Abstract
MicroRNAs (miRNAs) are 22-nucleotide single-stranded RNAs which regulate gene expression by targeting 3′ untranslated regions. Previous studies have suggested that miRNAs may be used as markers for investigating the molecular regulation of gene expression. In the present study, miRNA and mRNA expression profiles were investigated using a massively parallel next generation sequencing technique to compare herpes simplex virus (HSV)2-infected (n=3) and healthy (n=3) epithelial tissues from guinea pigs. Total RNA was isolated and RNA sequencing was performed using a HiSeq 2000 sequencing system. Differential expression of miRNA and mRNA was analyzed using two-tailed t-tests. A negative correlation was detected between the miRNAs and their predicted target genes. Following infection with HSV2, 205 and 159 miRNAs were demonstrated to be upregulated and downregulated, respectively. These differentially expressed miRNAs were associated with cellular and metabolic processes, biological regulation, response to stimuli and cellular components of the immune system, as determined by functional gene ontology analysis. Following HSV2 infection, 6 upregulated miRNAs including miR-592, miR-1245b-5p, miR-150, miR-342-5p, miR-1245b-3p and miR-124 were demonstrated to participate in the toll-like receptor (TLR) pathway by targeting related genes. These results suggested that the downregulated genes were associated with the TLR pathway after infection with HSV2. The results of reverse transcription-quantitative polymerase chain reaction analysis were consistent with RNA sequencing, indicating that the increased expression of these miRNAs downregulated the TLR pathway-associated genes, which may mediate the progression of HSV2-induced genital herpes.
Keywords: herpes simplex virus, genital herpes, micro RNA, sequence analysis, toll-like receptors
Introduction
Genital herpes is a common infection caused by herpes simplex virus (HSV), which is prevalent worldwide (1). As a ubiquitous enveloped virus, HSV is capable of transmitting through mucosal membranes and migrating into nerve tissues to induce prolonged infection (2). Two types of HSV have been described, including HSV-1 and HSV2, which are found in the trigeminal and lumbosacral ganglia, respectively, and have been demonstrated to infect orofacial areas and the genital tract (2). The worldwide prevalence of HSV2 has significantly increased since the late 1970s and incidence rates amongst adults are ~30% in the US (3). HSV infections are predominantly treated with antiviral drugs, including acyclovir, valacycolovir, famciclovir and penciclovir (4,5). Previous studies have suggested that both humoral and cellular immune responses are activated in response to HSV infection (6–8); and the recurrence of HSV has been associated with the depression of cellular immune responses in a guinea pig model of HSV (9,10), as demonstrated by HSV-specific T cells following HSV challenge (9). Furthermore, HSV-specific Th1 type CD41 T cells have also been demonstrated to contribute to immune defences against HSV infection (11,12); although the levels of the HSV-specific antibodies are not sufficient to prevent HSV-induced central nervous system damage (13).
Transcriptome studies have been used to investigate pathogen infection, and previous studies have demonstrated that noncoding microRNA (miRNA) have an important role in the immune response to pathogen infection (14,15). miRNAs, which are composed of ~22 nucleotides, regulate post-transcriptional gene silencing by selectively targeting mRNA in the 3′ untranslated region. Since a single miRNA is capable of regulating various genes, and it has been estimated that >60% of protein-coding genes are regulated by miRNA (16), miRNAs are regarded as potential markers for the investigation of coordinated gene expression.
To the best of our knowledge, the present study was the first to investigate whether miRNA expression profiles differ between normal and HSV2-infected tissues. Next-generation sequencing (NGS) was used to examine mRNA and miRNA expression profiles in guinea pigs following infection with HSV2. NGS provides unprecedented analysis of the expression levels of miRNAs and mRNAs. Compared with chromatin immunoprecipitation, the NGS platform facilitates the comparison of expression levels differences and can be used to locate novel miRNAs. Therefore, in the present study, NGS was used to elucidate miRNA-mRNA regulation following HSV2 infection in guinea pigs.
Thus, the current study aimed to investigate the gene expression and miRNA changes in HSV2-infected tissues, and to develop novel diagnostic criteria for HSV2 infection.
Materials and methods
Animals and virus
Animal experimental procedures were approved by the Animal Ethical Committee of Hunan University of Chinese Medicine (Changsha, China). Female Dunkin-Hartley guinea pigs (n=3 per group; age, 4 weeks; weight, 326.21±3.69 g; Jingfeng Laboratory Animal Breeding Co., Ltd., Jinan, China) were maintained in the Animal Culture Center at Hunan University of Chinese Medicine. The guinea pigs were maintained in 23°C, 50% humidity, with a 12-h dark:light cycle, and fed commercial diets at dawn and dusk from Sangon Biotech Co., Ltd. (Shanghai, China). Water was supplied using sipper tubes (Sangon Biotech Co., Ltd.). HSV2 strains were isolated from patients at the Hunan University of Chinese Medicine Affiliated Hospital. DNA sequencing was used to determine the virus stain as HSV2 SAV.
Construction of a guinea pig model of genital herpes
Prior to modeling, the guinea-pig vulva was cleaned with physiological saline. A total of 0.1 ml HSV-2 SAV (1%) was injected into the vagina (depth, 3–4 cm) of each guinea pig. Successful establishment of the guinea pig model of genital herpes via infection with HSV2 was confirmed after 7 days by polymerase chain reaction (PCR) and clinical characterization. Disease course was scored from 0–6 using a composite scale based on the clinical characterization of vaginitis, as follows: 0, no symptom of infection; 1, small vesicles on the vulva; 2, 10–50% of the vulva covered by vesicles; 3, 50–100% of the vulva covered by vesicles; 4, 10–50% of the vulva covered by small ulcers; 5, 50–100% of the vulva covered by severe ulcers; and 6, hind limb paralysis or mortality (17). Guinea pigs scoring 4 were used in the present study. Epithelial tissues were excised from the vulvae of guinea pigs in the control and infection groups (n=3 each) to perform the following experiments.
RNA extraction
Samples were homogenized by using a mortar and pestle with liquid nitrogen. Total RNA was extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's protocol and was subsequently digested with DNase I (Qiagen, Inc., Valencia, CA, USA) to remove any remaining DNA. RNA integrity was assessed using an Agilent BioAnalyzer 2100 (Agilent Technologies, Inc., Santa Clara, CA, USA).
RNA-sequencing (seq) and data analysis
Library preparation and sequencing was performed by the Beijing Genomics Institute (BGI; Shenzhen, China). Six libraries were constructed and RNA-seq was performed using an Illumina Hiseq 2000 sequencing system (Illumina, Inc., San Diego, CA, USA). Raw data were filtered in order to remove low quality base and adapter sequences. Reads were mapped to the human genome (ftp://ftp.ensembl.org/pub/release-84/fasta/homo_sapiens/dna/) using TopHat and Cufflinks software (version 2.2.1; http://cole-trapnell-lab.github.io/cufflinks/). In order to compare differential expression levels, the ‘significance of digital gene expression profiles’ method was used, as developed by the BGI (18). The false discovery rate (FDR) was controlled by adjusting the P-value with a Benjamini-Hochberg algorithm (19). Significant differences between the infection and control groups were assessed by Student's t-test or the paired t-test. Fold changes in expression levels were calculated using fragments per kilobase of exon model per million mapped reads using a DEGseq package in R (version 2.15.1; www.r-project.org).
Small RNA sequence and data analysis
Small RNAs were isolated and miRNA libraries were prepared using a TruSeq Small RNA Sample Prep kit (version 2; Illumina, Inc.), prior to sequencing on a HiSeq 2000 sequencer. Illumina HiSeq Control Software (version 2.0.12), Real-Time Analysis (version 1.12.4.2), and Consensus Assessment of Sequence And Variation (version 1.8.2; all Illumnia, Inc.) software were employed to process the raw data. Differential expression levels of miRNA were calculated and standardized to reads per million, as follows: Counts/total count of each sample × 1 million.
Interaction between mRNA and miRNA
Associations between the expression levels of mRNA and miRNA were estimated by correlation coefficient R analysis. miRNA-mRNA pairs were identified by these correlations and mRNA-miRNA interaction was analyzed using miRBase targets from the Wellcome Trust Sanger Institute (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/).
Functional gene ontology (GO) analysis
GO term enrichment was performed for the 573 target genes of the differentially expressed miRNAs using Blast2go software (version 3.0; www.blast2go.com). Default Blast2go parameters were used and only GO terms with P<0.001 were included in the present study.
Identification of miRNA-mRNA interaction with the toll-like receptors (TLRs) pathway
Associations between the miRNAs and the TLR pathway were analyzed using CluePedia (version 1.1.7; http://apps.cytoscape.org/apps/cluepedia) and miRBase. miRNA-mRNA pairs among the TLR pathway-related genes were investigated and miRNA-mRNA connections were plotted using R (version 2.15.1; www.r-project.org).
Reverse transcription-quantitative PCR (RT-qPCR)
Vulval epithelial samples were harvested from healthy and HSV2-infected guinea pigs (n=3 per group). Following total RNA isolation, cDNA were synthesized using a PrimeScript RT reagent kit with gDNA Eraser (Takara Biotechnology Co., Ltd., Dalian, China). Taqman miRNAs and mRNAs primers were purchased from Applied Biosystems (Thermo Fisher Scientific, Inc.). qPCR was performed on an ABI 7500 Real-Time PCR system (Applied Biosystems) to a total reaction volume of 50 µl, including SYBR Premix Ex Taq, according to the manufacturer's protocol. The reaction mixture contained TaqMan PCR, 5.5 µl SYBR Premix Ex Taq (Applied Biosystems), 0.5 µl primers and probes mix (Applied Biosystems), and 4.5 µl cDNA (1/20 dilution of cDNA). PCR thermal cycling was performed as follows: 1 cycle of 94°C for 2 min, followed by 40 cycles of 94°C for 30 sec, 60°C for 30 sec and 72°C for 45 sec, and 72°C for 5 min for 1 cycle. The primers were as follows: Forward, 5′-GCATGTTGTTTTCCCCACAAA-3′ and reverse, 5′-CTGCAGTGCTGACTGAAACATTC-3′ for TLR1; forward, 5′-GACTAGCAACTCCTTTGCCTCAGT-3′ and reverse, 5′-CCACCCTTCGGAGCATCA-3′ for TLR3; forward, 5′-CAGCTTTCCTGGCAATTGACT-3′ and reverse, 5′-CGACATCTTCCCTGGATGCT-3′ for TLR5; forward, 5′-GGCAAAGTGGGCGAGATG-3′ and reverse, 5′-GCTCTGCGTTTTGTCGAAGAC-3 for TLR9; forward, 5′-CACTTGGCACGACACCTACAA-3′ and reverse, 5′-CCTGGGCCAGCATTCTCA-3′ for TRAF6; forward, 5′-CAGGACGCCATAGACCACTCA-3′ and reverse, 5′-GGCGACAGTCGAAGTTGGA-3′ for TRIF; and forward, 5′-GCAACCCGAGACAAGATGGT-3′ and reverse, 5′-GCGTCCAATACGGCCAAAT-3′ for GAPDH. Dissociation curve analysis was used to verify the generation of single products. GAPDH was used as a housekeeping/reference gene, and all reactions were repeated three times. Data were analyzed using the ABI 7500 Prism sequence detection software (Applied Biosystems) and the relative expression was calculated using the 2−ΔΔCq method.
Statistical analysis
Statistical analysis was performed using SPSS statistical software (version 17.0; SPSS, Inc., Chicago, IL, USA The significant difference between the two groups was analyzed by one-way analysis of variance using paired t-test. Data are expressed as mean ± standard deviation. P<0.05 was considered as significant difference.
Results
mRNA and miRNA expression profiles
In the present study, three healthy and three HSV2-infected guinea pigs were investigated. The workflow of the mRNA and miRNA analyses employed in the present study is shown in Fig. 1. A total of 424 miRNAs and 9,336 genes were sequenced and 364 miRNAs and 4,130 genes were identified following data filtration and genome annotation (Fig. 2). A total of 105 miRNAs were upregulated and 141 were downregulated; whereas 798 genes were upregulated and 503 were downregulated in the HSV2-infected group, as compared with the control group. The differential expressions with the greatest degrees are listed in Table I.
Figure 1.
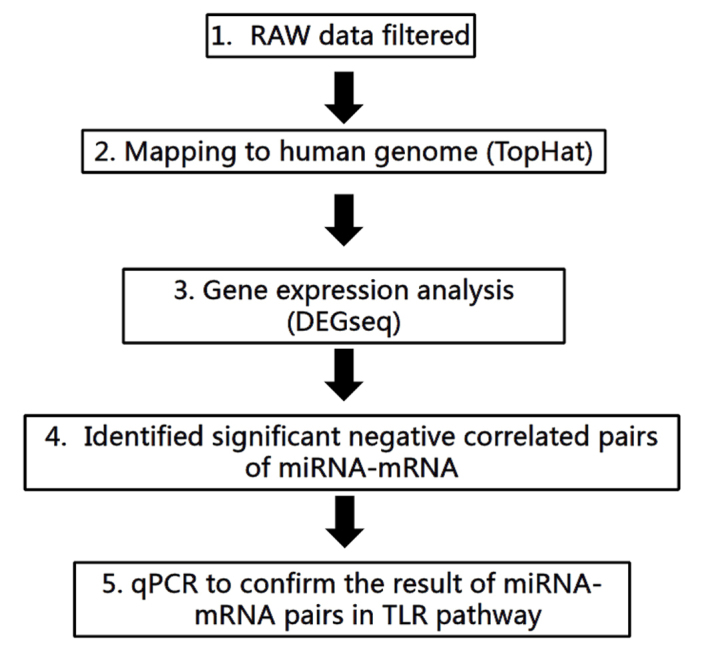
Workflow of the miRNA and mRNA analyses in the present study.
Figure 2.
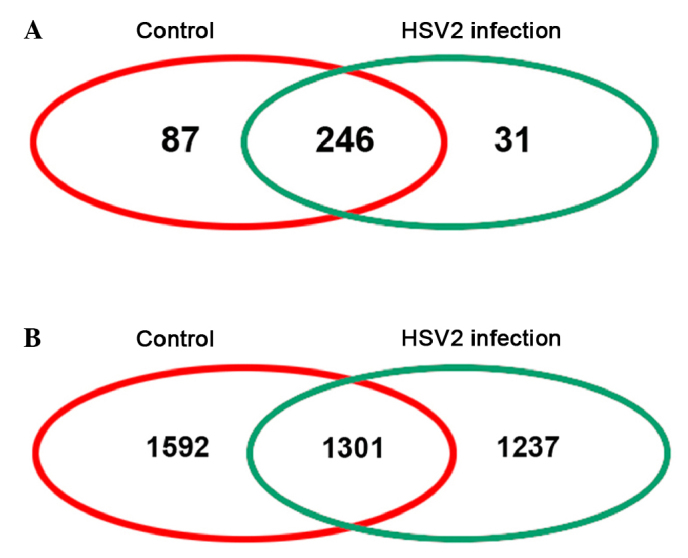
Venn plots of sequenced (A) miRNA and (B) mRNA from healthy control and herpes simplex virus 2-infected epithelial tissues harvested from guinea pigs.
Table I.
Top five upregulated and downregulated miRNAs and mRNA following infection with herpes simplex virus 2.
| Names | Log2 (infection/control) | P-value | |
|---|---|---|---|
| miRNA | |||
| Upregulated | |||
| miR-325 | 1.603 | 0.001 | |
| miR-401 | 1.618 | 0.010 | |
| miR-199 | 2.474 | 0.003 | |
| miR-195 | 2.893 | 0.001 | |
| miR-186 | 3.363 | 0.009 | |
| Downregulated | |||
| miR-153 | −3.381 | 0.003 | |
| miR-433 | −3.113 | 0.006 | |
| miR-131 | −2.844 | 0.001 | |
| miR-140-5p | −2.456 | 0.007 | |
| miR-427 | −2.127 | 0.002 | |
| mRNA | |||
| Upregulated | |||
| Dars | 11.946 | 0.004 | |
| PHF8 | 5.085 | 0.001 | |
| Mdk | 4.324 | 0.001 | |
| Brca1 | 3.773 | 0.006 | |
| PPP6C | 1.761 | 0.001 | |
| Downregulated | |||
| CCLC | −3.525 | 0.010 | |
| Acoxl | −4.662 | 0.003 | |
| TGF-β1 | −11.552 | 0.003 | |
| SFXN5 | −8.620 | 0.010 | |
| BCs | −3.662 | 0.001 |
Dars, Aspartyl-tRNA synthetase; PHF8, PHD Finger Protein 8; Mdk, midkine (neurite growth-promoting factor 2); Brca1, breast cancer susceptibility gene 1; PPP6C, protein phosphatase 6 catalytic subunit; CCLC, chemokine ligands gene; Acoxl, Acyl-CoA oxidase-like; TGF-β1, tumor growth factor-β1; SFXN5: sideroflexin 5; BCs, nematode protein-coding gene bcs.
GO analysis
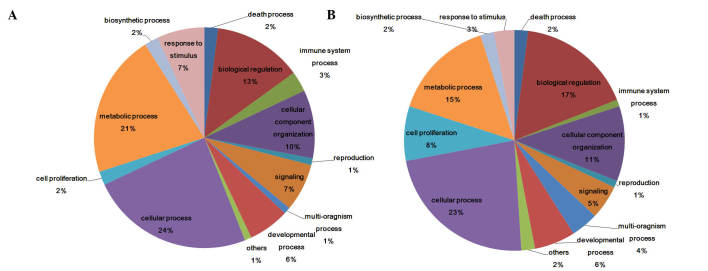
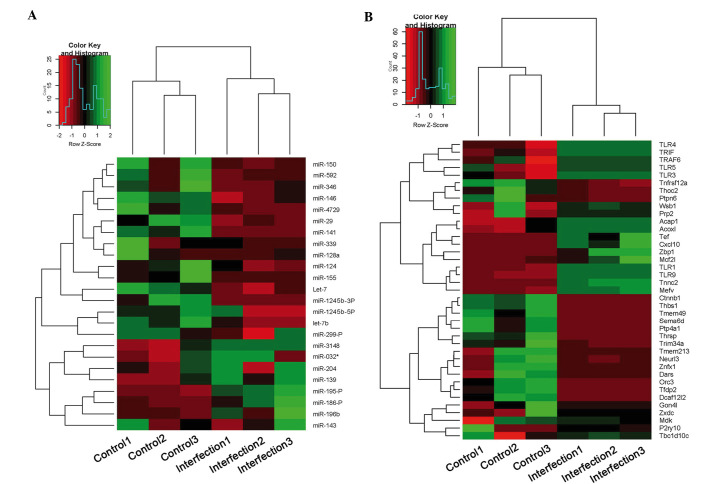
Differentially expressed genes and miRNAs were assigned GO terms (Fig. 3). The most relevant GO terms detected in the present study were ‘cellular process’, ‘metabolic process’, ‘biological regulation’ and ‘signalling’; whereas several differentially expressed genes were assigned into ‘response to stimulus’ and ‘immune system process’ terms. Similar results were detected in miRNAs and mRNAs (Fig. 3A and B). Although the most abundant GO terms for the miRNAs assessed in the present study were ‘cellular process’ (24%), ‘metabolic process’ (21%) and ‘biological regulation’ (13%), the ‘response to stimulus’ and ‘immune system process’ terms were also associated with several differentially expressed miRNAs (7 and 3%, respectively; Fig. 3B). The ‘signalling’ term was also high and different between miRNAs and mRNAs. Expression levels of immune-related miRNA and mRNAs are shown in Fig. 4. TLR1, TLR3, TLR5, TLR9, tumor necrosis factor receptor-associated factor (TRAF)6 and toll/interleukin-1 receptor-domain-containing adapter-inducing interferon-β (TRIF), which have been demonstrated to be key genes in TLR pathway, were in the ‘immune system process’.
Figure 3.
Biological processes of sequenced (A) miRNA and (B) mRNA from epithelial tissues in HSV2-infected and healthy control epithelial tissues harvested from guinea pigs, as determined by gene ontology analysis.
Figure 4.
Heatmap of immune-related (A) miRNA and (B) mRNAs from HSV2-infected and healthy control epithelial tissues harvested from guinea pigs.
Upregulated miRNAs depressed TLR-associated genes following HSV2 infection
It was observed that the TLR pathway was downregulated after HSV2 infection, and the negative correlation between miRNA and mRNA of these genes (Fig. 5A), as demonstrated by RT-qPCR analysis on epithelial samples from 10 normal and 10 HPV2-infected guinea pigs.. The results demonstrated that the expression levels of TLR1, TLR3, TLR5, TLR9, TRAF6 and TRIF significantly decreased (P<0.05; Fig. 5B) following infection with HSV2; whereas the expression levels of the miRNAs, including miR-592, miR-1245b-5p, miR-150, miR-342-5p, miR-1245b-3p and miR-124, significantly increased (P<0.05; Fig. 5C).
Figure 5.
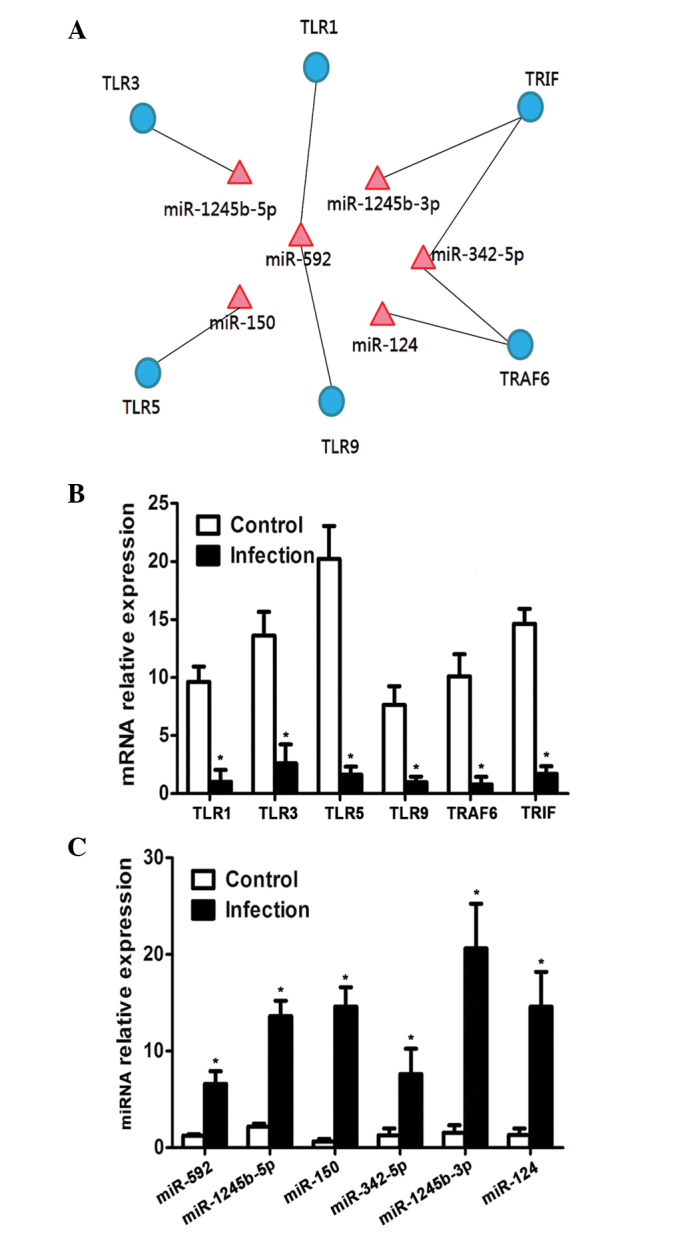
Aterations in the TLR pathway after infection with HSV2. (A) Genes related to the TLR pathway are under the regulation of miRNAs. Quantitative polymerase chain reaction demonstrated a (B) decrease in the expression levels of genes related to the TLR pathway after HSV infection (n=3); (C) whereas the expression levels of miRNAs related to TLR pathway increased after HSV infection (n=3). *P<0.05 vs. the control group. TLR, toll-like receptor; TRAF, tumor necrosis factor receptor-associated factor; TRIF, toll/interleukin-1 receptor-domain-containing adapter-inducing interferon-β.
miRNA-mRNA interaction
Negative correlations between putative miRNA and mRNA pairs were demonstrated (Fig. 5B and 5C) and miRNA target prediction was performed. A total of 13 miRNA with target genes in the transcriptome data were detected (Table II). Among these, 6 miRNA-mRNA pairs were demonstrated to be associated with TLR pathways. Furthermore, a negative correlation between mRNA and miRNA expression levels was detected in these miRNA-mRNA pairs (Table III).
Table II.
miRNAs with their putative target genes.
| miRNA | Log2 | P-value | P-value (hypergeometric) |
|---|---|---|---|
| miR-592 | 1.052 | 0.001 | <0.001 |
| miR-1245b-5p | 1.302 | 0.001 | <0.001 |
| miR-150 | 2.127 | 0.001 | <0.001 |
| miR-342-5p | 0.998 | 0.001 | <0.001 |
| miR-1245b-3p | 1.689 | 0.001 | <0.001 |
| miR-124 | 1.852 | 0.001 | <0.001 |
| miR-299-P | 1.452 | 0.001 | <0.001 |
| miR-365 | −1.618 | 0.001 | <0.001 |
| miR-321 | −1.603 | 0.001 | <0.001 |
| miR-339 | 1.020 | 0.001 | <0.001 |
| miR-346 | 1.478 | 0.001 | <0.001 |
| miR-424 | −1.300 | 0.001 | <0.001 |
| miR-4729 | 3.113 | 0.002 | <0.001 |
Log2 was calculated as infection/control.
Table III.
Differentially expressed miRNAs and their predicted target genes in the TLR pathway.
| miRNA | Log2 miRNA | Target gene | Log2 mRNA |
|---|---|---|---|
| miR-592 | 1.052 | TLR1 | −1.052 |
| TLR9 | −1.602 | ||
| miR-1245b-5p | 1.302 | TLR3 | −1.202 |
| miR-150 | 2.127 | TLR5 | −3.200 |
| miR-342-5p | 0.998 | TRAF6 | −2.690 |
| TRIF | −1.661 | ||
| miR-1245b-3p | 1.689 | TRIF | −1.661 |
| miR-124 | 1.852 | TRAF6 | −2.690 |
Log2 was calculated as infection/control. TLR, toll-like receptor; TRAF, tumor necrosis factor receptor-associated factor; TRIF, toll/interleukin-1 receptor-domain-containing adapter-inducing interferon-β.
Discussion
Previous studies have demonstrated that the abnormal expression of immune factors is mediated in HSV1 and HSV2-induced genital herpes (7–9). In particular, it has been demonstrated that HSV-specific T cells are triggered during HSV challenge (9), and various studies have focused on alterations in pro-inflammatory cytokines following infection (20–23). Furthermore, alterations in immune factor expression levels and hormone-triggered reproductive immune in vertebrates have been studied extensively (24–26). However, the molecular mechanism of immune dysregulation has not been demonstrated until now. The present study focused on mRNA and miRNA expression profiles following infection with HSV2. NGS was used to sequence mRNA and small RNA in order to elucidate the transcriptome and epigenetic alterations in tissues infected with HSV2 genital herpes.
Substantial progress has been made in understanding the TLR pathways involved in viral-infected organisms. It has previously been established that HSV1 and HSV2 trigger the expression of TLRs, which are stimulated by cytokines chemokines (21). These effects may be triggered by T- and B-cell-mediated immunity (27). It has been reported that the stimulation of TLR pathway induces remodeling of the lymph node (28). Furthermore, TLR recognition mediates the innate response to ensure an effective acquired immune response (29). Previous studies have demonstrated that TLR agonists could be used as potential therapies to treat pathogen infection (21,27). Therefore, novel treatment strategies which capitalize on the involvement of the TLR pathway in HSV infection are required.
Previous studies have suggested that both humoral and cellular immune responses are activated in response to HSV infection (6–8), and the recurrence of HSV has been associated with the depression of cellular immune responses in a guinea pig model of HSV (9,10). The present study investigated alterations in the expression levels of miRNA and mRNAs associated with the TLR pathway in HSV2-infected tissues. The results indicated that miR-592, miR-1245b-5p, miR-150, miR-342-5p, miR-1245b-3p and miR-124 expression levels were increased following infection with HSV2; whereas the following TLR pathway-associated genes were downregulated: TLR1, TLR3, TLR5, TLR9, TRAF6 and TRIF. These results demonstrated that miRNAs may downregulate these TLR pathway-associated genes during infection with HSV2. These results are consistent with a previous study by Li et al (30), which reported that HSV2 is capable of stimulating the expression of IL-1, IFN-β, TLR4 and TLR9 in cervical epithelial cells. There are numerous TLR pathway factors associated with various genes, and the present study demonstrated that the sequenced genes in this pathway were downregulated after infection with HSV2. Therefore, these results suggested network regulation of mRNA by miRNA in tissues infected with HSV2 genital herpes. NGS was used in the present study to investigate miRNA and mRNA expression profiles in the pithelial tissues of healthy and HSV2-infected guinea pigs. NGS technology provides an efficient and low cost method for the elucidation of the molecular mechanism underlying pathology. In the present study, a network of dysregulated miRNAs and target genes was constructed, which demonstrated that miR-592 and miR-342-5p share target genes in the TLR pathway. However, further studies are required in order to investigate the underlying mechanisms.
In conclusion, the present study used using NGS to elucidate the miRNA and mRNA expression profiles in the epithelial tissues of guinea pigs following infection with HSV2. A total of 105 and 141 miRNAs were upregulated and downregulated, respectively; whereas 798 upregulated and 503 downregulated genes were detected. Genes associated with the TLR pathway were downregulate, whereas their related miRNAs were highly expressed following in guinea pigs HSV2 infection. Enrichment analysis of the dysregulated genes demonstrated that biological and immune system processes were predominantly affected. Therefore, the results of the present study may contribute to the understanding of the molecular pathogenesis of HSV2-induced genital herpes.
Acknowledgements
The present study was supported by Chinese Nature Science Foundation (grant nos. 81202705 and 81373641) and by the Key Laboratory of Hunan Province for Integrated Traditional Chinese and Western Medicine on Prevention and Treatment of Cardio-Cerebral Diseases, and the Key Laboratory of Colleges and Universities in Hunan Province for Cytobiology and Molecular Biotechnology.
References
- 1.Delaney S, Gardella C, Daruthayan C, Saracino M, Drolette L, Corey L, Wald A. A prospective cohort study of partner testing for herpes simplex virus and sexual behavior during pregnancy. J Infect Dis. 2012;206:486–494. doi: 10.1093/infdis/jis403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rozenberg F, Deback C, Agut H. Herpes simplex encephalitis: From virus to therapy. Infect Disord Drug Targets. 2011;11:235–250. doi: 10.2174/187152611795768088. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong GL, Schillinger J, Markowitz L, Nahmias AJ, Johnson RE, McQuillan GM, St Louis ME. Incidence of herpes simplex virus type 2 infection in the United States. Am J Epidemiol. 2001;153:912–920. doi: 10.1093/aje/153.9.912. [DOI] [PubMed] [Google Scholar]
- 4.Tyring SK, Baker D, Snowden W. Valacyclovir for herpes simplex virus infection: Long-term safety and sustained efficacy after 20 years' experience with acyclovir. J Infect Dis. 2002;186:S40–S46. doi: 10.1086/342966. [DOI] [PubMed] [Google Scholar]
- 5.Zuckerman RA, Lucchetti A, Whittington WL, Sanchez J, Coombs RW, Zuñiga R, Magaret AS, Wald A, Corey L, Celum C. Herpes simplex virus (HSV) suppression with valacyclovir reduces rectal and blood plasma HIV-1 levels in HIV-1/HSV-2-seropositive men: A randomized, double-blind, placebo-controlled crossover trial. J Infect Dis. 2007;196:1500–1508. doi: 10.1086/522523. [DOI] [PubMed] [Google Scholar]
- 6.Bourne N, Milligan GN, Schleiss MR, Bernstein DI, Stanberry LR. DNA immunization confers protective immunity on mice challenged intravaginally with herpes simplex virus type 2. Vaccine. 1996;14:1230–1234. doi: 10.1016/S0264-410X(96)00027-8. [DOI] [PubMed] [Google Scholar]
- 7.Ashley R, Benedetti J, Corey L. Humoral immune response to HSV-1 and HSV-2 viral proteins in patients with primary genital herpes. J Med Virol. 1985;17:153–166. doi: 10.1002/jmv.1890170208. [DOI] [PubMed] [Google Scholar]
- 8.Corey L, Reeves WC, Holmes KK. Cellular immune response in genital herpes simplex virus infection. N Engl J Med. 1978;299:986–991. doi: 10.1056/NEJM197811022991805. [DOI] [PubMed] [Google Scholar]
- 9.Stanberry LR, Kern ER, Richards JT, Abbott TM, Overall JC. Genital herpes in guinea pigs: Pathogenesis of the primary infection and description of recurrent disease. J Infect Dis. 1982;146:397–404. doi: 10.1093/infdis/146.3.397. [DOI] [PubMed] [Google Scholar]
- 10.Skoberne M, Cardin R, Lee A, Kazimirova A, Zielinski V, Garvie D, Lundberg A, Larson S, Bravo FJ, Bernstein DI, et al. An adjuvanted herpes simplex virus 2 subunit vaccine elicits a T cell response in mice and is an effective therapeutic vaccine in guinea pigs. J Virol. 2013;87:3930–3942. doi: 10.1128/JVI.02745-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sin JI, Kim JJ, Zhang D, Weiner DB. Modulation of cellular responses by plasmid CD40L: CD40L plasmid vectors enhance antigen-specific helper T cell type 1 CD4+ T cell-mediated protective immunity against herpes simplex virus type 2 in vivo. Hum Gene Ther. 2001;12:1091–1102. doi: 10.1089/104303401750214302. [DOI] [PubMed] [Google Scholar]
- 12.BenMohamed L, Bertrand G, McNamara CD, Gras-Masse H, Hammer J, Wechsler SL, Nesburn AB. Identification of novel immunodominant CD4+ Th1-type T-cell peptide epitopes from herpes simplex virus glycoprotein D that confer protective immunity. J Virol. 2003;77:9463–9473. doi: 10.1128/JVI.77.17.9463-9473.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milligan GN, Bernstein DI, Bourne N. T lymphocytes are required for protection of the vaginal mucosae and sensory ganglia of immune mice against reinfection with herpes simplex virus type 2. J Immunol. 1998;160:6093–6100. [PubMed] [Google Scholar]
- 14.Xiao YL, Kash JC, Beres SB, Sheng ZM, Musser JM, Taubenberger JK. High-throughput RNA sequencing of a formalin-fixed, paraffin-embedded autopsy lung tissue sample from the 1918 influenza pandemic. J Pathol. 2013;229:535–545. doi: 10.1002/path.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papic N, Maxwell CI, Delker DA, Liu S, Heale BS, Hagedorn CH. RNA-sequencing analysis of 5′ capped RNAs identifies many new differentially expressed genes in acute hepatitis C virus infection. Viruses. 2012;4:581–612. doi: 10.3390/v4040581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1415. [DOI] [PubMed] [Google Scholar]
- 17.Hsiung GD, Mayo DR, Lucia HL, Landry ML. Genital herpes: Pathogenesis and chemotherapy in the guinea pig model. Rev Infect Dis. 1984;6:33–50. doi: 10.1093/clinids/6.1.33. [DOI] [PubMed] [Google Scholar]
- 18.Xiao S, Mo D, Wang Q, Jia J, Qin L, Yu X, Niu Y, Zhao X, Liu X, Chen Y. Aberrant host immune response induced by highly virulent PRRSV identified by digital gene expression tag profiling. BMC Genomics. 2010;11:544. doi: 10.1186/1471-2164-11-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferreira J, Zwinderman A. On the Benjamini–Hochberg method. Ann Stat. 2006;34:1827–1849. doi: 10.1214/009053606000000425. [DOI] [Google Scholar]
- 20.Morrison LA. The Toll of herpes simplex virus infection. Trends Microbiol. 2004;12:353–356. doi: 10.1016/j.tim.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 21.Nazli A, Yao XD, Smieja M, Rosenthal KL, Ashkar AA, Kaushic C. Differential induction of innate anti-viral responses by TLR ligands against Herpes simplex virus, type 2, infection in primary genital epithelium of women. Antiviral Res. 2009;81:103–112. doi: 10.1016/j.antiviral.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 22.Sainz B, Loutsch JM, Marquart ME, Hill JM. Stress-associated immunomodulation and herpes simplex virus infections. Med Hypotheses. 2001;56:348–356. doi: 10.1054/mehy.2000.1219. [DOI] [PubMed] [Google Scholar]
- 23.Bettahi I, Zhang X, Afifi RE, BenMohamed L. Protective immunity to genital herpes simplex virus type 1 and type 2 provided by self-adjuvanting lipopeptides that drive dendritic cell maturation and elicit a polarized Th1 immune response. Viral Immunol. 2006;19:220–236. doi: 10.1089/vim.2006.19.220. [DOI] [PubMed] [Google Scholar]
- 24.Zhong H, Zhou Y, Yu F, Xiao J, Gan X, Zhang M. Seasonal changes and human chorionic gonadotrophin (hCG) effects on innate immune genes expression in goldfish (Carassius auratus) Fish Shellfish Immunol. 2014;38:303–310. doi: 10.1016/j.fsi.2014.03.033. [DOI] [PubMed] [Google Scholar]
- 25.Norris K, Evans MR. Ecological immunology: Life history trade-offs and immune defense in birds. Behav Ecol. 2000;11:19–26. doi: 10.1093/beheco/11.1.19. [DOI] [Google Scholar]
- 26.McCallum ML, Trauth SE. Physiological trade-offs between immunity and reproduction in the northern cricket frog (Acris crepitans) Herpetologica. 2007;63:269–274. doi: 10.1655/0018-0831(2007)63[269:PTBIAR]2.0.CO;2. [DOI] [Google Scholar]
- 27.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 28.Soderberg KA, Payne GW, Sato A, Medzhitov R, Segal SS, Iwasaki A. Innate control of adaptive immunity via remodeling of lymph node feed arteriole. Proc Natl Acad Sci USA. 2005;102:16315–16320. doi: 10.1073/pnas.0506190102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeda K, Akira S. Toll-like receptors in innate immunity. Int Immunol. 2005;17:1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 30.Li H, Li X, Wei Y, Tan Y, Liu X, Wu X. HSV-2 induces TLRs and NF-κB-dependent cytokines in cervical epithelial cells. Biochem Biophys Res Commun. 2009;379:686–690. doi: 10.1016/j.bbrc.2008.12.150. [DOI] [PubMed] [Google Scholar]