y, Liu B-H, Wang Y-X, Lei C-W, Xiang R, Wang H-N. 2017. Colistin resistance gene mcr-1 and its variant in Escherichia coli isolates from chickens in China. Antimicrob Agents Chemother 61:e01204-16. doi: 10.1128/AAC.01204-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, Bhullar K, Canova MJ, De Pascale G, Ejim L, Kalan L, King AM, Koteva K, Morar M, Mulvey MR, O'Brien JS, Pawlowski AC, Piddock LJV, Spanogiannopoulos P, Sutherland AD, Tang I, Taylor PL, Thaker M, Wang W, Yan M, Yu T, Wright GD. 2013. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother 57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.McArthur AG, Wright GD. 2015. Bioinformatics of antimicrobial resistance in the age of molecular epidemiology. Curr Opin Microbiol 27:45–50. doi: 10.1016/j.mib.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 146.Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, Lago BA, Dave BM, Pereira S, Sharma AN, Doshi S, Courtot M, Lo R, Williams LE, Frye JG, Elsayegh T, Sardar D, Westman EL, Pawlowski AC, Johnson TA, Brinkman FSL, Wright GD, McArthur AG. 2017. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Liu B, Pop M. 2009. ARDB—antibiotic resistance genes database. Nucleic Acids Res 37:D443–D447. doi: 10.1093/nar/gkn656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Martínez-Martínez L, Pascual A, Jacoby GA. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797–799. doi: 10.1016/S0140-6736(97)07322-4. [DOI] [PubMed] [Google Scholar]
- 149.Boyd DA, Tyler S, Christianson S, McGeer A, Muller MP, Willey BM, Bryce E, Gardam M, Nordmann P, Mulvey MR. 2004. Complete nucleotide sequence of a 92-kilobase plasmid harboring the CTX-M-15 extended-spectrum beta-lactamase involved in an outbreak in long-term-care facilities in Toronto, Canada. Antimicrob Agents Chemother 48:3758–3764. doi: 10.1128/AAC.48.10.3758-3764.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.White AP, Sibley KA, Sibley CD, Wasmuth JD, Schaefer R, Surette MG, Edge TA, Neumann NF. 2011. Intergenic sequence comparison of Escherichia coli isolates reveals lifestyle adaptations but not host specificity. Appl Environ Microbiol 77:7620–7632. doi: 10.1128/AEM.05909-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhi S, Li Q, Yasui Y, Edge T, Topp E, Neumann NF. 2015. Assessing host-specificity of Escherichia coli using a supervised learning logic-regression-based analysis of single nucleotide polymorphisms in intergenic regions. Mol Phylogenet Evol 92:72–81. doi: 10.1016/j.ympev.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 154.Gardner SN, Slezak T, Hall BG. 2015. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31:2877–2878. doi: 10.1093/bioinformatics/btv271. [DOI] [PubMed] [Google Scholar]
- 155.PLoS One Staff. 2015. Correction, When whole-genome alignments just won't work: kSNP v2 software for alignment-free SNP discovery and phylogenetics of hundreds of microbial genomes. PLoS One 10:e0118258. doi: 10.1371/journal.pone.0118258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Köser CU, Holden MTG, Ellington MJ, Cartwright EJP, Brown NM, Ogilvy-Stuart AL, Hsu LY, Chewapreecha C, Croucher NJ, Harris SR, Sanders M, Enright MC, Dougan G, Bentley SD, Parkhill J, Fraser LJ, Betley JR, Schulz-Trieglaff OB, Smith GP, Peacock SJ. 2012. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N Engl J Med 366:2267–2275. doi: 10.1056/NEJMoa1109910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Enright AJ, Van Dongen S, Ouzounis CA. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Jolley KA, Maiden MCJ. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Jackson BR, Tarr C, Strain E, Jackson KA, Conrad A, Carleton H, Katz LS, Stroika S, Gould LH, Mody RK, Silk BJ, Beal J, Chen Y, Timme R, Doyle M, Fields A, Wise M, Tillman G, Defibaugh-Chavez S, Kucerova Z, Sabol A, Roache K, Trees E, Simmons M, Wasilenko J, Kubota K, Pouseele H, Klimke W, Besser J, Brown E, Allard M, Gerner-Smidt P. 2016. Implementation of nationwide real-time whole-genome sequencing to enhance listeriosis outbreak detection and investigation. Clin Infect Dis 63:380–386. doi: 10.1093/cid/ciw242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Karah N, Jolley KA, Hall RM, Uhlin BE, Lemarie C, Gaultier M. 2017. Database for the ampC alleles in Acinetobacter baumannii. PLoS One 12:e0176695. doi: 10.1371/journal.pone.0176695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kaas RS, Leekitcharoenphon P, Aarestrup FM, Lund O. 2014. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One 9:e104984. doi: 10.1371/journal.pone.0104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Price MN, Dehal PS, Arkin AP. 2009. Fasttree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Rosendahl Madsen AM, Holm A, Jensen TG, Knudsen E, Lundgaard H, Skov MN, Uldum SA, Kemp M. 2017. Whole-genome sequencing for identification of the source in hospital-acquired Legionnaires' disease. J Hosp Infect 96:392–395. doi: 10.1016/j.jhin.2017.04.020. [DOI] [PubMed] [Google Scholar]
- 165.Rasmussen LH, Dargis R, Højholt K, Christensen JJ, Skovgaard O, Justesen US, Rosenvinge FS, Moser C, Lukjancenko O, Rasmussen S, Nielsen XC. 2016. Whole genome sequencing as a tool for phylogenetic analysis of clinical strains of mitis group streptococci. Eur J Clin Microbiol Infect Dis 35:1615–1625. doi: 10.1007/s10096-016-2700-2. [DOI] [PubMed] [Google Scholar]
- 166.Sokal Michener C. 1958. A statistical method for evaluating systematic relationships. Univ Kansas Sci Bull 38:1409–1438. [Google Scholar]
- 167.Sahl JW, Lemmer D, Travis J, Schupp J, Gillece J, Aziz M, Driebe E, Drees K, Hicks N, Williamson C, Hepp C, Smith D, Roe C, Engelthaler D, Wagner D, Keim P. 2016. The Northern Arizona SNP Pipeline (NASP): accurate, flexible, and rapid identification of SNPs in WGS datasets. bioRxiv doi: 10.1101/037267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Mercante JW, Morrison SS, Desai HP, Raphael BH, Winchell JM. 2016. Genomic analysis reveals novel diversity among the 1976 Philadelphia Legionnaires' disease outbreak isolates and additional ST36 strains. PLoS One 11:e0164074. doi: 10.1371/journal.pone.0164074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Jiménez A, Castro JG, Silvia Munoz-Price L, De Pascale D, Shimose L, Mustapha MM, Spychala CN, Mettus RT, Cooper VS, Doi Y. 2017. Outbreak of Klebsiella pneumoniae carbapenemase-producing Citrobacter freundii at a tertiary acute care facility in Miami, Florida. Infect Control Hosp Epidemiol 38:320–326. doi: 10.1017/ice.2016.273. [DOI] [PubMed] [Google Scholar]
- 172.Moradigaravand D, Boinett CJ, Martin V, Peacock SJ, Parkhill J. 2016. Recent independent emergence of multiple multidrug-resistant Serratia marcescens clones within the United Kingdom and Ireland. Genome Res 26:1101–1109. doi: 10.1101/gr.205245.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.van Vliet AHM. 2017. Use of pan-genome analysis for the identification of lineage-specific genes of Helicobacter pylori. FEMS Microbiol Lett 364:fnw296. doi: 10.1093/femsle/fnw296. [DOI] [PubMed] [Google Scholar]
- 174.Cowley L, Dallman T, Fitzgerald S, Irvine N, Rooney P, McAteer S, Day M, Perry N, Bono J, Jenkins C, Gally D. 2016. Short-term evolution of Shiga toxin-producing Escherichia coli O157:H7 between two food-borne outbreaks. Microb Genomics 2:e000084. doi: 10.1099/mgen.0.000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Laing C, Buchanan C, Taboada EN, Zhang Y, Kropinski A, Villegas A, Thomas JE, Gannon VPJ. 2010. Pan-genome sequence analysis using Panseq: an online tool for the rapid analysis of core and accessory genomic regions. BMC Bioinformatics 11:461. doi: 10.1186/1471-2105-11-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol 5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sankarasubramanian J, Vishnu US, Sridhar J, Gunasekaran P, Rajendhran J. 2015. Pan-genome of Brucella species. Indian J Microbiol 55:88–101. doi: 10.1007/s12088-014-0486-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Laing C, Villegas A, Taboada EN, Kropinski A, Thomas JE, Gannon VPJ. 2011. Identification of Salmonella enterica species- and subgroup-specific genomic regions using Panseq 2.0. Infect Genet Evol 11:2151–2161. doi: 10.1016/j.meegid.2011.09.021. [DOI] [PubMed] [Google Scholar]
- 179.Katz LS, Griswold T, Williams-Newkirk AJ, Wagner D, Petkau A, Sieffert C, Van Domselaar G, Deng X, Carleton HA. 2017. A comparative analysis of the Lyve-SET phylogenomics pipeline for genomic epidemiology of foodborne pathogens. Front Microbiol 8:375. doi: 10.3389/fmicb.2017.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Tate H, Folster JP, Hsu C-H, Chen J, Hoffmann M, Li C, Morales C, Tyson GH, Mukerjee S, Brown AC, Green A, Wilson W, Dessai U, Abbott J, Joseph L, Haro J, Ayers S, McDermott PF, Zhao S. 2017. Comparative analysis of extended-spectrum-β-lactamase CTX-M-65-producing Salmonella enterica serovar Infantis isolates from humans, food animals, and retail chickens in the United States. Antimicrob Agents Chemother 61:e00488-17. doi: 10.1128/AAC.00488-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Haley BJ, Kim SW, Pettengill J, Luo Y, Karns JS, Van Kessel JAS. 2016. Genomic and evolutionary analysis of two Salmonella enterica serovar Kentucky sequence types isolated from bovine and poultry sources in North America. PLoS One 11:e0161225. doi: 10.1371/journal.pone.0161225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Sarovich DS, Price EP. 2014. SPANDx: a genomics pipeline for comparative analysis of large haploid whole genome re-sequencing datasets. BMC Res Notes 7:618. doi: 10.1186/1756-0500-7-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 https://arxiv.org/abs/1303.3997. [Google Scholar]
- 184.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Holder M, Lewis PO. 2003. Phylogeny estimation: traditional and Bayesian approaches. Nat Rev Genet 4:275–284. doi: 10.1038/nrg1044. [DOI] [PubMed] [Google Scholar]
- 186.Williams TL, Moret BME. 2003. An investigation of phylogenetic likelihood methods, p 79–86. In Proceedings of the Third IEEE Symposium on Bioinformatics and Bioengineering IEEE Computer Society, Washington, DC. [Google Scholar]
- 187.Posada D, Crandall KA. 2001. Selecting the best-fit model of nucleotide substitution. Syst Biol 50:580–601. doi: 10.1080/10635150118469. [DOI] [PubMed] [Google Scholar]
- 188.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Han MV, Zmasek CM. 2009. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics 10:356. doi: 10.1186/1471-2105-10-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Letunic I, Bork P. 2016. Interactive Tree of Life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Stamatakis A, Ludwig T, Meier H. 2005. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- 192.Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol 57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 193.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 194.Pattengale ND, Alipour M, Bininda-Emonds ORP, Moret BME, Stamatakis A. 2010. How many bootstrap replicates are necessary? J Comput Biol 17:337–354. doi: 10.1089/cmb.2009.0179. [DOI] [PubMed] [Google Scholar]
- 195.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Liu K, Linder CR, Warnow T. 2011. RAxML and FastTree: comparing two methods for large-scale maximum likelihood phylogeny estimation. PLoS One 6:e27731. doi: 10.1371/journal.pone.0027731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 199.Geyer CJ. 1991. Markov chain Monte Carlo maximum likelihood, p 156–163. In Computing science and statistics: proceedings of the 23rd Symposium of the Interface. Interface Foundation of North America, Fairfax Station, VA. [Google Scholar]
- 200.Kruskal JB. 1956. On the shortest spanning subtree of a graph and the traveling salesman problem. Proc Am Math Soc 7:48–50. [Google Scholar]
- 201.Prim RC. 1957. Shortest connection networks and some generalizations. Bell Syst Tech J 36:1389–1401. doi: 10.1002/j.1538-7305.1957.tb01515.x. [DOI] [Google Scholar]
- 202.Cormen TH, Leiserson CE, Rivest RL, Stein C. 2009. Introduction to algorithms. MIT Press, Cambridge, MA. [Google Scholar]
- 203.Mellmann A, Harmsen D, Cummings CA, Zentz EB, Leopold SR, Rico A, Prior K, Szczepanowski R, Ji Y, Zhang W, McLaughlin SF, Henkhaus JK, Leopold B, Bielaszewska M, Prager R, Brzoska PM, Moore RL, Guenther S, Rothberg JM, Karch H. 2011. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One 6:e22751. doi: 10.1371/journal.pone.0022751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Matanock A, Katz LS, Jackson KA, Kucerova Z, Conrad AR, Glover WA, Nguyen V, Mohr MC, Marsden-Haug N, Thompson D, Dunn JR, Stroika S, Melius B, Tarr C, Dietrich SE, Kao AS, Kornstein L, Li Z, Maroufi A, Marder EP, Meyer R, Perez-Osorio AC, Reddy V, Reporter R, Carleton H, Tweeten S, Waechter H, Yee LM, Wise ME, Davis K, Jackson BR. 2016. Two Listeria monocytogenes pseudo-outbreaks caused by contaminated laboratory culture media. J Clin Microbiol 54:768–770. doi: 10.1128/JCM.02035-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kingry LC, Rowe LA, Respicio-Kingry LB, Beard CB, Schriefer ME, Petersen JM. 2016. Whole genome multilocus sequence typing as an epidemiologic tool for Yersinia pestis. Diagn Microbiol Infect Dis 84:275–280. doi: 10.1016/j.diagmicrobio.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Cherry JL. 2017. A practical exact maximum compatibility algorithm for reconstruction of recent evolutionary history. BMC Bioinformatics 18:127. doi: 10.1186/s12859-017-1520-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Loman NJ, Quick J, Simpson JT. 2015. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat Methods 12:733–735. doi: 10.1038/nmeth.3444. [DOI] [PubMed] [Google Scholar]
- 208.Magi A, Semeraro R, Mingrino A, Giusti B, D’Aurizio R. 2017. Nanopore sequencing data analysis: state of the art, applications and challenges. Brief Bioinform 2017:bbx062. doi: 10.1093/bib/bbx062. [DOI] [PubMed] [Google Scholar]
- 209.van Belkum A, Dunne WM. 2013. Next-generation antimicrobial susceptibility testing. J Clin Microbiol 51:2018–2024. doi: 10.1128/JCM.00313-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Stoesser N, Batty EM, Eyre DW, Morgan M, Wyllie DH, Del Ojo Elias C, Johnson JR, Walker AS, Peto TEA, Crook DW. 2013. Predicting antimicrobial susceptibilities for Escherichia coli and Klebsiella pneumoniae isolates using whole genomic sequence data. J Antimicrob Chemother 68:2234–2244. doi: 10.1093/jac/dkt180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Tyson GH, McDermott PF, Li C, Chen Y, Tadesse DA, Mukherjee S, Bodeis-Jones S, Kabera C, Gaines SA, Loneragan GH, Edrington TS, Torrence M, Harhay DM, Zhao S. 2015. WGS accurately predicts antimicrobial resistance in Escherichia coli. J Antimicrob Chemother 70:2763–2769. doi: 10.1093/jac/dkv186. [DOI] [PubMed] [Google Scholar]
- 212.McDermott PF, Tyson GH, Kabera C, Chen Y, Li C, Folster JP, Ayers SL, Lam C, Tate HP, Zhao S. 2016. Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob Agents Chemother 60:5515–5520. doi: 10.1128/AAC.01030-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Aanensen DM, Feil EJ, Holden MTG, Dordel J, Yeats CA, Fedosejev A, Goater R, Castillo-Ramírez S, Corander J, Colijn C, Chlebowicz MA, Schouls L, Heck M, Pluister G, Ruimy R, Kahlmeter G, Åhman J, Matuschek E, Friedrich AW, Parkhill J, Bentley SD, Spratt BG, Grundmannj H, Krziwanek K, Stumvoll S, Koller W, Denis O, Struelens M, Nashev D, Budimir A, Kalenic S, Pieridou-Bagatzouni D, Jakubu V, Zemlickova H, Westh H, Larsen AR, Skov R, Laurent F, Ettienne J, Strommenger B, Witte W, Vourli S, Vatopoulos A, Vainio A, Vuopio-Varkila J, Fuzi M, Ungvári E, Murchan S, Rossney A, Miklasevics E, et al. . 2016. Whole-genome sequencing for routine pathogen surveillance in public health: a population snapshot of invasive Staphylococcus aureus in Europe. mBio 7:e00444-16. doi: 10.1128/mBio.00444-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Bradley P, Gordon NC, Walker TM, Dunn L, Heys S, Huang B, Earle S, Pankhurst LJ, Anson L, de Cesare M, Piazza P, Votintseva AA, Golubchik T, Wilson DJ, Wyllie DH, Diel R, Niemann S, Feuerriegel S, Kohl TA, Ismail N, Omar SV, Smith EG, Buck D, McVean G, Walker AS, Peto TEA, Crook DW, Iqbal Z. 2015. Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis. Nat Commun 6:10063. doi: 10.1038/ncomms10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Ellington MJ, Ekelund O, Aarestrup FM, Canton R, Doumith M, Giske C, Grundman H, Hasman H, Holden MTG, Hopkins KL, Iredell J, Kahlmeter G, Köser CU, MacGowan A, Mevius D, Mulvey M, Naas T, Peto T, Rolain J-M, Samuelsen Ø, Woodford N. 2017. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: report from the EUCAST Subcommittee. Clin Microbiol Infect 23:2–22. doi: 10.1016/j.cmi.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 216.Richter E, Brown-Elliott BA, Wallace RJ. 2011. Mycobacterium: laboratory characteristics of slowly growing mycobacteria, p 503–524. In Versalovic J, Carroll KC, Funke G, Jorgensen JH, Landry ML, Warnock DW (ed), Manual of clinical microbiology, 10th ed ASM Press, Washington, DC. [Google Scholar]
- 217.World Health Organization. 2016. Global tuberculosis report 2016. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 218.Eldholm V, Balloux F. 2016. Antimicrobial resistance in Mycobacterium tuberculosis: the odd one out. Trends Microbiol 24:637–648. doi: 10.1016/j.tim.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 219.Pankhurst LJ, del Ojo Elias C, Votintseva AA, Walker TM, Cole K, Davies J, Fermont JM, Gascoyne-Binzi DM, Kohl TA, Kong C, Lemaitre N, Niemann S, Paul J, Rogers TR, Roycroft E, Smith EG, Supply P, Tang P, Wilcox MH, Wordsworth S, Wyllie D, Xu L, Crook DW. 2016. Rapid, comprehensive, and affordable mycobacterial diagnosis with whole-genome sequencing: a prospective study. Lancet Respir Med 4:49–58. doi: 10.1016/S2213-2600(15)00466-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Votintseva AA, Bradley P, Pankhurst L, del Ojo Elias C, Loose M, Nilgiriwala K, Chatterjee A, Smith EG, Sanderson N, Walker TM, Morgan MR, Wyllie DH, Walker AS, Peto TEA, Crook DW, Iqbal Z. 2017. Same-day diagnostic and surveillance data for tuberculosis via whole-genome sequencing of direct respiratory samples. J Clin Microbiol 55:1285–1298. doi: 10.1128/JCM.02483-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Manson AL, Abeel T, Galagan JE, Sundaramurthi JC, Salazar A, Gehrmann T, Shanmugam SK, Palaniyandi K, Narayanan S, Swaminathan S, Earl AM. 2017. Mycobacterium tuberculosis whole genome sequences from southern India suggest novel resistance mechanisms and the need for region-specific diagnostics. Clin Infect Dis 64:1494–1501. doi: 10.1093/cid/cix169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Judge K, Harris SR, Reuter S, Parkhill J, Peacock SJ. 2015. Early insights into the potential of the Oxford Nanopore MinION for the detection of antimicrobial resistance genes. J Antimicrob Chemother 70:2775–2778. doi: 10.1093/jac/dkv206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Carter J-M, Hussain S. 2017. Robust long-read native DNA sequencing using the ONT CsgG nanopore system. Wellcome Open Res 2:23. doi: 10.12688/wellcomeopenres.11246.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Gargis AS, Kalman L, Lubin IM. 2016. Assuring the quality of next-generation sequencing in clinical microbiology and public health laboratories. J Clin Microbiol 54:2857–2865. doi: 10.1128/JCM.00949-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Moran-Gilad J, Sintchenko V, Pedersen SK, Wolfgang WJ, Pettengill J, Strain E, Hendriksen RS. 2015. Proficiency testing for bacterial whole genome sequencing: an end-user survey of current capabilities, requirements and priorities. BMC Infect Dis 15:174. doi: 10.1186/s12879-015-0902-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Duncavage EJ, Abel HJ, Pfeifer JD. 2017. In silico proficiency testing for clinical next-generation sequencing. J Mol Diagn 19:35–42. doi: 10.1016/j.jmoldx.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 227.Davies KD, Farooqi MS, Gruidl M, Hill CE, Woolworth-Hirschhorn J, Jones H, Jones KL, Magliocco A, Mitui M, O'Neill PH, O'Rourke R, Patel NM, Qin D, Ramos E, Rossi MR, Schneider TM, Smith GH, Zhang L, Park JY, Aisner DL. 2016. Multi-institutional FASTQ file exchange as a means of proficiency testing for next-generation sequencing bioinformatics and variant interpretation. J Mol Diagn 18:572–579. doi: 10.1016/j.jmoldx.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 228.Duncavage EJ, Abel HJ, Merker JD, Bodner JB, Zhao Q, Voelkerding KV, Pfeifer JD. 2016. A model study of in silico proficiency testing for clinical next-generation sequencing. Arch Pathol Lab Med 140:1085–1091. doi: 10.5858/arpa.2016-0194-CP. [DOI] [PubMed] [Google Scholar]
- 229.Hasman H, Hammerum AM, Hansen F, Hendriksen RS, Olesen B, Agersø Y, Zankari E, Leekitcharoenphon P, Stegger M, Kaas RS, Cavaco LM, Hansen DS, Aarestrup FM, Skov RL. 2015. Detection of mcr-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Euro Surveill 20:30085. doi: 10.2807/1560-7917.ES.2015.20.49.30085. [DOI] [PubMed] [Google Scholar]
- 230.Doumith M, Godbole G, Ashton P, Larkin L, Dallman T, Day M, Day M, Muller-Pebody B, Ellington MJ, de Pinna E, Johnson AP, Hopkins KL, Woodford N. 2016. Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J Antimicrob Chemother 71:2300–2305. doi: 10.1093/jac/dkw093. [DOI] [PubMed] [Google Scholar]
- 231.Ip CLC, Loose M, Tyson JR, de Cesare M, Brown BL, Jain M, Leggett RM, Eccles DA, Zalunin V, Urban JM, Piazza P, Bowden RJ, Paten B, Mwaigwisya S, Batty EM, Simpson JT, Snutch TP, Birney E, Buck D, Goodwin S, Jansen HJ, O'Grady J, Olsen HE, MinION Analysis and Reference Consortium. 2015. MinION Analysis and Reference Consortium: phase 1 data release and analysis. F1000Res 4:1075. doi: 10.12688/f1000research.7201.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Jain M, Fiddes IT, Miga KH, Olsen HE, Paten B, Akeson M. 2015. Improved data analysis for the MinION nanopore sequencer. Nat Methods 12:351–356. doi: 10.1038/nmeth.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Sugawara E, Nikaido H. 2014. Properties of AdeABC and AdeIJK efflux systems of Acinetobacter baumannii compared with those of the AcrAB-TolC system of Escherichia coli. Antimicrob Agents Chemother 58:7250–7257. doi: 10.1128/AAC.03728-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Chikhi R, Lavenier D. 2011. Localized genome assembly from reads to scaffolds: practical traversal of the paired string graph, p 39–48. In Algorithms in bioinformatics. Springer, New York, NY. [Google Scholar]