Abstract
It has been reported that FERM domain containing 7 (FRMD7) may cause X-linked idiopathic congenital nystagmus (ICN). A total of >40 mutations of the FRMD7 gene have been identified, however their pathogenic role remains unclear. In the present study, enhanced green fluorescent protein-tagged wild-type (WT) and mutant (MT) FRMD7 (c. C781>G) were expressed in stably expressing human neuroblastoma SH-SY5Y cells following viral transfection and antibiotic selection. Uniform expression of the FRMD7 fusion proteins was confirmed via fluorescence microscopy and western blotting. The expression profiles of neuron-specific proteins and Rho guanine triphosphatases (GTPases) differed significantly between the wild-type and mutant cell lines. Levels of Mtap2, NF-M, nestin, GAP43 and Rac1 mRNA were significantly increased in MT-FRMD7 cells compared with controls (P<0.01). However, the expression of Rac1 protein did not differ significantly among the two cell lines. Taken together, the results of the current study suggest that MT-FRMD7 influences the expression of neuron-specific genes and Rho GTPases, which may be involved in the pathogenesis of ICN. The FRMD7 stable expression cell line may facilitate future studies investigating the role of this protein in neuronal development.
Keywords: FERM domain containing 7, idiopathic congenital nystagmus, stable cell line, SH-SY5Y, neuron-specific genes
Introduction
It has been demonstrated that mutations in the FERM domain containing protein 7 (FRMD7) gene is one of the major causes of X-linked idiopathic congenital nystagmus (ICN) (1–5). ICN is an oculomotor disorder arising from a primary defect in the regions related to ocular motor control. It primarily presents with involuntary horizontal oscillations of both eyes and its onset usually develops within 6 months of birth (6). There are currently no effective treatments for ICN and patients experience a significant decrease in their quality of life over time. To date, >40 mutations of FRMD7 have been detected worldwide in people from various ethnic backgrounds (1–5,7–9). A previous study by the current authors identified two novel missense mutations (c.781C>G and c.886G>C) and a truncated mutation (c.1003C>T) of human FRMD7 in three X-linked ICN pedigrees (3).
The FRMD7 gene is comprised of 12 exons, encodes a 714-residue polypeptide, and contains the FERM domain (four-point-one, ezrin, radixin, moesin) at its amino terminal. It has been demonstrated that the FRMD7 protein co-localizes with actin in the primary neurites in differentiating mouse neuroblastoma (Neuro-2a) cells and that the overexpression of FRMD7 in Neuro-2a cells promotes neurite outgrowth (10), whereas FRMD7 knockdown causes a reduction in average neurite length (11). Previous studies by the current authors identified an association between FRMD7 and Rho-GDIα that is dependent on both the N- and C-terminals of FRMD7, and the latter is also important in subcellular localization (12,13). Furthermore, it was demonstrated that FRMD7 releases Rac1 from RhoGDIα in vitro activating Rac1 signalling, whereas an FRMD7 mutant failed to activate Rac1 signalling, which may lead to ICN (12). Watkins et al (13) identified an interaction between the FRMD7 protein and calcium/calmodulin-dependent serine protein kinase (CASK), which is known to link the plasma membrane and actin cytoskeleton in neurons. Mutations in FRMD7 may therefore disrupt the interaction with CASK that is essential to promote membrane extension during neurite outgrowth (13).
An in vivo study on FRMD7 knockout mice identified specific expression of the protein in starburst amacrine cells, which led to asymmetric inhibition of direction-selective retinal ganglion cells, indicating that FRMD7 is key in establishing neuronal circuit asymmetry (14). However, studies such as this are limited because neurons in mice are different from humans; thus, functional studies in human neuron-like cell lines are potentially more informative. In the present study, human wild-type (WT) and mutant-type (MT, C781>G) FRMD7 genes were cloned and the C-terminal was fused with enhanced green fluorescent protein (eGFP). Transfection of these genes into human neuroblastoma SH-SY5Y cells led to stable expression of WT- and MT-FRMD7-eGFP fusions, as confirmed by fluorescence microscopy and western blotting. The expression profiles of various neuron-specific genes during neuronal development in these cell lines were determined. The aim of the present study was to construct SH-SY5Y cell lines uniformly expressing FRMD7-eGFP fusions and to investigate the effect of the mutation on the expression of genes involved in neuronal development.
Materials and methods
Plasmid construction
The polymerase chain reaction (PCR) products of human WT and MT (c.781C>G) (3) FRMD7 [produced from the cDNA template (FRMD7 cDNA) indicated as previously described (15)] were confirmed by the Sangon company via direct sequencing (Sangon, Shanghai, China). The two FRMD7 cDNAs were fused with Flag-tag (sequence motif DYKDDDDK), digested with XhoI and BamHI and cloned into pLV-EF1a-EGFP-N (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Cell culture and plasmid transfection
The human neuroblastoma (SH-SY5Y) and the human renal epithelial cell lines (293T) were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The 293T cells and SH-SY5Y cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum (Invitrogen; Thermo Fisher Scientific., Inc.), 10% penicillin and streptomycin. Cells were maintained in 5% CO2 at 37°C and were passaged every 24–48 h.
The 293T cell line is a convenient source of recombinant virus. In the present study, 2.5×106 293T cells were seeded in 10 ml growth medium (DMEM supplemented with 10% FBS and 10% pencillin and streptomycin) in a 100-mm tissue culture dish 24 h prior to transfection. The pLV-EF1a-EGFP-FRMD7WT or pLV-EF1a-EGFP-FRMD7MT and two package carriers were used for transfection mediated by the Attractene Transfection kit following standard instructions (Qiagen Inc., Valencia, CA, USA). The virus titers were then detected using a QuickTiter™ Lentivirus Titer kit following the manufacturer's instructions (Cell Biolabs, Inc., San Diego, CA, USA).
Human neuroblastoma SH-SY5Y cells were cultivated in a 6-well plate (1×104 cells/well). When the cell density reached 70–80% confluence, the Lentivirus was used for cell transfection. Puromycin (A1113803, Invitrogen; Thermo Fisher Scientific, Inc.) selection (1 µg/ml) started 24 h after transfection. The medium was then changed with selection drug (1 µg/ml puromycin) every 2–3 days. Following 2–4 weeks, isolated colonies were selected and grown for later assays.
Western blotting and immunofluorescence
For western blot analysis, cultured SH-SY5Y cells were homogenized in radioimmunoprecipitation assay buffer with 1X protease inhibitor cocktail (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and 100 mg/ml phenylmethylsulfonyl fluoride at room temperature. Equal amounts of protein (50 µg) were separated using 8% SDS-PAGE and transferred to a PVDF membrane (Bio-Rad, Hercules, CA, USA). After blocking with 5% skim milk powder at 4°C 1 h, membranes were incubated with the primary mouse anti-flag antibody (F3040; Sigma-Aldrich; Merck KGaA) at a 1:4,000 dilution or mouse anti-Rac1 monoclonal antibody (05–389; EMD Millipore, Billerica, MA, USA) at a 1:1,000 dilution overnight at 4°C.
Then the membrane-bound antibody was incubated with horseradish peroxidase-conjugated secondary antibody at 1:2,000 dilution 2 h at 4°C (ab97040; Abcam, Shanghai, China). The membranes were then processed using an ECL advance western blotting detection kit (Qiagen, Inc.). β-actin (sc-58673; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) at 1:2,000 dilution was used as a loading control. Morphological features of eGFP positive cells were quantified using a confocal laser scanned microscope (Leica TCS SP5 X; Leica, Wetzlar, Germany). All experiments were performed in triplicate.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNAs were isolated from SH-SY5Y cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. RNA (5 µg) was reverse transcribed using oligo dT (SO131; Invitrogen; Thermo Fisher Scientific, Inc.) by reverse transcriptase. For PCR amplification, specific oligonucleotide primer pairs (10 pmol each) were incubated with 2 µl cDNA template in 25 µl PCR reaction mixtures containing 2.5 µl 10x PCR buffer, 1.5 mM MgSO4, mixed deoxynucleotides (1 mM each) and 0.5 U KOD PLUS polymerase (Toyobo Co., Ltd., Osaka, Japan). For amplification of full-length FRMD7, the following PCR conditions were followed: 35 cycles at 94°C for 2 min; 98°C for 15 sec; 58°C for 40 sec; 68°C for 2 min 20 sec; and a final extension step at 68°C for 7 min. Amplified PCR products were analyzed by 1% agarose gel electrophoresis and ethidium bromide staining in UVP (Gel Doc1000, Bio-Rad, USA). FRMD7 mutation c.781C>G was produced using overlap PCR. RT-qPCR was performed to assess the relative expression of neuron-specific genes [microtubule-associated protein 2 (Mtap2), NeuN, neurofilament light (NF-L), neurofilament medium (NF-M), neurofilament heavy (NF-H), Tuj-1, growth-associated protein 43 (GAP-43), microtubule assocaited protein tau (MAPT) and nestin] compared with the reference gene GAPDH. qPCR was performed using a custom-made RT-PCR kit with SYBR green chemistry (Toyobo, Co., Ltd.) according to the manufacturers instructions in an ABI Prism 7700 sequence detector (Applied Biosystems; Thermo Fisher Scientific, Inc.). For the qPCR, the following PCR conditions were followed: 95°C for 5 min, followed by 40 cycles of 95°C for 30 sec, 60°C for 30 min, 72°C for 1 min and a final extension step at 72°C for 10 min. The relative ratio of neuronal specific genes to the GAPDH gene concentrations was calculated using the 2−∆∆Cq method (16). The primer sequences of human neuron-specific genes are presented in Table I.
Table I.
Primer sequences used for reverse transcription-quantitative polymerase chain reaction.
| Gene | Direction | Primer sequence (5′-3′) | Product length (bp) |
|---|---|---|---|
| Mtap2 | Forward | CTGTCACAGTGGAGGAAGATTT | 87 |
| Mtap2 | Reverse | AAGGCTCAGCTGTAGAGGGA | |
| Nestin | Forward | GGCACAATCCGCGGACACCT | 156 |
| Nestin | Reverse | TGGCATCGGCCTGCCACGC | |
| NeuN | Forward | CTACAGCGACAGTTACGGCA | 123 |
| NeuN | Reverse | ATGGTCCGAGAAGGAAACGG | |
| NF-L | Forward | CTGGAAATCGAAGCATGCCG | 106 |
| NF-L | Reverse | TGATCGTGTCCTGCATAGCG | |
| NF-M | Forward | TCTCTACTTTTCCGGCAGTGA | 87 |
| NF-M | Reverse | GATGGTGTCCTGGCCATTCC | |
| NF-H | Forward | GGAGGCACTGAAAAGCACCA | 100 |
| NF-H | Reverse | CTGAATGGCTTCCTGGTAGGAG | |
| MAPT | Forward | GATTGGGTCCCTGGACAATA | 127 |
| MAPT | Reverse | GTGGTCTGTCTTGGCTTTGG | |
| Tuj-1 | Forward | ATCCATTTCTCGACTTTCCAAACT | 127 |
| Tuj-1 | Reverse | TTCCCAGAACTGTGGACGCC | |
| GAP-43 | Forward | TTACCTTGCCTGGGAGGCTT | 124 |
| GAP-43 | Reverse | CAATCTTTTGGTCGTCATCA | |
| GAPDH | Forward | GACAACAGCCTCAAGATCATCAG | 120 |
| GAPDH | Reverse | ATGGCATGGACTGTGGTCATGAG | |
| Rac1 | Forward | ATGCAGGCCATCAAGTGTG | 107 |
| Rac1 | Reverse | ACAGTAGGGATATATTCTC | |
| RhoA | Forward | TTCAGCAAGGACCAGTTCCC | 118 |
| RhoA | Reverse | CCTGCCCAGCTGTGTCCCAC | |
| Cdc42 | Forward | GCAGACAATTAAGTGTGTTG | 138 |
| Cdc42 | Reverse | CCAATCATAACTGTGACTGC |
bp, base pairs; Mtap2, microtubule-associated protein 2; NF-L, neurofilament light; NF-M, neurofilament medium; NF-H, neurofilament heavy; GAP-43, growth-associated protein 43; MAPT, microtubule assocaited protein tau; Cdc-42, cell division cycle 42.
Statistical analysis
Statistical analysis was performed by Student's t-test or Mann-Whitney U test using SPSS 16.0 software (SPSS, Inc., Chicago, IL, USA). All experiments were performed in triplicate as independent experiments and results were expressed as the mean ± standard error of the mean. P<0.05 was considered to indicate a statistically significant difference.
Results
Amplification of human FRMD7 and construction of pLV-EF1a-EGFP-FRMD7
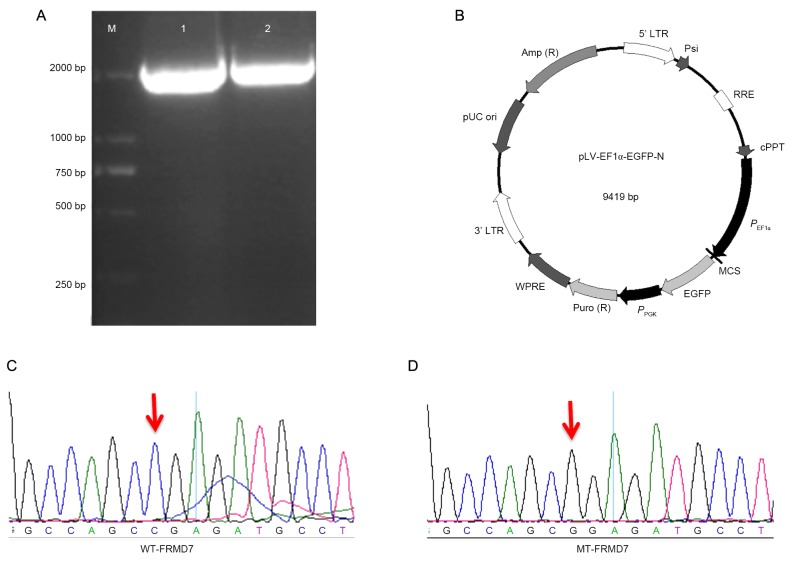
In order to enhance the transfection efficiency of SH-SY5Y cells, lentiviral vectors and a packaging virus were initially constructed. Human WT- and MT-FRMD7 were amplified by PCR, resulting in the expected 2.1 kb bands on an 1% agarose gel (Fig. 1A). Amplification products were digested with XhoI and BamHI and subcloned into the pLV-EF1a-EGFP-N vector (Fig. 1B). In addition, a Flag-tag was fused at the C-terminus of FMRD7 to facilitate expression analysis. Recombination plasmids were purified, sequenced and named pLV-EF1a-EGFP-FRMD7WT or pLV-EF1a-EGFP-FRMD7MT. Sequence alignment with the FRMD7 sequence obtained from GenBank (https://www.ncbi.nlm.nih.gov/genbank/) revealed 100% identity, confirming that genes had been successfully cloned (Fig. 1C and D).
Figure 1.
Construction of human wild type and mutant-type pLV-EF1a-EGFP-FRMD7. (A) 0.8% agarose gel presenting PCR amplification of human WT- (781C>G) and MT-FRMD7 (2.1 kb bands). Lane 1, WT and lane 2, MT. Human WT- and MT-FRMD7 were digested with XhoI and BamHI and (B) subcloned into the pLV-EF1a-EGFP-N vector. pLV-EF1a-EGFP-FRMD7WT and pLV-EF1a-EGFP-FRMD7MT were sequenced and compared with the GenBank FRMD7 sequence and the alignment (C) WT and (D) MT confirmed successful engineering of both constructs. The mutation is indicated by a red arrow. FMRD, FERM domain containing 7. WT, wild-type; MT, mutant type.
Construction of cell lines stably expressing WT- or MT-FRMD7-eGFP fusions
In order to engineer stably expressing SH-SY5Y cell lines, the virus was packaged, the virus titer tested and the cells were transfected and incubated in medium containing 1 mg/ml puromycin for 2–3 weeks. WT and MT clones were then selected and cultured, and it was demonstrated via immunofluorescence that they expressed eGFP (Fig. 2), unlike SH-SY5Y negative controls (cells transfected with empty pLV-EF1a-EGFP-N; Fig. 2). Furthermore, the expression of FRMD7 was determined by western blotting using the Flag-tag antibody (Fig. 3), with β-actin antibody used as an internal control. The results confirmed that the SH-SY5Y cell lines stably expressed WT- or MT-FRMD7-eGFP fusions.
Figure 2.
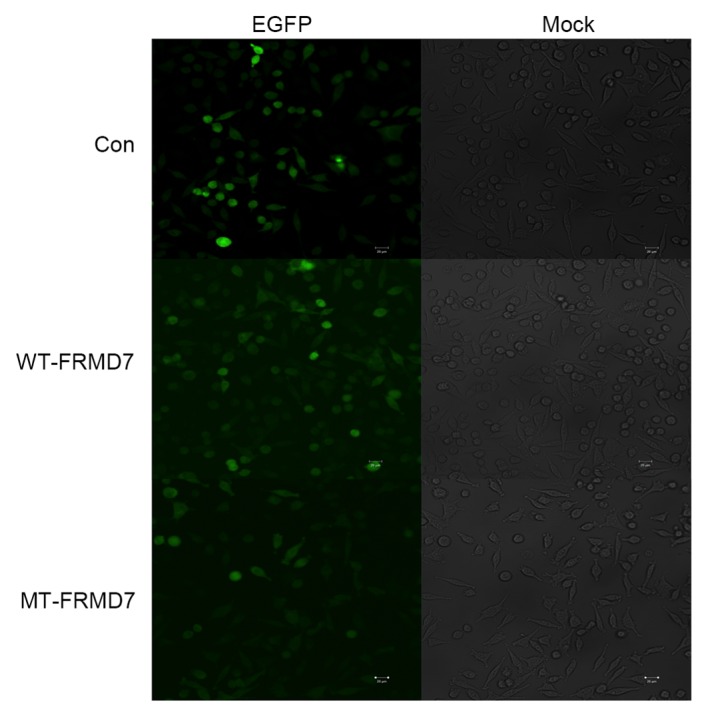
Generation of stable SH-SY5Y cell lines expressing FRMD7-eGFP fusion proteins. Left panels=normal-field fluorescence of SH-SY5Y cells stably expressing eGFP-tagged FRMD7 and bright-field fluorescence of SH-SY5Y cells. Right side=Control cells transfected with empty pLV-EF1a-EGFP-N vector. Scale bar=20 µm. Magnification, ×10. FMRD, FERM domain containing 7; eGFP, enhanced green fluorescent protein; WT, wild-type; MT, mutant type.
Figure 3.
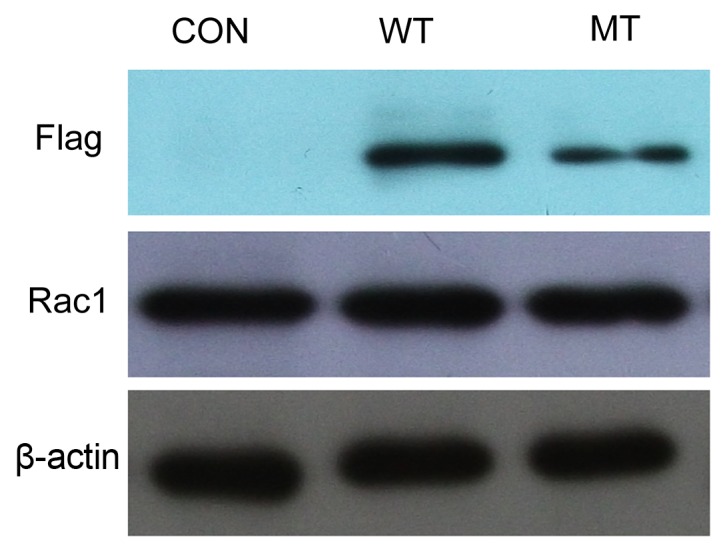
Western blotting of the expression of FRMD7 and Rac1 in stably expressing SH-SY5Y cells. Mouse anti-flag antibody and alkaline phosphatase-conjugated goat anti-mouse immunoglobulin G was used to confirm the stable expression of FRMD7. Rac1 expression was detected using Rac1 antibody and there was no apparent difference in its expression between the WT- or MT-FRMD7 cells and the controls. β-Actin was used as an internal control. Con, control; FMRD, FERM domain containing 7; eGFP, enhanced green fluorescent protein; WT, wild-type; MT, mutant type.
Mutation of FRMD7 affects the expression of neuron-specific genes
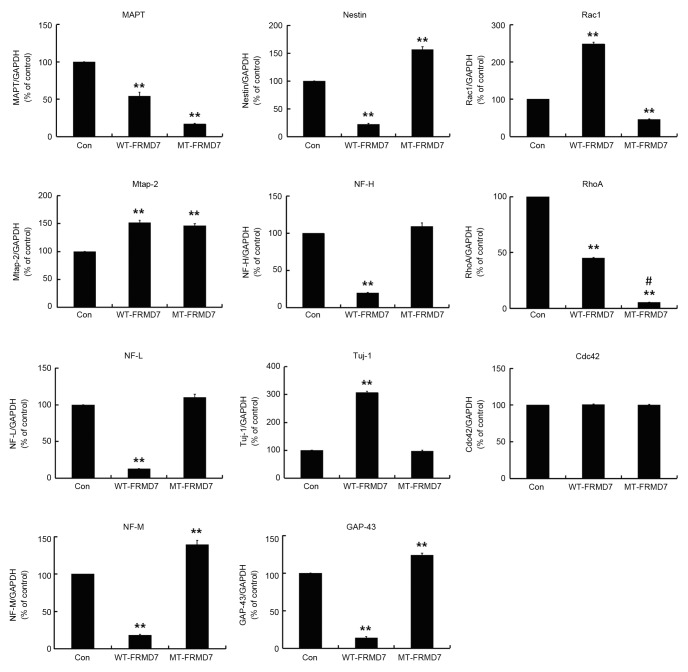
It has been demonstrated that FRMD7 regulates the expression of neuron-specific genes in mouse neuroblastoma Neuro-2a cells, including the cytoskeleton-associated genes Mtap2, NF-L and NF-M and the neuronal differentiation gene NeuN (11). However, it remains unclear whether FRMD7 serves a similar role in human neuronal cells and the effect of disease-related mutations is also unknown. To investigate these points, the mRNA levels of various neuron-specific genes associated with different stages of neuronal development were measured. More specifically, the expression of the cytoskeletal genes Mtap2, NF-L, NF-M, NF-M, Tuj-1 and MAPT, and neuronal differentiation markers GAP-43 and nestin were evaluated in SH-SY5Y cells stably expressing WT- and MT-FRMD7. There were signifcant differences between the two types of cells. In cells expressing WT-FRMD7, levels of Mtap2 and Tuj-1 mRNA were significantly increased compared with controls (P<0.01), whereas mRNA levels of NF-L, NF-M, NF-H, Nestin and GAP43 were significantly decreased (P<0.01). By contrast, expression of Mtap2, NF-M, Nestin and GAP43 were significantly increased in MT-FRMD7 cells compared with controls (P<0.01), whereas the NF-L, NF-H and Tuj-1 mRNA levels were unchanged (Fig. 4).
Figure 4.
Mutations in FRMD7 influence the expression of neuron-specific Rho GTPases in stably expressing SH-SY5Y cells. mRNA levels of Mtap2, NF-L, NF-M, NF-M, Tuj-1, MAPT, GAP-43 and nestin were measured via reverse-transcription quantitative polymerase reaction. In cells expressing WT-FRMD7, Mtap2 and Tuj-1 were significantly upregulated compared with the controls, whereas NF-L, NF-M, NF-H, nestin and GAP43 were downregulated. By contrast, transcription of Mtap2, NF-M, Nestin and GAP43 was clearly increased in cells expressing MT-FRMD7, whereas the expression of MAPT was downregulated. The expression of NF-L, NF-H and Tuj-1 were unaffected. Expression of Rho GTPase Rac1 was increased in WT-FRMD7 cells but decreased in MT-FRMD7 cells. Interestingly, both WT- and MT-FRMD7 evidently decreased the expression of Rho GTPase RhoA, but the effect was bigger with MT-FRMD7. Expression of the other tested Rho GTPases (Cdc-42) was not significantly different in the three cell lines. Cells transfected with empty vector served as a negative control (con group). All data are presented as the fold change in expression relative to the controls. All experiments were performed in triplicate, and the graph presents the average for the three measurements (columns, means; bars, standard error of the mean; **P<0.01 vs. Con; #P<0.01 vs. WT-FRMD7. Con, control; FMRD, FERM domain containing 7; eGFP, enhanced green fluorescent protein; WT, wild type; MT, mutant type; Mtap2, microtubule-associated protein 2; NF-L, neurofilament light; NF-M, neurofilament medium; NF-H, neurofilament heavy; GAP-43, growth-associated protein 43; MAPT, microtubule assocaited protein tau; Cdc-42, cell division cycle 42.
Mutation of FRMD7 affects the expression of Rho GTPases
A previous study by the present authors demonstrated that FRMD7 activates signalling of the Rho subfamily member Rac1, which regulates the reorganization of actin filaments and controls neuronal outgrowth. Mutations of FRMD7 affected Rac1 signalling, which may lead to abnormal neuronal outgrowth and the development of X-linked ICN (12). The present study tested whether mutations in FRMD7 influence the expression of the Rho GTPases Rac1, RhoA and cell division cycle 42 (Cdc42). In accordance with a previous result (11), WT-FRMD7 enhanced the expression of Rac1 compared with the controls, whereas MT-FRMD7 decreased expression of this protein, however these differences were not marked (Fig. 3). Interestingly, both WT- and MT-FRMD7 significantly decreased the mRNA expression of the Rho subfamily member RhoA (P<0.01), with a significantly greater decrease in MT-FRMD7 compared with WT-FRMD7 (P<0.01; Fig. 4). However, the mRNA levels of the other Rho GTPase, cell division cycle 42 (Cdc42), did not differ significantly among the three cell lines (Fig. 4).
Discussion
FRMD7 is a member of the 4.1 protein superfamily and has the characteristic four-point-one, ezrin, radixin, moesin (FERM) domain at its N-terminus (1). Mutations of FRMD7 are the major etiological contributor of ICN (1–5). A previous study by the current authors demonstrated that FRMD7 interacts with RhoGDI, whereby it specifically activates the Rac1 signalling involved in neuronal development. A missense (C781>G) mutation in human FRMD7 that replaces glycine 261 with an arginine (R261>G) reduces its ability to associate with RhoGDIα, which leads to a decreased release of Rac1 from the Rac1-RhoGDIα complex and decreased activation of Rac1 (12).
The development of FRMD7 knockout animals has helped to elucidate its role in neuronal development and represents progress from previous studies that utilized non-human neuronal cell lines (11,14). The development of genetically engineered stably expressing cell lines may also overcome this barrier. In the current study, human neuroblastoma SH-SY5Y cell lines that stably express WT and MT eGFP-tagged FRMD7 were enginerred using a lentiviral approach. A stable cell line was developed following puromycin antibiotic selection. Immunofluorescence measurements and western blotting confirmed the stable expression of FRMD7-eGFP fusions.
The engineered cell lines were used to investigate the expression of neuron-specific genes in the presence of WT- and MT-FRMD7 and revealed significant differences in their expression. Mtap2 and Tuj-1 were upregulated whereas NF-L, NF-M, NF-H, nestin and GAP43 were downregulated in WT-FRMD7 cells, compared with controls (P<0.01). However, in MT-FRMD7 cells, Mtap2, NF-M, nestin and GAP43 were significantly upregulated (P<0.01) compared with controls, whereas MAPT was downregulated (P<0.01), and levels of NF-L, NF-H and Tuj-1 were unaffected. The C781>G mutation present in MT-FRMD7 is known to lead to ICN (3), suggesting that the mutation is linked to pathogenesis. Furthermore, similar changes in expression were observed in a previous study involving FRMD7 overexpression and differentiation in Neuro-2a cells, however these changes were less pronounced (11). This raises the question as to why the results of the present study are not consistent with those of previous reports. It is hypothesized that human neuronal development is more complex than in other species such as mice, with intrinsic differences observed in human neurons. Alternatively, in prevous studies, investigations may have taken place during different developmental stages (15).
Both nestin and GAP43 are known markers of neuronal differentiation, and their expression is usually transient and does not persist into adulthood. Upon completion of differentiation, nestin is downregulated and replaced by tissue-specific intermediate filament proteins (17–20). Similarly, nestin and GAP43 expression is low in embryonic cortical neurons (17–20). It has been demonstrated that overexpression of WT-FRMD7 promotes neurite outgrowth in retinoic acid-mediated differentiation in Neuro-2a cells (11). The fact that nestin and GAP43 were downregulated in cells expressing WT-FRMD7 but upregulated in cells expressing MT-FRMD7 in the present study indicates that mutations in FRMD7 may influence the initiation of neuronal development.
Tuj-1 is a specific neuronal marker that is present in early stage neurons, whereas Mtap2 is a neuronal cytoskeletal protein that is expressed in mature neurons (21,22). These proteins are important in both early and late morphological events (21,23). Neurofilaments (NFs) are comprised of neuron-specific cytoskeletal intermediate filament proteins and undergo marked changes in their subunit compositions in both developing and adult neurons. Additionally, these changes influence both the physiology and morphology of axons (24). Furthermore, NFs determine axonal caliber and promote axonal growth over long distances (25). Although WT-FRMD7 increases the expression of Tuj-1 and Mtap2, NFs were not affected in the engineered SH-SY5Y cells and the expression of MT-FRMD7 only produced a weak influence on the expression on the neuron-specific proteins tested, with the exception of Mtap2. These results indicate that FRMD7 may be important in neuronal development by influencing microtubules and F-actin, but does not affect NFs. However, the exact mechanism behind this requires further investigation.
It was previously demonstrated by the current authors that human FRMD7 promotes the release of Rac1 from RhoGDIα, which activates the Rac1 signalling pathway (12). Therefore, the effect of WT- and MT-FRMD7 on the expression of Rho GTPases in the stable cell lines was investigated. It was observed that levels of Rac1 mRNA were higher in cells expressing WT-FRMD7 and lower in MT-FRMD7 cells, as expected. However, the expression of Rac1 protein did not markedly differ between the WT and MT cell lines, suggesting that FRMD7 activates Rac1 signalling via a mechanism that does not involve increasing Rac1 expression. The influence of FRMD7 on Rac1 and RhoA expression requires further study.
In conclusion, SY5Y neuronal cell lines that stably express WT- and MT-FRMD7 eGFP fusion proteins were successfully established in the current study and the resulting cell lines were used to investigate the role of FRMD7 in neuronal development. The results of the current study support a link between mutations in FRMD7 and the pathogenesis of ICN.
Acknowledgements
The present study was supported by the National Nature Science Foundation of China (grant no. 81400933) and Zhejiang Medical Science and Technology Plan project (grant no. 2016KYB119).
References
- 1.Tarpey P, Thomas S, Sarvananthan N, Mallya U, Lisgo S, Talbot CJ, Roberts EO, Awan M, Surendran M, McLean RJ, et al. Mutations in FRMD7, a newly identified member of the FERM family, cause X-linked idiopathic congenital nystagmus. Nat Genet. 2006;38:1242–1244. doi: 10.1038/ng1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schorderet DF, Tiab L, Gaillard MC, Lorenz B, Klainguti G, Kerrison JB, Traboulsi EI, Munier FL. Novel mutations in FRMD7 in X-linked congenital nystagmus. Mutation in brief #963. Online. Hum Mutat. 2007;28:525. doi: 10.1002/humu.9492. [DOI] [PubMed] [Google Scholar]
- 3.Zhang B, Liu Z, Zhao G, Xie X, Yin X, Hu Z, Xu S, Li Q, Song F, Tian J, et al. Novel mutations of the FRMD7 gene in X-linked congenital motor nystagmus. Mol Vis. 2007;13:1674–1679. [PubMed] [Google Scholar]
- 4.He X, Gu F, Wang Y, Yan J, Zhang M, Huang S, Ma X. A novel mutation in FRMD7 causing X-linked idiopathic congenital nystagmus in a large family. Mol Vis. 2008;14:56–60. [PMC free article] [PubMed] [Google Scholar]
- 5.Fingert JH, Roos B, Eyestone ME, Pham JD, Mellot ML, Stone E. Novel intragenic FRMD7 deletion in a pedigree with congenital X-linked nystagmus. Ophthalmic Genet. 2010;31:77–80. doi: 10.3109/13816810903584989. [DOI] [PubMed] [Google Scholar]
- 6.Lee AG, Brazis PW. Localizing forms of nystagmus: Symptoms, diagnosis, and treatment. Curr Neurol Neurosci Rep. 2006;6:414–420. doi: 10.1007/s11910-996-0022-y. [DOI] [PubMed] [Google Scholar]
- 7.Li N, Wang L, Cui L, Zhang L, Dai S, Li H, Chen X, Zhu L, Hejtmancik JF, Zhao K. Five novel mutations of the FRMD7 gene in Chinese families with X-linked infantile nystagmus. Mol Vis. 2008;14:733–738. [PMC free article] [PubMed] [Google Scholar]
- 8.Choi JH, Shin JH, Seo JH, Jung JH, Choi KD. A start codon mutation of the FRMD7 gene in two Korean families with idiopathic infantile nystagmus. Sci Rep. 2015;5:13003. doi: 10.1038/srep13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta S, Pathak E, Chaudhry VN, Chaudhry P, Mishra R, Chandra A, Mukherjee A, Mutsuddi M. A novel mutation in FRMD7 causes X-linked idiopathic congenital nystagmus in a North Indian family. Neurosci Lett. 2015;597:170–175. doi: 10.1016/j.neulet.2015.04.037. [DOI] [PubMed] [Google Scholar]
- 10.Betts-Henderson J, Bartesaghi S, Crosier M, Lindsay S, Chen HL, Salomoni P, Gottlob I, Nicotera P. The nystagmus-associated FRMD7 gene regulates neuronal outgrowth and development. Hum Mol Genet. 2010;19:342–351. doi: 10.1093/hmg/ddp500. [DOI] [PubMed] [Google Scholar]
- 11.Pu J, Lu X, Zhao G, Yan Y, Tian J, Zhang B. FERM domain containing protein 7 (FRMD7) upregulates the expression of neuronal cytoskeletal proteins and promotes neurite outgrowth in Neuro-2a cells. Mol Vis. 2012;18:1428–1435. [PMC free article] [PubMed] [Google Scholar]
- 12.Pu J, Mao Y, Lei X, Yan Y, Lu X, Tian J, Yin X, Zhao G, Zhang B. FERM domain containing protein7 interacts with the Rho GDP dissociation inhibitor and specifically activates Rac1 signaling. PloS One. 2013;8:e73108. doi: 10.1371/journal.pone.0073108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watkins RJ, Patil R, Goult BT, Thomas MG, Gottlob I, Shackleton S. A novel interaction between FRMD7 and CASK: Evidence for a causal role in idiopathic infantile nystagmus. Hum Mol Genet. 2013;22:2105–2118. doi: 10.1093/hmg/ddt060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yonehara K, Fiscella M, Drinnenberg A, Esposti F, Trenholm S, Krol J, Franke F, Scherf BG, Kusnyerik A, Müller J, et al. Congenital nystagmus gene FRMD7 is necessary for establishing a neuronal circuit asymmetry for direction selectivity. Neuron. 2016;89:177–193. doi: 10.1016/j.neuron.2015.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pu J, Li Y, Liu Z, Yan Y, Tian J, Chen S, Zhang B. Expression and localization of FRMD7 in human fetal brain, and a role for F-actin. Mol Vis. 2011;17:591–597. [PMC free article] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta DeltaC(T) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Hoffman RM. The potential of nestin-expressing hair follicle stem cells in regenerative medicine. Expert Opin Biol Ther. 2007;7:289–291. doi: 10.1517/14712598.7.3.289. [DOI] [PubMed] [Google Scholar]
- 18.Michalczyk K, Ziman M. Nestin structure and predicted function in cellular cytoskeletal organisation. Histol Histopathol. 2005;20:665–671. doi: 10.14670/HH-20.665. [DOI] [PubMed] [Google Scholar]
- 19.Jacobs KM, Neve RL, Donoghue JP. Neocortex and hippocampus contain distinct distributions of calcium-calmodulin protein kinase II and GAP43 mRNA. J Comp Neurol. 1993;336:151–160. doi: 10.1002/cne.903360112. [DOI] [PubMed] [Google Scholar]
- 20.Benowitz LI, Routtenberg A. GAP-43: An intrinsic determinant of neuronal development and plasticity. Trends Neurosci. 1997;20:84–91. doi: 10.1016/S0166-2236(96)10072-2. [DOI] [PubMed] [Google Scholar]
- 21.Roskams AJ, Cai X, Ronnett GV. Expression of neuron-specific beta-III tubulin during olfactory neurogenesis in the embryonic and adult rat. Neuroscience. 1998;83:191–200. doi: 10.1016/S0306-4522(97)00344-8. [DOI] [PubMed] [Google Scholar]
- 22.Matus A. Microtubule-associated proteins and neuronal morphogenesis. J Cell Sci Suppl. 1991;15:61–67. doi: 10.1242/jcs.1991.Supplement_15.9. [DOI] [PubMed] [Google Scholar]
- 23.Caceres A, Banker GA, Binder L. Immunocytochemical localization of tubulin and microtubule-associated protein 2 during the development of hippocampal neurons in culture. J Neurosci. 1986;6:714–722. doi: 10.1523/JNEUROSCI.06-03-00714.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lees JF, Shneidman PS, Skuntz SF, Carden MJ, Lazzarini RA. The structure and organization of the human heavy neurofilament subunit (NF-H) and the gene encoding it. EMBO J. 1988;7:1947–1955. doi: 10.1002/j.1460-2075.1988.tb03032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perrot R, Berges R, Bocquet A, Eyer J. Review of the multiple aspects of neurofilament functions and their possible contribution to neurodegeneration. Mol Neurobiol. 2008;38:27–65. doi: 10.1007/s12035-008-8033-0. [DOI] [PubMed] [Google Scholar]