Abstract
Background
Cervical cancer is the second leading cause of death among female patients with cancer in the world. High risk human papillomavirus has causal roles in cervical cancer initiation and progression by deregulating several cellular processes. However, HPV infection is not sufficient for cervical carcinoma development. Therefore, other genetic and epigenetic factors may be involved in this complex disease, and the identification of which may lead to better diagnosis and treatment. Our aim was to analyze the expression of microRNAs in cervical cancer cases positive or negative for HPV E6/E7 mRNA, and to assess their diagnostic usefulness and relevance.
Methods
The expression of three different microRNAs (miR-9, miR-21, and miR-155) in 52 formalin-fixed paraffin-embedded (FFPE) primary cervical cancer tissue samples and 50 FFPE normal cervical tissue samples were evaluated.
Results
MiR-9, miR-21, and miR-155 were significantly overexpressed in cervical cancer tissues compared to normal tissues (P < 0.001). MiR-21 and miR-155 expression combined with the HPV E6/E7 mRNA assay in HPV E6/E7 negative cervical cancer showed increased AUC of 0.7267 and 0.7000, respectively (P = 0.01, P = 0.04), demonstrating their potential as diagnostic tools. Moreover, miR-21 and miR-155 were predictors showing a 7 fold and 10.3 fold higher risk for HPV E6/E7 negative patients with cervical cancer (P = 0.024 and P = 0.017, respectively) while miR-155 was a predictor showing a 27.9 fold higher risk for HPV E6/E7 positive patients with cervical cancer (P < 0.0001).
Conclusions
There is a strong demand for additional, alternative molecular biomarkers for diagnosis and management of precancer patients. MiR-21 and miR-155 may be helpful in the prediction of both HPV positive and HPV negative cases of cervical cancer.
Keywords: Cervical cancer, microRNA, HPV E6/E7, RT-qPCR, Molecular diagnosis
Background
Cervical cancer is the third most common malignancy in women worldwide [1]. High risk human papillomavirus (HR-HPV) infection is recognized as the most important risk factor in cervical cancer. Persistent over-expression of the E6 and E7 oncogenes encoded in the HPV genome have a critical role in the development of cervical cancer by causing genetic and epigenetic instability [2]. HPV E6 leads to the degradation of p53, which is a critical tumor suppressor that regulates abrogation of cell growth arrest. Furthermore, HPV E7 binds and deactivates another important tumor suppressor, the retinoblastoma protein (pRb), thereby interfering with cell cycle regulation [3–6].
Recently, several studies reported the development of cervical cancers that are HPV negative despite increased sensitivity of HR-HPV detection methods. Through meta-analyses of HPV detection methods, both Tjalma, et al. and Giorgi, et al. found that 4.2 to 8.2% of cases were HPV negative in 574 invasive cervical cancers and 3162 invasive cervical cancers [7, 8]. A large international retrospective cross-sectional study including 10,575 cases with invasive cervical cancer found that 15% (1598 cases) were negative for HPV DNA [9]. Similarly, our previous study also found that 15% of patients with cervical cancer (100 cases) were HPV negative [10].
Epigenetic instability is affected by microRNAs (miRNA or miR-). MiRNAs are 19 to 25 nucleotides (nt) in length, and have a role in transcriptional and epigenetic regulation through binding the 3′-UTR of the target-mRNA [11, 12]. It is now widely known that miRNA dysregulation is associated with a wide variety of human malignancies, such as breast cancer, lung cancer, colon cancer, and gastric cancer [13–16].
Many miRNAs studies have tried to confirm the utility of each miRNAs in cervical cancers with different methods. Lui et al. used miRNA direct sequencing analysis with six human cervical carcinoma cell lines and frozen cervical tumor tissues [17]. Lee et al. had miRNA expression profiling with 157 panel analyses with frozen cervical tumor tissues [18]. Gocze et al. utilized quantitative real time polymerase chain reaction (RT-qPCR) of eight miRNAs (miR-21, miR-27a, miR-34a, miR-146a, miR-155, miR-196a, miR-203, miR-221) individually [19].
Among these several miRNAs, three miRNAs (miR-9, miR-21, and miR-155) having their revealed targets that might be related to cancer were selected. Ma et al. showed miR-9 increased cell motility and invasiveness by targeting Cadherin 1(CDH1) and lead to cancer metastasis [20]. Asangani et al. and Bumrungthai et al. showed miR-21 promoted invasion and cell proliferation targeting programmed cell death 4(PDCD4) [21, 22]. MiR-155 expression promotes the proliferation targeting liver kinase B1 (LKB1) [23, 24].
Although the roles of these three miRNAs (miR-9, miR-21, and miR-155) have been studied in cervical cancer, their potential diagnostic or prognostic value in a clinical setting has not been examined. In addition, it is not known whether there is an association between these three miRNAs and HR-HPV infection status in clinical tissue specimens. Therefore, the purpose of this study was to investigate miR-9, miR-21, and miR-155 expression levels in cervical cancer and normal tissue samples, and determine their possible relation to HR-HPV E6/E7 oncogene expression.
Methods
Clinical samples
A total of 52 FFPE cervical cancer tissue samples and 50 FFPE normal cervical tissue samples were used from the Department of Pathology, Yonsei University Wonju Severance Christian Hospital, Wonju, Republic of Korea, between January 2010 and December 2014 (Table 1). Institutional Ethics Committee at Yonsei University Wonju College of Medicine approved the study protocol (approval no. YWMR-12-4-010) and all subjects provided written informed consent. Cases with tissue biopsies available were reviewed by two pathologists. The 52 cervical cancer samples consisted of tissue samples from 50 squamous cell carcinomas and 2 adenocarcinomas.
Table 1.
Sample information in cervical cancer and normal
| Variables | Cancer, n (%) | Normal, n (%) |
|---|---|---|
| Age | ||
| < 50 years | 18 (34.6) | 31 (62.0) |
| ≥ 50 years | 34 (65.4) | 19 (38.0) |
| Histology | ||
| SCC | 50 (96.2) | |
| ADC | 2 (3.8) | |
| HPV E6/E7 mRNA expression | ||
| Positive | 37 (71.2) | 0 (0) |
| Negative | 15 (28.8) | 50 (100) |
| Total | 52 (100) | 50 (100) |
SCC Squamous cell carcinoma, ADC Adenocarcinoma
Deparaffinization of FFPE tissues and total RNA extraction
Three to four 10-μm thick sections of FFPE cervical tissue were used for total RNA extraction. To remove paraffin from FFPE tissue, 160 μL of Deparaffinization solution (Qiagen, Hilden, Germany) was added and vortexed, followed by incubation for 3 min at 56 °C. RNA extraction was performed using the Qiagen RNeasy FFPE kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. Total RNA purity and concentration were determined by measuring the ratio of the absorbance at 260 and 280 nm using an Infinite 200 spectrophotometer (Tecan, Salzburg, Austria). All preparation and handling procedures were conducted under RNase-free conditions. Isolated total RNA was stored at −70 °C until used.
cDNA synthesis
Complementary DNA (cDNA) was synthesized using a TaqMan microRNA Reverse Transcriptase kit (Applied Biosystems by Life Technologies, Foster City, CA, USA) according to manufacturer’s instructions. Briefly, 5 to 10 ng of total RNA was used for cDNA synthesis. The reverse transcriptase (RT) reaction mixture contained 0.15 μL of 100 mM dNTP mix (100 mM each dATP, dGTP, dCTP, and dTTP at a neutral pH), 1 μL of 50 U/μL reverse transcriptase, 1.5 μL of 10× reverse transcriptase buffer, 0.19 μL of 20 U/μL RNase inhibitor, and adjusted the total reaction volume to 15 μL with nuclease free water. The cDNA synthesis reaction was performed as follows: 16 °C for 30 min followed by 42 °C for 30 min, and 85 °C for 5 min.
MiRNA analysis using RT-qPCR
MiRNA expression was quantified by determining the cycle threshold (CT) which is the number of PCR cycles required for the fluorescence to exceed a value significantly higher than the background fluorescence, using the TaqMan small RNA assay (Applied Biosystems by Life Technologies) with miRNA specific primers according to manufacturer’s instructions. Briefly, 1.4 μL of cDNA was added to 10 μL of probe qPCR mix and 7.6 μL of nuclease free water. The following TaqMan small RNA assay (Applied Biosystems) primers were used: hsa-miR-9-5p, hsa-miR-21-5p, hsa-miR-155-5p, and RNU6B. All analyzed miRNAs are of human (Homo sapiens) origin and therefore, the prefix “hsa” is omitted throughout the text. RT-qPCR reactions were performed using a CFX96 Real-Time PCR System Detector (Bio-Rad, Hercules, CA, USA). Samples were run in duplicate for each experiment. Data were analyzed using the comparative Ct (2-ΔΔCT) method using the small nuclear RNA, RNU6B, as an endogenous control. To monitor reagent contamination, negative controls were included for each primer pair. PCR cycling conditions were as follows: 95 °C for 3 min 40 cycles of 95 °C for 15 s and 60 °C for 60 s.
HPV E6/E7 mRNA analysis using RT-qPCR
To detect HPV E6/E7 mRNA in FFPE cervical tissues, multiplex RT-qPCR was performed using the TaqMan assay with the OPTIMYGENE HPV E6/E7 mRNA RT-qDx assay kit (Optipharm, Osong, Republic of Korea). PCR primers and the corresponding TaqMan probes were designed for three different sets of HPV regions, with each set of probes targeting their conserved sequence (FAM: HPV genotypes 16, 31, 33, 35, 52, and 58; CY5: HPV genotypes 18, 39, 45, 51, 59, and 68; and HEX: HPV genotypes 53, 56, 66, and 69).
RT-qPCR reactions consisted of 10 μL of 2 × Thunderbird probe qPCR mix (Toyobo, Osaka, Japan), 5 μL of primers and TaqMan probe mixture, 2 μL of template cDNA, and distilled water for a final reaction volume of 20 μL. The multiplex RT-qPCR assay detected the HPV E6 and E7 genes simultaneously in a single tube by incorporating two targets (E6 and E7) using specific TaqMan probes, which were labeled with different fluorophores (FAM, HEX, and Cy5). Positive and negative controls were included throughout the procedure. PCR cycling conditions were as follows: 95 °C for 3 min 45 cycles of 95 °C for 20 s and 60 °C for 40 s. To avoid false negatives because of mRNA degradation, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an endogenous control.
Statistical analysis
Statistical analysis was performed using GraphPad Prism software version 5.02 (GraphPad, La Jolla, CA, USA) and MedCalc 9.0 software (MedCalc Software Inc., Mariakerke, Belgium). Student’s t-test and Mann Whitney U test were used to determine statistical significance between cervical cancer and normal cervical tissue samples as well as investigate miRNA expression in patients according to HPV infection status. Receiver operating characteristic (ROC) curves were generated to assess diagnostic accuracy of each miRNA, and the area under the ROC curve (AUC) was calculated to measure discriminatory capacity. The best sensitivity/specificity pair was selected based on the maximum likelihood ratio. Univariate and multivariate logistic regression by odds ratio (OR) and 95% confidential interval (95% CI) were performed to assess predictors for cervical cancer diagnosis using the XLSTAT software (Addinsoft, New York, USA). All statistical tests were two-sided, and a P value ≤0.05 was considered statistically significant.
Results
HPV E6/E7 mRNA expression in cervical cancer tissues
Prior to investigating miRNA expression levels, we first examined HPV E6/E7 mRNA expression in 52 FFPE cervical cancer tissue samples and 50 FFPE normal control samples. Fifteen (28.8%) of the 52 FFPE cervical cancer tissue samples were negative for HR-HPV E6/E7 expression (termed HR-HPV E6/E7-negative), while 37 (71.2%) samples were positive (termed HR-HPV E6/E7-positive). We found that all 50 FFPE normal cervical control samples were negative for HPV E6/E7 mRNA expression (Table 1).
MiRNA expression levels in cervical cancer and normal tissues
Expression levels of miR-9, miR-21, and miR-155 were investigated in our 52 FFPE cervical cancer tissue samples and 50 FFPE normal cervical tissue controls. All three miRNAs were significantly up regulated in FFPE cervical cancer tissues compared to FFPE normal cervical tissues (P < 0.0001) (Fig. 1a-c). The AUC was 0.7565 [95% confidence interval (CI) = 0.6624–0.8507] in miR-9, 0.8325 (95% CI = 0.7530–0.9120) in miR-21, and 0.8492 (95% CI = 0.7736–0.9249) in miR-155, all of which indicate these miRNAs may be used as potential biomarkers for cervical cancer (Fig. 1d-f).
Fig. 1.
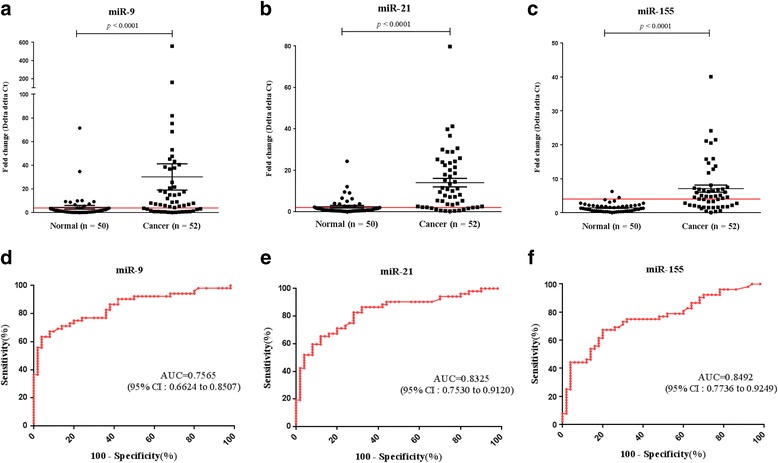
MiR-9, miR-21, and miR-155 expression levels in formalin-fixed paraffin-embedded (FFPE) cervical cancer and normal tissue samples. a MiR-9, b miR-21, and c miR-155 expression levels in 52 FFPE cervical cancer tissue samples were significantly different compared to that found in 50 FFPE normal cervical tissue samples (P < 0.0001 for all three comparisons). Receiver operating characteristic (ROC) curve analysis showed that d miR-9 had an area under the ROC curve (AUC) value of 0.7565 [95% confidence interval (CI) = 0.6624–0.8507], while e miR-21, and f miR-155 had AUC values of 0.8325 (95% CI = 0.7530–0.9120) and 0.8492 (95% CI = 0.7736–0.9249), respectively
Diagnostic value of miR-9, miR-21, and miR-155 miRNAs
To assess the potential diagnostic value of these three miRNAs, the performance characteristics sensitivity, specificity, positive predictive value, and negative predictive value were determined and evaluated. The cut-off values of miR-9, miR-21, and miR-155 as determined using the likelihood ratio, were 4.035, 1.975, and 3.880 respectively, for optimal sensitivity and specificity. The sensitivity of miR-9, miR-21, and miR-155 was 67.3% (95% CI = 52.9–79.7), 82.7% (95% CI = 69.7–91.8), and 65.4% (95% CI = 50.9–78.0) respectively, while the specificity was 80.0% (95% CI = 66.3–90.0) for miR-9, 72.0% (95% CI = 57.5–83.8) for miR-21, and 96.0% (95% CI = 86.3–99.5) for miR-155. Positive predictive values (PPVs) of miR-9, miR-21, and miR-155 were 77.8%, 75.4%, and 94.3% respectively, while the respective negative predictive values (NPVs) were 70.2%, 80.0%, and 71.6% (Table 2).
Table 2.
Clinical cut-off values, sensitivity, and specificity of miRNAs
| Cut-off value | Sensitivity, % (95% CI) | Specificity, % (95% CI) | PPV, % (95% CI) | NPV, % (95% CI) | Likelihood ratio | |
|---|---|---|---|---|---|---|
| miR-9 | >4.035 | 67.3 (52.9–79.7) | 80.0 (66.3–90.0) | 77.8 (62.9–88.8) | 70.2 (56.6–81.6) | 3.4 |
| miR-21 | >1.975 | 82.7 (69.7–91.8) | 72.0 (57.5–83.8) | 75.4 (62.2–85.9) | 80.0 (65.4–90.4) | 3.0 |
| miR-155 | >3.880 | 65.4 (50.9–78.0) | 96.0 (86.3–99.5) | 94.3 (80.8–90.6) | 71.6 (59.3–82.0) | 15.9 |
95% CI 95% confidence interval, PPV positive predictive value, NPV negative predictive value
MiR-9, miR-21, and miR-155 in HPV E6/E7-positive and -negative cervical cancer
To investigate the expression of miR-9, miR-21, and miR-155 with cervical cancer cases that were HPV E6/E7 mRNA-positive or -negative for HPV E6/E7 mRNA, expression levels of the three miRNAs were analyzed in three groups: HPV E6/E7-positive cancer samples, HPV E6/E7-negative cancer samples, and normal samples. We found that all three miRNAs were significantly up regulated in HR-HPV E6/E7-positive cancer tissue samples compared to normal tissue samples (P < 0.0001), while miR-21 and miR-155 were up regulated in HPV E6/E7-negative cancer tissue samples compared to normal controls (P = 0.0079 and P = 0.0384, respectively). We found no significant difference in miR-9 expression levels between HR-HPV E6/E7-negative cancer samples and normal cervical samples (Fig. 2).
Fig. 2.
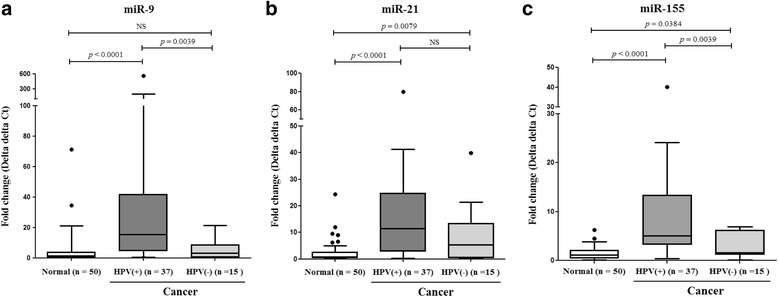
Box and whisker plots of comparisons between three miRNA expression levels in normal cervical tissues and cancer tissues with (+) or without (−) HPV E6/E7 mRNA expression. a MiR-9, b miR-21, and c miR-155 expression levels in high risk human papillomavirus (HR-HPV) E6/E7-positive cervical cancer tissues were significantly up regulated compared to that found in normal cervical tissue samples (P < 0.0001 for all three comparisons). MiR-21 and miR-155 expression levels in HR-HPV E6/E7-negative cervical cancer tissues were significantly up regulated compared to that found in normal cervical tissue samples (P = 0.0079 and P = 0.00384, respectively). NS, not significant
Diagnostic value of miR-9, miR-21, and miR-155 miRNAs in conjunction with the HPV E6/E7 mRNA assay
The effectiveness of each three miRNA combined with HPV E6/E7 mRNA assay was investigated by analysis of AUC. In all cases of cervical cancer, the value of AUC was 0.8558 (95% CI 0.7773–0.9343), 0.8135 (95% CI 0.7257–0.9013), 0.8215 (95% CI 0.7349–0.9082), 0.8935 (95% CI 0.8243–0.9626) in HPV E6/E7, HPV E6/E7 + miR-9, HPV E6/E7 + miR-21, and HPV E6/E7 + miR-155, respectively. Especially, in HPV negative cervical cancer, miR-21 and miR-155 in conjunction with the HPV E6/E7 mRNA was shown increased AUC value of 0.7267 (95% CI 0.5776–0.8757) and 0.7000 (95% CI 0.5152–0.8646) respectively, compared to HPV E6/E7 assay (Table 3).
Table 3.
Diagnostic values of miRNAs in conjunction with HPV E6/E7 for cervical cancer
| AUCa | 95% CI | P-value | |
|---|---|---|---|
| All cases | |||
| HPV E6/E7 | 0.8558 | 0.7773–0.9343 | <0.0001 |
| HPV E6/E7 + miR-9 | 0.8135 | 0.7257–0.9013 | <0.0001 |
| HPV E6/E7 + miR-21 | 0.8215 | 0.7349–0.9082 | <0.0001 |
| HPV E6/E7 + miR-155 | 0.8935 | 0.8243–0.9626 | <0.0001 |
| HPV negative cases | |||
| HPV E6/E7 | 0.5000 | 0.3321–0.6679 | 1.00 |
| HPV E6/E7 + miR-9 | 0.6000 | 0.4284–0.7716 | 0.24 |
| HPV E6/E7 + miR-21 | 0.7267 | 0.5776–0.8757 | 0.01 |
| HPV E6/E7 + miR-155 | 0.7000 | 0.5152–0.8648 | 0.04 |
a AUC an area under the ROC curve
MiRNA predictors for diagnosing cervical cancer in HPV E6/E7-negative cases
We analyzed the following risk predictors of cervical cancer: age, HPV E6/E7 mRNA-expressing status, and miR-9, miR-21, and miR-155 expression. We found that the highest risk factor was HPV E6/E7 mRNA expression (OR = 244.4, 95% CI = 13.6–4376.4) followed by expression of miR-155 (OR = 41.7, 95% CI = 9.1–191.2), miR-21 (OR = 12.3, 95% CI = 4.8–31.7), and miR-9 (OR = 8.2, 95% CI = 3.3–20.3). Ages below 60 years were not a significant risk factor but being over the age of 60 years did confer a higher risk to cervical cancer (Table 4).
Table 4.
The diagnostic utility of predictors for cervical cancer (univariate analysis)
| OR | 95% CI | P value | |
|---|---|---|---|
| Ages | |||
| < 30 years | 1 | ||
| 31–40 years | 1.4 | 0.4–5.1 | 0.580 |
| 41–50 years | 1.7 | 0.4–6.5 | 0.442 |
| 51–60 years | 3.5 | 0.8–16.4 | 0.109 |
| > 60 years | 35.2 | 3.6–344.2 | 0.002 |
| HPV E6/E7 | |||
| negative | 1 | ||
| positive | 244.4 | 13.6–4376.4 | <0.0001 |
| miR-9 | |||
| negative | 1 | ||
| positive | 8.2 | 3.3–20.3 | <0.0001 |
| miR-21 | |||
| negative | 1.0 | ||
| positive | 12.3 | 4.8–31.7 | <0.0001 |
| miR-155 | |||
| negative | 1.0 | ||
| positive | 41.7 | 9.1–191.2 | <0.0001 |
OR Odds ratio, 95% CI 95% confidential interval
To investigate the predictive power for diagnosing cervical cancer in HPV E6/E7 mRNA negative cases, the odds ratio of the miRNAs was determined based on HPV E6/E7 mRNA expression using multivariate analysis. We found that miR-21 and miR-155 in HPV E6/E7-negative cervical cancer patients conferred a 7.0 (95% CI = 1.3–37.6) and 10.3 (95% CI = 1.5–70.7) fold risk to cervical cancer, respectively (Table 5).
Table 5.
MiRNA predictors of cervical cancer according to HPV E6/E7 mRNA expression status in patients (multivariate analysis)
| OR | 95% CI | P value | |
|---|---|---|---|
| HPV positive | |||
| miR-9 | 3.3 | 0.6–18.7 | 0.173 |
| miR-21 | 1.8 | 0.3–11.1 | 0.515 |
| miR-155 | 27.9 | 5.0–155.7 | <0.0001 |
| HPV negative | |||
| miR-9 | 0.4 | 0.1–2.6 | 0.341 |
| miR-21 | 7.0 | 1.3–37.6 | 0.024 |
| miR-155 | 10.3 | 1.5–70.7 | 0.017 |
OR Odds ratio, 95% CI 95% confidential interval
Discussion
HPV DNA detection for cervical cancer screening is widely used as an early diagnostic guideline to prevent the progression of cervical cancer. Nevertheless, there is an ongoing need for studies investigating the molecular mechanisms related to cervical cancer carcinogenesis because most high-risk HPV infections that present without any symptoms go away within one to two years, and there have been reports of several HPV negative cases of cervical cancer [7–9, 25]. In our previous study, we found that 15 out of 100 FFPE cervical cancer tissue samples were HPV negative using an E6/E7 mRNA assay as well as testing for the HPV L1 genotype (data not shown).
Several oncogenic miRNAs are associated with cervical cancer tumorigenesis [17–23]. However, the results from those studies were not comprehensively evaluated using clinical specimens, and those studies have not tested for an association between miRNA expression levels and HR-HPV E6/E7 mRNA expression in clinical specimens. The aim of this study was to explore the potential clinical relevance of miR-9, miR-21, and miR-155 by investigating their expression levels in 52 FFPE cervical cancer tissue samples and in 50 FFPE normal cervical tissue samples. Evidence of association between these three miRNAs and HR-HPV E6/E7 mRNA expression was also investigated.
All three miRNAs (miR-9, miR-21, and miR-155) showed significantly higher expression in cervical cancer tissues compared to that found in normal cervical tissues (P < 0.0001) (Fig. 1). This finding supports the possibility that these three miRNAs may be implicated in cervical cancer development in clinical samples. Although four previous studies using cervical cancer cell lines and clinical samples found that miR-21, miR-155, and miR-9 were up regulated in cervical cancer, they did not validate these three miRNAs in terms of diagnostic value [13, 18, 26, 27]. This study is the first to assess these miRNAs as putative biomarkers and their possible discriminatory capacity in FFPE tissues.
Differences in expression levels of the three miRNAs between HR-HPV E6/E7-positive cervical cancer tissue samples and HR-HPV E6/E7–negative cervical cancer tissue samples showed the strongest association was between miR-9 expression and HR-HPV E6/E7-positive cancer cases compared to that found with the other miRNAs (Fig. 2). Similarly, Weijun Liu et al. found miR-9 and HPV E6 caused increased cell motility by down regulating follistatin like 1 (FSTL1) and activated leukocyte cell adhesion molecule (ALCAM) mRNAs, both of which are involved in cell migration [28].
Both miR-21 and miR-155 had reported other mechanism related to immune response as well as HR-HPV E6/E7 expression. Bumrungthai et al. found that miR-21 is correlated with increased expression of α-smooth muscle actin (α-SMA) and interleukin 6 (IL-6) and decreased expression of PDCD4 in cell proliferation and initiates inflammation-associated carcinogenesis via nuclear factor kappa-light chain-enhancer of activated B cells (NF-kB) and interleukin-6 (IL-6) signaling pathways in colon and cervical cancer cells and Asangani et al. found miR-21 down-regulates PDCD4 in colon cancer and functions as stimulating invasion, intravasation, and metastasis [21, 22, 29]. For miR-155, Guoying Lao et al. found that miR-155 regulates LKB1 expression, which functions as embryonic polarity, metabolism, and cell growth and up-regulation of miR155 promotes proliferation of cervical cancer cells [23].
In terms of diagnostic value, miR-21, and miR-155 expression in combination with the HPV E6/E7 mRNA assay may be useful in the diagnosis of cervical cancer independent of HPV infection because these miR-21 and miR-155 may have the discriminatory power to detect HPV negative cervical cancer cases (Table 3). In particular, miR-21 was independent of HPV status being consistently up regulated in cervical cancer (Fig. 2). We analyzed the odds ratios to assess the effects of the predictors age, HPV E6/E7 mRNA expression, and expression of miR-9, miR-21, and miR-155 on cervical cancer compared to normal controls. While we found that all of the predictors were significantly associated with cervical cancer, miR-21 and miR-155 expression were identified as predictors for high risk in HPV negative cervical cancer tissues compared to normal cervical tissues (Tables 4 and 5).
Some studies have investigated miRNAs for association with HPV infection as potential diagnostic and prognostic indicators. Xiaohong Wang et al. revealed that miR-92a and miR-378 expression was associated with cancer progression in HPV positive tissue samples [30]. Similarly, we found that miR-155 overexpression is associated with increased risk of cervical cancer in HPV E6/E7 mRNA positive tissues. Moreover, miR-21 and miR-155 overexpression in HPV E6/E7 mRNA negative tissue samples could complement approaches for cervical cancer diagnosis and prediction of progression.
Conclusions
Our findings showed that miRNA RT-qPCR assays for specific miRs may be useful tools in the diagnosis of cervical cancer and especially, HPV negative cases of cervical cancer. In addition, these findings are important towards determining the possible role of miRNA expression in cervical cancer development, and the relationship between miRNA and HPV infection. Further study is needed in pre-cancer lesions to understand the role of miRNAs in tumor carcinogenesis, and more tests using normal and cervical cancer samples will be necessary to clearly demonstrate the potential utility of these assays for cervical cancer screening and diagnosis.
Acknowledgments
Funding
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and future Planning (2015R1A2A2A04004455).
Availability of data and materials
All relevant materials are described in the manuscript. Additional data sets supporting the conclusions of this article are available at request from the corresponding author.
Abbreviations
- 95% CI
95% confidential interval
- ALCAM
Activated leukocyte cell adhesion molecule
- AUC
Area under the ROC curve
- FFPE
Formalin-fixed paraffin-embedded
- FSTL1
Follistatin like 1
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- HR-HPV
High risk human papillomavirus
- IL-6
Interleukin-6; LKB1: Liver kinase B1
- miRNA
microRNA
- NPV
Negative predictive value
- OR
Odds ratio
- PPV
Positive predictive value
- pRb
Retinoblastoma protein
- ROC
Receiver operating characteristic
- α-SMA
α-smooth muscle actin
Authors’ contributions
SP, KE, and JK participated in the design of the study and performed the statistical analysis and manuscript. KE and HJ and HW carried out the data extraction. HY and KP and HB conceived of the study, and participated in its design and coordination and helped to draft the manscripts. GK, DL, and SK helped to assemble the clinical data and performed the initial data interpretation together with HB and organize the experiment and sample collection. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Institutional Ethics Committee at Yonsei University Wonju College of Medicine approved the study protocol (approval no. YWMR-12-4-010) and all subjects provided written informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kwang Hwa Park, Phone: +82-33-741-1556, Email: abba@yonsei.ac.kr.
Hyeyoung Lee, Phone: +82-33-760-2740, Email: hyelee@yonsei.ac.kr.
References
- 1.McGuire S. World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press. 2015. Adv Nutr. 2016;7:418–419. doi: 10.3945/an.116.012211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crosbie EJ, Kitchener HC. Human papillomavirus in cervical screening and vaccination. Clin Sci (Lond) 2006;110:543–552. doi: 10.1042/CS20050230. [DOI] [PubMed] [Google Scholar]
- 3.Munger K, Werness BA, Dyson N, Phelps WC, Harlow E, Howley PM. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 1989;8:4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koivusalo R, Mialon A, Pitkanen H, Westermarck J, Hietanen S. Activation of p53 in cervical cancer cells by human papillomavirus E6 RNA interference is transient, but can be sustained by inhibiting endogenous nuclear export-dependent p53 antagonists. Cancer Res. 2006;66:11817–11824. doi: 10.1158/0008-5472.CAN-06-2185. [DOI] [PubMed] [Google Scholar]
- 5.Liu X, Clements A, Zhao K, Marmorstein R. Structure of the human Papillomavirus E7 oncoprotein and its mechanism for inactivation of the retinoblastoma tumor suppressor. J Biol Chem. 2006;281:578–586. doi: 10.1074/jbc.M508455200. [DOI] [PubMed] [Google Scholar]
- 6.Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 7.Giorgi Rossi P, Sideri M, Carozzi FM, Vocaturo A, Buonaguro FM, Tornesello ML, Burroni E, Mariani L, Boveri S, Zaffina LM, et al. HPV type distribution in invasive cervical cancers in Italy: pooled analysis of three large studies. Infect Agent Cancer. 2012;7:26. doi: 10.1186/1750-9378-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tjalma WA, Fiander A, Reich O, Powell N, Nowakowski AM, Kirschner B, Koiss R, O’Leary J, Joura EA, Rosenlund M, et al. Differences in human papillomavirus type distribution in high-grade cervical intraepithelial neoplasia and invasive cervical cancer in Europe. Int J Cancer. 2013;132:854–867. doi: 10.1002/ijc.27713. [DOI] [PubMed] [Google Scholar]
- 9.de Sanjose S, Quint WG, Alemany L, Geraets DT, Klaustermeier JE, Lloveras B, Tous S, Felix A, Bravo LE, Shin HR, et al. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol. 2010;11:1048–1056. doi: 10.1016/S1470-2045(10)70230-8. [DOI] [PubMed] [Google Scholar]
- 10.Wang HY, Kim G, Cho H, Kim S, Lee D, Park S, Park KH, Lee H. Diagnostic performance of HPV E6/E7, hTERT, and Ki67 mRNA RT-qPCR assays on formalin-fixed paraffin-embedded cervical tissue specimens from women with cervical cancer. Exp Mol Pathol. 2015;98:510–516. doi: 10.1016/j.yexmp.2015.03.036. [DOI] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 12.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 13.Guo J, Miao Y, Xiao B, Huan R, Jiang Z, Meng D, Wang Y. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol. 2009;24:652–657. doi: 10.1111/j.1440-1746.2008.05666.x. [DOI] [PubMed] [Google Scholar]
- 14.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 15.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 16.Michael MZ, O’Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 17.Lui WO, Pourmand N, Patterson BK, Fire A. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 2007;67:6031–6043. doi: 10.1158/0008-5472.CAN-06-0561. [DOI] [PubMed] [Google Scholar]
- 18.Lee JW, Choi CH, Choi JJ, Park YA, Kim SJ, Hwang SY, Kim WY, Kim TJ, Lee JH, Kim BG, Bae DS. Altered MicroRNA expression in cervical carcinomas. Clin Cancer Res. 2008;14:2535–2542. doi: 10.1158/1078-0432.CCR-07-1231. [DOI] [PubMed] [Google Scholar]
- 19.Gocze K, Gombos K, Juhasz K, Kovacs K, Kajtar B, Benczik M, Gocze P, Patczai B, Arany I, Ember I. Unique microRNA expression profiles in cervical cancer. Anticancer Res. 2013;33:2561–2567. [PubMed] [Google Scholar]
- 20.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bumrungthai S, Ekalaksananan T, Evans MF, Chopjitt P, Tangsiriwatthana T, Patarapadungkit N, Kleebkaow P, Luanratanakorn S, Kongyingyoes B, Worawichawong S, Pientong C. Up-regulation of miR-21 is associated with cervicitis and human papillomavirus infection in cervical tissues. PLoS One. 2015;10:5. [DOI] [PMC free article] [PubMed]
- 22.Asangani I, Rasheed S, Nikolova D, Leupold J, Colburn N, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 23.Lao G, Liu P, Wu Q, Zhang W, Liu Y, Yang L, Ma C. Mir-155 promotes cervical cancer cell proliferation through suppression of its target gene LKB1. Tumour Biol. 2014;35:11933–11938. doi: 10.1007/s13277-014-2479-7. [DOI] [PubMed] [Google Scholar]
- 24.Turner M, Vigorito E. Regulation of B- and T-cell differentiation by a single microRNA. Biochem Soc Trans. 2008;36:531–533. doi: 10.1042/BST0360531. [DOI] [PubMed] [Google Scholar]
- 25.Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2007;370:890–907. doi: 10.1016/S0140-6736(07)61416-0. [DOI] [PubMed] [Google Scholar]
- 26.Wang X, Tang S, Le SY, Lu R, Rader JS, Meyers C, Zheng ZM. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS One. 2008;3:e2557. doi: 10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pereira PM, Marques JP, Soares AR, Carreto L, Santos MA. MicroRNA expression variability in human cervical tissues. PLoS One. 2010;5:e11780. doi: 10.1371/journal.pone.0011780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu W, Gao G, Hu X, Wang Y, Schwarz JK, Chen JJ, Grigsby PW, Wang X. Activation of miR-9 by human papillomavirus in cervical cancer. Oncotarget. 2014;5:11620–11630. doi: 10.18632/oncotarget.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deftereos G, Corrie SR, Feng Q, Morihara J, Stern J, Hawes SE, Kiviat NB. Expression of mir-21 and mir-143 in cervical specimens ranging from histologically normal through to invasive cervical cancer. PLoS One. 2011;6:e28423. doi: 10.1371/journal.pone.0028423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Wang HK, Li Y, Hafner M, Banerjee NS, Tang S, Briskin D, Meyers C, Chow LT, Xie X, et al. microRNAs are biomarkers of oncogenic human papillomavirus infections. Proc Natl Acad Sci U S A. 2014;111:4262–4267. doi: 10.1073/pnas.1401430111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant materials are described in the manuscript. Additional data sets supporting the conclusions of this article are available at request from the corresponding author.


