Abstract
The present study aimed to evaluate the expression of microRNA (miR)-421 in gastric cancer and to investigate its biological function and underlying mechanism of action in the development of gastric cancer. The expression of miR-421 was measured in 60 pairs of clinically removed gastric cancer tissues and matched adjacent normal gastric tissues by reverse transcription-quantitative polymerase chain reaction. In addition, following transfection with an miR-421 inhibitor to suppress the expression of miR-421, the proliferation, migration and cell cycle distribution of human gastric carcinoma MKN28/MKN74 cells were determined by cell counting, Transwell and flow cytometry assays. The target gene of miR-421 was also predicted using bioinformatic analysis and verified by dual-luciferase reporter gene assay and western blot analysis. Furthermore, overexpression of the miR-421 target protein was induced in MKN28/MKN74 cells to determine its function. It was observed that miR-421 was significantly upregulated in gastric cancer tissues and that the expression of miR-421 was associated with lymph node metastasis and the clinical stage of gastric cancer (all P<0.05). Claudin11 (CLDN11) was predicted and verified as a direct target of miR-421. In vitro experiments demonstrated that inhibition of miR-421 expression suppressed the proliferation and metastasis of MKN28/MKN74 cells and induced G1/S-phase cell cycle arrest (all P<0.05). Analagous results were observed in MKN28/MKN74 cells following overexpression of the CLDN11 protein. Collectively, these data suggest that miR-421 may promote the proliferation, invasion and metastasis of gastric cancer by inhibiting the expression of CLDN11.
Keywords: gastric cancer, microRNA-421, claudin-11, metastasis
Introduction
Gastric cancer is among the most prevalent types of malignant tumor, and has the fourth and second highest rates of incidence and mortality, respectively, among malignant tumors (1,2). Until 2015, the incidence of gastric cancer has increased on a yearly basis and two thirds of all new cases were reported in developing countries, among which China accounted for 42% (3). As patients with gastric cancer are typically asymptomatic during the early stage of the disease, early diagnosis of gastric cancer is difficult and the five-year survival rate of all patients with gastric cancer is <20% (4). Thus, gastric cancer is a threat to global health. It has been documented that the invasion and metastasis of tumor cells is a major cause of mortality in patients with cancer (5). Clinical evidence has also indicated that the prognosis of gastric cancer is associated with the stage of disease at diagnosis, with early diagnosis of gastric cancer resulting in a lower incidence of metastasis and a five-year survival rate of >90% (6). Although a number of studies have focused on gastric cancer at the molecular level (7–9), the mechanism underlying the invasion and metastasis of gastric cancer remains unknown. Therefore further studies are warranted to determine the underlying molecular mechanisms of gastric cancer invasion and metastasis, in order to identify novel gene markers for the early diagnosis of gastric cancer.
Recent evidence has indicated that microRNAs (miRNAs or miRs), a class of post-transcriptional regulators, serve key roles in the development of gastric cancer (10). miRNAs are a conserved class of small RNAs (18–22 nucleotides long) that regulate gene expression by binding to complementary sequences at the 3′ untranslated region (3′UTR) of target mRNAs, which results in gene silencing through the translational repression or degradation of target mRNA (11). Recent studies have demonstrated that the miRNA expression profile is altered in gastric cancer tissues, and that miRNAs may participate in the development, proliferation and metastasis of gastric cancer as either oncogenes or tumor suppressors (11,12). For instance, miR-448 suppresses the metastasis of gastric cancer cells by targeting Disintegrin and metalloproteinase domain-containing protein 10, and was found to be downregulated in both gastric cancer tissues and cell lines (13). Furthermore, it has been demonstrated that miR-485 inhibits the development of gastric cancer by targeting flotillin-1, as observed in three gastric cancer cell lines (MKN45, BGC-823 and SGC7901) (14).
miR-421 is a recently identified miRNA typically expressed in tumors and is associated with the metastasis of multiple tumor types. It has been observed that the level of miR-421 expression in gastric cancer is associated with the prognosis of patients (15); however, the corresponding molecular mechanism is not well understood. Liu et al (15) demonstrated that higher levels of miR-421 expression were linked to poor patient prognosis, indicating that miR-421 may promote the development of gastric cancer. Zhang et al (16) also documented that the presence of miR-421 within gastric secretions may be a potential biomarker for gastric cancer. Furthermore, the higher positive detection rate of miR-421 than that of serum carcino-embryonic antigen in gastric cancer indicates that miR-421 may be a useful diagnostic marker for gastric cancer (17). In hepatocellular carcinoma, the human farnesoid X receptor has been implicated as a target of miR-421, as downregulation of the receptor promotes the proliferation and migration of hepatocellular carcinoma cells (18).
The present study aimed to elucidate the function of miR-421 in gastric cancer and its underlying mechanisms of action. Therefore, the expression of miR-421 in gastric cancer tissues was evaluated using reverse transcription-quantitative polymerase chain reaction (RT-qPCR). The roles of miR-421 in regulating the proliferation, migration and invasion of the gastric cancer cell line MKN28/MKN74 (19), were also investigated.
Materials and methods
Gastric cancer tissue collection
A total of 60 paired samples of human gastric cancer and matched adjacent noncancerous tissues were collected between December 2012 and October 2013 from patients with gastric cancer admitted to three hospitals in Lanzhou, China. A total of 26 patients were from the First Hospital Affiliated to Lanzhou University, 13 were from the Lanzhou Petrochemical General Hospital and 21 were from the Gansu Academy of Medical Science (all Lanzhou, China). Among these patients, 39 presented with lymph node metastasis (N1) and 20 were in stage I, 22 were in stage II, 11 were in stage III and 7 were in stage IV. This classification was in accordance with the TNM stage system (20). Gastric cancer tissues were histopathologically diagnosed and classified by two pathologists according to the 2003 World Health Organization standard of tumor classification (21). These tissues were frozen using liquid nitrogen immediately following surgery and stored at −80°C prior to use. Written informed consent was obtained from all patients prior to the current study and all protocols were approved by the ethics review board of the First Hospital Affiliated to Lanzhou University at the Lanzhou Petrochemical General Hospital and Gansu Academy of Medical Science.
Cell culture and transfection
The gastric cancer cell line MKN28 was purchased from Cobioer Biosciences (Nanjing, China). This cell line has been reported as cross-contaminated with MKN74, and as such is referred to as MKN28/MKN74 throughout the present study (19). Cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and penicillin-streptomycin (10,000 U/ml). Cultures were incubated in a humidified atmosphere with 5% CO2 at 37°C and passaged when a confluence of 80–90% was reached. When the cells were passaged, they were digested by trypsin for 3 min in RT. Cells were transfected with 25 pmol miR-421 inhibitor (TAG TTG TCT GTA ATT AAC CCG CG) or miR-421 mimic (ATC AAC AGA CAT TAA TTG GGC GC; Guangzhou Ribobio Co., Ltd., Guangzhou, China) using Lipofectamine® 2000 (Thermo Fisher Scientific Inc.), according to the manufacturer's instructions. As a control, MKN28/MKN74 cells were transfected with negative control (NC) miR-421 inhibitor (Guangzhou Ribobio Co., Ltd.) that did not target any human mature miRNA or with NC mimic (NC) that did not target any human gene products. To induce overexpression of CLDN11, a plasmid containing a CLDN11 coding sequence (ATG GTG GCCAC GTG CCT GCAG GTG GTG GGCT TCG TCA CGAGCTT CGT GGGC TGG ATC GGGG TCA TCG TGAC CAC CTCCACC AAT GAC TGGG TGG TGA CCTG CGG CTA CACC ATC CCC ACCT GCC GCA AGCT GGA TGA GCTG GGC TCCAAGG GGC TGT GGGC CGA CTG CGTC ATG GCC ACGG GGCTGT ACCA CTG CAA GCCC CTG GTG GACA TCC TCA TCCTGCC GGG CTAC GTG CAG GCCT GCC GCG CCCT GAT GATTGCT GCC TCG GTCC TGG GTC TGCC GGC CAT TTTACT G CTG CTGA CTG TTC TTCC CTG CAT CCGG ATG GGCCA GG AGC CCG TGT GGC TAA GTAC AGG CGG GCCC AGCTG GCTGG TGT TTT GCTC ATT CTG CTGG CTC TCT GCGCCC T TGT TGCC ACC ATC TGGT TCC CTG TGTG CGC CCACCG T GAG ACC ACCA TCG TGA GCTT TGG CTA CTCC CTG TATGCAG GCT GGA TTGG TGC TGT GCTG TGC CTC GTGGGTG GCT GTGT CAT CCT CTGC TGC GCT GGAG ATG CCCAGGC CTT TGG TGAA AAC CGT TTCT ACT ACA CTGC GGGCTC TAGC TCC CCG ACTC ATG CGA AGAG TGC CCA CGTAT AA) was provided by GeneChem Co., Ltd. (Shanghai, China), and empty plasmid was used as a negative control (plasmid NC; GeneChem Co., Ltd.). Plasmids were transfected into MKN28/MKN74 cells using Lipofectamine® 2000 and cells were cultured for 24 or 48 h at 37°C post-transfection before collection for subsequent experiments.
RNA extraction and RT-qPCR
Gastric cancer and matched normal tissues were ground in liquid nitrogen with a porcelain mortar and pestle. Total RNA was isolated using TRIzol® isolation reagent (Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. RNA was reverse transcribed into cDNA using a PrimeScript RT-PCR kit (Clontech Laboratories, Inc., Mountainview, CA, USA) following the addition of a poly (A) tail. qPCR was performed using a SYBR PrimeScript RT-PCR kit (Takara Biotechnology Co., Ltd., Dalian, China) on a Step One Plus Real-Time PCR system (Thermo Fisher Scientific, Inc.). Expression of U6 small nuclear (sn)RNA was used as an internal control. The forward and reverse primers were as follows: For miR-421, forward, 5′-ATCAACAGACAUUAATT-3′ and reverse, 5′-ATCAACAGACATTAATTGGG-3′ and for U6 snRNA, forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′. The mixture was amplified at 95°C for 10 min, followed by 40 cycles at 95°C for 1 min and 60°C for 30 sec. PCR was performed in triplicate on each sample, and relative expression levels were evaluated using the ΔΔCq method, as described previously (22).
Cell counting kit-8 (CCK-8) assay
MKN28/MKN74 cells, cultured as described above, were transfected with miR-421 mimic or NC mimic and seeded into 96-well plates at a density of 2×103 cells/well in triplicate. After 24 h incubation at 37°C to obtain adherent cells, a total of 20 µl CCK-8 solution (Beyotime Institute of Biotechnology, Haimen, China) was added to the wells every 24 h for 3 days and incubated for 30 min at 37°C. Data were not collected at 0 h while cells were suspended. The absorbance of each well was measured with a SpectraMax M5 microplate reader (Molecular Devices, LLC, Sunnyvale, CA, USA) at 450 nm and a proliferation curve was generated.
Migration and invasion assay
A total of 2×105 transfected MKN28/MKN74 cells in 200 µl RPMI-1640 medium without FBS were plated onto 8 µm pore filter inserts in the top chamber of a Transwell chamber (Corning Incorporated, NY, USA). RPMI-1640 supplemented with 10% FBS (600 µl) was added to the bottom chamber and incubated at 37°C and 5% CO2. To analyze the invasive capacity of cells, Matrigel (BD Biosciences, San Jose, CA, USA) was melted at 4°C overnight and diluted with RPMI-1640 in a 1:2 ratio in advance. The transwell migration chambers were coated with Matrigel on ice and incubated at 37°C for 60 min to allow the Matrigel to solidify. After 24 h (for migration assays) or 72 h (for invasion assays) incubation at 37°C, cells that did not pass through the chambers were removed with a cotton swab, while cells that had passed to the lower side of the chamber were fixed with 4% formaldehyde at room temperature for 30 min, stained with Giemsa and counted under an Olympus BX51/61 microscope (Olympus Corporation, Tokyo, Japan) at ×200 magnification. Migration and invasion data were calculated by counting the number of migrated or invaded cells in 5 randomly selected fields.
Flow cytometry cell cycle analysis
At 24 h after transfection with miR-421 inhibitor or NC inhibitor, or CLDN11 overexpression plasmid, 1×106 MKN28/MKN74 cells were seeded into separate plates, washed twice with cold phosphate-buffered saline (PBS) and stained with a BD Cycletest™ Plus DNA Reagent kit (BD Biosciences), according to the manufacturer's instructions. Briefly, 200 ul solution A from the kit was added and incubated at room temperature for 10 min, followed by addition of 120 µl solution B. Following incubation for 10 min at room temperature, 100 µl solution C was added and incubated for 15 min in the dark at room temperature. Following the addition of 200 µl PBS, cells were analyzed by flow cytometry using a BD FACSVerse™ flow cytometer (BD Biosciences) using Modfit software version 3.2 (Verity Software House, Inc., Topsham, ME, USA).
Western blotting
A total of 20 µg protein was extracted from MKN28/MKN74 cells transfected with miR-421 or NC inhibitor, or CLDN11 overexpression plasmid or control using radioimmunoprecipitation assay buffer (Bio-Rad Laboratories, Inc., Hercules, CA, USA) and separated by SDS-PAGE. Proteins were then transferred onto nitrocellulose membranes using the wet method. Following blocking with non-fat milk for 2 h at room temperature, membranes were incubated with polyclonal rabbit anti-CLDN11 antibody (sc-25711; 1:1,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) at 4°C overnight. Following washing with tris-buffered saline/Tween-20 3 times, membranes were incubated with monoclonal rabbit anti-mouse horseradish peroxidase-conjugated secondary antibody (sc-2004; 1:2,000; Santa Cruz Biotechnology, Inc.) at room temperature for 1 h. Membranes were then developed using the BeyoECL Plus enhanced chemiluminescence plus reagent (Beyotime Institute of Biotechnology, Haimen, China) and analyzed by Quantity One 4.62 software (Bio-Rad Laboratories, Inc.). Rabbit anti-GAPDH primary antibody (sc-367715; 1:1,000; Santa Cruz Biotechnology, Inc.) was used as an internal control, which was incubated at room temperature for 2 h. Western blotting was replicated 3 times and arepresentative result was shown.
Dual-luciferase reporter gene assay
According to results of the bioinformatics prediction, luciferase reporter plasmids were generated by inserting wildtype or mutant 3′UTR sequences of CLDN11 (23) into the multiple cloning site (Spe-1 and HindIII restriction sites) downstream of the luciferase reporter gene in the pMIR-REPORT™ Luciferase plasmid (Thermo Fisher Scientific, Inc.). The mutant 3′UTR was to completely mutate the binding sequence to seed sequence of miR-421. MKN28/MKN74 cells were transfected with 0.5 µg constructed luciferase reporter plasmid and 10 ng pMIR-REPORT™ β-gal Control plasmid (Thermo Fisher Scientific, Inc.) as an internal control to determine transfection efficiency. miR-421 mimic or NC mimic were also transfected. Luminescence was measured 24 h after transfection using a dual-luciferase detection kit (no. RG027; Beyotime Institute of Biotechnology), according to the manufacturer's instructions. Measurements of luminescence were performed on a GloMax 20/20 Luminometer (Promega Corporation, Madison, WI, USA).
Target prediction
To identify the target of miR-421, putative target genes were searched using Targetscan software (www.targetscan.org/). Has-miR-421 was entered into the searching bar and its predicted targets were obtained.
Statistical analysis
Data are expressed as the mean ± standard deviation of three independent experiments. Statistical significance was determined with a paired t-test for two groups and one-way analysis of variance with Student-Newman-Keuls post hoc test for three or more groups using SPSS 16.0 software (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
miR-421 expression is upregulated in gastric cancer tissue
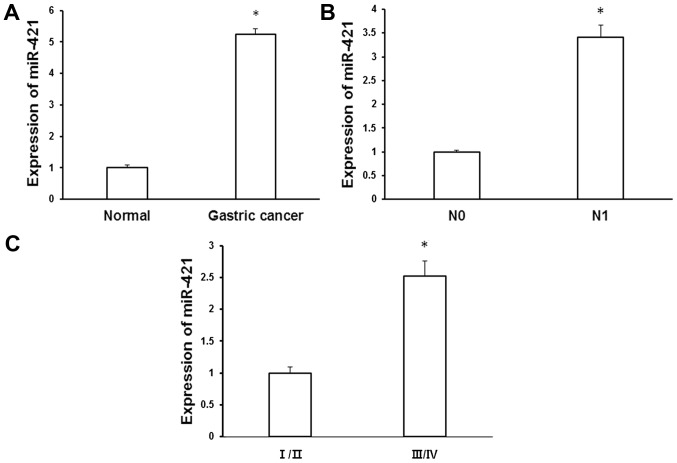
To evaluate the role of miR-421 in gastric cancer, levels of miR-421 expression were measured in gastric cancer tissues and matched adjacent normal tissues using RT-qPCR. It was determined that miR-421 was significantly upregulated in gastric cancer tissue relative to normal controls (5.24±0.19 vs. 1±0.09; P<0.05; Fig. 1A). To evaluate the expression pattern of miR-421 during the development of gastric cancer, the association between miR-421 expression and clinicopathological characteristics was evaluated. Patients with lymph node metastasis (N1) exhibited significantly increased expression of miR-421 relative to those without lymph node metastasis (3.41±0.26 vs. 1±0.04, P<0.05; Fig. 1B). In addition, miR-421 expression was significantly upregulated in stage III/IV gastric cancer relative to stage I/II cancer (2.52±0.24 vs. 1±0.1, P<0.05; Fig. 1C). These results suggest that upregulation in miR-421 may be associated with the development and metastasis of gastric cancer.
Figure 1.
Upregulation of miR-421 in gastric cancer tissue specimens. Total RNA was isolated from gastric cancer tissues and matched adjacent normal gastric tissue using TRIzol reagent. Levels of miR-421 expression were measured using reverse transcription-quantitative polymerase chain reaction and normalized to that of U6 (A) Quantification of miR-421 expression in gastric cancer tissues and matched adjacent normal tissues. (B) Quantification of miR-421 expression in N0 and N1 specimens. (C) Quantification of miR-421 expression in different clinical stages of gastric cancer. *P<0.05 vs. adjacent normal tissues. miR, microRNA; N0, negative for lymph node metastasis; N1, positive for lymph node metastasis; I/II and III/IV, clinical stage.
Inhibiton of miR-421 represses MKN28/MKN74 cell proliferation
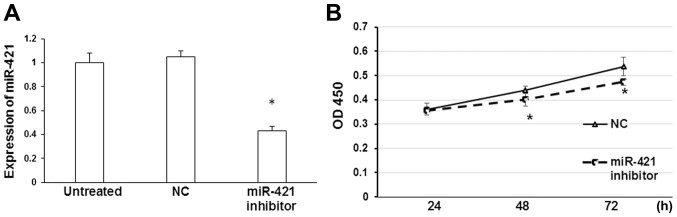
To determine the influence of miR-421 on gastric cancer cells, MKN28/MKN74 cells were transiently transfected with miR-421 inhibitor or NC. It was determined that transfection with miR-421 inhibitor significantly downregulated the expression of miR-421 in MKN28/MKN74 cells relative to the NC group (P<0.05; Fig. 2A). In addition, levels of miR-421 expression did not differ significantly between untreated and NC transfected cells (Fig. 2). Furthermore, as determined by a CCK-8 proliferation assay, inhibition of miR-421 significantly repressed the proliferation of MKN28/MKN74 cells 48 and 72 h post-transfection, relative to NC transfected cells (P<0.05; Fig. 2B). Therefore, miR-421 may promote the development of gastric cancer by inducing the proliferation of gastric cancer cells.
Figure 2.
CCK-8 assay of cell proliferation. (A) Reverse transcription-quantitative polymerase chain reaction analysis of miR-421 expression in MKN28/MKN74 cells following transfection with miR-421 inhibitor. (B) A CCK-8 assay was performed in MKN28/MKN74 cells transfected with miR-421 inhibitor or NC inhibitor. The optical density of each well was measured at 450 nm at the indicated time-points. *P<0.05 vs. NC. CCK-8, cell counting kit-8; miR, microRNA; NC, negative control; OD, optical density; h, hours.
Inhibition of miR-421 reduces the migration and invasion of gastric cancer cells
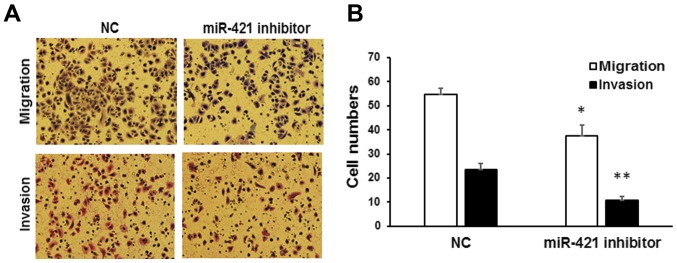
The influence of miR-421 on the migration and invasion of gastric cancer cells was subsequently determined. Using a Transwell assay, it was observed that the number of migrated MKN28/MKN74 cells was significantly reduced following the inhibition of miR-421, relative to cells transfected with NC inhibitor (37.5±4.60 vs. 54.8±2.37, P<0.05; Fig. 3). Similarly, the number of invaded cells following transfection with miR-421 inhibitor was significantly lower than that observed for cells transfected with NC (10.8±1.5 vs. 23.5±2.50, P<0.01; Fig. 3). Collectively these data suggest that miR-421 may serve a role in promoting tumor cell invasion in vitro.
Figure 3.
Migration and invasion of MKN28/MKN74 cells. Transwell migration and invasion assays of MKN28/MKN74 cells transfected with miR-421 or NC inhibitor. (A) Cells that had migrated through the filter into the lower wells were stained with Giemsa and viewed by fluorescence microscopy (magnification, ×200). (B) The numbers of migrated and invaded cells were expressed relative to the total number of cells in the lower wells. *P<0.05 and **P<0.01 vs. NCs. miR, microRNA; NC, negative control.
MiR-421 influences the cell cycle in MKN28/MKN74 cells
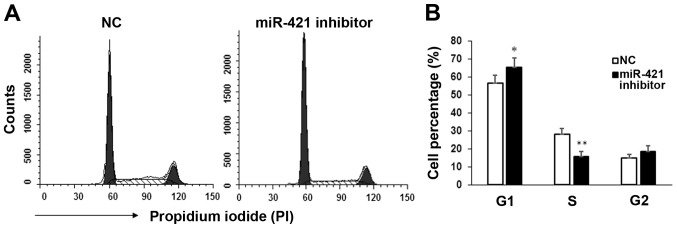
Cell cycle distribution in MKN28/MKN74 cells following transfection with miR-421 inhibitor was measured by flow cytometry. Significant inhibition (P<0.05) of the G1/S transition was observed in miR-421 inhibitor transfectants, relative to controls (Fig. 4), while the proportions of cells in the G2 stage were similar between the two groups. These results indicate that miR-421 may promote the proliferation of gastric cancer cells by regulating the G1/S transition.
Figure 4.
Flow cytometry analysis of the cell cycle. (A) The cell cycle distribution of MKN28/MKN74 cells was analyzed by flow cytometry 24 h after transfection with miR-421 or NC. (B) Percentages of cells at G1, S and G2 phase. *P<0.05 and **P<0.01 vs. NC at each phase. miR, microRNA; NC, negative control.
CLDN11 is a target of miR-421
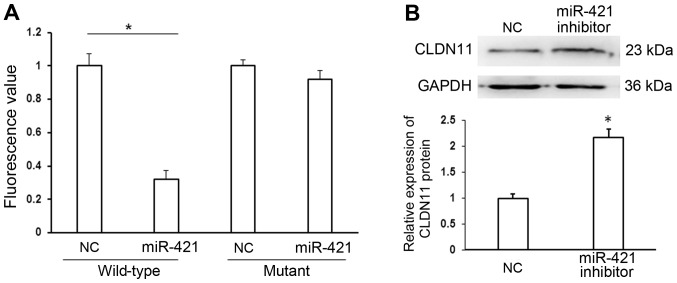
To identify the target of miR-421, putative target genes were searched using Targetscan software (www.targetscan.org/) and CLDN11 was identified as a target of miR-421. A wild-type putative binding site in the 3′UTR of CLDN11 or a mutated 3′UTR sequence was cloned into the pMIR-REPORT™ luciferase plasmid, and co-transfected with miR-421 or NC mimics into MKN28/MKN74 cells. Levels of luciferase activity were significantly lower in cells co-transfected with pMIR-REPORT-CLDN11 3′-UTR wild-type and miR-421 mimic, relative to cells co-transfected with pMIR-REPORT-CLDN11 3′-UTR wild-type and NC mimic (P<0.05). By contrast, levels of luciferase activity did not differ significantly between cells co-transfected with pMIR-REPORT-CLDN11 3′-UTR mutant and miR-421 mimic and cells co-transfected with pMIR-REPORT-CLDN11 3′-UTR mutant and NC mimic (Fig. 5A). The results of the dual luciferase reporter assay were verified by western blot analysis. Inhibition of miR-421 significantly increased the expression of CLDN11 protein compared with transfection with NC (P<0.05; Fig. 5B). Collectively these data indicate that CLDN11 may be a direct target of miR-421.
Figure 5.
CLDN11 is a direct target of miR-421. (A) Luciferase activity assays involving transfection with luciferase vectors encoding a wild-type or mutant 3′UTR of CLDN11 were performed following co-transfection with miR-421 or NC mimic. Luciferase activity was normalized to that of the pMIR-REPORT™ β-gal plasmid. (B) Western blot analysis of CLDN11 protein expression in MKN28/MKN74 cells transfected with miR-421 or NC inhibitor. GAPDH served as an internal control. *P<0.05 vs. NC. CLDN11, claudin-11; miR, microRNA; NC, negative control.
Overexpression of CLDN11 blocks the migration and invasion of gastric cancer cells
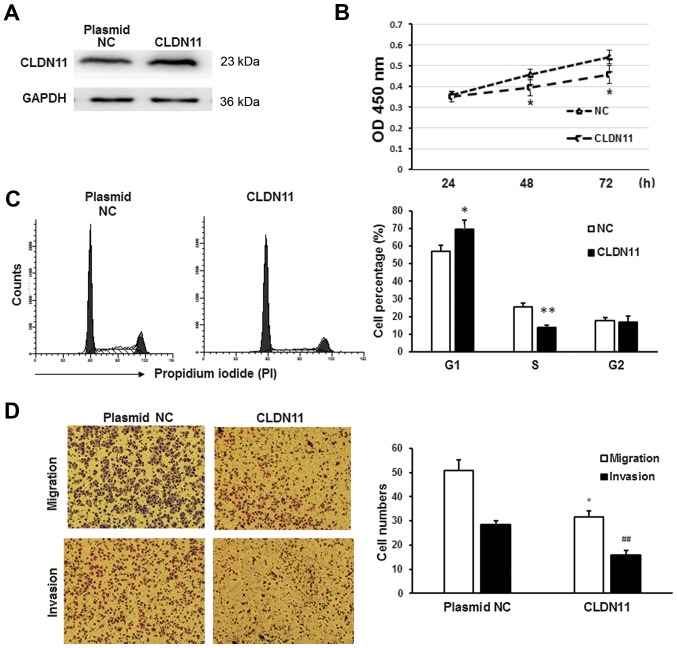
To determine whether miR-421 promotes the migration and metastasis of gastric cancer cells by downregulating CLDN11, CLDN11 was overexpressed in MKN28/MKN74 cells. Overexpression of CLDN11 was confirmed by western blot analysis (Fig. 6A). CLDN11 overexpression significantly reduced the proliferation rate of MKN28/MKN74 cells (P<0.05; Fig. 6B). Furthermore, flow cytometry indicated that CLDN11 inhibited the G1/S transition in MKN28 cells (P<0.05; Fig. 6C). In addition, CLDN11 overexpression significantly suppressed the migration and invasion of MKN28/MKN74 cells (P<0.05), relative to that in NC transfectants (Fig. 6D). These results were consistent with those obtained from the inhibition of miR-421, suggesting that downregulation of CLDN11 promotes the development of gastric cancer.
Figure 6.
Role of CLDN11 in MKN28/MKN74 cells. CLDN11 was overexpressed in MKN28/MKN74 cells using a CLDN11 overexpression plasmid. (A) Western blot analysis was used to confirm CLDN11 overexpression. GAPDH served as an internal control. (B) A Cell Counting Kit-8 assay was performed to measure the proliferation of MKN28/MKN74 cells following transfection. *P<0.05. (C) Flow cytometry analysis of cell cycle distribution 24 h after transfection. *P<0.05 and **P<0.01. (D) Transwell migration and invasion assays of MKN28/MKN74 cells. Cells that traversed to the lower wells were stained with Giemsa (red) and counted under a fluorescence microscope (magnification, ×200). *P<0.05 and ##P<0.01 vs. NC group. CLDN11, claudin-11; NC, negative control.
Discussion
In the present study, it was observed that miR-421 was significantly upregulated in gastric cancer tissues and that expression of miR-421 was associated with lymph node metastasis and clinical stage. In subsequent in vitro experiments, an miR-421 inhibitor was used to downregulate miR-421 in gastric cancer MKN28/MKN74 cells. It was observed that downregulation in miR-421 reduced the proliferation, migration and invasion of MKN28/MKN74 cells. Using bioinformatics, CLDN11 was also predicted to be a target of miR-421, and a dual luciferase report assay indicated that miR-421 may bind directly to the 3′ UTR of CLDN11 mRNA. Western blot analysis also indicated that miR-421 downregulated the expression of CLDN11 protein, while overexpression of CLDN11 inhibited the proliferation, migration and invasion of MKN28/MKN74 cells. Collectively, these data suggest that miR-421 may function as an oncogene in gastric cancer by targeting CLDN11 and that upregulation of miR-421 during gastric cancer may promote tumor development.
Mature miR-421 is related to the development of multiple tumors types. For instance, miR-421 may promote the development of neuroblastoma by targeting the tumor suppressor Menin (24). Wu et al (25) observed that miR-421 regulated cellular apoptosis in the undifferentiated gastric cancer cell line BGC-823 by targeting Caspase-3, and thus may be a novel diagnostic biomarker in gastric cancer (26). Analogous to previous results, the present study identified a potential relationship between elevated miR-421 expression and the development of gastric cancer, as indicated by the significant upregulation in miR-421 in patients presenting with lymph node metastasis and/or higher clinical stages of gastric cancer (III/IV). However, it has been demonstrated that in nasopharyngeal carcinoma, miR-421 may function as a tumor suppressor by downregulating the expression of forkhead box protein O4, leading to the inhibition of cell proliferation and metastasis (27). Thus, miR-421 may serve different biological functions in different types of tumors. In addition, the association between miR-421 expression and tumor development identified in the current study was inconsistent with previous reports by Zhang et al (16) and Jiang et al (17). This discrepancy may be due to variations in the samples studied, including variations in sex, age, environment and geographical location. Further studies are now warranted in larger sample sizes to verify the results.
CLDN11 is a member of the claudin protein family and is a critical component of tight junctions, where it serves a key role in regulating cell junctions and cell adhesion (28). Previous results have indicated that CLDN11 may suppress the proliferation, invasion and apoptosis of many tumor types. For instance, in hepatocellular carcinoma, downregulation of CLDN11 by miR-99 facilitated the metastasis of tumor cells (29), while in bladder cancer, expression of CLDN11 was associated with a reduced rate of distal metastasis (30). Agarwal et al (31) also observed that the metastatic ability of gastric cancer cells was increased following the silencing of CLDN11, indicating that CLDN11 may serve as a tumor suppressor in gastric cancer. Similarly, the present study demonstrated that overexpression of CLDN11, a potential target of miR-421, significantly decreased the proliferation, invasion and metastasis of gastric cancer cells.
In conclusion, miR-421 may promote the proliferation, invasion and metastasis of gastric cancer cells, and thus may be a potential diagnostic marker and therapeutic target in the treatment of gastric cancer.
Acknowledgements
The authors are thankful to Dr Weilin Yang at the Department of General Surgery, (The First Hospital of Lanzhou University, Lazhou, China) for his contributions to the current study.
References
- 1.Juzėnas S, Saltenienė V, Kupcinskas J, Link A, Kiudelis G, Jonaitis L, Jarmalaite S, Kupcinskas L, Malfertheiner P, Skieceviciene J. Correction: Analysis of deregulated microRNAs and their target genes in gastric cancer. PLoS One. 2015;10:e0135762. doi: 10.1371/journal.pone.0132327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang CM, Chen RF, Chen QY, Wei J, Zheng CH, Li P, Xie JW, Wang JB, Lin JX, Lu J, et al. Application value of a 6-type classification system for common hepatic artery absence during laparoscopic radical resections for gastric cancer: A large-scale single-center study. Medicine (Baltimore) 2015;94:e1280. doi: 10.1097/MD.0000000000001280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang C, Jia Z, Cao D, You L, Jin M, Wu X, Wen S, Cao X, Jiang J. Polymorphism of DNA methyltransferase 3b and association with development and prognosis in gastric cancer. PLoS One. 2015;10:e0134059. doi: 10.1371/journal.pone.0134059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu M, Li K, Maskey N, Xu Z, Peng C, Tian S, Li Y, Yang G. 15-PGDH expression as a predictive factor response to neoadjuvant chemotherapy in advanced gastric cancer. Int J Clin Exp Pathol. 2015;8:6910–6918. [PMC free article] [PubMed] [Google Scholar]
- 5.Matouk IJ, Halle D, Raveh E, Gilon M, Sorin V, Hochberg A. The role of the oncofetal H19 lncRNA in tumor metastasis: Orchestrating the EMT-MET decision. Oncotarget. 2016;7:3748–3765. doi: 10.18632/oncotarget.6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geredeli C, Dogru O, Omeroglu E, Yilmaz F, Cicekci F. Gastric metastasis of triple negative invasive lobular carcinoma. Rare Tumors. 2015;7:5764. doi: 10.4081/rt.2015.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim TH, Shivdasani RA. Stomach development, stem cells and disease. Development. 2016;143:554–565. doi: 10.1242/dev.124891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan L, Tan B, Li Y, Zhao Q, Liu Y, Wang D, Zhang Z. Silencing of ZNF139-siRNA induces apoptosis in human gastric cancer cell line BGC823. Int J Clin Exp Pathol. 2015;8:12428–12436. [PMC free article] [PubMed] [Google Scholar]
- 9.Tong GH, Tong WW, Qin XS, Lu LP, Liu Y. DBD-F induces apoptosis in gastric cancer-derived cells through suppressing HIF2α expression. Cell Oncol (Dordr) 2015;38:479–484. doi: 10.1007/s13402-015-0253-5. [DOI] [PubMed] [Google Scholar]
- 10.Karimi Kurdistani Z, Saberi S, Tsai KW, Mohammadi M. MicroRNA-21: Mechanisms of oncogenesis and its application in diagnosis and prognosis of gastric cancer. Arch Iran Med. 2015;18:524–536. [PubMed] [Google Scholar]
- 11.Assumpção MB, Moreira FC, Hamoy IG, Magalhães L, Vidal A, Pereira A, Burbano R, Khayat A, Silva A, Santos S, et al. High-throughput miRNA sequencing reveals a field effect in gastric cancer and suggests an epigenetic network mechanism. Bioinform Biol Insights. 2015;9:111–117. doi: 10.4137/BBI.S24066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imaoka H, Toiyama Y, Okigami M, Yasuda H, Saigusa S, Ohi M, Tanaka K, Inoue Y, Mohri Y, Kusunoki M. Circulating microRNA-203 predicts metastases, early recurrence, and poor prognosis in human gastric cancer. Gastric Cancer. 2016;19:744–753. doi: 10.1007/s10120-015-0521-0. [DOI] [PubMed] [Google Scholar]
- 13.Wu X, Tang H, Liu G, Wang H, Shu J, Sun F. miR-448 suppressed gastric cancer proliferation and invasion by regulating ADAM10. Tumour Biol. 2016;37:10545–10551. doi: 10.1007/s13277-016-4942-0. [DOI] [PubMed] [Google Scholar]
- 14.Kang M, Ren MP, Zhao L, Li CP, Deng MM. miR-485-5p acts as a negative regulator in gastric cancer progression by targeting flotillin-1. Am J Transl Res. 2015;7:2212–2222. [PMC free article] [PubMed] [Google Scholar]
- 15.Liu H, Gao Y, Song D, Liu T, Feng Y. Correlation between microRNA-421 expression level and prognosis of gastric cancer. Int J Clin Exp Pathol. 2015;8:15128–15132. [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Cui L, Ye G, Zheng T, Song H, Xia T, Yu X, Xiao B, Le Y, Guo J. Gastric juice microRNA-421 is a new biomarker for screening gastric cancer. Tumour Biol. 2012;33:2349–2355. doi: 10.1007/s13277-012-0497-x. [DOI] [PubMed] [Google Scholar]
- 17.Jiang Z, Guo J, Xiao B, Miao Y, Huang R, Li D, Zhang Y. Increased expression of miR-421 in human gastric carcinoma and its clinical association. J Gastroenterol. 2010;45:17–23. doi: 10.1007/s00535-009-0135-6. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Gong W, Dai S, Huang G, Shen X, Gao M, Xu Z, Zeng Y, He F. Downregulation of human farnesoid X receptor by miR-421 promotes proliferation and migration of hepatocellular carcinoma cells. Mol Cancer Res. 2012;10:516–522. doi: 10.1158/1541-7786.MCR-11-0473. [DOI] [PubMed] [Google Scholar]
- 19.Yokozaki H. Molecular characteristics of eight gastric cancer cell lines established in Japan. Pathol Int. 2000;50:767–777. doi: 10.1046/j.1440-1827.2000.01117.x. [DOI] [PubMed] [Google Scholar]
- 20.Shu P, Qin J, Shen K, Chen W, Liu F, Fang Y, Wang X, Wang H, Shen Z, Sun Y, Qin X. The IGCA staging system is more accurate than AJCC7 system in stratifying survival of patients with gastric cancer in stage III. BMC Cancer. 2017;17:238. doi: 10.1186/s12885-017-3235-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mezzetti M, Raveglia F, Panigalli T, Giuliani L, Lo Giudice F, Meda S, Conforti S. Assessment of outcomes in typical and atypical carcinoids according to latest WHO classification. Ann Thorac Surg. 2003;76:1838–1842. doi: 10.1016/S0003-4975(03)01194-9. [DOI] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Katsushima K, Shinjo K, Natsume A, Ohka F, Fujii M, Osada H, Sekido Y, Kondo Y. Contribution of microRNA-1275 to Claudin11 protein suppression via a polycomb-mediated silencing mechanism in human glioma stem-like cells. J Biol Chem. 2012;287:27396–27406. doi: 10.1074/jbc.M112.359109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Li W, Zhang JG, Li HY, Li YM. Downregulation of tumor suppressor menin by miR-421 promotes proliferation and migration of neuroblastoma. Tumour Biol. 2014;35:10011–10017. doi: 10.1007/s13277-014-1921-1. [DOI] [PubMed] [Google Scholar]
- 25.Wu JH, Yao YL, Gu T, Wang ZY, Pu XY, Sun WW, Zhang X, Jiang YB, Wang JJ. MiR-421 regulates apoptosis of BGC-823 gastric cancer cells by targeting caspase-3. Asian Pac J Cancer Prev. 2014;15:5463–5468. doi: 10.7314/APJCP.2014.15.13.5463. [DOI] [PubMed] [Google Scholar]
- 26.Wu J, Li G, Yao Y, Wang Z, Sun W, Wang J. MicroRNA-421 is a new potential diagnosis biomarker with higher sensitivity and specificity than carcinoembryonic antigen and cancer antigen 125 in gastric cancer. Biomarkers. 2015;20:58–63. doi: 10.3109/1354750X.2014.992812. [DOI] [PubMed] [Google Scholar]
- 27.Chen L, Tang Y, Wang J, Yan Z, Xu R. miR-421 induces cell proliferation and apoptosis resistance in human nasopharyngeal carcinoma via downregulation of FOXO4. Biochem Biophys Res Commun. 2013;435:745–750. doi: 10.1016/j.bbrc.2013.05.056. [DOI] [PubMed] [Google Scholar]
- 28.Zwanziger D, Badziong J, Ting S, Moeller LC, Schmid KW, Siebolts U, Wickenhauser C, Dralle H, Fuehrer D. The impact of CLAUDIN-1 in follicular thyroid carcinoma aggressiveness. Endocr Relat Cancer. 2015;22:819–830. doi: 10.1530/ERC-14-0502. [DOI] [PubMed] [Google Scholar]
- 29.Yang J, Liu X, Yuan X, Wang Z. miR-99b promotes metastasis of hepatocellular carcinoma through inhibition of claudin 11 expression and may serve as a prognostic marker. Oncol Rep. 2015;34:1415–1423. doi: 10.3892/or.2015.4104. [DOI] [PubMed] [Google Scholar]
- 30.Awsare NS, Martin TA, Haynes MD, Matthews PN, Jiang WG. Claudin-11 decreases the invasiveness of bladder cancer cells. Oncol Rep. 2011;25:1503–1509. doi: 10.3892/or.2011.1244. [DOI] [PubMed] [Google Scholar]
- 31.Agarwal R, Mori Y, Cheng Y, Jin Z, Olaru AV, Hamilton JP, David S, Selaru FM, Yang J, Abraham JM, et al. Silencing of claudin-11 is associated with increased invasiveness of gastric cancer cells. PLoS One. 2009;4:e8002. doi: 10.1371/journal.pone.0008002. [DOI] [PMC free article] [PubMed] [Google Scholar]