Abstract
The unfolded protein response (UPR) is an adaptive cellular program used by eukaryotic cells to cope with protein misfolding stress. During tumor development, cancer cells are facing intrinsic (oncogene activation) and extrinsic (limiting nutrient or oxygen supply) challenges, with which they must cope to survive. Moreover, chemotherapy represents an additional extrinsic challenge that cancer cells are facing and to which they adapt in the case of resistance. As of today, resistance to chemotherapy and targeted therapies is one of the important issues that oncologists have to deal with for treating cancer patients. In this review, we first describe the key molecular mechanisms controlling the UPR and their implication in solid cancers. Then, we review the literature that connects cancer chemotherapy resistance mechanisms and activation of the UPR. Finally, we discuss the possible applications of targeting the UPR to bypass drug resistance.
Introduction
The endoplasmic reticulum (ER) is the first intracellular compartment of the secretory pathway. It regulates calcium homeostasis, lipid biosynthesis and protein productive folding and quality control. About one-third of all the proteins transit through the ER1, 2, 3 towards their final cellular or extracellular location. The synthesis of these proteins occurs on the cytosolic side of the ER and productive protein folding is orchestrated by elaborated ER-resident molecular machines involving chaperones, foldases and quality control proteins. These molecular machines ensure protein biogenesis from their nascent form to their ER exportable form.4 However, in the course of this process, a significant proportion of proteins is not properly folded and fails ER protein quality control criteria.5 These misfolded proteins are therefore addressed to the ER-associated degradation (ERAD) system that targets them to the cytosol for ubiquitinylation and proteasomal degradation.1 If the ER faces an important protein folding demand or sees its folding and degradation capacity attenuated, is needed, ER capacity to handle protein biogenesis are overwhelmed, thereby leading to an accumulation of improperly folded proteins in this compartment and to a situation called ER stress. ER stress leads to the activation of an adaptive response, named the unfolded protein response (UPR) that aims at (i) limiting misfolded proteins accumulation in the ER by transiently attenuating protein translation; (ii) augmenting the ER folding capacity by increasing the transcription of ER-resident chaperones proteins; (iii) enhancing protein clearance from the ER by increasing its degradation capacity. If the ER stress persists, the UPR triggers cell death.6, 7
During cancer genesis, an acute demand of protein synthesis is needed to support different cellular functions such as tumor proliferation, migration and differentiation, often driven by oncogenic activation.3 Tumor microenvironment might also provide limited tumor growth/development conditions because of important tumor oxygen and nutrient demands and inadequate vascularization. Therefore, cancer cells have to adapt to such a selective milieu with hypoxia, pH variation and nutrient deprivation that leads to cellular stress,6, 8, 9, 10 by activating a range of cellular stress-response pathways including the UPR that will be described in the first part of this review.
Chemotherapy represents an additional source of cellular stress for cancer cells. Indeed, antitumor drugs emphasize the microenvironmental stress acting on the selection of drug-resistant cancer cells.11 Resistance to chemotherapy is a principal problem in treating the most commonly seen solid tumors. Chemotherapy efficacy is indeed exposed to the multiple intrinsic and acquired resistance mechanisms developed by tumor cells that will be presented in the second part of this review. Furthermore, we will discuss the involvement of the ER stress-induced UPR to anticancer drug resistance. Understanding the UPR mechanisms associated with cancer drug resistance will provide insights to open new therapeutic avenues in which the association of standard chemotherapy with drugs targeting the UPR could overtake cancer drug resistance.
UPR molecular mechanisms and their functions in cancers: the basics
The UPR is crucial for cells to adapt their ER folding capacity to selective conditions as such nutrients and oxygen privation.1 However, if environment-triggered ER stress cannot be resolved, prolonged UPR activation initiates cell death mechanisms. In this section, we will present the molecular actors of the UPR and describe its involvement in cancers.
UPR sensors and their downstream pathways
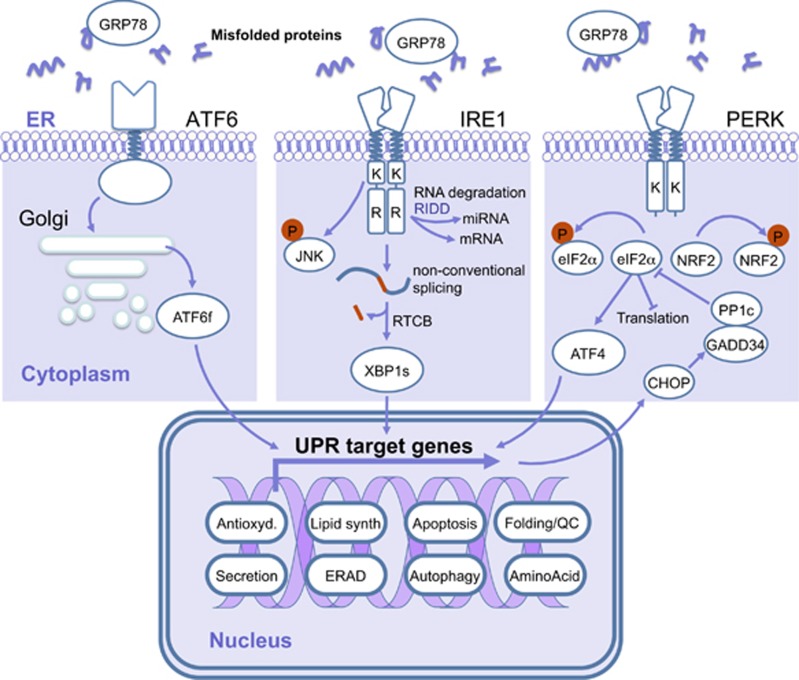
The three major mammalian UPR sensors were first described in the late 1990s: ATF6α (activating transcription factor 6α),12 IRE1α (inositol requiring enzyme 1α)13 and PERK (protein kinase RNA-activated-like ER kinase).14 The signaling pathways activated downstream of the three sensors lead to the reduction of protein misfolding, by slowing down de novo protein synthesis on the cytosolic side of the ER and by increasing protein folding and clearance in the ER (Figure 1). The activation of these three sensors is controlled by the ER-resident chaperone molecule GRP78/BiP (glucose-regulated protein 78/binding immunoglobulin protein). Indeed, under basal conditions, GRP78 constitutively associates with the luminal domains of the sensors through a noncanonical binding, thus preventing their activation.1, 2 Upon accumulation of misfolded proteins, GRP78 dissociates from the sensors when misfolded proteins accumulate in the ER, through mechanism depending on its substrate binding domain.15 This induces IRE1α and PERK oligomerization and autotransphosphorylation16 and the subsequent activation of the downstream signaling cascades. Moreover, BiP dissociation from AFT6α together with protein disulfide isomerase (PDI)-mediated disulfide bond modification17, 18 promotes ATF6α export to the Golgi complex.19, 20
Figure 1.
The UPR sensors and their downstream partners. During ER stress, GRP78 is released from IRE1α, PERK and ATF6 sensors allowing their dimerization/oligomerization or export to the Golgi apparatus. PERK activation leads to phosphorylation of NRF2 and eIF2α. Phosphorylation of eIF2α induces global translation attenuation and prompts that of AFT4. ATF4 and NRF2 induce expression of genes involved in antioxidant response, protein folding, amino-acid metabolism, autophagy and apoptosis. The negative feedback loop activated downstream of PERK dephosphorylates eIF2α to restore translation. IRE1α activation leads to c-Jun N-terminal protein kinase (JNK) phosphorylation, regulated IRE1-dependent decay (RIDD) activity and XBP1 splicing that induces expression of genes involved protein folding, secretion, ERAD and lipid synthesis. Activation of ATF6 leads to its export in the Golgi apparatus where its cytosolic domain is released to translocate to the nucleus and activate the transcription of genes involved in protein folding and ERAD. Antioxid, antioxidant response; Lipid synth, lipid synthesis; QC, quality control.
Activating transcription factor 6α
ER stress leads to ATF6α export from the ER to the Golgi apparatus where ATF6α proteolytic cleavage by S1P and S2P proteases releases an active membrane-free form ATF6f, which therefore translocates to the nucleus and induces the transcription of genes mainly involved in protein folding and ERAD.2, 3, 21, 22
Inositol requiring enzyme 1α
IRE1α is a type I ER-resident transmembrane protein. Its cytoplasmic domain presents two distinct molecular activities: a serine/threonine kinase and an endoribonuclease (RNase), resembling RNaseL. Upon ER stress, IRE1α dimerizes/oligomerizes and its trans-autophosphorylation induces a conformational change leading to endoribonuclease activation.1 The first substrate described for IRE1α RNase was X-box binding protein-1 (XBP1) mRNA that is processed together with the t-RNA ligase RTCB (RNA 2′,3′-cyclic phosphate and 5′-OH ligase) leading to a non-conventional mRNA splicing.23 The resulting open reading frame is shifted and leads to the translation of a stable and active transcription factor, XBP1s.24, 25 XBP1s activate the expression of genes involved in protein folding, secretion, ERAD and lipid synthesis.2, 26, 27 IRE1α RNase is also involved in ER-localized mRNA, ribosomal RNA and microRNAs degradation.28, 29, 30, 31, 32, 33, 34 This activity is named regulated IRE1-dependent decay. Importantly, regulated IRE1-dependent decay selectivity is highly dependent on IRE1α oligomerization state and the cell type, the precise mechanisms of regulated IRE1-dependent decay activation are still debated.35, 36, 37, 38
PKR-like ER kinase
As for IRE1α, PERK is a type I ER-resident transmembrane protein. Upon ER stress, PERK trans-autophosphorylates and phosphorylates the translation initiation factor eIF2α (eukaryotic initiation factor 2α) and the transcription factor NRF2 (nuclear respiratory factor 2). Activated eIF2α attenuates global protein translation, reducing the folding demand on the ER2, 3, 39, 40 whereas activated NRF2 controls the antioxidant response.2 PERK-mediated eIF2α phosphorylation also triggers the translational activation of the transcription factor ATF4 that induces expression of genes involved in protein folding, amino-acid metabolism, autophagy and apoptosis1, 2, 41, 42 such as the apoptosis-related gene CEBP (CCAAT/enhancer-binding protein) homologous protein CHOP (CEBP homologous protein/growth arrest and DNA-damaged-inductible protein 153 (GADD153)) that impacts on the control of cell death/survival outputs upon ER stress.43 Moreover, PERK/eIF2α activation is negatively controlled by a feedback mechanism involving the protein GADD34 induced by this PERK pathway, which, in association with the phosphatase PP1c (protein phosphatase 1c), is responsible for the dephosphorylation of eIF2α.44
UPR involvement in cancers
The role of ER stress signaling as a key actor in cancer development has been first proposed in 20048 and is now largely accepted by both the scientific and medical communities.45 For instance, increased expression levels of major actors of the UPR such as IRE1α, unspliced and spliced XBP1, PERK and ATF6 were observed in tissues sections from a variety of human tumors including brain, breast, gastric, kidney, liver, lung and pancreatic cancers (Table 1).46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67 Moreover, the chaperone GRP78 is also found overexpressed in many cancers46, 47, 48, 49, 50, 51, 52, 54, 56, 57, 58, 59, 60, 61, 62, 64, 65, 66 and is involved in the dissemination/metastasis of human tumors. GRP78 overexpression is associated with higher tumor grades and reduced patients’ survival.48, 53, 57, 59, 61, 65, 67 In experimental models including tumor cell lines and mouse tumor xenografts, GRP78 was also shown to have an important role in regulating cancer hallmarks (Table 2).46, 47, 48, 51, 54, 55, 56, 57, 59, 60, 61, 65, 66, 68, 69, 70, 71, 72, 73 For example, GRP78 regulates tumor cell proliferation and migration.47, 59, 65
Table 1. Clinical evidences of UPR involvement in solid cancers.
| Tumor origin | Materials | Methods | GRP78 | IRE1α | XBP1 | XBP1s | ATF6 | PERK | eIF2α | Others | Comments | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Brain | GBM | IHC, WB | + | + | + | + | (1) | 46 | ||||
| GBM | WB | + | 47 | |||||||||
| GBM, AAIII, AAII, ODG | Transcriptomic, IHC, WB | + | Increased in high-grade tumors | 48 | ||||||||
| Breast | Invasive (stages II and III) | IHC | + | 49 | ||||||||
| Ductal, lobular, stages II and III | NB, IHC, WB | + | 50 | |||||||||
| adenocarcinoma | IHC | + | + | Correlated with ERα expression | 51 | |||||||
| ERα+ invasive ductal carcinoma | transcriptomic | + | + | + | + | + | (2) | 52 | ||||
| ERα+ | IHC | + | Associated with poor prognosis | 53 | ||||||||
| Colorectal | stages II and III CRC | IHC | + | 54 | ||||||||
| Adenoma, CRC | RT–PCR, IHC | + | 55 | |||||||||
| CRC | IHC | + | No correlation with grade or metastases | 56 | ||||||||
| CRC | IHC | + | Increased in metastatic tumors | 57 | ||||||||
| Adenoma, adenocarcinoma | IHC | + | (3) | 58 | ||||||||
| Kidney | RCC (stages I– IV) | Q-PCR, IHC | + | Associated with high-stage tumors | 59 | |||||||
| Liver | HCC | IHC | + | 60 | ||||||||
| HCC | NB, Q-PCR, IHC | + | + | + | + | Associated with histologic grading | 61 | |||||
| HCC | IHC | + | Correlated with CD147 expression | 62 | ||||||||
| Lung | Adenocarcinoma | Q-PCR | + | + | + | (4) | Associated with low stages | 63 | ||||
| NSCLC | IHC | + | Correlated with RRBP1 expression | 64 | ||||||||
| Pancreas | PDAC | IHC | + | Associated with poor prognosis | 65 | |||||||
| PDAC | RT–PCR, IHC | + | + | + | + | + | (5) | Associated with MIA2 mutations | 66 | |||
| PDAC | IHC | (6) | Associated with poor prognossis correlated with decreased SMARCB1 expression | 67 | ||||||||
Abbreviations: AA, anaplastic astrocytoma; ATF, activating transcription factor; CRC, colorectal cancer; eIF2α, eukaryotic initiation factor 2α ERp, ER protein; GADD, growth arrest and DNA-damage-inducible protein; GBM, glioblastoma; HCC, hepatocellular carcinoma; IRE1α, inositol requiring enzyme 1α GRP, glucose-regulated protein; IHC, immunohistochemistry; NB, northern blot; NSCLC, non-small cell lung cancer; ODG, oligodendroglioma; PCR, polymerase chain reaction; PDAC, pancreatic ductal adenocarcinoma; PDI, protein disulfide isomerase; PERK, PKR-like endoplasmic reticulum kinase; Q-PCR, quantitative PCR; RCC, renal cell carcinoma; RT–PCR, reverse transcriptase–PCR; SERP, stress-associated ER protein; UPR, unfolded protein response; WB, western blot; XBP, X-box binding protein.
(1) Calreticulin(+), CHOP/GADD153(+), ERp72(+), GRP94(+), GRP170(+).
(2) CHOP(+), GADD34(+), GRP94(+), SERP1(+).
(3) Decreased CHOP.
(4) ERO1A.
(5) Calnexin(+), PDI(+).
(6) Phosphorylated ATF2.
Table 2. Cellular models demonstrating the importance of UPR in solid cancers.
| Tumor origin | Materials | Methods | GRP78 | IRE1α | XBP1 | XBP1s | ATF6 | PERK | eIF2α | ATF4 | Others | Comments | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Brain | U87 cell line | NB, WB | + | (1) | 46 | ||||||||
| U87 xenograft | NB, IHC, WB | + | + | + | (2) | ||||||||
| U87 and D245MG xenografts | NB, IHC, WB | + | + | + | (3) | ||||||||
| U87, U251, U138, A172, LN229 and T98G | WB, IHC | + | Associated with increased proliferation | 47 | |||||||||
| U87, U251, A172, LN229, LN443 and LNZ308 | WB | + | 48 | ||||||||||
| U251 | RT–PCR | + | + | + | + | + | (4) | Increased under arginine deprivation | 68 | ||||
| Breast | T47D cell line | WB | + | Increased under glucose privation increased under estrogen treatment | 51 | ||||||||
| Hs578T, MDA-MB-231 | + | + | + | + | + | (5) | Modulated by LOXL2 and associated with EMT | 69 | |||||
| Colorectal | Colo205, HCT116, SW480, SW626 | RT–PCR, WB | + | + | + | + | + | + | + | (6) | 54 | ||
| DLD1, HCT15, SW480, WiDr | RT–PCR | + | 55 | ||||||||||
| Colo205, HCT116, SW480, SW626 | RT–PCR, WB | + | + | + | + | + | + | (7) | 57 | ||||
| HT29 | WB | + | Increased under glucose deprivation or radiation | 56 | |||||||||
| HCT119 | RT–PCR, WB | + | + | + | + | + | (8) | Increased under arginine deprivation | 68 | ||||
| HT29 | RT–PCR, WB | + | + | + | + | + | (9) | ||||||
| HGC27 | WB | + | + | + | Increased under severe hypoxia | 70 | |||||||
| Kidney | 786-O, OS-RC-2 and Caki-1 | RT–PCR, WB | + | 59 | |||||||||
| 786-O, A498, ACHN, Caki, | RT–PCR, WB | + | Associated with increased proliferation | 71 | |||||||||
| Liver | HepG2 | WB | + | Increased under glucose privation | 60 | ||||||||
| HepG2, HuH7, HLF | NB, WB | + | + | + | + | 61 | |||||||
| HepG2, SMCC-7721, MHCC97-H | WB | + | + | (10) | 72 | ||||||||
| Ovary | SKOV3 | RT–PCR | + | + | + | + | (11) | Increased under arginine deprivation | 68 | ||||
| Pancreas | AsPC-1, BxPC-3, Capan-1, MIAPaCa-2, PCT-3 and SU.86.86 | WB | + | Associated with increased proliferation and migration | 65 | ||||||||
| Su86.86 | RT–PCR | + | Associated with MIA2 mutations | 66 | |||||||||
| Skin | A375, HMVII, WM9, WM3918 | RT–PCR, WB | + | + | + | + | + | + | (12) | Increased by HA15, a GRP78 inhibitor | 73 |
Abbreviations: ATF, activating transcription factor; EDEM, ER degradation enhancer, mannosidase α-like; eIF2α, eukaryotic initiation factor 2α EMT, epithelial-to-mesenchymal transition; ERp, ER protein; GRP, glucose-regulated protein; HERP, homocysteine-induced ER protein; IHC, immunohistochemistry; IRE1α, inositol requiring enzyme 1α LOXL2, lysyl oxidase like 2; NB, northern blot; PDI, protein disulfide isomerase; PERK, PKR-like endoplasmic reticulum kinase; UPR, unfolded protein response; WB, western blot; XBP, X-box binding protein.
(1) GRP94(+).
(2) CHOP(+).
(3) Calreticulin(+), CHOP(+), ERp72(+), GRP94(+), HERP(+), PDI(+).
(4) CHOP(+), EDEM1(+), GRP94(+).
(5) DDIT3(+), DNAJB9(+), EDEM1(+).
(6) Phosphorylated PERK and eIF2α.
(7) Phosphorylated eIF2α.
(8) CHOP(+), GRP94(+), phosphorylated eIF2α and GCN2.
(9) CHOP(+), EDEM1(+), phosphorylated eIF2α and GCN2.
(10) Phosphorylated IRE1α.
(11) CHOP+, GRP94+.
(12) CHOP(+), phosphorylated IRE1α, PERK and eIF2α.
Tumor progression is characterized by UPR activation induced by the challenging growth conditions associated with hypoxia and anticancers drugs.52 Furthermore, tumor cells develop specific metabolic processes to adapt to such environment,74 and examples of highly dynamic network between cancer cells’ adaptation and resistance to environmental stresses and UPR signaling pathways will be illustrated in the following section.
UPR linked to cancer initiation
In the normal gastrointestinal tract, a differential expression of GPR78 is observed and is lower in intestinal stem cells and higher in more differentiated transit amplifying cells.75 Interestingly, most of the colorectal cancers (CRCs) derive from transformed intestinal stem cell in which activation of the PERK/eIF2α axis is associated with the loss of stemness.76 This suggests that cancer initiation might be linked to ER stress in the gastrointestinal tract.3 Remarkably, in a colitis-associated cancer model, the IRE1α pathway appears to have an important role in mediating ER stress that induces intestinal stem cell expansion.77 Indeed, XBP1 loss in epithelial cells results in intestinal stem cell hyperproliferation, therefore promoting initiating phases of cancer development.3
UPR linked to tumor quiescence and aggressiveness
Cancer cells must cope with strict growth conditions forced by their intrinsic condition (oncogene expression) but also by the tumor environment including chemotherapy, nutrient starvation and in vivo microenvironmental challenges. They therefore develop adaptive mechanisms such as a metabolic resting state called quiescence/dormancy. Regulation of tumor cell dormancy has been associated with the activation of both ATF6α and PERK-eIF2α. Both pathways were identified as a survival factors for quiescent but not proliferative squamous carcinoma cells78 and under hypoxia,79 respectively. In triple-negative breast cancers, the IRE1α/XBP1s axis is found constitutively active, thereby conferring higher aggressiveness due to XBP1s-mediated hypoxia-inducible factor-1α activation.80 In glioblastoma (GBM), tumor migration/invasion is associated to aggressiveness. Interestingly, IRE1α endoribonuclease activity regulates the extracellular matrix protein SPARC (secreted protein acidic and rich in cysteine) itself involved in tumor invasion.81
UPR-linked ‘secretory switch’ in cancer cells
To sustain their own important metabolic demands and to adapt to their challenging environment, cancer cells reprogram their secretome and the associated secretory pathway needed to support tumor functions and necessary for cancer progression.3, 82 For instance, tumor invasion is facilitated by change in secreted extracellular matrix components and matrix metalloproteases.83, 84 Tumor cell proliferation and neoangiogenesis (see below) are sustained through the secretion of growth factors, cytokines and chemokines.3 As ER is the major site of protein production that also orchestrates their secretion, activation of the UPR strongly modulates tumor cells’ secretory switch during cancer development.
UPR linked to tumor epithelial-to-mesenchymal transition
Epithelial-to-mesenchymal transition (EMT) is a physiological process used by cancer cells to acquire critical oncogenic features such as migration/invasion, stemness and drug resistance.3 EMT is controlled by specific transcription factors involved in these cell functions and the UPR has been often involved in the expression of these transcription factors. For instance, in breast tumors, increased expression of XBP1s is observed in metastatic tumors, which correlates with the EMT inducer SNAIL (snail-related protein).85 LOXL2 (lysyl oxidase like 2)/GRP78 interaction in the ER also activates the IRE1-XBP1 signaling pathway thereby inducing the expression of several EMT-linked transcription factors including SNAI1 (snail family transcriptional repressor), SNAI2, ZEB2 (zinc-finger E-box-binding homeobox 2) and TCF3 (transcription factor 3).69 Moreover, the overexpression of the TWIST (twist-related protein) transcription factor correlates with PERK constitutive activation.86 The ‘secretory switch’ induced by UPR might also contribute to EMT.86, 87, 88 Indeed, overexpression of Serpin B3, a serine/cysteine protease inhibitor, is associated with chronic UPR induction leading to nuclear factor-κB activation and interleukin-6 production. This results in an EMT-like phenotype in mammary epithelial cells.89 In GBM, dominant-negative form of IRE1α modulates the expression molecules involved in extracellular matrix structures, angiogenesis and inflammatory chemokines, thus reflecting a mesenchymal drift.90
UPR-linked tumor angiogenesis
Expression of proangiogenic factors is affected by the UPR in cancer cells. For instance, vascular endothelial growth factor-A (VEGF-A), interleukin-1β and interleukin-6 are induced downstream of IRE1α signaling in GBM cells.90, 91 Moreover, IRE1α-mediated mRNA cleavage of the circadian gene PERIOD1,92 an important mediator of GBM infiltration, also supports tumor angiogenesis through the regulation of the CXCL3 chemokine.90 Furthermore, in response to hypoxia, VEGF is also upregulated by the PERK-ATF4 branch of the UPR to induce angiogenesis.2, 3, 74, 93 Interestingly, the UPR-regulated ER chaperone ORP150 (oxygen-regulated protein 150) controls tumor angiogenesis by promoting the secretion of VEGF in prostatic and glioma cancer cells.94, 95
UPR-linked tumor metabolic processes
Under nutrient deprivation, cancer cells adapt their metabolic demand in part through activation of the UPR. Downstream of IRE1α, XBP1s activates the expression of key enzymes of the hexosamine biosynthetic pathway that convert glucose to UDP-acetylglucosamine.96, 97 These are substrates for the O- and N-glycosylation of proteins, thereby improving global proteotasis. In addition, through hypoxia-inducible factor-1α activation, XBP1s also actively promotes glucose uptake in triple-negative breast cancer cells, which in turn upregulates the expression of several proteins involved in glycolytic processes including the glucose transporter 1.98
UPR linked to tumor autophagy
Autophagy is a cellular process that allows cancer cells to generate additional energy supplies through the selective or non-selective degradation of protein aggregates or damaged organelles. Under hypoxia, activation of the PERK/eIF2α/ATF4 pathway is protective for tumor cells through autophagy induction via LC3B (autophagy protein microtubule-associated protein 1 light chain 3b) and ATG5 (autophagy protein 5).99, 100, 101 Similarly, TNF receptor associated factor 2 (TRAF2)/IRE1α activates c-Jun N-terminal protein kinase that also induces autophagy.102
Chemotherapy resistance induced by UPR
General mechanisms of resistance to chemotherapy in cancer
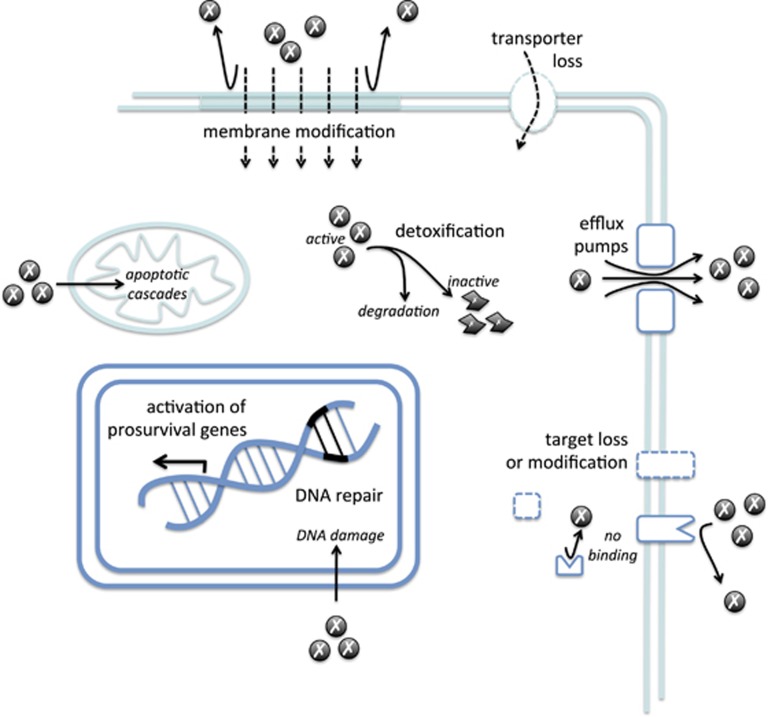
During the past decades, chemotherapy and targeted therapies have become the principal modes of treatment against cancers (Table 3), but their efficacy is confronted to the multiple intrinsic and acquired resistance mechanisms developed by tumor cells before and during the treatment. These resistance mechanisms can include the reduction of drug uptake, the alteration of the drug target, the induction of drug-detoxifying mechanisms, repair of drug-induced damages and insensitivity to drug-induced cell death (Figure 2).103, 104, 105
Table 3. Standard chemotherapy treatments and their targets in solid tumors.
| Drugs | Cancers | Targets |
|---|---|---|
| Alkylating agents | ||
| Carboplatin | Ovary | DNA alkylation |
| Cisplatin | Biliary, gastric, lung, urogenital | DNA alkylation |
| Cyclophosphamide | Urinary | DNA alkylation |
| Dacarbazine | Skin | DNA alkylation |
| Ifosfamide | Soft tissues | Guanine alkylation |
| Oxaliplatin | Biliary, colorectal, pancreas | DNA crosslinking |
| Temozolomide | Brain | Guanine alkylation |
| Antimetabolites | ||
| 5-Fluorouracil | Colorectal, gastric, pancreas | Pyrimidine analog, TS |
| Capecitabine | Breast, colorectal | Pyrimidine analog, TS |
| Gemcitabine | Biliary, lung, pancreas, urinary | Deoxycytidine analog |
| Methotrexate | Urinary | DHFR |
| Pemetrexed | Lung | TS, DHFR, GARFT |
| Antibiotics/intercalants | ||
| Doxorubicin | Endometrial, soft tissues, urinary | DNA intercalant |
| Camptothecin | Colorectal, pancreas | Topoisomerases I |
| Etoposide | Lung, urogenital | Topoisomerases II |
| Bleomycin | Genitourinary | DNA strand break inducer |
| Antimitotics/spindle poisons | ||
| Docetaxel | Breast, gastric, urinary | β-Tubulin |
| Paclitaxel | Breast, ovary | β-Tubulin |
| Vinblastin | Breast, kidney, urinary | Microtubules |
| Hormone therapy | ||
| Bicalutamide | Prostate | Androgen receptors |
| Goserelin | Prostate | GnRH agonist |
| Tamoxifen | Breast | Estrogen receptors |
| Targeted therapy | ||
| Erlotinib | Pancreas | EGFR |
| Bortezomib | Lymphoma, myeloma | Proteasome |
| Sorafenib | Kidney, liver | FLT3, c-KIT, PDFGRβ, c-RAF, b-RAF, VEGFRII and III |
| Sunitinib | Kidney | FLT3, c-KIT, PDGFRβ, RET, VEGFRI and II |
| Immunotherapy | ||
| Bevacizumab | Kidney, lung | VEGF |
| Trastuzumab | Breast | HER2/neu |
Abbreviations: DHFR, dihydrofolate reductase; EGFR, epidermal growth factor receptor; FLT, fms-like tyrosine kinase; GARFT, glycinamide ribonucleotide formyltransferase; GnRH, gonadotropin-releasing hormone; HER2/neu, human epidermal growth factor receptor; KIT, v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog; PDGFR, platelet-derived growth factor receptor; RAF, rapidly accelerated fibrosarcoma; RET, rearranged during transfection; TS, thymidylate synthase.
Figure 2.
General mechanisms involved in chemotherapy resistance. Tumor cells limit chemotherapy drugs accumulation by modifying their membrane composition, reducing drug transporters and increasing efflux pumps. Mechanisms of detoxification lead to drug inactivation. Drug target modification or loss also contributes to chemotherapy resistance. Finally, DNA damage and apoptosis induced by anticancer drugs are prevented by sophisticated DNA repair system and upregulation of prosurvival genes.
Resistance to anticancer drug accumulation
Drugs enter into tumor cells by three main routes: diffusion, active transport and endocytosis.103 However, tumor cells use several mechanisms to limit this entry by decreasing the uptake or increasing the efflux of the drug.103 For instance, the family of multidrug resistance proteins, acting as drug efflux pumps (reviewed in Chen and Tiwari106and Sodani et al.107), is the subject of intense research to characterize the role in chemotherapy resistance.11, 103 Expression of these proteins has been reported to correlate with resistance to chemotherapy in vitro.105 Modulation of their functions is also correlated to in vitro chemosensitivity to drugs such as cisplatin, doxorubicin, paclitaxel and vincristine in several cancer cell lines.108, 109 In addition, modulation of the expression of cell surface transporters or their mutations can reduce drug uptake. As such, in osteosarcoma, both decreased expression and mutations of the methotrexate transporter reduced folate carrier that reduce their drug affinity have been reported.103, 105, 110 Finally, cancer cell mutants that have defective endocytosis are resistant to immunotoxins that enter into tumor cells by endocytosis.103
Induction of drug-detoxifying mechanisms
Both drug inactivation and the absence of drug activation are specific for given classes of drugs.104 For instance, 5-fluorouracil (5-FU) is catabolized by dihydropyrimidine dehydrogenase that confers in vitro resistance to 5-FU once overexpressed in CRCs.105 Platinum drugs such as cisplatin, carboplatin and oxaliplatin can also be inactivated after covalent linkage to the thiol glutathione, decreasing the availability of the native drug to bind its target104, 108 and leading to drug efflux by ABC transporter proteins.105 High levels of glutathione have been found in tumor cells resistant to platinum drugs. Interestingly, expression of glutathione S-transferase-π, a member of the family of glutathione S-transferase that catalyzes glutathione conjugation, is linked to overall survival following cisplatin treatment of head and neck cancers and to cisplatin resistance of ovarian cancers.105, 108, 110
Modification of drug targets
Drug sensitivity is affected by alterations of the drug target, such as mutations and/or changes in expression level.104, 108 For instance, 5-FU and pemetrexed treatments inhibit translation of their target mRNA thymidylate synthase (TS),104 thus leading to increased TS expression level and increased 5-FU resistance.104, 105 Moreover, the overexpression and/or oncogenic mutations in many protein tyrosine kinases have been described in human cancers, rendering difficult the anti-protein tyrosine kinase targeting therapies. Indeed, efficacy of epidermal growth factor receptor (EGFR) inhibitors such as gefitinib and erlotinib is markedly reduced in non-small-cell lung cancers exhibiting the EGFR-T790M mutation.104 Amplification and mutations in anaplastic lymphoma kinase have been identified in pediatric neuroblastoma, but secondary mutations in the anaplastic lymphoma kinase tyrosine kinase domain or anaplastic lymphoma kinase fusion gene amplifications are observed after crizotinib treatment leading to the disease relapse.104
DNA-damage repair
Most chemotherapeutic drugs drive the induction of DNA damage in tumor cells either directly for platinum-based drugs or indirectly for 5-FU and topoisomerase inhibitors.104, 105 DNA topoisomerase-I mutations have been reported to affect camptothecin sensitivity.105 Similarly, DNA topoisomerase-II, a target of doxorubicin and etoposide, is mutated in resistant cancer cell lines.105 Reduction of DNA topoisomerase-II expression by post-transcriptional modifications such as ubiquitination and sumoylation also leads to drug resistance and reduction of DNA damage.6, 111 In normal cells, DNA lesions are quickly recognized by DNA-damage response factors, which activate cell cycle checkpoints and direct DNA repair.112 Consequently, the regulation of DNA repair systems in tumor cells is a critical factor for their response to chemotherapeutics.112 For instance platinum-induced DNA damage is repaired by the nucleotide excision repair pathway and in vitro correlation between enhanced nucleotide excision repair and resistance to cisplatin has been reported in many studies.108 High expression of ERCC1 (excision repair cross-complementing 1), one of the key components of nucleotide excision repair, is linked to poor response to chemotherapy in numerous cancer types.104 In addition, mutation and/or downregulation of key DNA mismatch repair proteins such as MLH1 (mutL homolog 1) is observed in cisplatin-resistant tumors.104, 108, 110
Activation of antiapoptotic and prosurvival pathways
Most tumors develop defects in the common cell death pathways that lead to chemotherapy resistance.104 For instance, levels of BIM (Bcl-2 interacting mediator of cell death), a proapoptotic protein of the Bcl-2 (B-cell lymphoma) family, predict clinical responsiveness to EGFR and ERBB2 inhibitors. Moreover, a germline deletion in BIM gene is significantly associated with resistance to protein tyrosine kinase inhibitors in patients with EGFR-mutant lung cancers.104 Expression levels of MCL1, another member of the Bcl-2 family, are important determinant of resistance to Bcl-2 inhibitor ABT-737 and other cytotoxic chemotherapeutics.104 Furthermore, under chemotherapy pressure, tumors develop novel survival signaling pathways that contribute to drug resistance.104 An important number of proteins is involved in these pathways: oncogenes such as RAS and AKT (v-Akt murine thymoma viral oncogene homolog); tumor suppressor genes such as TP53 (tumor protein 53) and PTEN (phosphatase and tensin homolog); and prosurvival factors as nuclear factor-κB and signal transducer and activator of transcription 3.104, 108 Mutations, amplifications, chromosomal translocations and overexpression of these genes are associated with various malignancies and linked to resistance to chemotherapy and targeted therapies.104
Other factors involved in drug resistance
The influence of the local tumor microenvironment is identified as important contributor to chemotherapy resistance.104 For instance, hypoxia enhances drug detoxification by interfering with the generation of oxygen radicals and by increasing hypoxia-inducible factor-1-mediated activation of survival signals.108 Furthermore tumor heterogeneity at the genetic, molecular and cellular levels contributes substantially to chemotherapy resistance. For instance, the presence of cancer stem cells with robust intrinsic drug resistance capabilities reduces the chemotherapy efficacy.104 In solid tumors, the stroma (extracellular matrix, cancer-associated fibroblasts, immune and inflammatory cells and blood vessels) protects cancer cells from cytotoxic agents, thus allowing them to evade apoptosis and to develop acquired resistance leading to disease relapse.11, 104, 108 Recently, EMT has been associated with chemotherapy and targeted therapy resistance.104 Finally, as most anticancer drugs are primarily targeted against proliferating cancer cells, a significant proportion of cancer cells are in a dormancy/quiescent state, thereby exhibiting a degree of drug resistance linked to their decreased ability to proliferate.11, 108
Chemotherapy resistance induced by the UPR
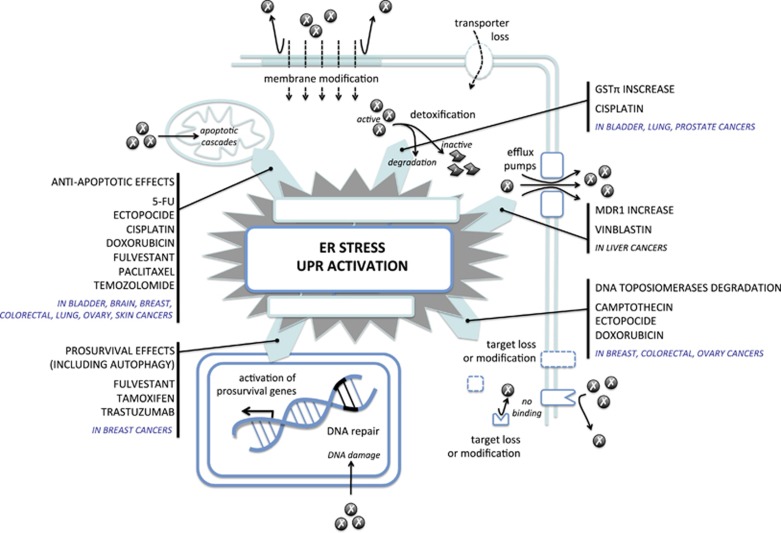
UPR activation is commonly observed in various tumor specimens (see UPR involvement in cancers) and correlates with drug resistance. Clinical evidences and in vitro demonstrations of tight link between UPR activation and drug resistance will be first reviewed in this section. The link between UPR and cellular adaptation of cancer cells including autophagy and hypoxia that also contributes to antidrug resistance will be presented in the next paragraphs (Figure 3).
Figure 3.
The UPR intervention in chemotherapy resistance. UPR activation contributes to chemotherapy drug resistance by increasing drug detoxification and efflux pump expression, by modulating drug targets and activating antiapoptotic and prosurvival genes expression. Examples of anticancer drugs used several cancer types described in the literature are indicated.
Clinical relevance of the UPR activation and chemotherapy resistance
Clinical evidences of such phenomenon are almost exclusively limited to breast cancers (Table 4).49, 52, 113, 114, 115 Indeed, expression of the UPR sensors and their downstream partners are correlated with resistance to tamoxifen, thereby leading to decreased time to recurrence and poor survival.52 Interestingly, opposite effects are observed with the expression of XBP1u and XBP1s. XBP1u is associated with longer survival of breast patients treated with tamoxifen, whereas XBP1s is associated with shorter survival.113 This underlines IRE1α involvement in tamoxifen resistance. In contrast, GRP78 involvement seems to be more complex. High GRP78 expression in breast cancer specimens predicts a shorter recurrence-free survival in patients who received doxorubicin-based adjuvant chemotherapy. However, the opposite effect is observed in patients treated with doxorubicin and cyclophosphamide, followed by taxane (paclitaxel or docetaxel) on a clinical trial, where GRP78-positive staining predicts a better recurrence-free survival.114 These results underline the possibility of use combined anticancer drugs to overcome cancer resistance (Figure 3).
Table 4. Clinical evidences of UPR involvement in cancer chemotherapy resistance.
| Tumor origin | Materials | Chemotherapy | Methods | GRP78 | IRE1α | XBP1 | XBP1s | ATF6 | PERK | Others | Comments | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Breast | Ductal/lobular (stages II and III) | Doxorubicin | IHC | + | Associated with reduced time to recurrence | 49 | ||||||
| ERα+ | Tamoxifen | Transcriptomic | + | + | + | (1) | Associated with poor prognosis | 52 | ||||
| Invasive ductal (stages I–III) | Tamoxifen | Q-PCR | + | + | Associated with high or poor survival respectively | 113 | ||||||
| Invasive ductal (stages II and III) | Doxorubicin, cyclophosphamide+ taxane (paclitaxel or docetaxel) | IHC | + | Associated with longer survival | 114 | |||||||
| Colorectal | Rectal cancer | 5-FU | WB | (2) | Associated with poor response to therapy | 115 |
Abbreviations: ATF, activating transcription factor; eIF2α, eukaryotic initiation factor 2α ER, estrogen receptor; ERO1L, ER oxidoreduction 1-like; 5-FU, 5-fluorouracil; GADD, growth arrest and DNA-damage-inducible protein; GRP, glucose-regulated protein; HERPUD, HERP ubiquitin-like domain; IHC, immunohistochemistry; IRE1α, inositol requiring enzyme 1α PERK, PKR-like endoplasmic reticulum kinase; Q-PCR, quantitative PCR; RT–PCR, reverse transcriptase–PCR; SERP1, stress-associated ER protein 1; SYNV, synoviolin; UPR, unfolded protein response; XBP, X-box binding protein.
(1) 18 genes: ATF4, ATF6α, CHOP, DNAJB9, DNAJC3, EDEM1, eIF2α, ERO1L, ERO1LB, GADD34, GRP78, GRP94, HERPUD1, IRE1α, PERK, XBP1, SERP1, SYNV1.
(2) Calnexin(+).
Induction of UPR-dependent chemotherapy resistance in vitro
Correlations between UPR activation and chemotherapy resistance were mainly demonstrated in cellular models in many types of cancer (Table 5).46, 47, 48, 51, 52, 53, 54, 57, 60, 62, 64, 71, 72, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130 A vast number of these studies demonstrate the impact of GRP78 expression on drug resistance mainly involving a reduced effect of drug-induced apoptosis.47, 48, 54, 60, 64, 116, 117, 120, 123, 125, 128, 129 However, the precise molecular mechanisms involved remain to be discovered. In chemotherapy-resistant breast cancer cells, GRP78 suppresses doxorubicin-mediated apoptosis in part through inhibition of BAX (Bcl-2-associated X protein) and caspase-7 activation.49 GRP78 also forms complexes with BIK (Bcl-2-interacting killer), an apoptotic BH3-only protein, and blocks its apoptotic activity under estrogen starvation.120 Finally, the PDIA5/ATF6α activation loop was described to be essential to confer imatinib resistance in K562 leukemia cells.17 The direct involvement of the UPR sensors in other mechanisms associated with cancer resistance to chemotherapy (i.e. reduction of anticancer drug accumulation, drug-detoxifying mechanisms, modification of drug targets and DNA-damage repair) is up to now rather limited. For instance, a role for PERK in chemotherapy-resistant HT29 colon cancer cells has been involved in the upregulation of MDR related protein 1 through the regulation of NRF2.131
Table 5. Cellular models demonstrating the importance of UPR in cancer chemotherapy resistance.
| Tumor origin | Materials | Chemotherapy | Methods | GRP78 | IRE1α | XBP1 | XBP1s | ATF6 | PERK | eIF2α | ATF4 | Others | Comments | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bladder | T24/83 | Etoposide, doxorubicin, camptothecin | WB | + | Associated with resistance to apoptosis | 116 | ||||||||
| Bone | MG-63, SaOS-2 | Cisplatin | WB | + | (1) | Associated with resistance to apoptosis | 117 | |||||||
| Brain | U87 | Temozolomide | WB | + | Increased with ER stress (DTT) | 46 | ||||||||
| U87 and U251 | Temozolomide | WB | + | (1) | 47 | |||||||||
| LN229 | Temozolomide, camptothecin, 5-FU | WB | + | Associated with resistance to apoptosis | 47 | |||||||||
| A172 and LNZ308 | Etoposide, cisplatin | IHC | + | Associated with resistance to apoptosis | 48 | |||||||||
| U87 and U251 | Temozolomide | + | + | (2) | Associated with radicol-induced apoptosis | 118 | ||||||||
| Breast | MCF-7 | Doxorubicin | WB | + | (3) | 119 | ||||||||
| T47D | Estrogen | Q-PCR, WB | + | + | + | (4) | 52 | |||||||
| MCF-7 | Estrogen | Q-PCR, WB | + | + | + | + | + | + | + | (5) | 52 | |||
| MCF-7 xenograft | Estrogen | Q-PCR | + | − | − | + | − | − | + | (6) | 52 | |||
| 293T, MCF-7 | Etoposide | WB | + | Associated with BIK interaction | 120 | |||||||||
| MCF-7, T47D | Fulvestrant | WB | + | + | 121 | |||||||||
| LCC1, LCC9 | Fulvestrant | WB | + | + | + | (7) | Associated with autophagy | 122 | ||||||
| LCC9, MCF-7 | Fulvestrant | WB | + | + | Associated with resistance to apoptosis | 123 | ||||||||
| MDA-M35, T47D, MCF-7 | Quercetin | Q-PCR, WB | + | + | (8) | 124 | ||||||||
| MCF-7 | Paclitaxel | WB | + | Associated with resistance to apoptosis | 125 | |||||||||
| T47D | Tamoxifen | WB | + | 51 | ||||||||||
| MCF-7, T47D MCF-7 xenograft | Tamoxifen Tamoxifen | RT–PCR, WB WB | + + | (9) | Decreased resistance with IRE1 inhibitor decreased with IRE1 inhibitor | 53 | ||||||||
| MCF-7, T47D | Tamoxifen | WB | + | + | 121 | |||||||||
| MCF-7 xenograft | Tamoxifen | WB | + | + | 121 | |||||||||
| Rat DMBA-induced mammary tumors | Tamoxifen | WB | + | + | + | (1) | Associated with autophagy | 126 | ||||||
| SKBr3 | Trastuzumab | Q-PCR, ELISA | + | (10) | Increased with ER stress (Tg) | 127 | ||||||||
| Cervix | SiHa-derived stem-like cells | Cisplatin | RT–PCR, WB | + | + | (11) | increased apoptosis with IRE1 inhibitor | 128 | ||||||
| Colorectal | Colo205, HCT116, SW480, SW626 | Cisplatin, 5-FU | WB | + | Associated with resistance to apoptosis | 54 | ||||||||
| HCT116 HT29 | 5-FU | (12) | Associated with resistance to apoptosis | 57 | ||||||||||
| Kidney | A498, ACHN | Doxorubicin, 5-FU | IHC | + | Associated to cell cycle control | 71 | ||||||||
| Liver | HepG2 | Doxorubicin | RT–PCR, WB | + | Increased survival under glucose privation | 60 | ||||||||
| 7741, HepG2 and 7741 xenograft | Doxorubicin, VP-16 | IHC, WB | + | Correlated with CD147 expression | 62 | |||||||||
| HepG2, MHCC97 | Sorafenib | + | (9) | Associated with resistance to apoptosis-dependent of RACK expression | 72 | |||||||||
| Lung | PC13, PC14 | Doxorubicin | WB | + | Associated with resistance to apoptosis | 64 | ||||||||
| Ovary | PEO4 | Estrogen | Q-PCR, WB | + | + | 52 | ||||||||
| OVCAR-3 | Paclitaxel | WB | + | Associated with resistance to apoptosis | 125 | |||||||||
| Skin | Hep3 (dormant versus tumorigene) | Etoposide, doxorubicin | Q-PCR, WB | + | (13) | Associated with resistance to apoptosis | 129 | |||||||
| Others | CHO (hamster) | Etoposide, doxorubicin, camptothecin | WB | + | Associated with resistance to apoptosis | 116 | ||||||||
| CHO (hamster) | Etoposide | WB | + | Increased under ER stress (tg) | 130 | |||||||||
| NIH3T3 | Etoposide | WB | + | Increased under ER stress (tg) | 130 |
Abbreviations: ATF, activating transcription factor; BIK, Bcl-2-interacting killer; DTT, dithiothreitol; eIF2α, eukaryotic initiation factor 2α ERO1L, ER oxidoreduction 1-like; 5-FU, 5-fluorouracil; FRP, glucose-regulated protein; HSP, heat-shock protein; IHC, immunohistochemistry; IRE1α, inositol requiring enzyme 1α JNK, c-Jun N-terminal protein kinase; LCN2, lipocalin 2; PDI, protein disulfide isomerase; PERK, PKR-like endoplasmic reticulum kinase; Q-PCR, quantitative PCR; RT–PCR, reverse transcriptase–PCR; Tg, thapsigargin; UPR, unfolded protein response; WB, western blot; XBP, X-box binding protein.
(1) CHOP(+).
(2) calnexin(+), calreticulin(+), CHOP(+), GRP94(+), PDI(−), phosphorylated IRE1α, PERK and eIF2α(+).
(3) CHOP(+), phosphorylated PERK.
(4) Decreased CHOP, cleaved ATF6, phosphorylated PERK and eIF2α.
(5) DNAJC3, ERO1LB, GRP94.
(6) CHOP(+), DNAJC3(−), ERO1Lb(−), GADD34(+).
(7) CHOP(+), GRP94(+), cleaved ATF6, phosphorylated eIF2α.
(8) CHOP(+), phosphorylated eIF2α and JNK.
(9) Phosphorylated IRE1α.
(10) CHOP(+), LCN2(+).
(11) Phosphorylated eIF2α.
(12) Calnexin(+).
(13) HSP47(+), PDI(+), phosphorylated PERK and eIF2.
UPR and cellular adaptation links to cancer chemotherapy resistance
Different anticancer treatments, including those that stimulate ER stress, activate autophagy in tumor cells, which has been proposed to either enhance cancer cell death or act as a mechanism of resistance to chemotherapy.104, 132 Indeed, autophagy is a lysosome-dependent degradation pathway that degrades cellular components to maintain cellular biosynthesis and viability during metabolic stresses such as nutrient deprivation. During chemotherapy, autophagy facilitates cancer cell survival to cope with metabolic stresses caused by anticancer drugs.104 For instance, in breast cancer cell models, resistance to endocrine therapy such as tamoxifen and fulvestrant is the result of activation and interactions between different cellular mechanisms including UPR activation, autophagy and apoptosis in breast cancers.122, 123, 125, 126, 133 Indeed, antiestrogen-resistant breast cancer cells display higher levels of basal autophagy than sensitive cells.123 In addition, XBP1s-overexpressing MCF-7 cells displayed much higher basal levels of autophagy as demonstrated with increased basal LC3II levels and decreased p62 levels.123 Autophagy induced by XBP1s overexpression protects the cells against apoptosis. Furthermore, XBP1s-overexpressing cells become sensitive to tamoxifen when autophagy is blocked.123
Hypoxia is known to confer cancer cells with resistance to chemotherapy and to modulate UPR during ER stress.134, 135, 136 In breast cancers, taxol rapidly induces UPR activation including ATF6α, IRE1α and PERK pathways. However, hypoxia modulates taxol-induced UPR activation acting specifically on the UPR branches PERK, ATF6α and IRE1α.137 Indeed, ATF4 activation leads to taxol-induced autophagy completion and cell death resistance. Finally, ATF4 expression in association with hypoxia-induced genes, such as adrenomedullin, is a biomarker of a poor prognosis for human breast cancer patients.137 Intratumoral hypoxia is one predominant feature of GBM and is associated with resistance to temozolomide (TMZ), the standard chemotherapy for GBM.138 TMZ sensitivity of both sensitive and resistant GBM cells is significantly enhanced under hyperoxia in vitro through the induction of caspase-dependent pathways.138 In addition, elevated PDIA1 expression also occurs in hypoxic brain tumor cells. PDIA1, which belongs to the protein disulfide isomerase superfamily, is the key foldase that has been found to be significantly dysregulated during the development of TMZ resistance in GBM cells.139 Hyperoxia resensitizes TMZ-resistant GBM cells to TMZ by abrogating the hypoxia-induced UPR-related protective mechanisms. Hyperoxia, alone or synergistically with TMZ, activates the UPR in sensitive and resistant cell lines.139 Hyperoxia impairs protein folding that in turn induces UPR-mediated apoptosis. Its reduces survival benefit of cancer cells with PDIA1 overexpression through the UPR by decreasing GRP78 and PDIA1 expression and consequently triggering cell death via downregulation of the ER stress chaperone protectors.139 Interestingly, TMZ increases galectin-1 expression in glioma cells.134 Galectin-1 increases the expression of genes implicated in chemotherapy resistance such as GRP78, ORP150, HERP (homocysteine-induced ER protein), transcription associated factor 1 (TRA1), BNIP3L (Bcl-2/adenovirus E1B 19 kDa protein-interacting protein 3-like), GADD45B and CYR61 (cysteine-rich angiogenic inducer 61), some of which are located in the ER and modified by hypoxia.134 Additionally, under severe hypoxia and chemotherapy, UPR activation occurs in hypopharyngeal carcinomas leading to increased expression of GRP78 associated with hypoxia-induced chemotherapy resistance.136 Diminution of GRP78 inhibits cell proliferation and promotes apoptosis under cisplatin treatment with severely hypoxic conditions, indicating that GRP78 confers cancer cell resistance to cisplatin in response to severe hypoxia. This phenomenon involves increased CHOP and BAX expression levels and decreased Bcl-2 expression levels with simultaneous increased apoptosis under severely hypoxic conditions.136 A number of studies indicated that improving oxygenation inside the tumor could serve as a potential strategy to target hypoxia-induced chemotherapy resistance.135 In liver cancers, hypoxia increases cisplatin resistance. The use of a hemoglobin-based oxygen carrier (OC89) enhances the efficacy of cisplatin-based transarterial chemoembolization in rat liver cancer model. OC89 delivery knocks down the balance of UPR pathway by decreasing GRP78 expression and increasing that of CHOP. This leads to increase tumor apoptosis and to inhibit tumor cell proliferation.135
Interestingly, UPR activation is also observed in non-tumoral cells that compose the tumor microenvironment.140 Indeed, UPR markers GRP78, ATF4 and CHOP are significantly upregulated in endothelial cells from oral squamous cell carcinomas. Furthermore, under severe acidic conditions and hypoxia, which recapitulate the tumor microenvironment, microvascular endothelial cells increase GRP78 expression, acquire antiapoptosis capacities and resist to sunitinib, an antiangiogenic drug.140 GRP78 knockdown resensitizes endothelial cells to drug treatment.140
Conclusion and perspectives: targeting the UPR to bypass resistance
The UPR is a physiological mechanism developed by cells to cope with misfolded protein accumulation induced by challenging conditions. As observed for other cellular mechanisms, tumor cells hijack the UPR to allow drug resistance, through the activation of the UPR sensors ATF6, IRE1α and PERK, and their master regulator GRP78. As presented above, the involvement of the UPR in chemotherapy resistance is complex and not fully covered yet. This is in part due to the links between the UPR and other tumor adaptive mechanisms as such antiapoptotic mechanisms, autophagy or dormancy. Therefore, a global understanding of the molecular mechanisms controlling UPR-mediated drug resistance is highly needed.
Small-molecule UPR inhibitors that directly target the UPR sensors ATF6α, IRE1α, PERK and their regulators or effectors such as PDIA1 and eIF2α, respectively, have been recently identified.141 Their potential use in combination with chemotherapeutics might greatly improve anticancer drug efficacy. For instance, ISRIB, a drug that reverses the effects of eIF2α phosphorylation, increased gemcitabine-induced death of pancreatic cancer cells.142 Recent evidences have also been provided from leukemic tumors. The PDI inhibitor 16F16 reverses leukemia cell resistance to imatinib linked to the ATF6α pathway most likely by blocking PDIA5.17 Finally, MKC-3946, an IRE1α RNase inhibitor, synergizes bortezomib or arsenic trioxide induced toxicity of acute myeloid leukemia cells.143
Alternatively, modulating UPR with pharmacological drugs has shown promising results in vitro. For instance, epigallocatechin gallate, which specifically targets GRP78, resensitizes glioma cells to TMZ.47, 144 Although targeting GRP78 might be an attractive therapeutic approach, the challenge will be to minimize systemic toxicity in normal organs in which GRP78 is essential for the survival and functions of various cellular subtypes.145 This implies that GRP78-targeting drugs should selectively target tumor cells that require a high level of GRP78 and spare normal organs. Bortezomib, a proteasome inhibitor that amplifies the protein misfolding burden, confers a chemosensitizing effect to cisplatin, doxorubicin or camptothecin in various tumor types including breast, colon pancreatic cancers.146 Sorafenib, a potent multikinase inhibitor, induces both apoptosis and autophagy in human hepatocellular carcinoma cells through an ER stress-dependent mechanism and the alteration of normal secretory functions. Furthermore, the combination of sorafenib with the autophagy inhibitor chloroquine leads to enhance liver cancer suppression.147 Verteporfin, a YAP1 (Yes-associated protein 1) inhibitor, has been recently involved in the oligomerized protein accumulation in CRC cells, leading in part to tumor apoptosis. Furthermore, hypoxic or nutrient-deprived conditions amplify verteporfin-mediated CRC cell death.148 Resistance of melanoma cells to vemurafenib or PLX4032, two BRAFV600E kinase inhibitors, is bypassed in the presence of thapsigargin, an inhibitor of the SERCA pumps or in the presence of HA15, which targets GRP78, respectively, by inducing tumor apoptosis.73, 149
In conclusion, future challenges will certainly lead to the development of combined therapeutic approaches with new drugs that specifically target the UPR sensors and downstream partners and will to bypass anticancer drug resistance.
Acknowledgments
This work was supported by grants from Institut National du Cancer (INCa_5869, INCa_7981 and PLBIO: 2015-111), la Ligue Contre le Cancer (comités 35, 37 and 56) and EU H2020 MSCA ITN-675448 (TRAINERS).
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The authors declare no conflict of interest.
References
- Hetz C, Chevet E, Oakes SA. Proteostasis control by the unfolded protein response. Nat Cell Biol 2015; 17: 829–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevet E, Hetz C, Samali A. Endoplasmic reticulum stress-activated cell reprogramming in oncogenesis. Cancer Discov 2015; 5: 586–597. [DOI] [PubMed] [Google Scholar]
- Dejeans N, Barroso K, Fernandez-Zapico ME, Samali A, Chevet E. Novel roles of the unfolded protein response in the control of tumor development and aggressiveness. Semin Cancer Biol 2015; 33: 67–73. [DOI] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science 2008; 319: 916–919. [DOI] [PubMed] [Google Scholar]
- Schubert U, Anton LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 2000; 404: 770–774. [DOI] [PubMed] [Google Scholar]
- Mann MJ, Hendershot LM. UPR activation alters chemosensitivity of tumor cells. Cancer Biol Ther 2006; 5: 736–740. [DOI] [PubMed] [Google Scholar]
- Urra H, Dufey E, Lisbona F, Rojas-Rivera D, Hetz C. When ER stress reaches a dead end. Biochim Biophys Acta 2013; 1833: 3507–3517. [DOI] [PubMed] [Google Scholar]
- Ma Y, Hendershot LM. The role of the unfolded protein response in tumour development: friend or foe? Nat Rev Cancer 2004; 4: 966–977. [DOI] [PubMed] [Google Scholar]
- Cubillos-Ruiz JR, Bettigole SE, Glimcher LH. Tumorigenic and immunosuppressive effects of endoplasmic reticulum stress in cancer. Cell 2017; 168: 692–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Kaufman RJ. How does protein misfolding in the endoplasmic reticulum affect lipid metabolism in the liver? Curr Opin Lipidol 2014; 25: 125–132. [DOI] [PubMed] [Google Scholar]
- Tsuruo T, Naito M, Tomida A, Fujita N, Mashima T, Sakamoto H et al. Molecular targeting therapy of cancer: drug resistance, apoptosis and survival signal. Cancer Sci 2003; 94: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell 1999; 10: 3787–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev 1998; 12: 1812–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 1999; 397: 271–274. [DOI] [PubMed] [Google Scholar]
- Carrara M, Prischi F, Nowak PR, Kopp MC, Ali MM. Noncanonical binding of BiP ATPase domain to Ire1 and Perk is dissociated by unfolded protein CH1 to initiate ER stress signaling. Elife 2015; 4: 3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol 2000; 2: 326–332. [DOI] [PubMed] [Google Scholar]
- Higa A, Taouji S, Lhomond S, Jensen D, Fernandez-Zapico ME, Simpson JC et al. Endoplasmic reticulum stress-activated transcription factor ATF6alpha requires the disulfide isomerase PDIA5 to modulate chemoresistance. Mol Cell Biol 2014; 34: 1839–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadanaka S, Okada T, Yoshida H, Mori K. Role of disulfide bridges formed in the luminal domain of ATF6 in sensing endoplasmic reticulum stress. Mol Cell Biol 2007; 27: 1027–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell 2002; 3: 99–111. [DOI] [PubMed] [Google Scholar]
- Lu Y, Liang FX, Wang X. A synthetic biology approach identifies the mammalian UPR RNA ligase RtcB. Mol Cell 2014; 55: 758–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H et al. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell 2007; 13: 365–376. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 2001; 107: 881–891. [DOI] [PubMed] [Google Scholar]
- Jurkin J, Henkel T, Nielsen AF, Minnich M, Popow J, Kaufmann T et al. The mammalian tRNA ligase complex mediates splicing of XBP1 mRNA and controls antibody secretion in plasma cells. EMBO J 2014; 33: 2922–2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T et al. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev 2002; 16: 452–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 2002; 415: 92–96. [DOI] [PubMed] [Google Scholar]
- Hetz C, Martinon F, Rodriguez D, Glimcher LH. The unfolded protein response: integrating stress signals through the stress sensor IRE1alpha. Physiol Rev 2011; 91: 1219–1243. [DOI] [PubMed] [Google Scholar]
- Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C et al. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell 2007; 27: 53–66. [DOI] [PubMed] [Google Scholar]
- Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 2006; 313: 104–107. [DOI] [PubMed] [Google Scholar]
- Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol 2009; 186: 323–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D, Lerner AG, Vande Walle L, Upton JP, Xu W, Hagen A et al. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell 2009; 138: 562–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwawaki T, Hosoda A, Okuda T, Kamigori Y, Nomura-Furuwatari C, Kimata Y et al. Translational control by the ER transmembrane kinase/ribonuclease IRE1 under ER stress. Nat Cell Biol 2001; 3: 158–164. [DOI] [PubMed] [Google Scholar]
- Lerner AG, Upton JP, Praveen PV, Ghosh R, Nakagawa Y, Igbaria A et al. IRE1alpha induces thioredoxin-interacting protein to activate the NLRP3 inflammasome and promote programmed cell death under irremediable ER stress. Cell Metab 2012; 16: 250–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton JP, Wang L, Han D, Wang ES, Huskey NE, Lim L et al. IRE1alpha cleaves select microRNAs during ER stress to derepress translation of proapoptotic caspase-2. Science 2012; 338: 818–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel M, Chevet E, Tavernier J, Gerlo S. Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem Sci 2014; 39: 245–254. [DOI] [PubMed] [Google Scholar]
- Ghosh R, Wang L, Wang ES, Perera BG, Igbaria A, Morita S et al. Allosteric inhibition of the IRE1alpha RNase preserves cell viability and function during endoplasmic reticulum stress. Cell 2014; 158: 534–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam AB, Koong AC, Niwa M. Ire1 has distinct catalytic mechanisms for XBP1/HAC1 splicing and RIDD. Cell Rep 2014; 9: 850–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchecareilh M, Higa A, Fribourg S, Moenner M, Chevet E. Peptides derived from the bifunctional kinase/RNase enzyme IRE1alpha modulate IRE1alpha activity and protect cells from endoplasmic reticulum stress. FASEB J 2011; 25: 3115–3129. [DOI] [PubMed] [Google Scholar]
- Han D, Lerner AG, Vande Walle L, Upton J-P, Xu W, Hagen A et al. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell 2009; 138: 562–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P et al. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell 2001; 7: 1165–1176. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell 2000; 5: 897–904. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 2003; 11: 619–633. [DOI] [PubMed] [Google Scholar]
- Ye J, Koumenis C. ATF4, an ER stress and hypoxia-inducible transcription factor and its potential role in hypoxia tolerance and tumorigenesis. Curr Mol Med 2009; 9: 411–416. [DOI] [PubMed] [Google Scholar]
- Han J, Back SH, Hur J, Lin YH, Gildersleeve R, Shan J et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol 2013; 15: 481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa I, Zeng H, Harding HP, Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J Cell Biol 2001; 153: 1011–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obacz J, Avril T, Le Reste PJ, Urra H, Quillien V, Hetz C et al. Endoplasmic reticulum proteostasis in glioblastoma—from molecular mechanisms to therapeutic perspectives. Sci Signal 2017; 10: eaal2323. [DOI] [PubMed] [Google Scholar]
- Epple LM, Dodd RD, Merz AL, Dechkovskaia AM, Herring M, Winston BA et al. Induction of the unfolded protein response drives enhanced metabolism and chemoresistance in glioma cells. PLoS ONE 2013; 8: e73267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrko P, Schonthal AH, Hofman FM, Chen TC, Lee AS. The unfolded protein response regulator GRP78/BiP as a novel target for increasing chemosensitivity in malignant gliomas. Cancer Res 2007; 67: 9809–9816. [DOI] [PubMed] [Google Scholar]
- Lee HK, Xiang C, Cazacu S, Finniss S, Kazimirsky G, Lemke N et al. GRP78 is overexpressed in glioblastomas and regulates glioma cell growth and apoptosis. Neuro-oncology 2008; 10: 236–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Nichols P, Spicer D, Groshen S, Yu MC, Lee AS. GRP78 as a novel predictor of responsiveness to chemotherapy in breast cancer. Cancer Res 2006; 66: 7849–7853. [DOI] [PubMed] [Google Scholar]
- Fernandez PM, Tabbara SO, Jacobs LK, Manning FC, Tsangaris TN, Schwartz AM et al. Overexpression of the glucose-regulated stress gene GRP78 in malignant but not benign human breast lesions. Breast Cancer Res Treat 2000; 59: 15–26. [DOI] [PubMed] [Google Scholar]
- Scriven P, Coulson S, Haines R, Balasubramanian S, Cross S, Wyld L. Activation and clinical significance of the unfolded protein response in breast cancer. Br J Cancer 2009; 101: 1692–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andruska N, Zheng X, Yang X, Helferich WG, Shapiro DJ. Anticipatory estrogen activation of the unfolded protein response is linked to cell proliferation and poor survival in estrogen receptor alpha-positive breast cancer. Oncogene 2015; 34: 3760–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming J, Ruan S, Wang M, Ye D, Fan N, Meng Q et al. A novel chemical, STF-083010, reverses tamoxifen-related drug resistance in breast cancer by inhibiting IRE1/XBP1. Oncotarget 2015; 6: 40692–40703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhaidat NM, Alzoubi KH, Almomani N, Khabour OF. Expression of glucose regulated protein 78 (GRP78) determines colorectal cancer response to chemotherapy. Cancer Biomarkers 2015; 15: 197–203. [DOI] [PubMed] [Google Scholar]
- Fujimoto T, Yoshimatsu K, Watanabe K, Yokomizo H, Otani T, Matsumoto A et al. Overexpression of human X-box binding protein 1 (XBP-1) in colorectal adenomas and adenocarcinomas. Anticancer Res 2007; 27: 127–131. [PubMed] [Google Scholar]
- Drake TM, Ritchie JE, Kanthou C, Staves JJ, Narramore R, Wyld L. Targeting the endoplasmic reticulum mediates radiation sensitivity in colorectal cancer. Exp Mol Pathol 2015; 98: 532–539. [DOI] [PubMed] [Google Scholar]
- Mhaidat NM, Alzoubi KH, Khabour OF, Banihani MN, Al-Balas QA, Swaidan S. GRP78 regulates sensitivity of human colorectal cancer cells to DNA targeting agents. Cytotechnology 2016; 68: 459–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piton N, Wason J, Colasse E, Cornic M, Lemoine F, Le Pessot F et al. Endoplasmic reticulum stress, unfolded protein response and development of colon adenocarcinoma. Virchows Archiv 2016; 469: 145–154. [DOI] [PubMed] [Google Scholar]
- Fu W, Wu X, Li J, Mo Z, Yang Z, Huang W et al. Upregulation of GRP78 in renal cell carcinoma and its significance. Urology 2010; 75: 603–607. [DOI] [PubMed] [Google Scholar]
- Al-Rawashdeh FY, Scriven P, Cameron IC, Vergani PV, Wyld L. Unfolded protein response activation contributes to chemoresistance in hepatocellular carcinoma. Eur J Gastroenterol Hepatol 2010; 22: 1099–1105. [DOI] [PubMed] [Google Scholar]
- Shuda M, Kondoh N, Imazeki N, Tanaka K, Okada T, Mori K et al. Activation of the ATF6, XBP1 and grp78 genes in human hepatocellular carcinoma: a possible involvement of the ER stress pathway in hepatocarcinogenesis. J Hepatol 2003; 38: 605–614. [DOI] [PubMed] [Google Scholar]
- Tang J, Guo YS, Zhang Y, Yu XL, Li L, Huang W et al. CD147 induces UPR to inhibit apoptosis and chemosensitivity by increasing the transcription of Bip in hepatocellular carcinoma. Cell Death Differ 2012; 19: 1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakatani T, Maemura K, Hiyama N, Amano Y, Watanabe K, Kage H et al. High expression of IRE1 in lung adenocarcinoma is associated with a lower rate of recurrence. Jpn J Clin Oncol 2017; 47: 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HY, Yang YF, Wu AT, Yang CJ, Liu YP, Jan YH et al. Endoplasmic reticulum ribosome-binding protein 1 (RRBP1) overexpression is frequently found in lung cancer patients and alleviates intracellular stress-induced apoptosis through the enhancement of GRP78. Oncogene 2013; 32: 4921–4931. [DOI] [PubMed] [Google Scholar]
- Niu Z, Wang M, Zhou L, Yao L, Liao Q, Zhao Y. Elevated GRP78 expression is associated with poor prognosis in patients with pancreatic cancer. Scientific Rep 2015; 5: 16067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong B, Wu W, Valkovska N, Jager C, Hong X, Nitsche U et al. A common genetic variation of melanoma inhibitory activity-2 labels a subtype of pancreatic adenocarcinoma with high endoplasmic reticulum stress levels. Scientific Rep 2015; 5: 8109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese G, Carugo A, Tepper J, Robinson FS, Li L, Svelto M et al. Synthetic vulnerabilities of mesenchymal subpopulations in pancreatic cancer. Nature 2017; 542: 362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobak Y, Kurlishchuk Y, Vynnytska-Myronovska B, Grydzuk O, Shuvayeva G, Redowicz MJ et al. Arginine deprivation induces endoplasmic reticulum stress in human solid cancer cells. Int J Biochem Cell Biol 2016; 70: 29–38. [DOI] [PubMed] [Google Scholar]
- Cuevas EP, Eraso P, Mazon MJ, Santos V, Moreno-Bueno G, Cano A et al. LOXL2 drives epithelial–mesenchymal transition via activation of IRE1-XBP1 signalling pathway. Scientific Rep 2017; 7: 44988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Xue Y, Si Y, Wang Q, Wang Z, Yuan J et al. The unfolded protein response potentiates epithelial-to-mesenchymal transition (EMT) of gastric cancer cells under severe hypoxic conditions. Med Oncol 2015; 32: 447. [DOI] [PubMed] [Google Scholar]
- Lin JA, Fang SU, Su CL, Hsiao CJ, Chang CC, Lin YF et al. Silencing glucose-regulated protein 78 induced renal cell carcinoma cell line G1 cell-cycle arrest and resistance to conventional chemotherapy. Urol Oncol 2014; 32: 29.e1–11. [DOI] [PubMed] [Google Scholar]
- Zhou T, Lv X, Guo X, Ruan B, Liu D, Ding R et al. RACK1 modulates apoptosis induced by sorafenib in HCC cells by interfering with the IRE1/XBP1 axis. Oncol Rep 2015; 33: 3006–3014. [DOI] [PubMed] [Google Scholar]
- Cerezo M, Lehraiki A, Millet A, Rouaud F, Plaisant M, Jaune E et al. Compounds triggering ER stress exert anti-melanoma effects and overcome BRAF inhibitor resistance. Cancer Cell 2016; 29: 805–819. [DOI] [PubMed] [Google Scholar]
- Manie SN, Lebeau J, Chevet E. Cellular mechanisms of endoplasmic reticulum stress signaling in health and disease. 3. Orchestrating the unfolded protein response in oncogenesis: an update. Am J Physiol Cell Physiol 2014; 307: C901–C907. [DOI] [PubMed] [Google Scholar]
- Heijmans J, van Lidth de Jeude JF, Koo BK, Rosekrans SL, Wielenga MC, van de Wetering M et al. ER stress causes rapid loss of intestinal epithelial stemness through activation of the unfolded protein response. Cell Rep 2013; 3: 1128–1139. [DOI] [PubMed] [Google Scholar]
- Vermeulen L, Snippert HJ. Stem cell dynamics in homeostasis and cancer of the intestine. Nat Rev Cancer 2014; 14: 468–480. [DOI] [PubMed] [Google Scholar]
- Niederreiter L, Fritz TM, Adolph TE, Krismer AM, Offner FA, Tschurtschenthaler M et al. ER stress transcription factor Xbp1 suppresses intestinal tumorigenesis and directs intestinal stem cells. J Exp Med 2013; 210: 2041–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schewe DM, Aguirre-Ghiso JA. ATF6alpha-Rheb-mTOR signaling promotes survival of dormant tumor cells in vivo. Proc Natl Acad Sci USA 2008; 105: 10519–10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fels DR, Koumenis C. The PERK/eIF2alpha/ATF4 module of the UPR in hypoxia resistance and tumor growth. Cancer Biol Ther 2006; 5: 723–728. [DOI] [PubMed] [Google Scholar]
- Chen X, Iliopoulos D, Zhang Q, Tang Q, Greenblatt MB, Hatziapostolou M et al. XBP1 promotes triple-negative breast cancer by controlling the HIF1alpha pathway. Nature 2014; 508: 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejeans N, Pluquet O, Lhomond S, Grise F, Bouchecareilh M, Juin A et al. Autocrine control of glioma cells adhesion and migration through IRE1alpha-mediated cleavage of SPARC mRNA. J Cell Sci 2012; 125(Part 18): 4278–4287. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674. [DOI] [PubMed] [Google Scholar]
- Bissell MJ, Hines WC. Why don't we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med 2011; 17: 320–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung CT, Brugge JS. Outgrowth of single oncogene-expressing cells from suppressive epithelial environments. Nature 2012; 482: 410–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Chen X, Gao Y, Wu J, Zeng F, Song F. XBP1 induces snail expression to promote epithelial-to-mesenchymal transition and invasion of breast cancer cells. Cell Signal 2015; 27: 82–89. [DOI] [PubMed] [Google Scholar]
- Feng YX, Sokol ES, Del Vecchio CA, Sanduja S, Claessen JH, Proia TA et al. Epithelial-to-mesenchymal transition activates PERK-eIF2alpha and sensitizes cells to endoplasmic reticulum stress. Cancer Discov 2014; 4: 702–715. [DOI] [PubMed] [Google Scholar]
- Ulianich L, Garbi C, Treglia AS, Punzi D, Miele C, Raciti GA et al. ER stress is associated with dedifferentiation and an epithelial-to-mesenchymal transition-like phenotype in PC Cl3 thyroid cells. J Cell Sci 2008; 121(Part 4): 477–486. [DOI] [PubMed] [Google Scholar]
- Del Vecchio CA, Feng Y, Sokol ES, Tillman EJ, Sanduja S, Reinhardt F et al. De-differentiation confers multidrug resistance via noncanonical PERK-Nrf2 signaling. PLoS Biol 2014; 12: e1001945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheshadri N, Catanzaro JM, Bott AJ, Sun Y, Ullman E, Chen EI et al. SCCA1/SERPINB3 promotes oncogenesis and epithelial–mesenchymal transition via the unfolded protein response and IL6 signaling. Cancer Res 2014; 74: 6318–6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auf G, Jabouille A, Guerit S, Pineau R, Delugin M, Bouchecareilh M et al. Inositol-requiring enzyme 1alpha is a key regulator of angiogenesis and invasion in malignant glioma. Proc Natl Acad Sci USA 2010; 107: 15553–15558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drogat B, Auguste P, Nguyen DT, Bouchecareilh M, Pineau R, Nalbantoglu J et al. IRE1 signaling is essential for ischemia-induced vascular endothelial growth factor-A expression and contributes to angiogenesis and tumor growth in vivo. Cancer Res 2007; 67: 6700–6707. [DOI] [PubMed] [Google Scholar]
- Pluquet O, Dejeans N, Bouchecareilh M, Lhomond S, Pineau R, Higa A et al. Posttranscriptional regulation of PER1 underlies the oncogenic function of IREalpha. Cancer Res 2013; 73: 4732–4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais JD, Addison CL, Edge R, Falls T, Zhao H, Wary K et al. Perk-dependent translational regulation promotes tumor cell adaptation and angiogenesis in response to hypoxic stress. Mol Cell Biol 2006; 26: 9517–9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa K, Tsukamoto Y, Hori O, Kitao Y, Yanagi H, Stern DM et al. Regulation of tumor angiogenesis by oxygen-regulated protein 150, an inducible endoplasmic reticulum chaperone. Cancer Res 2001; 61: 4206–4213. [PubMed] [Google Scholar]
- Miyagi T, Hori O, Koshida K, Egawa M, Kato H, Kitagawa Y et al. Antitumor effect of reduction of 150-kDa oxygen-regulated protein expression on human prostate cancer cells. Int J Urol 2002; 9: 577–585. [DOI] [PubMed] [Google Scholar]
- Wang ZV, Deng Y, Gao N, Pedrozo Z, Li DL, Morales CR et al. Spliced X-box binding protein 1 couples the unfolded protein response to hexosamine biosynthetic pathway. Cell 2014; 156: 1179–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzel MS, Storm NJ, Gutschmidt A, Baddi R, Hinze Y, Jarosch E et al. Hexosamine pathway metabolites enhance protein quality control and prolong life. Cell 2014; 156: 1167–1178. [DOI] [PubMed] [Google Scholar]
- Ferrer CM, Lynch TP, Sodi VL, Falcone JN, Schwab LP, Peacock DL et al. O-GlcNAcylation regulates cancer metabolism and survival stress signaling via regulation of the HIF-1 pathway. Mol Cell 2014; 54: 820–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais JD, Filipenko V, Bi M, Harding HP, Ron D, Koumenis C et al. Activating transcription factor 4 is translationally regulated by hypoxic stress. Mol Cell Biol 2004; 24: 7469–7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi M, Naczki C, Koritzinsky M, Fels D, Blais J, Hu N et al. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J 2005; 24: 3470–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouschop KM, van den Beucken T, Dubois L, Niessen H, Bussink J, Savelkouls K et al. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Invest 2010; 120: 127–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol 2006; 26: 9220–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman MM. Mechanisms of cancer drug resistance. Annu Rev Med 2002; 53: 615–627. [DOI] [PubMed] [Google Scholar]
- Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer 2013; 13: 714–726. [DOI] [PubMed] [Google Scholar]
- Longley DB, Johnston PG. Molecular mechanisms of drug resistance. J Pathol 2005; 205: 275–292. [DOI] [PubMed] [Google Scholar]
- Chen ZS, Tiwari AK. Multidrug resistance proteins (MRPs/ABCCs) in cancer chemotherapy and genetic diseases. FEBS J 2011; 278: 3226–3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodani K, Patel A, Kathawala RJ, Chen ZS. Multidrug resistance associated proteins in multidrug resistance. Chin J Cancer 2012; 31: 58–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal R, Kaye SB. Ovarian cancer: strategies for overcoming resistance to chemotherapy. Nat Rev Cancer 2003; 3: 502–516. [DOI] [PubMed] [Google Scholar]
- Borst P, Evers R, Kool M, Wijnholds J. A family of drug transporters: the multidrug resistance-associated proteins. J Natl Cancer Inst 2000; 92: 1295–1302. [DOI] [PubMed] [Google Scholar]
- Chou AJ, Gorlick R. Chemotherapy resistance in osteosarcoma: current challenges and future directions. Expert Rev Anticancer Ther 2006; 6: 1075–1085. [DOI] [PubMed] [Google Scholar]
- Mann MJ, Pereira ER, Liao N, Hendershot LM. UPR-induced resistance to etoposide is downstream of PERK and independent of changes in topoisomerase IIalpha levels. PLoS ONE 2012; 7: e47931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman P, Jonkers J. The effects of deregulated DNA damage signalling on cancer chemotherapy response and resistance. Nat Rev Cancer 2012; 12: 587–598. [DOI] [PubMed] [Google Scholar]
- Davies MP, Barraclough DL, Stewart C, Joyce KA, Eccles RM, Barraclough R et al. Expression and splicing of the unfolded protein response gene XBP-1 are significantly associated with clinical outcome of endocrine-treated breast cancer. Int J Cancer 2008; 123: 85–88. [DOI] [PubMed] [Google Scholar]
- Lee E, Nichols P, Groshen S, Spicer D, Lee AS. GRP78 as potential predictor for breast cancer response to adjuvant taxane therapy. Int J Cancer 2011; 128: 726–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan D, Carberry S, Murphy AC, Lindner AU, Fay J, Hector S et al. Calnexin, an ER-induced protein, is a prognostic marker and potential therapeutic target in colorectal cancer. J Transl Med 2016; 14: 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy RK, Mao C, Baumeister P, Austin RC, Kaufman RJ, Lee AS. Endoplasmic reticulum chaperone protein GRP78 protects cells from apoptosis induced by topoisomerase inhibitors: role of ATP binding site in suppression of caspase-7 activation. J Biol Chem 2003; 278: 20915–20924. [DOI] [PubMed] [Google Scholar]
- Yan M, Ni J, Song D, Ding M, Huang J. Activation of unfolded protein response protects osteosarcoma cells from cisplatin-induced apoptosis through NF-kappaB pathway. Int J Clin Exp Pathol 2015; 8: 10204–10215. [PMC free article] [PubMed] [Google Scholar]
- Li ZY, Zhang C, Chen L, Chen BD, Li QZ, Zhang XJ et al. Radicol, a novel trinorguaiane-type sesquiterpene, induces temozolomide-resistant glioma cell apoptosis via ER stress and Akt/mTOR pathway blockade. Phytother Res 2017; 31: 729–739. [DOI] [PubMed] [Google Scholar]
- Chakravarty G, Mathur A, Mallade P, Gerlach S, Willis J, Datta A et al. Nelfinavir targets multiple drug resistance mechanisms to increase the efficacy of doxorubicin in MCF-7/Dox breast cancer cells. Biochimie 2016; 124: 53–64. [DOI] [PubMed] [Google Scholar]
- Fu Y, Li J, Lee AS. GRP78/BiP inhibits endoplasmic reticulum BIK and protects human breast cancer cells against estrogen starvation-induced apoptosis. Cancer Res 2007; 67: 3734–3740. [DOI] [PubMed] [Google Scholar]
- Gomez BP, Riggins RB, Shajahan AN, Klimach U, Wang A, Crawford AC et al. Human X-box binding protein-1 confers both estrogen independence and antiestrogen resistance in breast cancer cell lines. FASEB J 2007; 21: 4013–4027. [DOI] [PubMed] [Google Scholar]
- Cook KL, Clarke PA, Parmar J, Hu R, Schwartz-Roberts JL, Abu-Asab M et al. Knockdown of estrogen receptor-alpha induces autophagy and inhibits antiestrogen-mediated unfolded protein response activation, promoting ROS-induced breast cancer cell death. FASEB J 2014; 28: 3891–3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R, Warri A, Jin L, Zwart A, Riggins RB, Fang HB et al. NF-kappaB signaling is required for XBP1 (unspliced and spliced)-mediated effects on antiestrogen responsiveness and cell fate decisions in breast cancer. Mol Cell Biol 2015; 35: 379–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Wang J, Jing J, Hua H, Luo T, Xu L et al. Synergistic promotion of breast cancer cells death by targeting molecular chaperone GRP78 and heat shock protein 70. J Cell Mol Med 2009; 13: 4540–4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung BH, Kwan BW, He QY, Lee AS, Liu J, Wong AS. Glucose-regulated protein 78 as a novel effector of BRCA1 for inhibiting stress-induced apoptosis. Oncogene 2008; 27: 6782–6789. [DOI] [PubMed] [Google Scholar]
- Zhang X, Cook KL, Warri A, Cruz IM, Rosim M, Riskin J et al. Lifetime genistein intake increases the response of mammary tumors to tamoxifen in rats. Clin Cancer Res 2017; 23: 814–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumandan S, Mahadevan NR, Chiu K, DeLaney A, Zanetti M. Activation of the unfolded protein response bypasses trastuzumab-mediated inhibition of the PI-3K pathway. Cancer Lett 2013; 329: 236–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto A, Kawana K, Taguchi A, Adachi K, Sato M, Nakamura H et al. Inhibition of endoplasmic reticulum (ER) stress sensors sensitizes cancer stem-like cells to ER stress-mediated apoptosis. Oncotarget 2016; 7: 51854–51864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan AC, Zhang L, Adam AP, Aguirre-Ghiso JA. Functional coupling of p38-induced up-regulation of BiP and activation of RNA-dependent protein kinase-like endoplasmic reticulum kinase to drug resistance of dormant carcinoma cells. Cancer Res 2006; 66: 1702–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MD, Mann M, Nitiss JL, Hendershot LM. Activation of the unfolded protein response is necessary and sufficient for reducing topoisomerase IIalpha protein levels and decreasing sensitivity to topoisomerase-targeted drugs. Mol Pharmacol 2005; 68: 1699–1707. [DOI] [PubMed] [Google Scholar]
- Salaroglio IC, Panada E, Moiso E, Buondonno I, Provero P, Rubinstein M et al. PERK induces resistance to cell death elicited by endoplasmic reticulum stress and chemotherapy. Mol Cancer 2017; 16: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar M, Hernandez-Tiedra S, Torres S, Lorente M, Guzman M, Velasco G. Detecting autophagy in response to ER stress signals in cancer. Methods Enzymol 2011; 489: 297–317. [DOI] [PubMed] [Google Scholar]
- Parmar JH, Cook KL, Shajahan-Haq AN, Clarke PA, Tavassoly I, Clarke R et al. Modelling the effect of GRP78 on anti-oestrogen sensitivity and resistance in breast cancer. Interface Focus 2013; 3: 20130012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Mercier M, Lefranc F, Mijatovic T, Debeir O, Haibe-Kains B, Bontempi G et al. Evidence of galectin-1 involvement in glioma chemoresistance. Toxicol Appl Pharmacol 2008; 229: 172–183. [DOI] [PubMed] [Google Scholar]
- Liu XB, Cheng Q, Geng W, Ling CC, Liu Y, Ng KT et al. Enhancement of cisplatin-based TACE by a hemoglobin-based oxygen carrier in an orthotopic rat HCC model. Artif Cells Nanomed Biotechnol 2014; 42: 229–236. [DOI] [PubMed] [Google Scholar]
- Pi L, Li X, Song Q, Shen Y, Lu X, Di B. Knockdown of glucose-regulated protein 78 abrogates chemoresistance of hypopharyngeal carcinoma cells to cisplatin induced by unfolded protein in response to severe hypoxia. Oncol Lett 2014; 7: 685–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notte A, Rebucci M, Fransolet M, Roegiers E, Genin M, Tellier C et al. Taxol-induced unfolded protein response activation in breast cancer cells exposed to hypoxia: ATF4 activation regulates autophagy and inhibits apoptosis. Int J Chem Cell Biol 2015; 62: 1–14. [DOI] [PubMed] [Google Scholar]
- Sun S, Lee D, Lee NP, Pu JK, Wong ST, Lui WM et al. Hyperoxia resensitizes chemoresistant human glioblastoma cells to temozolomide. J Neuro-oncol 2012; 109: 467–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Sun S, Ho AS, Kiang KM, Zhang XQ, Xu FF et al. Hyperoxia resensitizes chemoresistant glioblastoma cells to temozolomide through unfolded protein response. Anticancer Res 2014; 34: 2957–2966. [PubMed] [Google Scholar]
- Visioli F, Wang Y, Alam GN, Ning Y, Rados PV, Nor JE et al. Glucose-regulated protein 78 (Grp78) confers chemoresistance to tumor endothelial cells under acidic stress. PLoS ONE 2014; 9: e101053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doultsinos D, Avril T, Lhomond S, Dejeans N, Guedat P, Chevet E. Control of the unfolded protein response in health and disease. SLAS Discov 2017; 22: 787–800. [DOI] [PubMed] [Google Scholar]
- Palam LR, Gore J, Craven KE, Wilson JL, Korc M. Integrated stress response is critical for gemcitabine resistance in pancreatic ductal adenocarcinoma. Cell Death Dis 2015; 6: e1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Lin DC, Guo X, Kharabi Masouleh B, Gery S, Cao Q et al. Inhibition of IRE1alpha-driven pro-survival pathways is a promising therapeutic application in acute myeloid leukemia. Oncotarget 2016; 7: 18736–18749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee R, Devi A, Mishra S. Molecular docking and molecular dynamics studies reveal structural basis of inhibition and selectivity of inhibitors EGCG and OSU-03012 toward glucose regulated protein-78 (GRP78) overexpressed in glioblastoma. J Mol Model 2015; 21: 272. [DOI] [PubMed] [Google Scholar]
- Lee AS. Glucose-regulated proteins in cancer: molecular mechanisms and therapeutic potential. Nat Rev Cancer 2014; 14: 263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams J. The development of proteasome inhibitors as anticancer drugs. Cancer Cell 2004; 5: 417–421. [DOI] [PubMed] [Google Scholar]
- Shi YH, Ding ZB, Zhou J, Hui B, Shi GM, Ke AW et al. Targeting autophagy enhances sorafenib lethality for hepatocellular carcinoma via ER stress-related apoptosis. Autophagy 2011; 7: 1159–1172. [DOI] [PubMed] [Google Scholar]
- Zhang H, Ramakrishnan SK, Triner D, Centofanti B, Maitra D, Gyorffy B et al. Tumor-selective proteotoxicity of verteporfin inhibits colon cancer progression independently of YAP1. Sci Signal 2015; 8: ra98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck D, Niessner H, Smalley KS, Flaherty K, Paraiso KH, Busch C et al. Vemurafenib potently induces endoplasmic reticulum stress-mediated apoptosis in BRAFV600E melanoma cells. Sci Signal 2013; 6: ra7. [DOI] [PMC free article] [PubMed] [Google Scholar]