Summary
Living cells have evolved to detect and process various signals and can self‐replicate, presenting an attractive platform for engineering scalable and affordable biosensing devices. Microbes are perfect candidates: they are inexpensive and easy to manipulate and store. Recent advances in synthetic biology promise to streamline the engineering of microbial biosensors with unprecedented capabilities. Here we review the applications of microbially‐derived biosensors with a focus on environmental monitoring and healthcare applications. We also identify critical challenges that need to be addressed in order to translate the potential of synthetic microbial biosensors into large‐scale, real‐world applications.
Part of achieving the UN sustainable development goal #3 (‘Good health and well being’) relies on using biosensing technologies for the detection of environmental hazards or early diagnostics of diseases. As the majority of target populations live in developing countries, the next generation of biosensors need to solve the conundrum of being cost‐effective and easy to operate, while remaining highly sensitive and specific. Microbes could help address this challenge by providing a robust and inexpensive self‐manufacturing platform capable of integrating various physical and chemical signals. Here, we focus on how microbially‐derived biosensors can provide a solution to problems in environmental monitoring of harmful substances and in medical diagnostics. We present developments based on bacterial‐ as well as yeast‐based whole‐cell biosensors and conclude with recently developed cell‐free biosensors.
Whole‐cell biosensors for environmental and food monitoring
The widespread use of chemicals in industry and agriculture has led to an increased environmental release of toxic compounds and subsequent food contamination. Consequently, sensitive, rapid, reliable and cost‐effective tools are needed to detect these toxic compounds and contribute to pollution mitigation programmes and treatment strategies. Traditionally, environmental pollutants have been measured by chromatographic methods (Liu et al., 2010). However, these technologies are expensive and require specialized equipments and well‐trained users. In addition, important parameters such as bioavailability, toxicity and genotoxicity can only be assayed using living cells (Harmsen, 2007).
Microbial biosensors are naturally occurring or engineered microorganisms producing a detectable signal upon environmental stimuli (Yagi, 2007; van der Meer and Belkin, 2010). Most of the sensors were designed based on bacteria in which a promoter induced by a molecule of interest drives expression of a reporter gene, producing a colorimetric, luminometric or fluorimetric output signal. In addition to the biotechnological workhorse E. coli, different soil‐borne bacteria such as P. fluorescens, P. putida or S. aureus have been engineered as whole‐cell biosensors to reduce the influence of native soil constituents (Renella and Giagnoni, 2016). To date, a variety of target analytes such as organic xenobiotics (naphthalene, BTEX [benzene, toluene, ethylbenzene and xylene], alkylsulphonates, polychlorinated biphenyls), heavy metals and metalloids (As, Cd, Zn, Ni, Cu, Cr, Cu), or nutrients and physiologically active molecules, can be detected by different kinds of whole‐cell biosensors (for in‐depth review, see (Yagi, 2007; van der Meer and Belkin, 2010), and references therein). Additionally, soil monitoring by whole‐cell biosensors that can detect molecules such as galactose, galactoside (Bringhurst et al., 2001) or nitrate (DeAngelis et al., 2005) also provide information about plant–microbe interaction and rhizosphere ecology for sustainable agriculture development. Genotoxins, chemical compounds causing harmful DNA damage, can be detected using the umu‐test, which is based on microbial DNA repair system (Biran et al., 2010).
For food monitoring, bacterial biosensors have been used to detect residual environmental pollutants, metabolizable products (e.g. ethanol, urea) and macro‐ and micronutrients (sugars, short‐chain fatty acids, amino acids, or vitamins). Bacterial biosensors also have been applied to the detection and identification of antibiotic residues to prevent allergies, toxicological effects and the emergence of antibiotic‐resistant bacteria. Engineered bacteriophages expressing a reporter gene upon infection are also a promising platform for identifying contamination in food or beverages by pathogenic bacteria and their toxins (Smartt and Ripp, 2010).
Beyond prokaryotic biosensors, yeast‐based biosensors, mostly using Saccharomyces cerevisiae, present several advantages including robustness, easy genetic manipulation and higher‐eukaryotic sensing modalities (Shimomura‐Shimizu and Karube, 2010). Yeast biosensors detecting environmental pollutants took advantage of yeast changes in respiration activity that can be monitored using a dissolved oxygen electrode that functions as an index of the level of degradable organic compounds present in the sample (Shimomura‐Shimizu and Karube, 2010; Jarque et al., 2016). These biosensors can detect biodegradable organics (Yudina et al., 2015), toxic heavy metals such as Cu2+ (Lehmann et al., 2000) or endocrine disrupting compounds (EDCs) (Schwartz‐Mittelman et al., 2005). The utilization of transcription factors activated by a molecule of interest and controlling a reporter gene are also a common strategy in yeast biosensor engineering (Shimomura‐Shimizu and Karube, 2010; Jarque et al., 2016). The similarity of DNA repair mechanisms between yeast and higher eukaryotes has also been used to expand yeast biosensors by detecting methylation‐based DNA damage (Moser et al., 2013) or general genotoxicity indicators (Benton et al., 2007).
Whole‐cell biosensors for medical diagnostics and epidemiology
The gold standard method for detecting infectious agents remains culture isolation (Yager et al., 2008), which requires significant knowledge, training and time. Strategies for amplifying pathogens' nucleic acids are not adapted for low‐cost, point‐of‐care (POC) testing in low‐resource settings (Yager et al., 2008). Enzyme‐linked immunosorbent assay (ELISA)‐based tests are expensive and not well suited to use outside of state‐of‐the‐art laboratories (Fu et al., 2011). The majority of testing for infectious diseases in resource‐limited settings is thus performed by microscopy or agglutination tests.
For the development of POC in vitro diagnostics, bacteria able to sense quorum‐sensing molecules were engineered for detecting infections (Kumari et al., 2008). Whole‐cell biosensors have faced hurdles to operate in clinical samples due to unreliable operation and low signal‐to‐noise ratio in complex and heterogenous samples. In addition, WCBs usually have a limited signal processing capability precluding integration of several signals (e.g. multiple biomarkers) for accurate diagnosis. Recently, however, synthetic genetic circuits capable of signal amplification and multiplex processing have allowed the detection of pathological biomarkers in human clinical samples including abnormal glucosuria in the urine of diabetic patients (Courbet et al., 2015). Bacterial biosensors could further be extended to in vivo diagnosis (Slomovic et al., 2015). For example, engineered E. coli were tested in mouse models to indicate liver metastasis by producing detectable signals in urine (Danino et al., 2015), or to target cancer cells via cell‐surface expression of synthetic adhesins (Piñero‐Lambea et al., 2015). Another effective method is yeast‐based antibody display which has been applied to design highly sensitive POC technology for biosensing devices (Colby et al., 2004; Venkatesh et al., 2015; Aronoff‐Spencer et al., 2016). Antibody display was used to perform electrochemical detection of Salmonella or Hepatitis C virus (Aronoff‐Spencer et al., 2016). Another approach for yeast biosensor design uses conditionally stable ligand‐binding domains degraded in the absence of a cognate ligand to sense different small molecules (Feng et al., 2015). The use of the novo‐designed binders opens up the possibility of generating yeast biosensors for ligands with relevance in POC diagnostics.
Cell‐free systems: going beyond living cells
While microbial biosensors hold a great deal of promise, they still have several limitations. For example, cell growth phase influences genetic circuit behaviour, adding noise to system and complicating reproducibility. Additionally, many compounds cannot cross the cellular membrane and are therefore not detectable using sensing machinery in the cytoplasm. Finally, many hazardous environmental contaminants remain toxic to the biosensor host organisms (Pellinen et al., 2004). One potential workaround is the use of cell‐free protein synthesis‐based biosensors. Cell‐free protein synthesis has long been used as a research tool and for recombinant protein expression (Nirenberg and Matthaei, 1961) and is now a common tool in synthetic biology (Perez et al., 2016). Briefly, cell‐free systems use the cellular machinery from harvested cell extracts, or individually purified compounds (Shimizu et al., 2005), to produce protein from DNA without the need for a living system. Unlike WCBs in which a large percentage of cellular resources is devoted to cell survival and replication, cell‐free systems allow all extant resources to be used in the biosensor. Cell‐free systems are more tolerant to a wide range of toxins and can detect mercury and antibiotics at higher concentrations than in vivo biosensors (Pellinen et al., 2004). Interestingly, recent work has shown that cell extracts containing these synthetic gene networks could be freeze‐dried on cellulose and stored more than a year at room temperature, while still remaining active when rehydrated. These paper‐based systems were used to engineer biosensors for Ebola and Zika viruses for POC (Pardee et al., 2014, 2016). CRISPR‐based systems were also used for highly sensitive detection of nucleic acid biomarkers (Gootenberg et al., 2017). Among the other advantages of paper‐based cell‐free system is the lack of a membrane, widening the range of detectable molecules.
Challenges faced by microbially‐derived biosensors
While microbial‐derived biosensors are useful tools for the detection of a wide range of analytes, these biosensors face several technical and societal challenges that have limited their widespread adoption and use. First, several technical limitations need to be solved. The natural promoters used in many biosensors can exhibit off‐target reactivity, responding not only to the molecule of interest but also to a group of compounds which interfere with promoter function (Cases and de Lorenzo, 2005). This is even more true for systems operating in complex, noisy samples like physiological fluids. In addition, the long response time for cell growth and reporter gene production complicates WCB usability for real‐time clinical monitoring (Yagi, 2007). Many applications, in particular clinical diagnosis, require multiplexed detection and processing of several biomarkers. Synthetic biology is providing tools that could address these problems (Fig. 1) (Courbet et al., 2016; Wei and Cheng, 2016). Also, using biosensors in low‐resource settings and harsh environments requires the development of convenient and adequate encapsulation formats, for example based on hydrogels (van der Meer and Belkin, 2010). Finally, a critical hurdle is that many ligands of interest cannot be detected because no receptor for them exists in nature. Therefore, current research should push towards developing synthetic receptors that could be easily tailored to detect many ligands of interest, ideally using versatile and well‐established antibody technologies.
Figure 1.
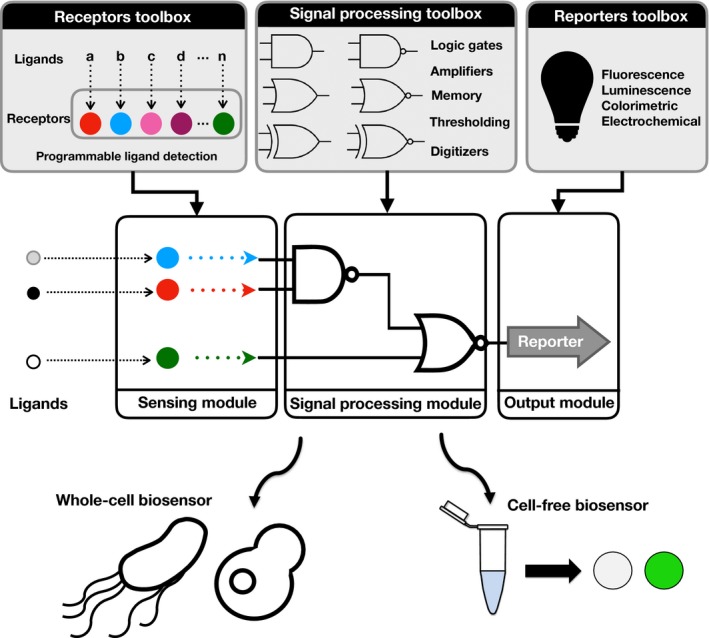
Next‐generation microbially derived biosensors. Synthetic biology research is providing an increasing number of biological parts enabling custom ligand detection, advanced signal processing and reporter output. These parts can be differentially composed into corresponding modules according to design specifications dictated by the envisioned application. Depending on the application constraints, the synthetic system obtained can be implemented either in a whole‐cell biosensor or in a cell‐free system operating on paper.
Field release of microbial biosensors also faces several regulatory hurdles, resulting in long periods of waiting time before their use validation. Recently developed kill switches (Caliando and Voigt, 2015) and synthetic auxotrophies (Malyshev et al., 2014; Mandell et al., 2015) should support tighter control of microorganism spread. As an alternative, biosensors engineered using abiotic, cell‐free systems could help bypass these issues by providing a more controlled and non‐proliferating platform. Ethical issues are even more important for in vivo clinical applications, for which long and costly clinical assays and safety assessment need to be performed. Open and constructive debates need to be regularly conducted to define the societal and cultural context in which these technologies can be deployed (Webb et al., 2017).
Conflict of interest
None declared.
Microbial Biotechnology (2017) 10(5), 1031–1035
Funding information European Research Council (ERC) Grant number: ERC‐Stg_2014_637579‐COMPUCELL
References
- Aronoff‐Spencer, E. , Venkatesh, A.G. , Sun, A. , Brickner, H. , Looney, D. , and Hall, D.A. (2016) Detection of Hepatitis C core antibody by dual‐affinity yeast chimera and smartphone‐based electrochemical sensing. Biosens Bioelectron 86: 690–696. [DOI] [PubMed] [Google Scholar]
- Benton, M.G. , Glasser, N.R. , and Palecek, S.P. (2007) The utilization of a Saccharomyces cerevisiae HUG1P‐GFP promoter‐reporter construct for the selective detection of DNA damage. Mutat Res 633: 21–34. [DOI] [PubMed] [Google Scholar]
- Biran, A. , Yagur‐Kroll, S. , Pedahzur, R. , Buchinger, S. , Reifferscheid, G. , Ben‐Yoav, H. , et al (2010) Bacterial genotoxicity bioreporters. Microb Biotechnol 3: 412–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringhurst, R.M. , Cardon, Z.G. , and Gage, D.J. (2001) Galactosides in the rhizosphere: utilization by Sinorhizobium meliloti and development of a biosensor. Proc Natl Acad Sci USA 98: 4540–4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliando, B.J. , and Voigt, C.A. (2015) Targeted DNA degradation using a CRISPR device stably carried in the host genome. Nat Commun 6: 6989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases, I. , and de Lorenzo, V. (2005) Promoters in the environment: transcriptional regulation in its natural context. Nat Rev Microbiol 3: 105–118. [DOI] [PubMed] [Google Scholar]
- Colby, D.W. , Kellogg, B.A. , Graff, C.P. , Yeung, Y.A. , Swers, J.S. , and Wittrup, K.D. (2004) Engineering antibody affinity by yeast surface display. Methods Enzymol 388: 348–358. [DOI] [PubMed] [Google Scholar]
- Courbet, A. , Endy, D. , Renard, E. , Molina, F. and Bonnet, J. (2015) Detection of pathological biomarkers in human clinical samples via amplifying genetic switches and logic gates. Sci Transl Med 7, 289ra83. [DOI] [PubMed] [Google Scholar]
- Courbet, A. , Renard, E. , and Molina, F. (2016) Bringing next‐generation diagnostics to the clinic through synthetic biology. EMBO Mol Med 8: 987–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danino, T. , Prindle, A. , Kwong, G.A. , Skalak, M. , Li, H. , Allen, K. , et al (2015) Programmable probiotics for detection of cancer in urine. Sci Transl Med 7, 289ra84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAngelis, K.M. , Ji, P. , Firestone, M.K. , and Lindow, S.E. (2005) Two novel bacterial biosensors for detection of nitrate availability in the rhizosphere. Appl Environ Microbiol 71: 8537–8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, J. , Jester, B.W. , Tinberg, C.E. , Mandell, D.J. , Antunes, M.S. , Chari, R. , et al (2015) A general strategy to construct small molecule biosensors in eukaryotes. Elife 4: e10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, E. , Yager, P. , Floriano, P.N. , Christodoulides, N. , and McDevitt, J.T. (2011) Perspective on diagnostics for global health. IEEE Pulse, 2: 40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg, J.S. , Abudayyeh, O.O. , Lee, J.W. , Essletzbichler, P. , Dy, A.J. , Joung, J. , et al (2017) Nucleic acid detection with CRISPR‐Cas13a/C2c2. Science 356: 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmsen, J. (2007) Measuring bioavailability: from a scientific approach to standard methods. J Environ Qual 36: 1420–1428. [DOI] [PubMed] [Google Scholar]
- Jarque, S. , Bittner, M. , Blaha, L. , and Hilscherova, K. (2016) Yeast biosensors for detection of environmental pollutants: current state and limitations. Trends Biotechnol 34: 408–419. [DOI] [PubMed] [Google Scholar]
- Kumari, A. , Pasini, P. , and Daunert, S. (2008) Detection of bacterial quorum sensing N‐acyl homoserine lactones in clinical samples. Anal Bioanal Chem 391: 1619–1627. [DOI] [PubMed] [Google Scholar]
- Lehmann, M. , Riedel, K. , Adler, K. , and Kunze, G. (2000) Amperometric measurement of copper ions with a deputy substrate using a novel Saccharomyces cerevisiae sensor. Biosens Bioelectron 15: 211–219. [DOI] [PubMed] [Google Scholar]
- Liu, X. , Germaine, K.J. , Ryan, D. , and Dowling, D.N. (2010) Whole‐cell fluorescent biosensors for bioavailability and biodegradation of polychlorinated biphenyls. Sensors 10: 1377–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyshev, D.A. , Dhami, K. , Lavergne, T. , Chen, T. , Dai, N. , Foster, J.M. , et al (2014) A semi‐synthetic organism with an expanded genetic alphabet. Nature 509: 385–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell, D.J. , Lajoie, M.J. , Mee, M.T. , Takeuchi, R. , Kuznetsov, G. , Norville, J.E. , et al (2015) Biocontainment of genetically modified organisms by synthetic protein design. Nature 518: 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer, J.R. , and Belkin, S. (2010) Where microbiology meets microengineering: design and applications of reporter bacteria. Nat Rev Microbiol 8: 511–522. [DOI] [PubMed] [Google Scholar]
- Moser, F. , Horwitz, A. , Chen, J. , Lim, W. , and Voigt, C.A. (2013) Genetic sensor for strong methylating compounds. ACS Synth Biol 2: 614–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirenberg, M.W. , and Matthaei, J.H. (1961) The dependence of cell‐free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proc Natl Acad Sci USA 47: 1588–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee, K. , Green, A.A. , Ferrante, T. , Cameron, D.E. , DaleyKeyser, A. , Yin, P. , and Collins, J.J. (2014) Paper‐based synthetic gene networks. Cell 159: 940–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee, K. , Green, A.A. , Takahashi, M.K. , Braff, D. , Lambert, G. , Lee, J.W. , et al (2016) Rapid, low‐cost detection of Zika virus using programmable biomolecular components. Cell 165: 1255–1266. [DOI] [PubMed] [Google Scholar]
- Pellinen, T. , Huovinen, T. , and Karp, M. (2004) A cell‐free biosensor for the detection of transcriptional inducers using firefly luciferase as a reporter. Anal Biochem 330: 52–57. [DOI] [PubMed] [Google Scholar]
- Perez, J.G. , Stark, J.C. , and Jewett, M.C. (2016) Cell‐free synthetic biology: engineering beyond the cell. Cold Spring Harbor Perspectives in Biology 8: a023853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñero‐Lambea, C. , Bodelón, G. , Fernández‐Periáñez, R. , Cuesta, A.M. , Álvarez‐Vallina, L. , and Fernández, L.Á. (2015) Programming controlled adhesion of E. coli to target surfaces, cells, and tumors with synthetic adhesins. ACS Synth Biol 4: 463–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renella, G. , and Giagnoni, L. (2016) Light dazzles from the black box: whole‐cell biosensors are ready to inform on fundamental soil biological processes. Chem Biol Technol Agric 3: 8. [Google Scholar]
- Schwartz‐Mittelman, A. , Baruch, A. , Neufeld, T. , Buchner, V. , and Rishpon, J. (2005) Electrochemical detection of xenoestrogenic and antiestrogenic compounds using a yeast two‐hybrid‐17‐beta‐estradiol system. Bioelectrochemistry 65: 149–156. [DOI] [PubMed] [Google Scholar]
- Shimizu, Y. , Kanamori, T. , and Ueda, T. (2005) Protein synthesis by pure translation systems. Methods 36: 299–304. [DOI] [PubMed] [Google Scholar]
- Shimomura‐Shimizu, M. , and Karube, I. (2010) Yeast based sensors. Adv Biochem Eng Biotechnol 117: 1–19. [DOI] [PubMed] [Google Scholar]
- Slomovic, S. , Pardee, K. , and Collins, J.J. (2015) Synthetic biology devices for in vitro and in vivo diagnostics. Proc Natl Acad Sci USA 112: 14429–14435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smartt, A.E. , and Ripp, S. (2010) Bacteriophage reporter technology for sensing and detecting microbial targets. Anal Bioanal Chem 400: 991–1007. [DOI] [PubMed] [Google Scholar]
- Venkatesh, A.G. , Sun, A. , Brickner, H. , Looney, D. , Hall, D.A. , and Aronoff‐Spencer, E. (2015) Yeast dual‐affinity biobricks: progress towards renewable whole‐cell biosensors. Biosens Bioelectron 70: 462–468. [DOI] [PubMed] [Google Scholar]
- Webb, A.J. , Kelwick, R. , and Freemont, P.S. (2017) Opportunities for applying whole‐cell bioreporters towards parasite detection. Microb Biotechnol 10: 244–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, T.‐Y. , and Cheng, C.‐M. (2016) Synthetic biology‐based point‐of‐care diagnostics for infectious disease. Cell Chem Biol 23: 1056–1066. [DOI] [PubMed] [Google Scholar]
- Yager, P. , Domingo, G.J. , and Gerdes, J. (2008) Point‐of‐Care Diagnostics for Global Health. Annual Review of Biomedical Engineering 10: 107–144. [DOI] [PubMed] [Google Scholar]
- Yagi, K. (2007) Applications of whole‐cell bacterial sensors in biotechnology and environmental science. Appl Microbiol Biotechnol 73: 1251–1258. [DOI] [PubMed] [Google Scholar]
- Yudina, N.Y. , Arlyapov, V.A. , Chepurnova, M.A. , Alferov, S.V. , and Reshetilov, A.N. (2015) A yeast co‐culture‐based biosensor for determination of waste water contamination levels. Enzyme Microb Technol 78: 46–53. [DOI] [PubMed] [Google Scholar]


