Abstract
Mesenchymal stem cells (MSCs) are able to differentiate into hepatocytes, promote the regeneration of hepatic cells and inhibit the progression of hepatic fibrosis. Transforming growth factor (TGF)-β1 is one of the key factors in the development of liver fibrosis, which also promotes extracellular matrix (ECM) formation. Drosophila mothers against decapentaplegic 7 (Smad7) is an essential negative regulator in the TGF-β1/Smad signaling pathway. In the present study, bone mesenchymal stem cells (BMSCs) were isolated from rat bone marrow and transfected with lentiviral vectors carrying the Smad7 gene. Smad7-enhanced green fluorescent protein (EGFP)-BMSCs stably expressing Smad7 were subsequently co-cultured with hepatic stellate cells (HSCs) for 48 h. Smad7 and TGF-β1 levels in the culture medium were detected using ELISA, and the levels of collagen (Col) I, Col III, laminin (LN) and hyaluronic acid (HA) were measured using immunoassays. The early apoptosis rates of HSCs were determined via flow cytometry. Reverse transcription-quantitative polymerase chain reaction and western blotting were performed to evaluate the mRNA and protein expression profiles, respectively. The results indicated that Smad7-EGFP-BMSCs stably expressing Smad7 were successfully constructed. Upon co-culturing with rat Smad7-EGFP-BMSCs, the early apoptotic rate of HSCs was significantly increased (P<0.05). Levels of Smad7 in the culture medium were also significantly increased (P<0.05), whereas the levels of TGF-β1, Col I, Col III, LN and HA were significantly decreased (P<0.05). Furthermore, the mRNA and protein levels of Smad7 and matrix metalloproteinase 1 were significantly increased (P<0.05), whereas those of TGF-β1, α-SMA, Smad2, smad3, TGF-β receptor I, Col I, tissue inhibitors of metalloproteinase-1 and Col III were significantly decreased. The results of the present study suggest that rat BMSCs overexpressing Smad7 may inhibit the fibrosis of HSCs by regulating the TGF-β1/Smad signaling pathway. This provides a novel insight into future treatments for liver fibrosis.
Keywords: mesenchymal stem cells, Drosophila mothers against decapentaplegic 7, hepatic stellate cells, fibrotic, transforming growth factor-β
Introduction
Stem cells that are capable of self-renewal and differentiation are thought to have therapeutic potential for the treatment of liver cirrhosis (1). There are two types of stem cells, embryonic and adult; adult stem cells are the focus of clinical research (2). Mesenchymal stem cells (MSCs) are multipotent adult stem cells with low immunogenicity that are able to differentiate into liver cells. MSCs are able to promote liver regeneration and suppress liver fibrosis, and thus may partially recover liver function and reverse liver cirrhosis (3). Transforming growth factor (TGF)-β1 is one of the key factors in the development of liver fibrosis, which also promotes extracellular matrix (ECM) formation (4). Drosophila mothers against decapentaplegics (SMADs) are intracellular proteins that transduce extracellular signals from TGF-β1 to the nucleus and further activate downstream gene transcription. Smad7 protein is an essential negative regulator in the TGF-β1/Smad7 signaling pathway (5).
Hepatic stellate cells (HSCs) have a primary role in the pathogenesis of liver fibrosis (6). A previous experimental study suggested that activated HSCs are an attractive target for antifibrotic therapy (7). Potential therapeutic strategies include downregulating HSC activation and neutralizing the antiproliferative, fibrogenic, and contractile responses of HSCs (8). It has been reported that migrated MSCs are present in the portal areas and fibrous bands in the liver, where activated HSCs are localized (9). This suggests that suppressing TGF-β1 function may improve the therapeutic effect of treatment with MSCs.
In the present study, rat BMSCs with stably expressed Smad7 that were able to suppress TGF-β were constructed and co-cultured with HSCs. The aim was to investigate the inhibitory effects of Smad7-enhanced green fluorescent protein (EGFP)-BMSCs on the fibrosis of HSCs and the mechanisms involved in order to provide experimental evidence for the development of future liver cirrhosis treatment strategies.
Materials and methods
Isolation and purification of rat BMSCs
A total of 10 6-week-old male Wistar rats (each weighing 160 to 200 g) were purchased from the Laboratory Animal Center of Sun Yat-sen University (Guangzhou, China). Rats received ordinary feed for 1 week and had free access to food and water, and were exposed to a 12-h light/dark cycle (humidity, 40–70%; temperature, 37°C). All animal experiments were performed under the ethical guidelines of the Ethics Committee of The Second Affiliated Hospital of Jinan University (Shenzhen, China). Wistar rats were sacrificed and their femurs were harvested. Bone marrow cells were harvested by carefully washing the marrow cavity with PBS. The cells were isolated using density gradient centrifugation (×400 g for 10 min at 37°C) and an adherence screening approach (10). Subsequently, cells were cultured in Dulbecco's modified Eagle's medium (DMEM)/F12 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) at a density of 1×106 cells/ml. Half of the culture medium was replenished after 3 days, and thereafter the whole culture medium was replaced every 3 days. When cells reached 80% confluence, they were digested with 0.05% trypsin and passaged at 30% confluence. Following the third passage, BMSCs were stained with green fluorescent protein, and surface markers of MSCs, including cluster of differentiation (CD)31, CD34, CD45, CD73, CD105 and CD106, were detected by flow cytometry (ALTRA; Beckman Coulter, Inc., Brea, CA, USA).
Construction of the lentiviral vectors carrying the Smad7 gene and Smad7-EGFP-BMSCs
Total RNA was extracted from the cerebral cortex, and reverse transcription/amplification was performed to obtain the Smad7 DNA fragments. Reverse transcription was performed using a reverse transcription kit (205310; Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Reverse transcription reaction conditions included 10 min at 40°C and 95°C for 2 min. The PCR reaction conditions were as follows: Pre-denaturation at 94°C for 5 min and 94°C 30 sec; annealing at 55°C for 45 sec, 72°C for 60 sec for a total of 35 cycles, followed by an extension of 72°C for 10 min. The following primers were used: Smad7, forward 5′-GGGTTTACAACCGCAGCAGT-3′ and reverse: 5′-GCCTTGATGGAGAAACCAGG-3′. Upon cleaving the lentiviral vector, pCDH-CMV-MCS-EF1-copEGFP (Shanghai GenePharma Co., Ltd., Shanghai, China), with restriction enzymes (EcoRI and BamHI; Shanghai GenePharma Co., Ltd.), the Smad7 DNA fragment was inserted and co-expressed with EGFP. The resultant recombinant plasmid was Smad7-EGFP.
BMSCs after the third passage were seeded in a 6-well plate at a density of 1×106 cells/ml. When the cells reached 70% confluence, Smad7-EGFP recombinant plasmid and Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) were added into each well and the culture medium was replenished after 10 h. Neomycin was added to the culture medium to screen and select for the recombinants. The transfection efficiency was evaluated under a fluorescence microscope and cells with stably expressed Smad7-EGFP were subcultured at 37°C for 24 h in vitro.
Cell culture
Hepatic stellate cells (HSCs) line T6 were purchased from the Shanghai Institute of Cell Biology (Shanghai, China). Cell lines were cultured in DMEM supplemented with 10% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin. Cells were maintained in a humidified incubator at 37°C in an atmosphere containing 5% CO2. All cell lines were passaged for <6 months.
Cells were divided into the following four groups: i) Smad7-EGFP-BMSCs + HSCs; ii) BMSCs + HSCs; iii) PBS + HSCs; and iv) Smad7-EGFP recombinant plasmid + HSCs. For the Smad7-EGFP-BMSCs + HSCs experimental group, the two cells lines were co-cultured as previously described (11). Following 48 h of cultivation, the Smad7 and TGF-β1 levels in the culture medium were detected using ELISA, according to the manufacturer's instruction, and the levels of Col I (cat. no. CX20063), Col III (cat. no. CX20064), LN (cat. no. KT20202) and HA (cat. no. 23610) were measured using an immunoassay according to manufacturer's instruction (Merck, New Jersey, CA, USA).
Flow cytometry
Upon co-cultivation with Smad7-EGFP-BMSCs for 48 h, HSCs were washed twice with PBS, fixed with 70% ethanol, and treated with RNase A (1 mg/ml (Invitrogen; Thermo Fisher Scientific, Inc.). Cells were double-stained with fluorescein isothiocyanate-conjugated Annexin V and propidium iodide (PI) solution (50 µg/ml; Invitrogen, Thermo Fisher Scientific, Inc.). For each sample, data from ~10,000 cells were recorded in the list mode on logarithmic scales. Apoptosis and necrosis were analyzed using a flow cytometer (Altra; Beckman Coulter, Inc., Brea, CA, USA) and quadrant statistics on double negative, Annexin V-positive/PI-negative, Annexin V-negative/PI-positive, and double positive cells were applied.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA samples were extracted from the transfected HSCs using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. qPCR analysis was performed using a kit (4389986; Invitrogen; Thermo Fisher Scientific, Inc). The following cycling conditions were performed: A total of 10 min at 40°C, 95°C for 2 min followed by 40 cycles of denaturation at 50°C for 2 min, annealing at 95°C for 2 min, elongation at 95°C for 15 sec and extension at 60°C for 32 sec. containing SYBR-Green according to the manufacturer's instructions and an Applied Biosystems 7500 Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). GAPDH was used as an internal control for mRNAs. The level of expression was calculated using the 2−ΔΔCq method (12). Primers used in qPCR analysis are provided in Table I.
Table I.
Primers used for reverse transcription-quantitative polymerase chain reaction.
| Gene | Direction | Primer sequence |
|---|---|---|
| Smad7 | F | 5′-GGGTTTACAACCGCAGCAGT-3′ |
| R | 5′-GCCTTGATGGAGAAACCAGG-3′ | |
| TGFβ1 | F | 5′-AAACGGAAGCGCATCGAA-3′ |
| R | 5′-GGGACTGGCGAGCCTTAGTT-3′ | |
| Smad2 | F | 5′-GTGTTTGCCGAGTGCCTAAGT-3′ |
| R | 5′-TTACAGCCTGGTGGGATTTTG-3′ | |
| Smad3 | F | 5′-GGACGCAGGCTCTCCAAAC-3′ |
| R | 5′-AGGAGATGGAGCACCAAAAGG-3′ | |
| TIMP-1 | F | 5′-GATTCGACGCTGTGGGAAAT-3′ |
| R | 5′-AAACTCCTCGCTGCGGTTCT-3′ | |
| Collagen I | F | 5′-CTCCCAGCGGTGGTTATGAC-3′ |
| R | 5′-TGCTGGCTCAGGCTCTTGA-3′ | |
| Collagen | F | 5′-CCCTCTCTTATTTTGGCACAGC-3′ |
| III | R | 5′-TGACATGGTTCTGGCTTCCAG-3′ |
| α-SMA | F | 5′-AATGGCTCCGGGCTCTGTA-3′ |
| R | 5′-TTCCAACCATCACTCCCTGG-3′ | |
| TβRI | F | 5′-GCTGTGGTTGGTGTCGGATT-3′ |
| R | 5′-CGTGGACAGAGCGAGTTTGAT-3′ | |
| MMP1 | F | 5′-CACAACAATCCTCGTTGGACC-3′ |
| R | 5′-AAGGCTCTCTCGATGGCGTT-3′ | |
| GAPDH | F | 5′-AGGGCTGCCTTCTCTTGTGA-3′ |
| R | 5′-AACTTGCCGTGGGTAGAGTCA-3′ |
Smad, Drosophila mothers against decapentaplegic; F, forward; R, reverse; TGF-β1, transforming growth factor-β1; TIMP-1, tissue inhibitors of metalloproteinase-1; α-SMA, α-smooth muscle actin; TβRI, TGF-β receptor I; MMP-1, matrix metalloproteinase 1.
Western blot analysis
HSCs were lysed in radioimmunoprecipitation assay buffer containing a proteinase inhibitor cocktail (Shenergy Biocolor BioScience & Technology, Shanghai, China). Protein concentration was determined using bicinchoninic acid (BIOSS, Beijing, China). Proteins were separated by 10% SDS-PAGE at 30 µg/gel lane and electrotransferred to nitrocellulose membranes. The resolved proteins were transferred onto a nitrocellulose membrane (EMD Millipore; Billerica, MA, USA), which was then exposed to 5% non-fat dried milk in Tris-buffered saline-Tween buffer for 1 at room temperature prior to incubation overnight at 4°C with primary antibodies. Membranes were incubated with rabbit polyclonal primary antibodies against Smad7 (SC-11392), Smad2 (:5339), Smad3 (9513), TGF-β receptor I (TβRI; Ab31013), matrix metalloproteinase 1 (MMP-1; SC-241561), α-smooth muscle actin (α-SMA; 14968), collagen (Col) I (Ab7778), Col III (Ba0290), TGF-β1 (SC-5538) and tissue inhibitors of metalloproteinase-1 (TIMP-1; all 1:1,000; all Shunho Cell Biotech Co., Ltd., Tianjin, China) at 4°C overnight. Following washing with 0.1% Tween-20 in PBS, membranes were subsequently incubated with a secondary antibody conjugated to horseradish peroxidase (1:1,000; Ab7076; BD Pharmingen; BD Biosciences, San Jose, CA, USA) at room temperature for 3 h. Immunolabeling was visualized using an enhanced chemiluminescence system (GE Healthcare Life Sciences, Little Chalfont, UK).
Statistical analysis
Data are presented as the means ± standard deviation of at least three independent experiments from separately treated and transfected cultures. Statistical comparisons were made by one-way analysis of variance and the Student-Newmnan-Keuls test was applied. P<0.05 was considered to indicate a statistically significant difference.
Results
Isolation and purification of rat BMSCs
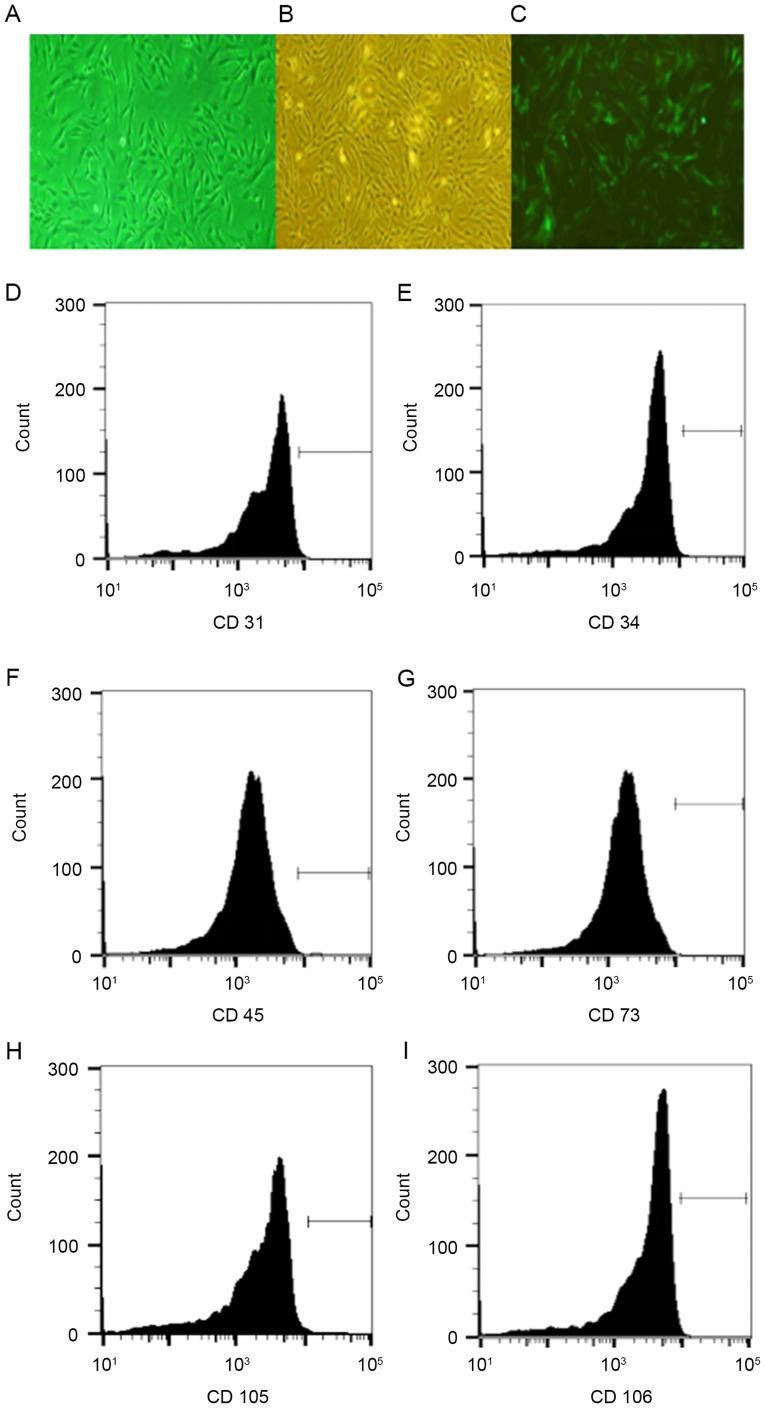
Rat BMSCs were isolated via density gradient centrifugation and an adherence screening approach, followed by confirmation using flow cytometry. Results indicated that the purity of the isolated Wistar rat BMSCs was 98%, and the transfection efficiency of lentiviral vector pCDH-CMV-MCS-EF1-copEGFP was 90% (Fig. 1A-C). Surface markers of BMSCs (CD31, CD34, CD45, CD73, CD105 and CD106) were successfully detected by flow cytometry, which further confirmed that the isolated cells were BMSCs (Fig. 1D-I).
Figure 1.
Isolation and purification of rat BMSCs. Representative images of (A) isolated rat BMSCs, (B) purified BMSCs (C) enhanced green fluorescent protein-positive BMSCs (magnification, ×40). (D) BMSCs with CD31 staining; (E) BMSCs with CD34 staining; (F) BMSCs with CD45 staining; (G) BMSCs with CD73 staining; (H) BMSCs with CD105 staining; and (I) BMSCs with CD106 staining. BMSCs, bone marrow stem cells; EGFP; CD, cluster of differentiation.
Successful construction of lentiviral vectors carrying the Smad7 gene and Smad7-EGFP-BMSCs
Total RNA was extracted from the cerebral cortex, and reverse transcription/amplification was performed to obtain Smad7 DNA fragments. Upon cleaving the lentiviral vector pCDH-CMV-MCS-EF1-copEGFP with restriction enzymes (EcoRI and BamHIs), the Smad7 DNA fragment was then inserted and co-expressed with EGFP. The resultant recombinant plasmid was subjected to DNA sequencing for further confirmation (Fig. 2A). Plasmids carrying the Smad7 gene were subsequently transfected into BMSCs, and the transfection efficiency was revealed to be ~90% when observed under a fluorescence microscope (Fig. 2B). These results indicated that the lentiviral vectors carrying the Smad7 gene and Smad7-EGFP-BMSCs were successfully constructed.
Figure 2.
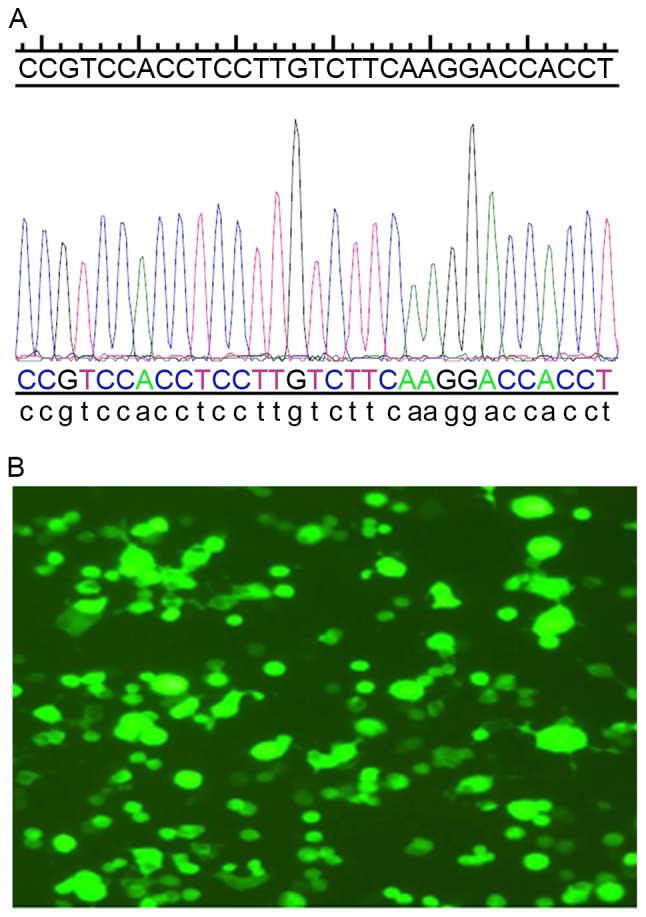
Construction of lentiviral vectors carrying the Smad7 gene. (A) Correct coding sequence of Drosophila mothers against decapentaplegic 7 as confirmed by DNA sequencing. (B) Lentiviral titers observed under fluorescence microscope (magnification, ×400).
Smad7-EGFP-BMSCs affect the expression of TGF-β1, Smad7, Col I, Col III, LN and HA in HSCs
Upon co-cultivation with Smad7-EGFP-BMSC, Smad7 and TGF-β1 levels in the culture medium were detected using ELISA. The results suggested that, in the Smad7-EGFP-BMSCs + HSCs group, the Smad7 level was significantly increased but the TGF-β1 level was significantly reduced compared with the other groups (P<0.05; Table II). Furthermore, the immunoassay results indicated that in the Smad7-EGFP-BMSCs + HSCs group, Col I, Col III, LN and HA levels were significantly decreased when compared with the other groups (P<0.05; Table III). These results suggest that Smad7-EGFP-BMSCs affects the expression of TGF-β1 and Smad7, which contributes to HSC fibrosis.
Table II.
Levels of TGF-β1 and Smad7 in the co-culture medium of different groups (pg/ml).
| Group | TGF-β1 | Smad7 |
|---|---|---|
| Smad7-EGFP-BMSCs + HSCs | 17.91±1.23a | 457.51±12.12a |
| BMSCs + HSCs | 83.71±5.62 | 222.82±18.64 |
| PBS + HSCs | 174.62±11.23 | 38.33±2.65 |
| Smad7 + HSCs | 56.91±1.78 | 300.21±9.68 |
P<0.05 vs. all other groups. TGF-β1, transforming growth factor-β1; Smad7, Drosophila mothers against decapentaplegic 7; EGFP, enhanced green fluorescent protein; BMSCs, bone marrow stem cells; HSCs, hepatic stellate cells.
Table III.
Levels of Col I, Col III, LN and HA in the co-culture medium of different groups (µg/ml).
| Group | Col I | Col III | LN | HA |
|---|---|---|---|---|
| Smad7-EGFP-BMSCs + HSCs | 32.11±4.36a | 40.9±5.98a | 23.89±2.65a | 13.67±0.96a |
| BMSCs + HSCs | 102.1±2.96 | 98.76±5.69 | 90.67±7.63 | 56.78±6.02 |
| PBS + HSCs | 198.67±7.54 | 234.56±13.23 | 309.78±12.6 | 114.5±5.873 |
| Smad7 + HSCs | 289.92±5.69 | 179.65±9.86 | 213.22±10.36 | 118.9±7.45 |
P<0.05 vs. all other groups. Col, collagen; LN, laminin; HA, hyaluronic acid; Smad7, Drosophila mothers against decapentaplegic 7; EGFP, enhanced green fluorescent protein; BMSCs, bone marrow stem cells; HSCs, hepatic stellate cells.
Smad7-EGFP-BMSC promotes the early apoptosis of HSCs
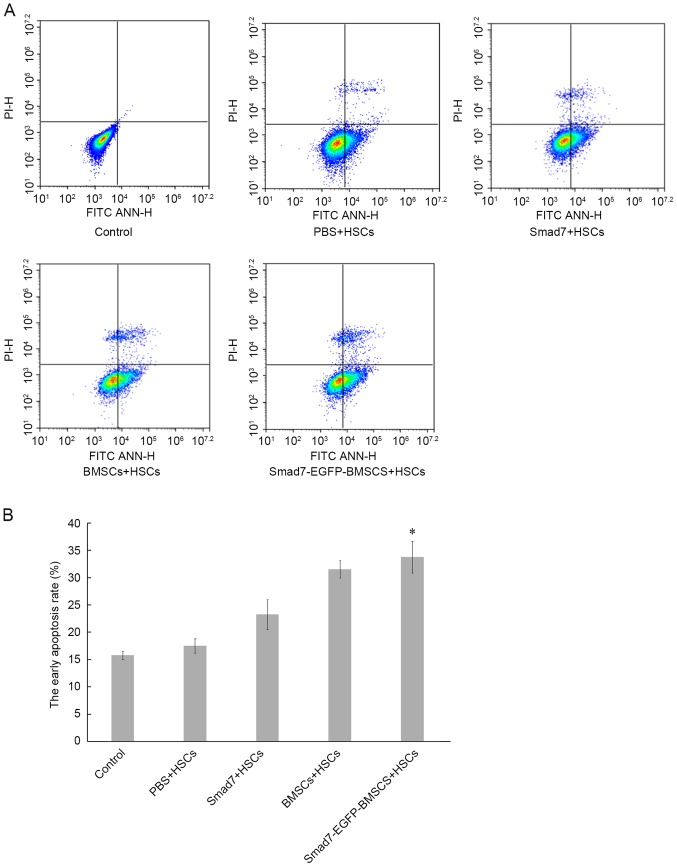
Following co-cultivation with Smad7-EGFP-BMSC, the early apoptotic rate of HSCs was detected by flow cytometry. The results indicated that the early apoptotic rate of HSCs in the Smad7-EGFP-BMSCs + HSCs group was significantly increased compared with the control, PBS + HSCs and Smad7 + HSCs groups (P<0.05; Fig. 3). Flow cytometry results indicated that co-cultivation with Smad7-EGFP-BMSCs promotes early apoptosis of HSCs and inhibits fibrosis.
Figure 3.
Early apoptosis of HSCs, as determined by flow cytometry. (A) ANN-V/PI staining of HSCs cells in different groups. (B) Histogram showing apoptotic HSC rates in different groups. *P<0.05 vs. BMSCs + HSCs, PBS + HSCs and Smad7 + HSCs groups.
Smad7-EGFP-BMSCs affect the expression of Smad7, Smad2, Smad3, TβRI, MMP-1, α-SMA, Col I, Col III, TGF-β1 and TIMP-1 mRNA in HSCs
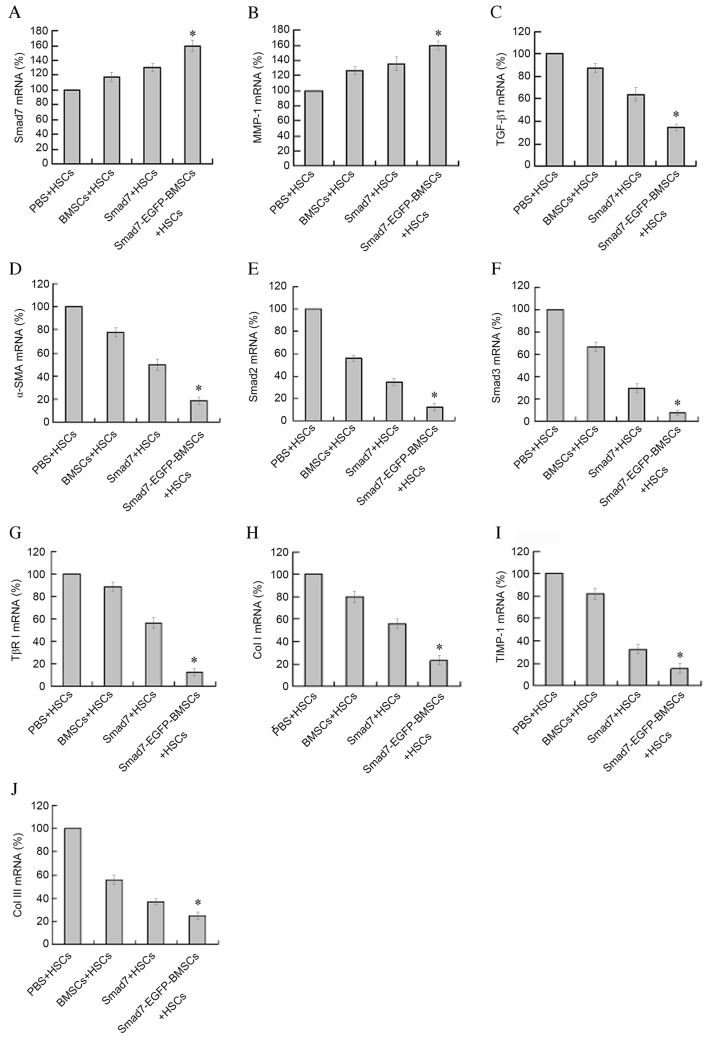
Following co-cultivation with Smad7-EGFP-BMSC, the mRNA levels of Smad7 and MMP-1 were significantly increased when compared with BMSCs + HSCs, PBS + HSCs and Smad7 + HSCs groups (P<0.05; Fig. 4A and B). However, the mRNA levels of TGF-β1, α-SMA, Smad2, Smad3, TβRI, Col I, TIMP-1 and Col III were decreased when compared with BMSCs + HSCs, PBS + HSCs and Smad7 + HSCs groups. (P<0.01; Fig. 4C-J). These results suggest that Smad7-EGFP-BMSC affects the mRNA expression levels of Smad7, Smad2, Smad3, TβRI, MMP-1, α-SMA, Col I, Col III, TGF-β1 and TIMP-1 in HSCs, which are essential in HSC fibrosis (13).
Figure 4.
mRNA expression detected by reverse transcription-quantitative polymerase chain reaction. mRNA levels of (A) Smad7, (B) MMP-1, (C) TGF-β1, (D) α-SMA, (E) Smad2, (F) Smad3, (G) TβRI (H) Col I, (I) TIMP-1 and (J) Col III. *P<0.05 vs. BMSCs + HSCs, PBS + HSCs and Smad7 + HSCs groups.
Smad7-EGFP-BMSCs affect the protein expression of Smad7, Smad2, Smad3, TβRI, MMP-1, α-SMA, Col I, Col III, TGF-β1 and TIMP-1 in HSCs
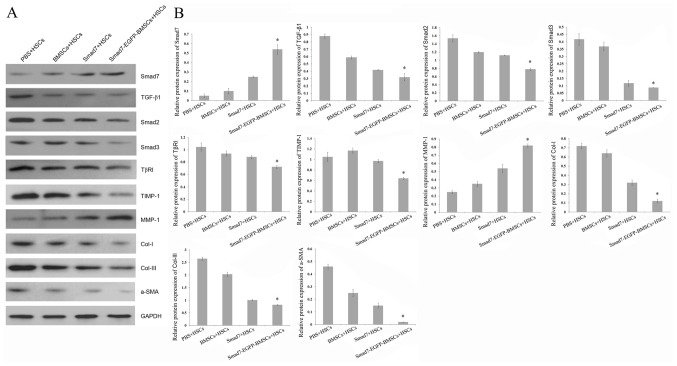
Western blotting revealed that, following co-cultivation with Smad7-EGFP-BMSC, the protein levels of Smad7 and MMP-1 were significantly increased when compared with BMSCs + HSCs, PBS + HSCs and Smad7 + HSCs groups. (P<0.05; Fig. 5). However, the protein levels of TGF-β1, α-SMA, Smad2, Smad3, TβRI, Col I, TIMP-1 and Col III were significantly decreased following co-cultivation with Smad7-EGFP-BMSC when compared with BMSCs + HSCs, PBS + HSCs and Smad7 + HSCs group (P<0.05; Fig. 5), indicating that Smad7-EGFP-BMSC affects these protein expressions, contributing to HSCs fibrosis.
Figure 5.
Protein expression levels, as detected by western blot analysis. (A) Representative image of western blots. (B) Relative quantitative of protein expression. *P<0.05 vs. BMSCs + HSCs, PBS + HSCs and Smad7 + HSCs groups.
Discussion
BMSCs were first isolated by Friedenstein et al (14), and since then have been considered as the progenitor cells for the skeletal tissues. BMSCs are clonogenic, fibroblastic in shape, and are capable of differentiating into multiple lineages, such as osteoblasts, chondrocytes, adipocytes, and hematopoiesis-supportive stroma (15). Furthermore, BMSCs are able to migrate and attach to the damaged area, and help to repair the damaged organ (6). In the present study, BMSCs were isolated and purified from rat bone marrow cells and identified by detecting BMSC surface markers (CD31, CD34, CD45, CD73, CD105 and CD106) using flow cytometry. The results indicated that rat BMSCs were successfully isolated.
Smad7 was initially identified as an inhibitor of TGF-β due mainly to its ability to bind TβRI and prevent TGF-β-associated Smad signaling (16). Recently it has been demonstrated that Smad7 interacts with other intracellular proteins and regulates TGF-β-independent signaling pathways, thus making a valid contribution to neoplastic processes in various organs (17). In the present study, lentiviral vectors carrying the Smad7 gene were successfully constructed and transfected into BMSCs. The results suggest that Smad7-EGFP-BMSCs affect TGF-β1 and Smad7 expression in HSCs following co-cultivation. Since liver fibrosis is characterized by the excessive deposition of ECM, serum markers representing ECM components are widely employed to assess the development of liver fibrosis. HA, LN, Col I, and Col IV are major serum markers of liver fibrosis (18). In the present study, Smad7-EGFP-BMSCs significantly induced early apoptosis of HSCs and suppressed the expression levels of Col I, Col III, LN and HA, suggesting that Smad7 may inhibit HSC fibrosis.
As liver fibrosis is characterized by the excessive deposition of ECM, the key step is fibrosis of HSCs. Activation of HSCs transforms them into myofibroblasts and fibroblasts, resulting in increased ECM synthesis and α-SMA expression, thus accelerating liver fibrosis (19). TGF-β1 is a key factor in the development of liver fibrosis and also promotes ECM formation (7). Smad7 is an essential negative regulator in the TGF-β1/Smad signaling pathway (20). TGF-β1 is able to activate HSCs via the TGF-β1/Smad signaling pathway, upregulate the expression of Col I, Col III, Col IV, LN and fibronectin, and promote ECM synthesis and deposition. Furthermore TGF-β1 is also able to upregulate the expression of TIMP-1 by suppressing the synthesis of MMP-1, reducing ECM degradation and by increasing ECM deposition in the damaged tissue or organ (21). TGF-β1 activates HSC with a positive feedback effect, resulting in increased TGF-β1 expression and rapid progression of liver fibrosis (22). In the present study, co-cultivation with Smad7-EGFP-BMSC resulted in a significant increase in mRNA and protein levels of Smad7 and MMP-1, whereas the mRNA and protein levels of TGF-β1, α-SMA, Smad2, Smad3, TβRI, Col I, TIMP-1 and Col III were decreased. A potential reason for this may be Smad7 and Smad2/3 competing to bind TβRI, triggering an upregulation in the expression of the downstream effector, MMP-1, and downregulating the expression levels of α-SMA, Col I, TIMP-1 and Col III. According to Pan et al (23), the Dlk1/Notch signaling pathway may also be involved in the process by which BMSCs inhibit HSC fibrosis.
The results of the present study indicate that rat BMSCs with Smad7 overexpression may inhibit the fibrosis of HSCs by regulating the TGF-β1/Smad signaling pathway. Hence, Smad7-EGFP-BMSC may be used for the treatment of liver cirrhosis, as it has the potential to transform into liver cells, and also may suppress liver fibrosis by releasing Smad7 and inhibiting the function of TGF-β1. To further evaluate these therapeutic effects, an in vivo study should be conducted on a rat liver cirrhosis model with direct Smad7-EGFP-BMSC transplantation to provide further insights into the future treatment of liver cirrhosis.
Acknowledgements
The present study was supported by the Shenzhen Municipal Science and Technology Innovation Committee Project: Study on the Treatment Effect and Mechanism of Mesenchymal Stem Cells Modified by Smad7 Gene in Liver Cirrhosis (grant no. JCYJ 20140416122812003).
Glossary
Abbreviations
- MSCs
mesenchymal stem cells
- ECM
extracellular matrix
- HSCs
hepatic stellate cells
- TGF-β1
transforming growth factor-β1
- Smad7
Drosophila mothers against decapentaplegic 7
- MMP-1
matrix metalloproteinase 1
- α-SMA
α-smooth muscle actin
- TIMP-1
tissue inhibitors of metalloproteinase-1
- HA
hyaluronic acid
- LN
laminin
References
- 1.Merion RM. Current status and future of liver transplantation. Semin Liver Dis. 2010;30:411–421. doi: 10.1055/s-0030-1267541. [DOI] [PubMed] [Google Scholar]
- 2.Ren HY, ZHao QJ, Xing W, Yang SG, Lu SH, Ren Q, Zhang L, Han ZC. Differention of human umbilical cord derived mesenchylmal stem cells into low immunogenic and functional hepatocye-like cells in vitro. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2010;32:190–194. doi: 10.3881/j.issn.1000-503X.2010.02.013. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 3.Li GC, Zhang HW, Zhao QC, Sun LI, Yang JJ, Hong L, Feng F, Cai L. Mesenchymal stem cells promote tumor angiogenesis via the action of transforming growth factor β1. Oncol Lett. 2016;11:1089–1094. doi: 10.3892/ol.2015.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dooley S, ten Dijke P. TGF-β in progression of liver disease. Cell Tissue Res. 2012;347:245–256. doi: 10.1007/s00441-011-1246-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Zhang L, Lei R, Shen Y, Shen H, Wu Z, Xu J. Effects of blocking two sites of transforming growth factor-β/Smads signaling on the formation of scar-related proteins in human skin fibroblasts. Zhonghua Shao Shang Za Zhi. 2015;31:372–377. (In Chinese) [PubMed] [Google Scholar]
- 6.Yin M, Wu L. Effect of mast cell chymase on activation, proliferation and transdifferentiation of hepatic stellate cells. Hepatogastroenterology. 2015;62:1007–10010. [PubMed] [Google Scholar]
- 7.Wang SL, Wu HM, He CZ, Yang L, Gao HJ, Yang CQ. Aldosterone and TGF-β1 synergistically increase PAI-1 expression in hepatic stellate cells of rats. Int J Clin Exp Pathol. 2015;8:9845–9853. [PMC free article] [PubMed] [Google Scholar]
- 8.Pan Q, Wang YQ, Li GM, Duan XY, Fan JG. Fuzheng huayu recipe ameliorates liver fibrosis by restoring balance between Epithelial-to-Mesenchymal transition and Mesenchymal-to-Epithelial transition in hepatic stellate cells. Biomed Res Int. 2015;2015:935903. doi: 10.1155/2015/935903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ravichandran R, Venugopal JR, Sundarrajan S, Mukherjee S, Ramakrishna S. Cardiogenic differentiation of mesenchymal stem cells on elastomeric poly (glycerol sebacate)/collagen core/shell fibers. World J Cardiol. 2013;5:28–41. doi: 10.4330/wjc.v5.i3.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu X, Liu Z, Deng W, Zhang Z, Liu Y, Wei L, Zhang Y, Zhou L, Wang Y. Derivation and characterization of sheep bone marrow-derived mesenchymal stem cells induced with telomerase reverse transcriptase. Saudi J Biol Sci. 2017;24:519–525. doi: 10.1016/j.sjbs.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su DN, Wu SP. Effect of Smad7 gene modified BMSCs to TGF-B signal conduction in hepatic stellate cells. Chin J Immunol. 2016;32:692–696. [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Chen S, Xu L, Lin N, Pan W, Hu K, Xu R. Activation of Notch1 signaling by marrow-derived mesenchymal stem cells through cell-cell contact inhibits proliferation of hepatic stellate cells. Life Sci. 2011;89:975–981. doi: 10.1016/j.lfs.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 14.Friedenstein AJ, Latzinik NV, Gorskaya YuF, Luria EA, Moskvina IL. Bone marrow stromal colony formation requires stimulation by haemopoietic cells. Bone Miner. 1992;18:199–213. doi: 10.1016/0169-6009(92)90807-P. [DOI] [PubMed] [Google Scholar]
- 15.Li X, Yuan Z, Wei X, Li H, Zhao G, Miao J, Wu D, Liu B, Cao S, An D, et al. Application potential of bone marrow mesenchymal stem cell (BMSCs) based tissue-engineering for spinal cord defect repair in rat fetuses with spina bifida aperta. J Mater Sci Mater Med. 2016;27:77. doi: 10.1007/s10856-016-5684-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Argentou N, Germanidis G, Hytiroglou P, Apostolou E, Vassiliadis T, Patsiaoura K, Sideras P, Germenis AE, Speletas M. TGF-β signaling is activated in patients with chronic HBV infection and repressed by SMAD7 overexpression after successful antiviral treatment. Inflamm Res. 2016;65:355–365. doi: 10.1007/s00011-016-0921-6. [DOI] [PubMed] [Google Scholar]
- 17.Zhao Z, Xi H, Xu D, Li C. Transforming growth factor β receptor signaling restrains growth of pancreatic carcinoma cells. Tumour Biol. 2015;36:7711–7716. doi: 10.1007/s13277-015-3466-3. [DOI] [PubMed] [Google Scholar]
- 18.Zhu C, Qi X, Li H, Peng Y, Dai J, Chen J, Xia C, Hou Y, Zhang W, Guo X. Correlation of serum liver fibrosis markers with severity of liver dysfunction in liver cirrhosis: A retrospective cross-sectional study. Int J Clin Exp Med. 2015;8:5989–5998. [PMC free article] [PubMed] [Google Scholar]
- 19.Chen YX, Weng ZH, Zhang SL. Notch3 regulates the activation of hepatic stellate cells. World J Gastroenterol. 2012;18:1397–1403. doi: 10.3748/wjg.v18.i12.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seki N, Toh U, Kawaguchi K, Ninomiya M, Koketsu M, Watanabe K, Aoki M, Fujii T, Nakamura A, Akagi Y, et al. Tricin inhibits proliferation of human hepatic stellate cells in vitro by blocking tyrosine phosphorylationof PDGF receptor and its signaling pathways. J Cell Biochem. 2012;113:2346–2355. doi: 10.1002/jcb.24107. [DOI] [PubMed] [Google Scholar]
- 21.Xie WL, Jiang R, Shen XL, Chen ZY, Deng XM. Diosgenin attenuates hepatic stellate cell activation through transforming growth factor-β/Smad signaling pathway. Int J Clin Exp Med. 2015;8:20323–20329. [PMC free article] [PubMed] [Google Scholar]
- 22.You SP, Ma L, Zhao J, Zhang SL, Liu T. Phenylethanol glycosides from cistanche tubulosa suppress hepatic stellate cell activation and block the conduction of signaling pathways in TGF-β1/smad as potential anti-hepatic fibrosis agents. Molecules. 2016;21:102. doi: 10.3390/molecules21010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan RL, Wang P, Xiang LX, Shao JZ. Delta-like 1 serves as a new target and contributor to liver fibrosis down-regulated by mesenchymal stem cell transplantation. J Biol Chem. 2011;286:12340–12348. doi: 10.1074/jbc.M110.194498. [DOI] [PMC free article] [PubMed] [Google Scholar]