Abstract
Hepatitis C virus (HCV) envelope proteins are essential not only for maintaining the viral life cycle, but also for evading the host's immune response and in clinical intervention. A thorough understanding of HCV envelope proteins depends on the availability of detailed structural information. Two crystal structures of the E2 core portion and of the E2 ectodomain, and one structure of the N-terminus of E1 ectodomain have shed new light on the complexity of HCV envelope proteins. In addition, the full-length E1-E2 complex has recently been modeled. The present review focuses on these advancements, introduces the recently solved structures and their biological implications and proposes novel ideas for studying the full-length E1-E2 complex.
Keywords: hepatitis C virus, envelope proteins, glycoprotein, structure
1. Introduction
Hepatitis C virus (HCV), a member of the Hepacivirus genus within the Flaviviridae family, is an enveloped virus consisting of positive-sense single-stranded RNA with approximately 9,600 nucleotides (1). HCV is a notable pathogen that primarily infects human liver, leading to chronic liver disease; this may include liver steatosis, fibrosis, cirrhosis and hepatocellular carcinoma (2). Approximately 3 to 4 million individuals are newly infected with HCV each year, whereas 170 million are chronically infected and the mortality rate from HCV-related causes is 350,000 each year (3). The early standard therapy (before 2011) was limited to the combination of PEG-interferon and ribavirin, which was not effective in treating HCV genotype 1 (4), in addition to causing side effects, such as pruritus and thrombocytopenia (5). When the structures of the viral protease (6) and RNA polymerase (7) were clearly resolved, the development of small molecule drugs against the vital proteins involved in HCV replication was accelerated; several specific antivirals (including direct acting antivirals, telaprevir, boceprevir and sofosbuvir) have now been approved and have significantly improved the antiviral efficacy against HCV, highlighting the critical contribution of resolved viral protein structures to HCV therapy (8–11). It is expected that a complete understanding of the structures of E1 and E2 glycoproteins will further contribute to glycoprotein-targeted drug development, in addition to therapeutic and preventive vaccines.
The HCV RNA genome encodes a single polyprotein that is 3,000 amino acids (aa) long. This precursor polyprotein is subsequently processed by cellular and viral proteases into 10 proteins; this includes four structural proteins, E1, E2, p7 and Core, and six nonstructural proteins (NS2, NS3, NS4A, NS4B, NS5A and NS5B) (12–15).
E1 and E2 (aa192-383 and aa384-746, respectively), which assemble to cover the core of the virus particle, consist of a highly glycosylated N-terminal ectodomain and a short C-terminal transmembrane domain (16). The N-terminal ectodomain is further divided into two parts: A receptor-binding domain and a C-terminal membrane-proximal stem region. (17). The short C-terminal transmembrane domain contains retention signals for the endoplasmic reticulum (18) and anchors each glycoprotein in the cytoplasmic membrane (19,20). E1 and E2 from the JFH-1 HCV strain isolate contain 8 and 18 cysteine residues in their ectodomains, respectively (21,22), which form disulfide bonds that stabilize the large covalent envelope proteins complex on the surface of the virion (23,24). However, the complex is initially intracellularly assembled as a noncovalent heterodimer (23,24) (Fig. 1).
Figure 1.

Schematic representation of hepatitis C virus polyprotein, highlighting E1 and E2 structure organization. Cysteines, glycans and residues involved in CD81 binding are indicated. The construct design of Protein Data Bank IDs 4UOI, 4MWF and 4NX3 are presented. TMD, transmembrane domain; ter, terminus.
The individual functions of these glycoproteins have not been fully elucidated; it is understood that E2 consists of conserved sequences for receptors binding and of multiple epitopes for inducing protective immune response (25–27). Less is known about the function of E1 due to the challenge in purifying E1 protein in its native form.
HCV entry into cells is associated with several host-generated molecules including low density lipoprotein receptor, glycosaminoglycans, high density lipoprotein receptor scavenger receptor class B type I, tetraspanin CD81, and two tight junction proteins, claudin-1 and occludin (28). The HCV life cycle is illustrated in Fig. 2. It remains unclear which cellular proteins mediate the fusion step in the host cell entry process. Knowledge of E1 and E2 structures is required to fully understand membrane fusion.
Figure 2.
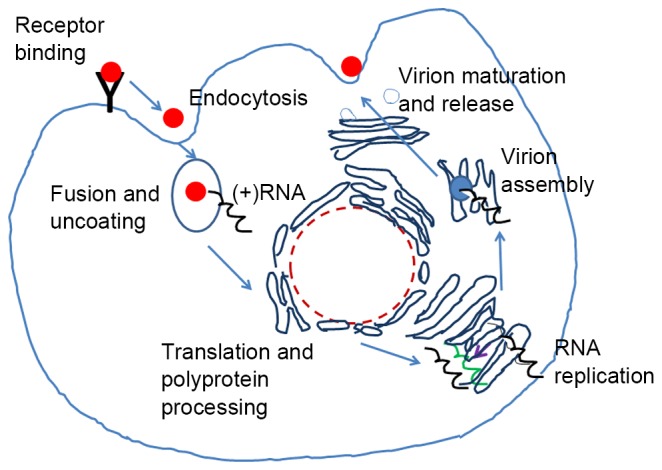
Brief overview of hepatitis C virus life cycle in hepatocytes. The virus entry involves several receptors at the cell surface. The HCV genome is released following fusion with the endosomal membrane. The genome is then translated directly into a single polyprotein, which is processed into 10 individual mature proteins, including glycoproteins E1 and E2. RNA replication and assembly take place on the intracellular membrane. The viral particles, budding from intracellular membrane and travelling through the Golgi apparatus, are secreted by the classical secretory pathway. HCV, hepatitis C virus.
Significant progress in understanding E1 and E2 structures has recently been made: Two crystal structures of the core portion (E2c) of E2 ectodomain (E2e) and one structure of the N-terminal of E1 ectodomain have been successfully established through single-crystal X-ray diffraction methods (29–31). Furthermore, a full-length E1-E2 complex has also been modeled in silico (16). In the current review, the recent structural progress in E1 and E2 proteins is summarized, and possible explanations for the inconsistent previous findings are proposed.
2. The unexpected 3D structures of E1 and E2
Unexpected tertiary structures of E2 glycoprotein
Despite previous challenges, the structure of E2c from E2e (Protein Data Bank IDs: 4MWF and 4NX3; rcsb.org/pdb/home/home.do) has recently been solved by two antibody studies, via single-crystal X-ray diffraction (30,31) (Figs. 3 and 4). E2 protein was truncated and modified in both studies. In 4MWF (aa412-645) (31), the engineered E2c construct expressing genotype 1a E2 (H77 isolate) not only deleted the two termini and two glycosylation sites (N448 and N576), but also replaced HRV2 (aa460-485) with a Gly-Ser-Ser-Gly linker. In 4NX3 (aa456-656) (30), the construct that expressed genotype 2a (J6 isolate) deleted 80 residues at the N-terminus, which contains the implicated cellular receptor binding sites and neutralizing epitopes. The antibodies used by two studies were also different. 4MWF employed an antibody specific to antigenic region 3 (AR3) of E2, which blocked CD81 receptor binding (31). A different antibody (2A12) recognizing a linear epitope at the C-terminus of the ectodomain was utilized in 4NX3, which does not prevent E2 from binding to receptors (30).
Figure 3.

Comparison of the structures of hepatitis C virus E2c from Protein Data Bank ID 4NX3 and 4MWF constructs. Glycans are reported as yellow regions and CD81 binding sites are indicated as red spheres.
Figure 4.
Cartoon presentation of the N-terminal of hepatitis C virus E1 ectodomain from Protein Data Bank ID 4UOI. (A) The N- and C termini of the construct. (B) The six monomers of the construct are indicated in six colors; disulfide bonds are represented with spheres.
As expected, the two structures demonstrated overall similarity; E2c was a monomer containing a central immunoglobin-like domain, which was formed by β-sheets surrounded by short α-helices dispersed in loops. However, regions within E2 that contained no regular secondary structure were not uncommon, and the majority of the preserved N-linked glycans were in a flexible pattern (30,31). The absent glycosylation in 4NX3 accounted for roughly a third of the expressed protein mass (30).
Like other viruses belonging to the Flaviviridae family, HCV contains class II fusion protein, which exhibits an extended architecture (32). Unexpectedly, two studies have revealed that the E2c architecture, similarly to E1, was compact (30,31). These studies also demonstrated the structural interaction between E2 and CD81, which may facilitate the development of therapeutic and preventive drugs or vaccines by interrupting receptor binding. In 4MWF (31), several key residues for CD81 binding were identified in the C-terminal lobe that is adjacent to the front layer, which has been known to contain key CD81 binding residues. Surprisingly, CD81 bound to the same exposed hydrophobic surface as AR3C and other broad neutralizing antibodies. As the surface of HCV particles is covered with E2 glycoproteins primarily consisting of relatively conserved residues, and is free of N-linked glycans, this represents an ideal choice for immunogen design. However, it may also contain a few non-conserved residues that provide a basis for immune evasion. In the 4NX3 model (30), the epitope including the N7 site, which binds the AR3C antibody and is competitively inhibitory of CD81 binding, lies at the interface of the basic and hydrophobic planes. Notably, CD81 binding improved following deletion of the N6 site, which is only 7 residues away from N7, and which represents the AR3C binding site; this possibly occurs via increased reliability of CD81 binding.
Unexpected crystallographic structure of the N-terminal of E1 ectodomain (nE1)
Structural scientists face enormous challenges while studying the E1 structure as the traditional methods, including soaking with heavy atoms and SeMet phasing (as E1 contains no methionines), are not suitable for deriving the structure of HCV E1 (33). There was no significant progress until recently, when an unexpected fold of nE1 (aa1-79) of HCV was successfully revealed by X-ray crystallography (29) (Figs. 1 and 4). The elucidated structure highlighted three unexpected findings. Firstly, that nE1 was arranged as a covalent homodimer with domains potentially swapped, as it is possible that the β-hairpin may fold back in monomeric E1 to replace the dimer interface. Overall, it consisted of a β-hairpin (residues 1–14) followed by an α-helix (residues 17–32), which was between a two and three-stranded antiparallel β-sheet. The β-hairpin was associated with the domain swap and homodimer formation. Unexpectedly, it showed that E1 was not a fusion protein, which is discussed in detail in a subsequent section.
Secondly, nE1 contained 6 monomers that were stabilized by disulfide bonds. These disulfide bonds are biologically important as the presence of disulfide bonds may improve lateral interactions between E1 and E2 on the surface and facilitate the budding of HCV particles (34).
In addition, another study indicated that these glycoproteins harbor the reduced cysteines that form couplings following virus-host interactions (35). In this respect, the covalent nE1 dimer may be relevant to the post-attachment conformation, for example if disulfide bonds are present, but these reduced cysteines have not been identified.
Lastly, the nE1 dimer is homologous to phosphatidylcholine transfer protein (number of matches, 74). Structurally, the steroidogenic acute regulatory protein-related transfer domain of this protein possesses partial similarity to the extended β-sheet of nE1Notably, this transfer domain is responsible for binding sterols and lipid-like hydrophobic molecules, just as N-terminal domain of E1 is hypothesized to be responsible for binding to apolipoproteins.
Putative structure of E1-E2 complex
E1 and E2 form a heterodimer on the surface of the HCV particle and together they facilitate viral entry (19,36). Most previous studies that analyzed either the structure of E1 or E2 were required to separate the appropriate glycoprotein from the heterodimer, which may impact the conformation of both glycoproteins (30,31,37–39). However, the amino acids located between E1 and E2 are partially inserted into the membrane, resulting in great difficulty in acquiring a single full-length crystal of either glycoprotein heterodimer that is free of cellular elements (16).
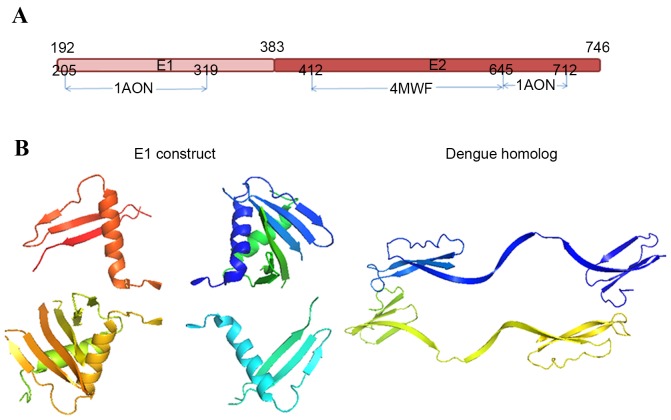
In order to attenuate the difficulty in obtaining the full-length envelope protein (E1 plus E2), a 3D structure of E1-E2 complex monomer ([E1.E2]) of HCV genotype 1 has recently been predicted (16). Dengue virus, another member of the Flaviviridae family, shares many structural similarities with HCV (40). The model of [E1.E2] monomer was assembled using the solved E2c (4MWF) (31), loops that were missed in the E2c (generated by MODELLER; salilab.org/modeller) and two fragments of E1 (residues 205–319) and E2 (residues 646–716), and using the dengue crystal structure 1OAN as a template (Fig. 5A). Most glycosylation sites in E1 and E2 are preserved in this model.
Figure 5.
Model of the HCV E1-E2 complex. (A) Organization of the HCV E1-E2 model. The model is made of two homologous fragments from dengue protein Protein Data Bank ID 1OAN, and one reported crystal structure of E2c from Protein Data Bank ID 4MWF. (B) Comparison of E1 construct (aa205-271) from Protein Data Bank ID 4UOI with dengue homolog fragment from Protein Data Bank ID 1AON. HCV, hepatitis C virus.
In the model of E1 and E2 monomer, 35% of the residues formed extended/β-strands, while <6% were helical (16). Although the percentage for extended/β-strand was 1% higher than in the structure predicted by the GOR4 program (41), it was predominantly consistent with the data of the solved structure.
The novel contribution made by this model was that it offers an overview of the appearance of both HCV envelope proteins, and additionally a prediction of several exposed residues and epitopes as priority targets for subsequent study. Unexpectedly, the present model suggests that E2 is not only a central component in fulfilling a fundamental function in the HCV life cycle, but also helps to maintain correct E1 folding. The model predicts that the N-terminus of E1, with a length of 115 residues, forms a portion of domains I and II.
However, this prediction may not be correct as the homology model of dengue did not resemble the crystallized short peptide of HCV E1 (Fig. 5B). This inconsistency may be caused by variation in crystallization procedures. A method by which this model could be verified would be to express the overlapping epitopes within the N-terminus of E1, which could then be used to compete for the antibody binding sites with the corresponding epitopes in E1-E2.
3. Modifications may impact protein structure
A native fold of 4MWF E2c was verified by binding anti-E2 monoclonal antibodies (mAbs) and the CD81 receptor, and by inhibiting HCV cell entry through receptor competition (31). Several previous studies that discussed the two recently solved structures indicated that regions of the structure were in conflict with previous reports of crystallized fragments, and contradicted each other in many respects (42–44). That the two E2c constructs used by Kong et al (31) and Khan et al (30) were expressed in the absence of E1, and had to be modified before crystallization, may be relevant to this. It is known that a small modification may affect the resultant conformation and aggregation, particularly the cysteine binding (16). Evidence for this concern is apparent. All cysteines form disulfide bonds when E2e was expressed as a soluble, secreted protein (45). However, the data could not be confirmed with other functional studies that explored E1-E2 assembly, interaction with CD81 receptors and viral infectivity subsequent to replacement of cysteine residues (45–47). In addition, Fraser et al (35) expressed E2 on the virion surface and used alkylation experiments to demonstrate that E2 comprised at least one free cysteine, and that this was necessary in E2 rearrangement following attachment. These data imply that the soluble and truncated E2 was not in a native form of folding, and that the resultant cysteine coupling was altered compared with the native folding (48).
Taken together, the recently solved E2 core structures had limitations and possible biases. Furthermore, a large, highly disordered portion of the structure remains to be elucidated, and nearly half of the glycoprotein is not included in the present structural analysis. Novel techniques are therefore required to refine and extend these structures (42–44).
Overall, the obstacles to obtain the correct E2e structure point to the form-dependent flexible cysteine coupling (for instance, soluble or truncated), the variable glycosylation state and the necessary E1 involvement in maintaining the correct conformation of E2e. In fact, the disulfide bonds and glycosylation sites are crucial in maintaining the correct protein folding, but the challenge is to understand how their flexibility results in such a high degree of E2 structural variability (42,49–51); this hampers the homogenous crystal arrangement and influences the reconstruction of the E2e density maps in the measurement of E2 structure by SAXS and cryoelectron microscopy (cryoEM), leading to low-resolution maps (30,31).
4. Novel techniques for better characterizing E2 tertiary structure under native conditions
Experimental modifications preceding use of X-ray crystallography may impact the native structure of E2 and markedly alter the protein function, leading to artificial structural data (52,53). This necessitates developing novel techniques to solve these current problems.
Alanine-scanning
One such novel technique is alanine-scanning. This may be performed on the full-length protein expressed under a near-native environment, and even in the context of whole viral particles, reducing potential biases derived from non-native conformations (42). The principle of alanine scanning is to generate X (residue in the target protein) to alanine variants, and subsequently compare the reactivity pattern of mAbs recognizing the conformational epitopes of mutants and wild-type protein. Therefore, this method may be used to infer the native structure using the antibody binding profile and may be used to map epitopes, in addition to identifying key residues that trigger changes to correct protein conformation (42). For example, cysteine or histidine single mutations within E2 may be introduced to study their effects on viral entry and assembly (45,47,54). Furthermore, a recent study using alanine scanning achieved the native E2 structure by expressing full-length E1 and E2 in mammalian cells. This study suggested a novel cysteine coupling between C486-C589 that was not included in the truncated E2c, but was critical for correct E2 folding (42).
Computation modeling
Another possible technique is computation modeling. Over last 10 years, a wide range of computational methods and algorithms have been developed to model the 3D structure of proteins (55–58). Among these, a co-evolution-based algorithm, based on the principle of coordinated changes that occur in pairs within organisms or biomolecules, appears to be an alternative approach for deducing the structure of E2, as this glycoprotein varies considerably in sequence and as the majority of sequences are available. Significant advances in disentangling the indirect and direct co-evolution signals have been made, which may be used to more accurately predict the contact regions between protein residues, or to identify residue pairs in close proximity. A de novo remodeling of the entire 3D protein has also been performed, implying that unknown regions of E2e can be rebuilt de novo and in silico (55–58). Therefore, in this distance-based algorithm, the structural data from all direct and indirect studies and the predicted unknown regions may be pooled to remodel the structure of the entire E2e. The accuracy of this structural prediction will be structurally assessed by the secondary structures, fitting them into the E2e maps revealed by cryoEM and SAXS and comparing the experimentally determined structures (using nuclear magnetic resonance or X-ray diffraction). It is of note that the prediction will be more valid if mapped to functional and immunogenic regions that may then be validated by mAbs (42,59). However, it is also key to recognize that many technical limitations of the structural prediction in silico remain unsolved for further details see (59).
5. Proposals to ascertain fusion and other steps of viral entry by analyzing the E1-E2 complex
An increase of correct E2 structural data is likely to contribute to a better understanding of protein function. E2 shares similarities to envelope proteins within Flavividae family, and was previously suggested to be a fusion protein (31,32,60). Lavillette et al (60) and Zhu et al (61) proposed that E1 and E2 coordinate in the fusion process, as the two glycoproteins both harbor a putative fusion peptide. This hypothesis was supported by E1-E2 mutagenesis of both proteins together and by functional fusion assays (62–66). In the identification of additional E2 protein structures, it appears unlikely that E2 is independently responsible for completion of the fusion step. The E2 tertiary structure from bovine viral diarrhea virus (another member of the Flaviviridae family) does not support a role of this glycoprotein as a fusion peptide (61). Furthermore, the crystal structure of E2c displayed a unique compact globular structure, which was markedly different from the extended class II fusion protein (16). However, E1 was previously implicated in HCV receptor attachment and membrane fusion via mutation of cysteine (34). Vieyres et al (44) confirmed E1 as a fusion protein, by integrating previously published data. However, a recent report on E1 N-terminal architecture, suggested that nE1 was not comparable to class II fusion fold (31). In fact, neither glycoprotein was alleged to resemble any of the three classes of fusion proteins, suggesting that HCV may adopt a novel fusion mechanism that differs from that of other Flaviviridae family members (30,31,67). As the HCV E1-E2 heterodimer is indispensable for viral fusion, expression of E1-E2 as a complex protein may help in the task of displaying the potential fusion domain, in addition to the possible alternate steps of viral entry.
6. Conclusions
Recently published E1 and E2 structures represent breakthroughs in the structural analysis of E1 and E2 glycoproteins. For the first time, the crystal structures of a large portion of the ectodomain of these glycoproteins were revealed by X-ray diffraction. This novel structural information both helps to better understand the fusion mechanism and the conformational epitopes, and to facilitate novel vaccine design and therapeutic drug development. However, the structures of both proteins have questionable reliability as they were prepared under non-native conditions. To compensate for such deficiencies, the [E1.E2] model was re-established in silico with the help of the currently available structural data of both glycoproteins, and the homology data of dengue viral protein. With increasingly available structural and functional data, it is predicted that a refined 3D model for the entire E1-E2 complex is likely to be generated soon. Such a high quality model may then be used for predicting antigenic epitopes and drug targets.
Acknowledgements
Meijiedeng, an English Language Service, helped in preparation of the current manuscript. The present work was supported by the National Natural Science Foundation of China (grant no. 81201291), the National Science and Technology Major Project (grant no. 2012ZX10002003), the Science and Technology Major Projects of Zhejiang Province (grant no. 2009C03011-2) and Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry (grant no. J20140017).
References
- 1.Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 2.Bartenschlager R, Penin F, Lohmann V, André P. Assembly of infectious hepatitis C virus particles. Trends Microbiol. 2011;19:95–103. doi: 10.1016/j.tim.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 3.Perz JF, Armstrong GL, Farrington LA, Hutin YJ, Bell BP. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol. 2006;45:529–538. doi: 10.1016/j.jhep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 4.Asselah T, Marcellin P. Interferon free therapy with direct acting antivirals for HCV. Liver Int. 2013;33:S93–S104. doi: 10.1111/liv.12076. (Suppl 1) [DOI] [PubMed] [Google Scholar]
- 5.Conteduca V, Sansonno D, Russi S, Pavone F, Dammacco F. Therapy of chronic hepatitis C virus infection in the era of direct-acting and host-targeting antiviral agents. J Infect. 2014;68:1–20. doi: 10.1016/j.jinf.2013.08.019. [DOI] [PubMed] [Google Scholar]
- 6.Lorenz IC, Marcotrigiano J, Dentzer TG, Rice CM. Structure of the catalytic domain of the hepatitis C virus NS2-3 protease. Nature. 2006;442:831–835. doi: 10.1038/nature04975. [DOI] [PubMed] [Google Scholar]
- 7.Appleby TC, Perry JK, Murakami E, Barauskas O, Feng J, Cho A, Fox D, III, Wetmore DR, McGrath ME, Ray AS, et al. Viral replication. Structural basis for RNA replication by the hepatitis C virus polymerase. Science. 2015;347:771–775. doi: 10.1126/science.1259210. [DOI] [PubMed] [Google Scholar]
- 8.Wyles DL. Antiviral resistance and the future landscape of hepatitis C virus infection therapy. J Infect Dis. 2013;207:S33–S39. doi: 10.1093/infdis/jis761. (Suppl 1) [DOI] [PubMed] [Google Scholar]
- 9.deLemos AS, Chung RT. Hepatitis C treatment: An incipient therapeutic revolution. Trends Mol Med. 2014;20:315–321. doi: 10.1016/j.molmed.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Rose L, Bias TE, Mathias CB, Trooskin SB, Fong JJ. Sofosbuvir: A Nucleotide NS5B Inhibitor for the Treatment of Chronic Hepatitis C Infection. Ann Pharmacother. 2014;48:1019–1029. doi: 10.1177/1060028014534194. [DOI] [PubMed] [Google Scholar]
- 11.Scheel TK, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med. 2013;19:837–849. doi: 10.1038/nm.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xue J, Zhu H, Chen Z. Therapeutic vaccines against hepatitis C virus. Infect Genet Evol. 2014;22:120–129. doi: 10.1016/j.meegid.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 13.Reed KE, Rice CM. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr Top Microbiol Immunol. 2000;242:55–84. doi: 10.1007/978-3-642-59605-6_4. [DOI] [PubMed] [Google Scholar]
- 14.Grakoui A, Wychowski C, Lin C, Feinstone SM, Rice CM. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993;67:1385–1395. doi: 10.1128/jvi.67.3.1385-1395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang H, Grise H. Cellular and molecular biology of HCV infection and hepatitis. Clin Sci (Lond) 2009;117:49–65. doi: 10.1042/CS20080631. [DOI] [PubMed] [Google Scholar]
- 16.Nayak A, Pattabiraman N, Fadra N, Goldman R, Kosakovsky Pond SL, Mazumder R. Structure-function analysis of hepatitis C virus envelope glycoproteins E1 and E2. J Biomol Struct Dyn. 2015;33:1682–1694. doi: 10.1080/07391102.2014.967300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helle F, Goffard A, Morel V, Duverlie G, McKeating J, Keck ZY, Foung S, Penin F, Dubuisson J, Voisset C. The neutralizing activity of anti-hepatitis c virus antibodies is modulated by specific glycans on the E2 envelope protein. J Virol. 2007;81:8101–8111. doi: 10.1128/JVI.00127-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cocquerel L, Duvet S, Meunier JC, Pillez A, Cacan R, Wychowski C, Dubuisson J. The transmembrane domain of hepatitis C virus glycoprotein E1 is a signal for static retention in the endoplasmic reticulum. J Virol. 1999;73:2641–2649. doi: 10.1128/jvi.73.4.2641-2649.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goffard A, Callens N, Bartosch B, Wychowski C, Cosset FL, Montpellier C, Dubuisson J. Role of N-linked glycans in the functions of hepatitis C virus envelope glycoproteins. J Virol. 2005;79:8400–8409. doi: 10.1128/JVI.79.13.8400-8409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartosch B, Dubuisson J, Cosset FL. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J Exp Med. 2003;197:633–642. doi: 10.1084/jem.20021756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cocquerel L, Wychowski C, Minner F, Penin F, Dubuisson J. Charged residues in the transmembrane domains of hepatitis C virus glycoproteins play a major role in the processing, subcellular localization, and assembly of these envelope proteins. J Virol. 2000;74:3623–3633. doi: 10.1128/JVI.74.8.3623-3633.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kato T, Furusaka A, Miyamoto M, Date T, Yasui K, Hiramoto J, Nagayama K, Tanaka T, Wakita T. Sequence analysis of hepatitis C virus isolated from a fulminant hepatitis patient. J Med Virol. 2001;64:334–339. doi: 10.1002/jmv.1055. [DOI] [PubMed] [Google Scholar]
- 23.Vieyres G, Thomas X, Descamps V, Duverlie G, Patel AH, Dubuisson J. Characterization of the envelope glycoproteins associated with infectious hepatitis C virus. J Virol. 2010;84:10159–10168. doi: 10.1128/JVI.01180-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenberg S. Recent advances in the molecular biology of hepatitis C virus. J Mol Biol. 2001;313:451–464. doi: 10.1006/jmbi.2001.5055. [DOI] [PubMed] [Google Scholar]
- 25.Keck ZY, Xia J, Wang Y, Wang W, Krey T, Prentoe J, Carlsen T, Li AY, Patel AH, Lemon SM, et al. Human monoclonal antibodies to a novel cluster of conformational epitopes on HCV E2 with resistance to neutralization escape in a genotype 2a isolate. PLoS Pathog. 2012;8:e1002653. doi: 10.1371/journal.ppat.1002653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wahid A, Dubuisson J. Virus-neutralizing antibodies to hepatitis C virus. J Viral Hepat. 2013;20:369–376. doi: 10.1111/jvh.12094. [DOI] [PubMed] [Google Scholar]
- 27.Kong L, Giang E, Robbins JB, Stanfield RL, Burton DR, Wilson IA, Law M. Structural basis of hepatitis C virus neutralization by broadly neutralizing antibody HCV1. Proc Natl Acad Sci USA. 2012;109:9499–9504. doi: 10.1073/pnas.1202924109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ploss A, Evans MJ. Hepatitis C virus host cell entry. Curr Opin Virol. 2012;2:14–19. doi: 10.1016/j.coviro.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.El Omari K, Iourin O, Kadlec J, Sutton G, Harlos K, Grimes JM, Stuart DI. Unexpected structure for the N-terminal domain of hepatitis C virus envelope glycoprotein E1. Nat Commun. 2014;5:4874. doi: 10.1038/ncomms5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khan AG, Whidby J, Miller MT, Scarborough H, Zatorski AV, Cygan A, Price AA, Yost SA, Bohannon CD, Jacob J, et al. Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature. 2014;509:381–384. doi: 10.1038/nature13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kong L, Giang E, Nieusma T, Kadam RU, Cogburn KE, Hua Y, Dai X, Stanfield RL, Burton DR, Ward AB, et al. Hepatitis C virus E2 envelope glycoprotein core structure. Science. 2013;342:1090–1094. doi: 10.1126/science.1243876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krey T, d'Alayer J, Kikuti CM, Saulnier A, Damier-Piolle L, Petitpas I, Johansson DX, Tawar RG, Baron B, Robert B, et al. The disulfide bonds in glycoprotein E2 of hepatitis C virus reveal the tertiary organization of the molecule. PLoS Pathog. 2010;6:e1000762. doi: 10.1371/journal.ppat.1000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.El Omari K, Iourin O, Kadlec J, Fearn R, Hall DR, Harlos K, Grimes JM, Stuart DI. Pushing the limits of sulfur SAD phasing: De novo structure solution of the N-terminal domain of the ectodomain of HCV E1. Acta Crystallogr D Biol Crystallogr. 2014;70:2197–2203. doi: 10.1107/S139900471401339X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wahid A, Helle F, Descamps V, Duverlie G, Penin F, Dubuisson J. Disulfide bonds in hepatitis C virus glycoprotein E1 control the assembly and entry functions of E2 glycoprotein. J Virol. 2013;87:1605–1617. doi: 10.1128/JVI.02659-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fraser J, Boo I, Poumbourios P, Drummer HE. Hepatitis C virus (HCV) envelope glycoproteins E1 and E2 contain reduced cysteine residues essential for virus entry. J Biol Chem. 2011;286:31984–31992. doi: 10.1074/jbc.M111.269605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lavie M, Goffard A, Dubuisson J. HCV Glycoproteins: Assembly of a Functional E1-E2 Heterodimer. In: Tan SL, editor. Source Hepatitis C Viruses: Genomes and Molecular Biology. Norfolk (UK): Horizon Bioscience; 2006. Chapter 4. [PubMed] [Google Scholar]
- 37.Albecka A, Montserret R, Krey T, Tarr AW, Diesis E, Ball JK, Descamps V, Duverlie G, Rey F, Penin F, Dubuisson J. Identification of new functional regions in hepatitis C virus envelope glycoprotein E2. J Virol. 2011;85:1777–1792. doi: 10.1128/JVI.02170-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng L, Zhong L, Struble E, Duan H, Ma L, Harman C, Yan H, Virata-Theimer ML, Zhao Z, Feinstone S, et al. Structural evidence for a bifurcated mode of action in the antibody-mediated neutralization of hepatitis C virus. Proc Natl Acad Sci USA. 2013;110:7418–7422. doi: 10.1073/pnas.1305306110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kong L, Giang E, Nieusma T, Robbins JB, Deller MC, Stanfield RL, Wilson IA, Law M. Structure of hepatitis C virus envelope glycoprotein E2 antigenic site 412 to 423 in complex with antibody AP33. J Virol. 2012;86:13085–13088. doi: 10.1128/JVI.01939-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuiken C, Hraber P, Thurmond J, Yusim K. The hepatitis C sequence database in Los Alamos. Nucleic Acids Res. 2008;36:D512–D516. doi: 10.1093/nar/gkm962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gibrat JF, Garnier J, Robson B. Further developments of protein secondary structure prediction using information theory. New parameters and consideration of residue pairs. J Mol Biol. 1987;198:425–443. doi: 10.1016/0022-2836(87)90292-0. [DOI] [PubMed] [Google Scholar]
- 42.Castelli M, Clementi N, Sautto GA, Pfaff J, Kahle KM, Barnes T, Doranz BJ, Dal Peraro M, Clementi M, Burioni R, Mancini N. HCV E2 core structures and mAbs: Something is still missing. Drug Discov Today. 2014;19:1964–1970. doi: 10.1016/j.drudis.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sabahi A, Uprichard SL, Wimley WC, Dash S, Garry RF. Unexpected structural features of the hepatitis C virus envelope protein 2 ectodomain. J Virol. 2014;88:10280–10288. doi: 10.1128/JVI.00874-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vieyres G, Dubuisson J, Pietschmann T. Incorporation of hepatitis C virus E1 and E2 glycoproteins: The keystones on a peculiar virion. Viruses. 2014;6:1149–1187. doi: 10.3390/v6031149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang W, Guan M, Liu Y, Xu Q, Peng H, Liu X, Tang Z, Zhu Y, Wu D, Ren H, et al. Alanine scanning mutagenesis of hepatitis C virus E2 cysteine residues: Insights into E2 biogenesis and antigenicity. Virology. 2014;448:229–237. doi: 10.1016/j.virol.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 46.McCaffrey K, Boo I, Poumbourios P, Drummer HE. Expression and characterization of a minimal hepatitis C virus glycoprotein E2 core domain that retains CD81 binding. J Virol. 2007;81:9584–9590. doi: 10.1128/JVI.02782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCaffrey K, Boo I, Tewierek K, Edmunds ML, Poumbourios P, Drummer HE. Role of conserved cysteine residues in hepatitis C virus glycoprotein e2 folding and function. J Virol. 2012;86:3961–3974. doi: 10.1128/JVI.05396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Owsianka A, Clayton RF, Loomis-Price LD, McKeating JA, Patel AH. Functional analysis of hepatitis C virus E2 glycoproteins and virus-like particles reveals structural dissimilarities between different forms of E2. J Gen Virol. 2001;82:1877–1883. doi: 10.1099/0022-1317-82-8-1877. [DOI] [PubMed] [Google Scholar]
- 49.Brazzoli M, Helenius A, Foung SK, Houghton M, Abrignani S, Merola M. Folding and dimerization of hepatitis C virus E1 and E2 glycoproteins in stably transfected CHO cells. Virology. 2005;332:438–453. doi: 10.1016/j.virol.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 50.Bräutigam J, Scheidig AJ, Egge-Jacobsen W. Mass spectrometric analysis of hepatitis C viral envelope protein E2 reveals extended microheterogeneity of mucin-type O-linked glycosylation. Glycobiology. 2013;23:453–474. doi: 10.1093/glycob/cws171. [DOI] [PubMed] [Google Scholar]
- 51.Iacob RE, Perdivara I, Przybylski M, Tomer KB. Mass spectrometric characterization of glycosylation of hepatitis C virus E2 envelope glycoprotein reveals extended microheterogeneity of N-glycans. J Am Soc Mass Spectrom. 2008;19:428–444. doi: 10.1016/j.jasms.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quick M, Shi L, Zehnpfennig B, Weinstein H, Javitch JA. Experimental conditions can obscure the second high-affinity site in LeuT. Nat Struct Mol Biol. 2012;19:207–211. doi: 10.1038/nsmb.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rashin AA, Domagalski MJ, Zimmermann MT, Minor W, Chruszcz M, Jernigan RL. Factors correlating with significant differences between X-ray structures of myoglobin. Acta Crystallogr D Biol Crystallogr. 2014;70:481–491. doi: 10.1107/S1399004713028812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clementi N, Mancini N, Criscuolo E, Cappelletti F, Clementi M, Burioni R. Epitope mapping by epitope excision, hydrogen/deuterium exchange, and peptide-panning techniques combined with in silico analysis. Methods Mol Biol. 2014;1131:427–446. doi: 10.1007/978-1-62703-992-5_26. [DOI] [PubMed] [Google Scholar]
- 55.de Juan D, Pazos F, Valencia A. Emerging methods in protein co-evolution. Nat Rev Genet. 2013;14:249–261. doi: 10.1038/nrg3414. [DOI] [PubMed] [Google Scholar]
- 56.Baldassi C, Zamparo M, Feinauer C, Procaccini A, Zecchina R, Weigt M, Pagnani A. Fast and accurate multivariate Gaussian modeling of protein families: Predicting residue contacts and protein-interaction partners. PLoS One. 2014;9:e92721. doi: 10.1371/journal.pone.0092721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones DT, Buchan DW, Cozzetto D, Pontil M. PSICOV: Precise structural contact prediction using sparse inverse covariance estimation on large multiple sequence alignments. Bioinformatics. 2012;28:184–190. doi: 10.1093/bioinformatics/btr638. [DOI] [PubMed] [Google Scholar]
- 58.Marks DS, Colwell LJ, Sheridan R, Hopf TA, Pagnani A, Zecchina R, Sander C. Protein 3D structure computed from evolutionary sequence variation. PLoS One. 2011;6:e28766. doi: 10.1371/journal.pone.0028766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dorn M, Silva EMB, Buriol LS, Lamb LC. Three-dimensional protein structure prediction: Methods and computational strategies. Comput Biol Chem. 2014;53PB:251–276. doi: 10.1016/j.compbiolchem.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 60.Lavillette D, Pécheur EI, Donot P, Fresquet J, Molle J, Corbau R, Dreux M, Penin F, Cosset FL. Characterization of fusion determinants points to the involvement of three discrete regions of both E1 and E2 glycoproteins in the membrane fusion process of hepatitis C virus. J Virol. 2007;81:8752–8765. doi: 10.1128/JVI.02642-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu YZ, Qian XJ, Zhao P, Qi ZT. How hepatitis C virus invades hepatocytes: The mystery of viral entry. World J Gastroenterol. 2014;20:3457–3467. doi: 10.3748/wjg.v20.i13.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Douam F, Dao Thi VL, Maurin G, Fresquet J, Mompelat D, Zeisel MB, Baumert TF, Cosset FL, Lavillette D. Critical interaction between E1 and E2 glycoproteins determines binding and fusion properties of hepatitis C virus during cell entry. Hepatology. 2014;59:776–788. doi: 10.1002/hep.26733. [DOI] [PubMed] [Google Scholar]
- 63.Flint M, Thomas JM, Maidens CM, Shotton C, Levy S, Barclay WS, McKeating JA. Functional analysis of cell surface-expressed hepatitis C virus E2 glycoprotein. J Virol. 1999;73:6782–6790. doi: 10.1128/jvi.73.8.6782-6790.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Drummer HE, Boo I, Poumbourios P. Mutagenesis of a conserved fusion peptide-like motif and membrane-proximal heptad-repeat region of hepatitis C virus glycoprotein E1. J Gen Virol. 2007;88:1144–1148. doi: 10.1099/vir.0.82567-0. [DOI] [PubMed] [Google Scholar]
- 65.Li HF, Huang CH, Ai LS, Chuang CK, Chen SS. Mutagenesis of the fusion peptide-like domain of hepatitis C virus E1 glycoprotein: Involvement in cell fusion and virus entry. J Biomed Sci. 2009;16:89. doi: 10.1186/1423-0127-16-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Russell RS, Kawaguchi K, Meunier JC, Takikawa S, Faulk K, Bukh J, Purcell RH, Emerson SU. Mutational analysis of the hepatitis C virus E1 glycoprotein in retroviral pseudoparticles and cell-culture-derived H77/JFH1 chimeric infectious virus particles. J Viral Hepat. 2009;16:621–632. doi: 10.1111/j.1365-2893.2009.01111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li Y, Modis Y. A novel membrane fusion protein family in Flaviviridae? Trends Microbiol. 2014;22:176–182. doi: 10.1016/j.tim.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]