Abstract
The aim of the present study was to investigate the changes in retinal gene expression at three time points and assess the underlying molecular mechanisms of diabetic retinopathy (DR) in a streptozotocin (STZ)-induced diabetes rat model using bioinformatics analysis. The gene expression profile of GSE28831 was extracted from the Gene Expression Omnibus database and differentially expressed genes (DEGs) were identified at three different time points (1, 4 and 12 weeks) using the limma package in R language. Gene ontology (GO) enrichment analysis of DEGs was performed followed by a principal component and pathway enrichment analysis of the selected DEGs along with protein-protein interaction network construction at the three time points. A total of 402, 105 and 213 DEGs were screened at 1, 4 and 12 weeks, respectively. In addition, the expression of 8 genes was identified to be significantly different at different time points, including cytochrome P450 2B2 (CYP2B2; downregulated gene; P=0.048; at 1 week), mannan binding lectin-associated serine protease-2 (MASP2; downregulated gene; P=0.044), lecithin retinol acyltransferase (LRAT; downregulated gene; P=0.015), retinal pigment epithelium (RPE)-specific protein 65 kDa (RPE65; downregulated gene; P=0.025), 11-cis-retinoldehydrogenase (RDH5; downregulated gene; P=0.04; at 4 weeks), mitogen-activated protein kinase 13 (MAPK13; upregulated gene; P=0.036), LRAT (downregulated gene; P=0.01) and RPE65 (downregulated gene; P=0.009; at 12 weeks). Furthermore, pathway enrichment and GO enrichment analyses revealed that DEGs at 4 weeks were primarily enriched in retinol metabolism and processes associated with visual functions, including ‘visual perception’ and ‘retinol metabolism’. DEGs, including CYP2B2, MASP2, LRAT, RPE65, RDH5 and MAPK13 may be potential targets for the diagnosis and treatment of DR. Thus, the current study demonstrated that abnormal visual functions occur at 4 weeks in STZ-induced diabetic rats. This may provide a scientific basis for the diagnosis and treatment of DR because DEGs may be used to facilitate the development of novel therapeutic strategies to diagnose and treat DR.
Keywords: diabetic retinopathy, differentially expressed gene, principal component analysis, pathway enrichment analysis, function enrichment analysis
Introduction
Diabetic retinopathy (DR) is a microvascular complication and is the most common cause of vision impairment and blindness among adults aged 20–74 years (1). The incidence of DR is associated with a number of risk factors such as hyperglycemia, hyperlipidemia and hypertension (2) and is characterized by signs of retinal ischemia and/or signs of increased retinal vascular permeability (3). Furthermore, DR is a progressive disease that endangers all retinal layers via insults of metabolic and neurological inflammation, which may contribute to vascular disruptions over time (4). Although clinical trials have demonstrated that effective treatments for diabetic retinopathy may reduce severe vision loss by 90% (5–7), these studies have emphasized the critical requirement for periodic eye examinations for all patients with diabetes. Thus, it is necessary to improve the means by which retinopathy is identified and prevented in its earliest stages rather than wait for the onset of vision-threatening lesions (8).
Over the past decade, studies have demonstrated that numerous factors may be involved in the pathogenesis of DR (9,10) and it has been determined that chronic hyperglycemia is a factor involved in the development and progression of DR (11–13). Chronic hyperglycemia may induce multiple cellular changes that lead to diabetic complications due to its toxic effects and/or the effects of its pathophysiological derivatives that directly act on tissues (14,15). Other factors including non-enzymatic glycation and glycoxidation may also induce DR. Furthermore, a number of candidate genes associated with the visual cycle, including lecithin retinol acyltransferase (LRAT) (16), retinal pigment epithelium (RPE)-specific protein 65 kDa (RPE65) (17) and 11-cis-retinoldehydrogenase (RDH5) (18) have been implicated in DR. Although early changes in retinal gene expression have been evaluated by analyzing retinal gene expression in streptozotocin (STZ)-induced diabetic rats (19), the specific underlying molecular mechanisms that occur during DR development and progression remain to be elucidated.
In the present study, bioinformatics was used to analyze the changes in retinal gene expression over a period of 12 weeks. Differentially expressed genes were analyzed at three different time points (1, 4 and 12 weeks) in the gene expression profile of retinal samples. The present study assessed the changes in retinal gene expression at the three time points with the aim of identifying the underlying molecular mechanisms of DR in diabetes.
Materials and methods
Affymetrix microarray data of retina gene
The gene expression profile of GSE28831 was downloaded from the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE28831). The dataset of GSE28831 was provided by a previous study by Kirwin et al (19), which obtained retinal samples of Long Evans rats, sacrificed 7 days, 4 weeks and 3 months after the induction of diabetes. A total of 21 retinal samples including 12 control samples and 9 diabetic samples were eligible for further analysis. Control samples consisted of 9 normal samples and 3 samples that received an injection of streptozotocin (STZ), but did not develop hyperglycemia. Diabetes samples included 9 retina samples which were obtained from STZ-induced diabetic rats. Retinal samples were collected from the retinas of control (n=3) and STZ-induced diabetic rats (n=3) at the three time points (1, 4 and 12 weeks after the induction of diabetes). In addition, 3 other samples were obtained from rats that were normoglycemic 1 week following STZ treatment that were used as normal controls.
The platform used was the GPL7294 Agilent-014879 Whole Rat Genome Microarray 4× 44K G4131F (Agilent Technologies, Inc., Santa Clara, CA, USA). The raw file and the annotation information file of the platform were also downloaded.
Data preprocessing and differential expression analysis
Each probe was mapped with one or multiple gene names labeled in an annotation platform. To make statistical inferences, each gene was normalized between microarrays based on quartile normalization (20). When multiple probes were mapped to the same gene ID, the mean expression of those probes was calculated. When one probe was mapped to multiple gene sets, information about the probe was deleted. The linear models of microarray data (limma) package (Version 3.30.3; Bioconductor, Seattle, WI, USA) in R language was used to identify genes that were differentially expressed between control and disease samples at the three time points (21). The P values of genes were calculated using student's t test and adjusted by multiple testing to circumvent the false-positive results. Subsequently, the log2-fold-change (logFC) was calculated. Only genes with an adjusted P<0.05 and |log2FC|>1 were selected as differentially expressed genes (DEGs) (22), which were the signature genes of DR.
Comparison of DEGs between control and disease samples
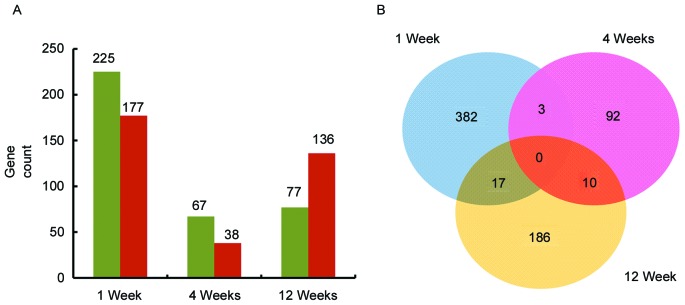
The DEGs between control and disease samples at the three time points were classified into upregulated and downregulated gene sets for further analysis. Subsequently, the contrast between the DEGs at the three time points was assessed according to the number of DEGs. Then gene sets at the three time points were compared and illustrated using a Venn diagram (Fig. 1) (23).
Figure 1.
Comparision of differentially expressed genes at the three timepoints. (A) Historgram graph demonstrating the number of upregulated and downregulated genes at three time points. Green represents upregulated genes and red represents downregulated genes. (B) Venn diagram of the number of genes at each timepoint. Blue represents 1 week, pink represents 4 weeks and yellow represents 12 weeks.
Principal component analysis (PCA) of DEGs
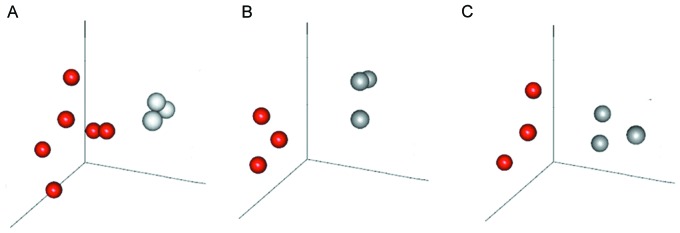
PCA, a multivariate regression analysis (24), was used to distinguish samples with multiple measurements (25). A PCA of DEGs was conducted in the present study using prcomp algorithm in R language (26,27). A 3D graph was then obtained, in which DEGs were considered as variables and the difference between control and disease samples at the three time points were observed.
Pathway enrichment analysis of DEGs
Pathway enrichment analysis is widely used to analyze high-throughput data and help link individual genes or proteins, which are differentially expressed under specific conditions, in order to improve understanding regarding biological pathways (28). To identify the pathways associated with DEGs selected at the three time points, the software tool KOBAS [kyoto encyclopedia of genes and genomes (KEGG) Orthology Based Annotation System] version 2 (http://kobas.cbi.pku.edu.cn), which was used for the annotation and identification of enriched pathways and diseases (29) based on the cumulative hypergeometric distribution algorithm (a discrete probability distribution), which was used for drawing from >2 selections in a population, again without replacement (30) with P<0.05 denoting a significant difference.
Protein-Protein interaction (PPI) network construction
Numerous physical activities of the cells were evaluated based on protein integration and dissociation. A variety of cellular physiological activities and the reaction of cells to the external and internal environment are regulated by PPI networks (31). Thus, a more thorough investigation of protein interactions is required to understand this biological phenomenon (32).
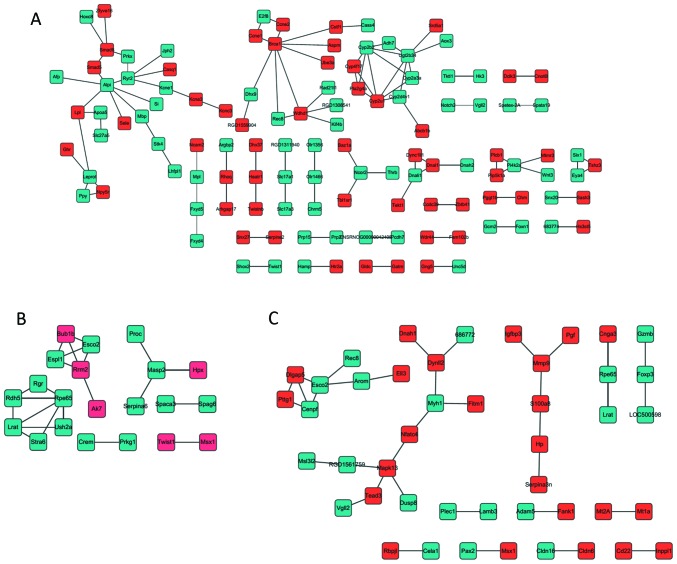
STRING (search tool for retrieving interacting genes/proteins; http://string-db.org/) (33) provides uniquely comprehensive coverage and ease of access to experimental and predicted interaction information. A PPI network was constructed, in which the interactions of DEGs between control and disease samples at the three time points were mapped to STRING based on information including the sequence characters and structures. Interaction patterns with a combined score >0.4 were selected. Subsequently, the PPI network at the three time points was visualized using Cytoscape software (version 3.4.0; Cytoscape Consortium, San Diego, CA, USA) (34). The nodes with higher degree served an important role in the PPI network, and may be the key nodes. Nodes represent the proteins encoded by DEGs, and a degree is considered to be the numbers of proteins that interact with that protein.
Functional analysis of the DEGs at the three time points
Gene ontology enrichment analysis has been extensively used to provide a detailed view for understanding the molecular mechanism and identifying a deeply complicated network, which consisted of a set of genes with similar functions (35).
DAVID (a database for annotation, visualization, and integrated discovery; version 6.8; https://david.ncifcrf.gov/home.jsp) (36), a comprehensive set of functional annotation tools, was used for systematic and integrative analyses of large gene/protein lists. It evaluated the functions of the DEGs at the three time points and defined significant function enrichment of these genes in multiple gene ontology (GO) categories when P<0.05.
Results
Identification of DEGs
A total of 402, 105 and 213 DEGs with the cut-off criteria of an adjusted value of significance (P<0.05) and |log2FC|>1 were selected as the thresholds to screen DEGs in control and disease samples at 1, 4 and 12 weeks.
Comparison of DEGs at the three time points
As presented in Fig. 1A, a total of 720 DEGs including 369 upregulated and 351 downregulated DEGs were screened. There were a greater number of downregulated than upregulated DEGs, with the exception of at 12 weeks following STZ-induced diabetes where the converse was true.
As presented in Fig. 1B, there were no common DEGs at all three time points. However, there were 3 common DEGs in weeks 1 and 4, 17 common DEGs in weeks 1 and 12 and 10 common DEGs in weeks 4 and 12.
PCA of DEGs
As presented in Fig. 2, disease and control samples were completely separated by the selected DEGs at the three time points, meaning that the expression patterns of screened DEGs were specific at different time points and could be used to completely distiguish between disease and control samples.
Figure 2.
Principal component analysis of DEGs at the three timepoints. Analysis of DEGs at (A) 1, (B) 4 and (C) 12 weeks. Red pellets, STZ-induced rat retinal sample; gray pellets, normal rat retinal sample; DEGs, differentially expressed genes.
Pathway enrichment analysis of the DEGs
The significantly enriched pathways for the up- and downregulated DEGs are presented in Table I. There were two KEGG pathways (Neuroactive ligand-receptor interaction and Arachidonic acid metabolism) associated with DEGs at 1 week and one KEGG pathway (Retinol metabolism) associated with DEGs at 4 weeks. The retinol metabolism pathway included three downregulated genes (LRAT, RPE65 and RDH5) and the LRAT and RPE65 genes were also downregulated in DEGs at 12 weeks (Fig. 3).
Table I.
DEGs enriched in the KEGG pathway at the three time points.
| A, Week 1 | |||
|---|---|---|---|
| Term | Count | Genes | P-value |
| rno04080: Neuroactive ligand-receptor interaction | 13 | THRB, LEPR, VIPR1, NPY5R, CHRM5, GPR35, CHRM4, PTGDR, ADRA1B, MTNR1B, ADRA2B, HTR2A, GHR | 0.005083 |
| rno00590: Arachidonic acid metabolism | 5 | CYP2U1, CYP2B2, PLA2G4A, CYP4F17, GPX3 | 0.048328 |
| B, Week 4 | |||
| Term | Count | Genes | P-value |
| rno00830: Retinol metabolism | 3 | LRAT, RPE65, RDH5 | 0.041873 |
DEGs, differentially expressed genes; KEGG, kyoto encyclopedia of genes and genomes.
Figure 3.
Protein-protein interaction network at the three timepoints. Network at (A) 1, (B) 4 and (C) 12 weeks. Green nodes represent proteins expressed by downregulated differentially expressed genes; red nodes represent proteins expressed by upregulated differentially expressed genes.
PPI network construction
A total of 104, 22 and 35 PPI pairs at 1, 4 and 12 weeks, respectively were obtained. These PPI pairs were then used to construct the PPI network by integrating these associations in the retina (Fig. 3). In this network, the proteins cytochrome P450 2B2 (CYP2B2; 1 week), mannan binding lectin-associated serine protease-2 (MASP2), LRAT, RPE65, RDH5 (4 weeks), mitogen-activated protein kinase 13 (MAPK13), LRAT and RPE65 (12 weeks) with high degrees form a local network and these proteins served an important role in the PPI network, and may be the key proteins in the development of DR.
GO enrichment analysis of DEGs involved in PPI network
As presented in Table II, the DEGs in the PPI network at 1, 4 and 12 weeks were enriched in 12, 7 and 10 GO categories, respectively. Furthermore the DEGs in PPI network at 1 week were primarily enriched in processes associated with metabolism and response, including ‘fatty acid metabolic processes’, ‘responses to drug’ and ‘responses to endogenous stimulus’. The DEGs in the PPI network at 4 weeks were enriched in processes associated with visual functions, such as ‘visual perception’, ‘sensory perception of light stimulus’ and ‘vitamin A metabolic process’. The DEGs in the PPI network at 12 weeks were primarily enriched in regulatory processes including ‘positive regulation of transcription, DNA-dependent’, ‘positive regulation of RNA metabolic process’ and ‘negative regulation of cell proliferation’.
Table II.
Enriched GO terms in the gene interaction network.
| A, Week 1 | ||
|---|---|---|
| Term | Genes | P-value |
| GO:0010033 response to organic substance | CYP2B2, GATM, SI, RHOQ, ADH7, BRCA1, AFP, CCNE1, PLA2G4A, ABCB1B, APOA5, CYP2D4V1, RYR2, GNG5, CASQ1, SELE, NCOR2, PGGT1B, GHR | <0.0001 |
| GO:0006631 fatty acid metabolic process | CYP2B2, LPL, PLA2G4A, CYP2D4V1, ADH7, BRCA1, SLC27A5, GHR | <0.0001 |
| GO:0009725 response to hormone stimulus | CCNE1, CYP2B2, PLA2G4A, GATM, ABCB1B, APOA5, SI, RHOQ, GNG5, BRCA1, NCOR2, GHR | <0.0001 |
| GO:0009719 response to endogenous stimulus | CCNE1, CYP2B2, PLA2G4A, GATM, ABCB1B, APOA5, SI, RHOQ, GNG5, BRCA1, NCOR2, GHR | 0.001969 |
| GO:0022602 ovulation cycle process | AFP, CCNE1, PLA2G4A, NPY5R, NCOR2 | 0.002192 |
| GO:0042698 ovulation cycle | AFP, CCNE1, PLA2G4A, NPY5R, NCOR2 | 0.002986 |
| GO:0042592 homeostatic process | LPL, JPH2, SMAD5, MBP, GCM2, PLA2G4A, HAMP, APOA5, KCNE1, RYR2, NCOR2, SASH3, HTR2A | 0.003682 |
| GO:0019369 arachidonic acid metabolic process | CYP2B2, PLA2G4A, CYP2D4V1 | 0.004721 |
| GO:0048878 chemical homeostasis | LPL, PLA2G4A, GCM2, JPH2, HAMP, APOA5, KCNE1, RYR2, NCOR2, MBP | 0.006974 |
| GO:0042493 response to drug | CCNE1, CYP2B2, LPL, SNX27, ABCB1B, RYR2, SRD5A1, HTR2A | 0.00758 |
| GO:0042304 regulation of fatty acid biosynthetic process | PLA2G4A, APOA5, BRCA1 | 0.008354 |
| GO:0031099 regeneration | AFP, CCNE1, NOTCH2, GATM, APOA5 | 0.008868 |
| B, 4 weeks | ||
| Term | Genes | P-value |
| GO:0007601 visual perception | LRAT, RPE65, RGR, USH2A, RDH5 | <0.0001 |
| GO:0050953 sensory perception of light stimulus | LRAT, RPE65, RGR, USH2A, RDH5 | <0.0001 |
| GO:0007600 sensory perception | LRAT, RPE65, RGR, USH2A, RDH5 | 0.0199 |
| GO:0050890 cognition | LRAT, RPE65, RGR, USH2A, RDH5 | 0.029055 |
| GO:0006776 vitamin A metabolic process | LRAT, RPE65 | 0.030184 |
| GO:0016101 diterpenoid metabolic process | LRAT, RPE65 | 0.030184 |
| GO:0001523 retinoid metabolic process | LRAT, RPE65 | 0.030184 |
| C, 12 weeks | ||
| Term | Genes | P-value |
| GO:0045893 positive regulation of transcription, DNA-dependent | MSX1, VGLL2, TEAD3, FOXP3, ELL3, PAX2, RBPJL | 0.001781 |
| GO:0051254 positive regulation of RNA metabolic process | MSX1, VGLL2, TEAD3, FOXP3, ELL3, PAX2, RBPJL | 0.001862 |
| GO:0045941 positive regulation of transcription | MSX1, VGLL2, TEAD3, FOXP3, ELL3, PAX2, RBPJL | 0.003924 |
| GO:0010628 positive regulation of gene expression | MSX1, VGLL2, TEAD3, FOXP3, ELL3, PAX2, RBPJL | 0.004415 |
| GO:0045944 positive regulation of transcription from RNA polymerase II promoter | MSX1, VGLL2, TEAD3, FOXP3, ELL3, PAX2 | 0.004659 |
| GO:0008285 negative regulation of cell proliferation | MSX1, INPPL1, PTTG1, FOXP3, IGFBP3 | 0.006831 |
| GO:0045935 positive regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | MSX1, VGLL2, TEAD3, FOXP3, ELL3, PAX2, RBPJL | 0.007352 |
| GO:0051173 positive regulation of nitrogen compound metabolic process | MSX1, VGLL2, TEAD3, FOXP3, ELL3, PAX2, RBPJL | 0.008442 |
| GO:0010557 positive regulation of macromolecule biosynthetic process | MSX1, VGLL2, TEAD3, FOXP3, ELL3, PAX2, RBPJL | 0.008569 |
| GO:0010273 detoxification of copper ion | MT1A, MT2A | 0.008659 |
GO, gene ontology.
Discussion
Due to the increasing number of individuals diagnosed with diabetes, blindness caused by DR may become more common unless improvements are made in the diagnosis and treatment of DR (7). To improve understanding regarding the mechanisms involved in DR formation, the present study investigated the biological processes and signaling pathways associated with DR. The analysis of gene expression profiling identified the abnormally expressed genes associated with DR and enabled the identification of novel targets for therapeutic strategies. In the current study, a total of 402, 105 and 213 DEGs were screened at 1, 4 and 12 weeks, respectively, and eight significant genes were identified including CYP2B2 (at 1 week), MASP2, LRAT, RPE65, RDH5 (at 4 weeks), MAPK13, LRAT and RPE65 (at 12 weeks). Pathway enrichment and GO enrichment analyses revealed that DEGs were primarily enriched in processes associated with visual functions, including ‘visual perception’ and ‘retinol metabolism’ at 4 weeks.
CYP2B2 encodes an enzyme that controls arachidonic acid metabolism (37). Arachidonic acid is a precursor of inflammatory mediators, and early stage DR is regarded as a low-grade chronic inflammatory condition (38). Pathway enrichment analysis in the present study revealed that DEGs at 1 week were enriched in arachidonic acid metabolism. However, early inflammation of the eyes does not lead to any clinical manifestations. Therefore, inflammation in the retina may be an important component of early DR without clinical manifestations and the associated genes, including CYP2B2, may serve important roles in the evaluation of these functions. This is in accordance with a previous study by Joussen et al (39), which suggested that the response of the retina to the diabetic challenge consists of an inflammatory component.
In the present study, MASP2 was downregulated at 4 weeks. MASP2 encodes a novel serine protease that acts in the mannan-binding lectin (MBL)-lectin complement fixation pathway (40). The MBL-lectin complement fixation pathway is widely thought to serve a major role in host defense against infection and the expression of MBL is upregulated during inflammation (41). Østergaard et al (42) suggested that the complement system, specifically the MBL pathway, serves an important role in the pathogenesis of diabetic vascular complications such as DR. The decreased expression of MASP2 leads to MBL deficiency, which increases the overall susceptibility of an individual to infectious diseases (43). Thus, the present study suggests that the downregulation of MASP2 may stimulate the progression of DR and promote the appearance of other symptoms associated with DR.
LRAT, RPE65 and RDH5 are involved in retinol metabolism, and these genes are all part of the visual cycle that recycles all-trans-retinal from bleached photoreceptors to the 11-cis-retina required for phototransduction (20). The visual cycle is critical for visual function. Previous studies have reported that mutations in three different genes involved in the visual cycle, RPE65, LRAT and RDH12 cause early onset retinal dystrophy (44). Furthermore, RPE65 is essential for the maintenance of normal vision (45,46). Thus, the downregulation of LRAT, RPE65 and RDH5 may lead to abnormal visual function. In addition, LRAT and RPE65 were included in the downregulated DEGs at 12 weeks and GO enrichment analysis revealed that he majority of DEGs at 4 weeks were associated with visual functions, including ‘visual perception’, ‘sensory perception of light stimulus’ and ‘vitamin A metabolic process’. It has therefore been suggested that abnormal visual functions occur at 4 weeks in STZ-induced diabetic rats, which may be caused by the downregulation of genes associated with visual functions, including LRAT, RPE65 and RDH5. However, considering the present study is a bioinformatic study, further long-term studies are required to verify the association between gene expression and visual function.
MAPK13 encodes an enzyme that is included in the MAPK family. In the PPI network at 12 weeks, MAPK13 with a high degree formed a local network, suggesting that MAPK13 may serve a major role in DR. MAPKs have been implicated in a number of diseases including cancer, inflammatory disease, obesity and diabetes (47,48). Previous studies have reported that MAPKs are involved in regulating insulin secretion and β cell survival (47,49,50) and MAPK inhibitors contribute to the prevention of cardiovascular diseases such as retinopathy (51). Therefore, MAPK13 may be involved in the pathogenesis of DR, although further studies are required to confirm this observation.
In conclusion, the data in the present study provide comprehensive bioinformatic analysis of DEGs, which may be involved in DR. It was also demonstrated that abnormalities in visual functions occur in STZ-induced diabetic rats at 4 weeks. DEGs, including CYP2B2, MASP2, LRAT, RPE65, RDH5 and MAPK13 may be potential targets for DR diagnosis and treatment. The results of the current study may provide a scientific basis for the diagnosis and targeted therapy of DR.
References
- 1.Fong DS, Aiello L, Gardner TW, King GL, Blankenship G, Cavallerano JD, Ferris FL, III, Klein R. American Diabetes Association: Retinopathy in diabetes. Diabetes Care. 2004;27:S84–S87. doi: 10.2337/diacare.27.2007.S84. (Suppl 1) [DOI] [PubMed] [Google Scholar]
- 2.Malone JI, Morrison AD, Pavan PR, Cuthbertson DD. Diabetic Control and Complications Trial: Prevalence and significance of retinopathy in subjects with type 1 diabetes of less than 5 years' duration screened for the diabetes control and complications trial. Diabetes Care. 2001;24:522–526. doi: 10.2337/diacare.24.3.522. [DOI] [PubMed] [Google Scholar]
- 3.Kempen JH, O'Colmain BJ, Leske MC, Haffner SM, Klein R, Moss SE, Taylor HR, Hamman RF. Eye Diseases Prevalence Research Group: The prevalence of diabetic retinopathy among adults in the united states. Arch Ophthalmol. 2004;122:552–563. doi: 10.1001/archopht.122.4.552. [DOI] [PubMed] [Google Scholar]
- 4.Ciulla TA, Amador AG, Zinman B. Diabetic retinopathy and diabetic macular edema: Pathophysiology, screening, and novel therapies. Diabetes Care. 2003;26:2653–2664. doi: 10.2337/diacare.26.9.2653. [DOI] [PubMed] [Google Scholar]
- 5.Photocoagulation treatment of proliferative diabetic retinopathy: The second report of diabetic retinopathy study findings. Ophthalmology. 1978;85:82–106. doi: 10.1016/S0161-6420(78)35693-1. [DOI] [PubMed] [Google Scholar]
- 6.Early photocoagulation for diabetic retinopathy. ETDRS report number 9. Early treatment diabetic retinopathy study research group. Ophthalmology. 1991;98:S766–S785. (5 Suppl) [PubMed] [Google Scholar]
- 7.Wilkinson C, Ferris FL, III, Klein RE, Lee PP, Agardh CD, Davis M, Dills D, Kampik A, Pararajasegaram R, Verdaguer JT. Global Diabetic Retinopathy Project Group: Proposed international clinical diabetic retinopathy and diabetic macular edema disease severity scales. Ophthalmology. 2003;110:1677–1682. doi: 10.1016/S0161-6420(03)00475-5. [DOI] [PubMed] [Google Scholar]
- 8.Antonetti DA, Barber AJ, Bronson SK, Freeman WM, Gardner TW, Jefferson LS, Kester M, Kimball SR, Krady JK, LaNoue KF, et al. Diabetic retinopathy: seeing beyond glucose-induced microvascular disease. Diabetes. 2006;55:2401–2411. doi: 10.2337/db05-1635. [DOI] [PubMed] [Google Scholar]
- 9.Sulochana KN, Ramakrishnan S, Rajesh M, Coral K, Badrinath SS. Diabetic retinopathy: Molecular mechanisms, present regime of treatment and future perspectives. Curr Sci. 2001;80:133–142. [Google Scholar]
- 10.Takagi H, Oh H, Otani A, Suzuma K, Suzuma I, Ohashi H, Wantanabe D, Kemmochi S, Ojima T, Suganami E, et al. Molecular mechanisms of retinal neovascularization in diabetic retinopathy. Int Congress Ser. 2004;1262:160–163. doi: 10.1016/j.ics.2003.11.017. [DOI] [Google Scholar]
- 11.Klein R, Klein BK, Moss SE, Cruickshanks KJ. Relationship of hyperglycemia to the long-term incidence and progression of diabetic retinopathy. Arch Intern Med. 1994;154:2169–2178. doi: 10.1001/archinte.154.19.2169. [DOI] [PubMed] [Google Scholar]
- 12.Engerman RL, Kern TS. Hyperglycemia as a cause of diabetic retinopathy. Metabolism. 1986;35:S20–S23. doi: 10.1016/0026-0495(86)90182-4. (4 Suppl 1) [DOI] [PubMed] [Google Scholar]
- 13.Balasubramanyam M, Rema M, Premanand C. Biochemical and molecular mechanisms of diabetic retinopathy. Curr Sci. 2002;83:1506–1514. [Google Scholar]
- 14.Sheetz MJ, King GL. Molecular understanding of hyperglycemia's adverse effects for diabetic complications. JAMA. 2002;288:2579–2588. doi: 10.1001/jama.288.20.2579. [DOI] [PubMed] [Google Scholar]
- 15.Nishikawa T, Edelstein D, Brownlee M. The missing link: A single unifying mechanism for diabetic complications. Kidney Int Suppl. 2000;77:S26–S30. doi: 10.1046/j.1523-1755.2000.07705.x. [DOI] [PubMed] [Google Scholar]
- 16.Larsen LM. Prevention of retinopathy by inhibition of the visual cycle. US Patent 9421175 B2. 2016 Filed March 16, 2005; issued August 23. [Google Scholar]
- 17.Jin M, Li S, Moghrabi WN, Sun H, Travis GH. Rpe65 is the retinoid isomerase in bovine retinal pigment epithelium. Cell. 2005;122:449–459. doi: 10.1016/j.cell.2005.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duester G. Families of retinoid dehydrogenases regulating vitamin A function: Production of visual pigment and retinoic acid. Eur J Biochem. 2000;267:4315–4324. doi: 10.1046/j.1432-1327.2000.01497.x. [DOI] [PubMed] [Google Scholar]
- 19.Kirwin SJ, Kanaly ST, Hansen CR, Cairns BJ, Ren M, Edelman JL. Retinal gene expression and visually evoked behavior in diabetic long evans rats. Invest Ophthalmol Vis Sci. 2011;52:7654–7663. doi: 10.1167/iovs.10-6609. [DOI] [PubMed] [Google Scholar]
- 20.Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seyednasrollah F, Laiho A, Elo LL. Comparison of software packages for detecting differential expression in RNA-seq studies. Brief Bioinform. 2015;16:59–70. doi: 10.1093/bib/bbt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheng WH, Sheng KT, Zhao YX, Li H, Zhou JL, Yao HY, Li XH. Identifying the biomarkers of multiple sclerosis based on non-coding RNA signature. Eur Rev Med Pharmacol Sci. 2015;19:3635–3642. [PubMed] [Google Scholar]
- 23.Hulsen T, de Vlieg J, Alkema W. BioVenn-a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vyas S, Kumaranayake L. Constructing socio-economic status indices: How to use principal components analysis. Health Policy Plan. 2006;21:459–468. doi: 10.1093/heapol/czl029. [DOI] [PubMed] [Google Scholar]
- 25.Hild M, Beckmann B, Haas SA, Koch B, Solovyev V, Busold C, Fellenberg K, Boutros M, Vingron M, Sauer F, et al. An integrated gene annotation and transcriptional profiling approach towards the full gene content of the Drosophila genome. Genome Biol. 2003;5:R3. doi: 10.1186/gb-2003-5-2-p3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stafford R, Goodenough AE, Slater K, Carpenter W, Collins L, Cruickshank H, Downing S, Hall S, McDonald K, McDonnell H, et al. Inferential and visual analysis of ethogram data using multivariate techniques. Animal Behaviour. 2012;83:563–569. doi: 10.1016/j.anbehav.2011.11.020. [DOI] [Google Scholar]
- 27.Venables W, Ripely B. Modern Applied Statistics with S. 4th. Springer; New York, NY: 2002. [DOI] [Google Scholar]
- 28.Mao X, Zhang Y, Xu Y. SEAS: A system for SEED-based pathway enrichment analysis. PLoS One. 2011;6:e22556. doi: 10.1371/journal.pone.0022556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–W322. doi: 10.1093/nar/gkr483. (Web Server Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agner F. Calculation Methods for Wallenius' Noncentral Hypergeometric Distribution. Commun Stat. 2008;37:258–273. doi: 10.1080/03610910701790269. [DOI] [Google Scholar]
- 31.Giot L, Bader J, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 32.Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan GS, Gao J, Zhang ZM, Du J, Mu GQ, Pan GT. Gene-Ontology analysis on the differentially expressed genes in maize (Zea mays L.) Ear Rot. J Life Sci. 2013;7:219–226. [Google Scholar]
- 36.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 37.Zordoky BN, Aboutabl ME, El-Kadi AO. Modulation of cytochrome P450 gene expression and arachidonic acid metabolism during isoproterenol-induced cardiac hypertrophy in rats. Drug Metab Dispos. 2008;36:2277–2286. doi: 10.1124/dmd.108.023077. [DOI] [PubMed] [Google Scholar]
- 38.Chen W, Jump DB, Grant MB, Esselman WJ, Busik JV. Dyslipidemia, but not hyperglycemia, induces inflammatory adhesion molecules in human retinal vascular endothelial cells. Invest Ophthalmol Vis Sci. 2003;44:5016–5022. doi: 10.1167/iovs.03-0418. [DOI] [PubMed] [Google Scholar]
- 39.Joussen AM, Huang S, Poulaki V, Camphausen K, Beecken WD, Kirchhof B, Adamis AP. In vivo retinal gene expression in early diabetes. Invest Ophthalmol Vis Sci. 2001;42:3047–3057. [PubMed] [Google Scholar]
- 40.Jensenius JC, Thiel S. MASP 2, a complement-fixing enzyme and uses for it. US Patent 20020082208 A1. 2002 Filed June 4, 2001; issued June 27. [Google Scholar]
- 41.Schwaeble HW, Stover CM, Tedford CE, Parent JB, Fujita T. Methods for treating conditions associated with MASP-2 dependent complement activation. WO Patent 2005123128 A2. 2005 Filed June 9, 2005; issued December 29. [Google Scholar]
- 42.Østergaard J, Hansen TK, Thiel S, Flyvbjerg A. Complement activation and diabetic vascular complications. Clin Chim Acta. 2005;361:10–19. doi: 10.1016/j.cccn.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 43.Turner MW. The role of mannose-binding lectin in health and disease. Mol Immunol. 2003;40:423–429. doi: 10.1016/S0161-5890(03)00155-X. [DOI] [PubMed] [Google Scholar]
- 44.Janecke AR, Thompson DA, Utermann G, Becker C, Hübner CA, Schmid E, McHenry CL, Nair AR, Rüschendorf F, Heckenlively J, et al. Mutations in RDH12 encoding a photoreceptor cell retinol dehydrogenase cause childhood-onset severe retinal dystrophy. Nat Genet. 2004;36:850–854. doi: 10.1038/ng0904-1024c. [DOI] [PubMed] [Google Scholar]
- 45.Moiseyev G, Chen Y, Takahashi Y, Wu BX, Ma JX. RPE65 is the isomerohydrolase in the retinoid visual cycle. Proc Natl Acad Sci USA. 2005;102:12413–12418. doi: 10.1073/pnas.0503460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campochiaro PA. Seeing the light: New insights into the molecular pathogenesis of retinal diseases. J Cell Physiol. 2007;213:348–354. doi: 10.1002/jcp.21213. [DOI] [PubMed] [Google Scholar]
- 47.Huang P, Han J, Hui L. MAPK signaling in inflammation-associated cancer development. Protein Cell. 2010;1:218–226. doi: 10.1007/s13238-010-0019-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lawrence MC, Jivan A, Shao C, Duan L, Goad D, Zaganjor E, Osborne J, McGlynn K, Stippec S, Earnest S, et al. The roles of MAPKs in disease. Cell Res. 2008;18:436–442. doi: 10.1038/cr.2008.37. [DOI] [PubMed] [Google Scholar]
- 49.Cuenda A, Nebreda AR. p38delta and PKD1: Kinase switches for insulin secretion. Cell. 2009;136:209–210. doi: 10.1016/j.cell.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 50.Sumara G, Formentini I, Collins S, Sumara I, Windak R, Bodenmiller B, Ramracheya R, Caille D, Jiang H, Platt KA, et al. Regulation of PKD by the MAPK p38delta in insulin secretion and glucose homeostasis. Cell. 2009;136:235–248. doi: 10.1016/j.cell.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Osman N, Ballinger ML, Dadlani HM, Getachew R, Burch ML, Little PJ. p38 MAP kinase mediated proteoglycan synthesis as a target for the prevention of atherosclerosis. Cardiovasc Hematol Disord Drug Targets. 2008;8:287–292. doi: 10.2174/187152908786786205. [DOI] [PubMed] [Google Scholar]