Abstract
The Runt-related transcription factor (RUNX) gene family consists of three members, RUNX1, −2 and −3, which heterodimerize with a common protein, core-binding factor β, and contain the highly conserved Runt-homology domain. RUNX1 and −2 have essential roles in hematopoiesis and osteogenesis. Runx3 protein regulates cell lineage decisions in neurogenesis and thymopoiesis. The aim of the present study was to determine the expression features of the Runx3 protein in a murine asthma model. In vivo, Runx3 protein and mRNA were found to be almost equivalently expressed in the murine lung tissue of the control, ovalbumin (OVA) and genistein groups; however, the nuclear Runx3 protein was abated in lung tissue in OVA-immunized and challenged mice. Following treatment with genistein, which is a flavonoid previously demonstrated to decrease airway inflammation in asthma, the allergic airway inflammation and airway hyper-responsiveness were attenuated and the Runx3 protein tended to augment in the nucleus. These results were further determined in vitro. These results indicated that the mislocalization of Runx3 protein is a molecular mechanism of allergic inflammation and airway hyper-responsiveness in a murine asthma model.
Keywords: asthma, airway inflammation, runx3, genistein, interleukin-2-inducible T cell kinase inhibitor
Introduction
Asthma is a chronic allergic inflammatory disease of airways that lymphocytes, eosinophils, macrophages and mast cells are associated with. In the majority of patients with asthma, allergen-specific T helper-type (Th)2 cells have been previously reported to be present in the lungs (1). Th2 cells produce cytokines that regulate allergen-specific synthesis of immunoglobulin E (IgE), eosinophil recruitment, mast cell growth and recruitment and airway hyper-reactivity (AHR), which is the primary symptom of asthma.
The function and differentiation of each T-cell subset are controlled by a subset-specific master transcription factor. As is evidenced by previous in vitro experiments (2–5), Runt-related transcription factor 3 (Runx3) interplays with these regulatory factors in various aspects of T cell-mediated immunity. Furthermore, previous studies have demonstrated that Runx3 acts as a Th1/Th2 determinant that activates the Th1 and represses the Th2 pathway (2,3,6).
Previous findings have suggested that Runx3 may be associated with the pathogenesis of asthma. Runx3 knockout mice have been demonstrated to develop spontaneous eosinophilic lung inflammation (7) and physiological conditions typically associated with asthma, including hypersensitivity to inhaled lipopolysaccharide, increased levels of serum IgE and AHR (8). In humans, the gene encoding Runx3 is located in a region of chromosome 1p36 (9), which also contains genes that have been suggested to induce asthma and hypersensitivity to environmental antigens (10,11).
However, the molecular mechanism of Runx3 in asthma remains elusive. The regulation of Runx3 is dependent on its substrate level tyrosine phosphorylation (12). Inducible T-cell kinase (ITK), a member of the Tec family of tyrosine kinases, is specifically expressed in T-cells (13); therefore, Runx3 may be a downstream transcription factor of ITK in signaling pathways that mediate asthma. Genistein, which is an interleukin-2-ITK inhibitor, has been previously demonstrated to decrease airway inflammation in allergic asthma (14), which was used to support the hypothesis that Runx3 is a downstream transcription of ITK signaling pathways.
Materials and methods
Mice
A total of 24 male Balb/c mice were purchased from Shanghai Laboratory Animal Centre (Shanghai, China). Mice (age, 6–8 weeks old; weight, 20–25 g) and were housed under specific pathogen-free conditions (humidity, 50–60%; temperature, 18–22°C) and exposed to a 12 h light-dark cycle. Mice had free access to food and water and all procedures were in accordance with the guidelines of the Animal Care Committee of Nanjing Medical University (Nanjing, China). The present study was approved by the Ethics Committee of Wuxi People's Hospital Affiliated to Nanjing Medical University.
Ovalbumin (OVA) sensitization, challenge and treatment
Balb/c mice were randomly divided into the control, asthmatic and genistein groups (n=8 in each) using a random number table. Mice in the asthmatic and genistein groups were sensitized on days 0 and 7 by intraperitoneal injection of 100 µg OVA (grade II; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) with 100 µl of 10% aluminum hydroxide gel. Two weeks post-sensitization, mice were exposed to aerosolized 1% OVA for 30 min/day in an exposure chamber (15×20×15 cm) to sensitize the airways, for seven successive days. For inhalation, OVA was dissolved in saline and aerosolized at 1 ml/min using an ultrasonic nebulizer (NE-U11B; Omron Co., Kyoto, Japan). Mice in the genistein group were administered 40 mg/kg genistein (Rongli Pharmaceutical Research Institute; Shijiazhuang, China) by interperitoneal injection 30 min prior to OVA exposure. The control group received an intraperitoneal injection of 0.1 ml saline and inhaled aerosolized saline.
Assessment of airway responsiveness to methacholine
Whole-body plethysmography was performed to measure airway hyper-responsiveness 24 h following the final OVA exposure. Mice were anesthetized by intraperitoneal injection with 70 mg/kg pentobarbital sodium (Sigma-Aldrich; Merck KGaA) and tracheostomized. Mice were subsequently placed in a whole-body plethysmograph and mechanically ventilated using an animal ventilator (AniRes2003; Beijing SYNOL High-Tech Co., Ltd., Beijing, China) at a rate of 120–150 breaths/min, with a tidal volume of 0.2 ml. Following detection of stable baseline lung resistance, increasing methacholine doses (3.125, 6.25, 12.5, 25 and 50 mg/ml) were administered by aerosol for 10 sec using an ultrasonic nebulizer, and lung resistance was recorded for 5 min. Maximum values of lung resistance were recorded.
Lung histology
The left lungs of mice were inflated with and fixed by immersion in 10% neutral buffered formalin. The tissue was routinely processed, embedded in paraffin and cut to 5-µm slices. The sections were stained with hematoxylin and eosin (H&E; Beyotime Institute of Biotechnology, Haimen, China) and observed under a microscope (Olympus-IX71; Olympus, Tokyo, Japan).
Isolation of splenic mononuclear cells and culture conditions
Two weeks post-immunization, mice were sacrificed via cervical dislocation and their spleens were aseptically excised and placed in RPMI 1640 medium (Thermo Fisher Scientific, Inc., Waltham MA, USA). The spleens were coarsely chopped and single-cell suspensions were prepared by passing the tissue through a 75-µm nylon tissue strainer. Cells were subsequently washed in RPMI 1640 medium and erythrocytes were lysed with ammonium chloride reagent (BD Pharmingen, San Diego, CA, USA). Cells were cultured in RPMI 1640 supplemented with 10% fetal calf serum, 2 mM L-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin (all from Thermo Fisher Scientific, Inc.) at a density of 1×106/ml (6 ml/well) in 6-well culture plates. Cells were cultured with 100 µg/ml OVA alone (OVA group), 100 µg/ml OVA and 1.2 µm 5-aminomethylbenzimidazole (ITK inhibitor group; Boehringer Ingelheim International GmbH; Ingelheim, Germany) or not cultured with OVA or aminomethylbenzimidazole (Control group). Subsequently, all cells were incubated in a humidified atmosphere containing 5% CO2 at 37°C for 24 h. Supernatants and cell pellets were subsequently harvested and subject to assessment of interleukin (IL)-4 and interferon (IFN)-γ release, and Runx3 protein and mRNA expression, respectively, as described below.
Reverse-transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the lung tissues from the three groups and cultured splenic mononuclear cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Equal amounts of total RNA (1 µg), as determined using an ultraviolet spectrophotometer, were used in a 20-µl complementary (c)DNA synthesis reaction system (Takara Bio, Inc., Otsu, Japan) that consisted of SYBR Premix Ex Taq II (2x, 10.0 µl), PCR forward primer (10 µM, 0.8 µl), PCR reverse primer (10 µM, 0.8 µl), ROX Reference Dye II (50x, 0.4 µl), DNA template (2.0 µl), dH2O (6.0 µl). PCR cycling conditions were as follows: 95.0°C for 30 sec; 40 cycles of 95°C (denaturation) for 5 sec, 60°C (annealing) for 34 sec; and a dissociation stage of 95°C for 15 sec, 60°C for 60 sec and 95°C for 15 sec. Control reactions without reverse transcriptase, were run in parallel. Prior to cDNA synthesis, residual genomic DNA was removed from total RNA using a DNA Eraser (Takara Bio, Inc.). qPCR was performed using a 7500 Real-time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) with SYBR-Green. PCR reactions were performed in triplicate, and the threshold cycle numbers were averaged. Gene expression levels were normalized to murine β-actin (ACTB). The relative mRNA expression levels were calculated according to the comparative Cq method in which the relative expression equals 2−ΔΔCq (15). PCR primers were designed and synthetized by Takara Bio, Inc., as follows: Murine Runx3 forward, 5′-GAACGGTGGTGACCGTGATG-3′ and reverse, 5′-TTGGTGAACACGGTGATTGTGA-3′; and murine ACTB forward, 5′-CATCCGTAAAGACCTCTATGCCAAC-3′ and reverse, 5′-ATGGAGCCACCGATCCACA-3′ as an internal control.
Western blot analysis
Lung tissues and splenic mononuclear cells were lysed in lysis buffer containing 1% Triton X-100, 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% sodium deoxycholate, 0.1% SDS, sodium orthovanadate, sodium fluoride, EDTA, leupeptin and 1 mM phenylmethylsulfonylfluoride for extraction of total protein. Nuclear protein was isolated from lung tissue and splenic mononuclear cells using a nuclei protein extraction kit (CW Biotech Co., Ltd., Beijing, China; cat. no. 07142011) according to the manufacturer's instructions. Protein concentration was determined using the bicinchoninic acid assay. A total of 40–60 µg protein lysates were separated by 10% SDS-PAGE and transferred to polyvinylidenedifluoride membranes (EMD Millipore, Billerica, MA, USA). Unspecific binding was blocked with 5% milk in Tris-buffered saline and Tween (TBST), and membranes were subsequently incubated with a 1:5,000 dilution of mouse anti-Runx3 (R 3-1E10; cat. no. D236-3; Medical and Biological Laboratories Co., Ltd., Nagoya, Japan), a 1:5,000 dilution of mouse anti-actin (5A7; cat. no. M20010; Abmart Inc., Shanghai, China) or a 1:1,000 dilution of rabbit anti-histone H3 (D1H2; cat. no. 4499; Cell Signaling Technology, Inc., Danvers, MA, USA) in blocking buffer containing 5% milk in TBST overnight at 4°C. The membranes were washed three times with TBST. Blots were developed with a 1:5,000 dilution of anti-mouse (cat. no. 0102s; CW Biotech Co., Ltd.) or a 1:5,000 dilution of anti-rabbit (cat. no. 0103s; CW Biotech Co., Ltd.) IgG horseradish peroxidase-labeled monoclonal antibody at room temperature for 1.5 h. Following incubation, the blots were washed three times using TBST. Immune complexes were detected using an enhanced chemiluminescence system (EMD Millipore) and blots were analyzed using Image J v2.1.4.7 (National Institutes of Health, Bethesda, MA, USA).
ELISA
IL-4 and IFN-γ levels in the supernatant fluid of cultured cells were detected using Mouse IL-4 ELISA kit and Mouse IFN-γ ELISA kit according to the manufacturer's protocol (eBioscience, Inc., San Diego, CA, USA).
Statistical analysis
Experiments were performed at least three times and statistical analysis was conducted using SPSS software (version 19.0; IBM SPSS, Armonk, NY, USA). Values are presented in bar graphs as the mean with error bars indicating the standard error of the mean. The differences between means were tested for statistical significance using one-way analysis of variance followed by a least-significant difference test. P<0.05 was considered to indicate a statistically significant difference.
Results
Genistein attenuates allergic airway inflammation in mice
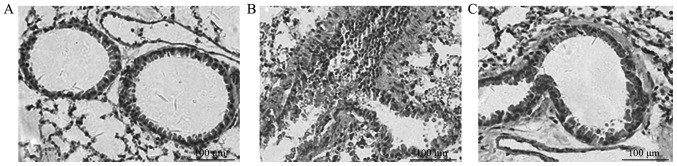
Lung specimens from the murine model of OVA-induced asthma were stained with H&E and, compared with those from control mice (Fig. 1A), showed a marked infiltration of inflammatory cells, particularly eosinophils and lymphocytes, in the interstitium surrounding blood vessels and airways (Fig. 1B). In severe cases, the inflammatory cells filled the alveolar spaces. Hyperplasia, disorders of airway epithelium and shedding of cilia from the epithelial surface were also evident in asthmatic mice. In contrast to asthmatic mice, mice pre-treated with genistein exhibited a marked reduction in the severity of allergic airway disease (Fig. 1C) and were histologically similar to the normal control mice (Fig. 1A). Detailed statistics have been previously provided by a previous study (16). These observations indicated that a murine model of asthma was successfully established and genistein was able to reduce the severity of allergic airway disease.
Figure 1.
Relief of allergic airway inflammation in mice treated with genistein. Hematoxylin and eosin staining of lung sections obtained from mice immunized and exposed to OVA and mice administered with genistein or saline. (A) Normal lungs from mice of the control group demonstrating airways and blood vessels surrounded by alveoli. (B) Lungs from OVA-induced mice. Infiltrating inflammatory cells, predominantly eosinophils, were accumulating in the interstitium surrounding the blood vessels and airways. Hyperplasia and disorders of the airway epithelium and shedding of cilia from the epithelial surface were also observed. (C) Lungs in mice that were injected with genistein, showing that damage was markedly reduced. Scale bars, 100 µm. OVA, ovalbumin. Magnification, ×200.
Genistein reduces airway hyper-responsiveness in asthmatic mice
Anesthetized tracheotomized mice were exposed to nebulized methacholine and airway resistance was measured using invasive plethysmography. Methacholine-induced airway hyper-responsiveness was elevated in the OVA group compared with the normal control group, with statistical significance for methacholine doses of 3.125–50 mg/ml (P<0.05; Fig. 2). Compared with the asthma group, the genistein group showed a significant reduction (P<0.05) of airway hyper-responsiveness at each dose of methacholine used. These results suggested that genistein treatment is able to suppress airway hyper-responsiveness in mice with OVA-induced asthma.
Figure 2.

Airway hyper-responsiveness in response to methacholine administration in mice. Plethysmography was performed 24 h following the final treatment and nebulization. Increasing doses of methacholine were administered and maximal resistance values (cm H2O/ml/sec) were recorded after 1 min. The ovalbumin-immunized mice exhibited elevated airway resistance with all methacholine doses compared with control and genistein mice (n=6/group). *P<0.05 vs. control and genistein groups. OVA, ovalbumin-immunized group; genistein, genistein-treated group; Re, responsiveness.
Runx3 mRNA and protein expression in the murine asthma model
Previous studies (2–7,17) have identified Runx3 as a Th1/Th2 determinant that activates the Th1 and represses the Th2 pathway. To determine whether Runx3 has a role in the development of asthma, Runx3 mRNA levels in the lung tissue of OVA-exposed mice were analyzed. Of note, no significant difference was observed in the expression of Runx3 mRNA in whole lung tissue between asthmatic and normal mice. Furthermore, Runx3 mRNA was expressed in asthmatic and genistein-injected mice (Fig. 3A).
Figure 3.
Expression levels of Runx3 mRNA and protein in ovalbumin-immunized and exposed mice. Relative mRNA expression was measured using real-time polymerase chain reaction with SYBR-Green. (A) Runx3 mRNA relative to murine β-actin in lung tissue of ovalbumin-immunized mice compared with that in normal controls and genistein-injected mice (n=8/group). (B) Runx3 protein expression was measured in lung tissue from control, OVA and genistein-injected mice (n=6/group). (C) Nuclear expression of Runx3 protein was determined in lung tissue from control, OVA and genistein-injected (n=6/group) mice. *P<0.05 vs. Control group; **P<0.05 vs. OVA group. Runx3, runt-related transcription factor 3; OVA, ovalbumin-immunized group; Genistein, genistein-treated group.
To evaluate Runx3 protein expression levels, western blot analysis was performed on lung tissue. Similar to the measured mRNA levels, Runx3 protein was equally expressed among the three groups (Fig. 3B).
These findings suggested that the overall expression levels of Runx3 mRNA and protein do not contribute to the development of OVA-induced asthma in mice.
Genistein augments the nuclear expression of Runx3 in the murine asthma model
As nuclear expression of Runx3 is necessary for the expression of downstream target genes, it was reasoned that there may be a difference in the nuclear content Runx3 between groups. To evaluate this potential difference, the nuclear protein levels of Runx3 were measured in each group via western blotting. The results demonstrated that the nuclear Runx3 protein levels were significantly reduced in lung tissue in asthmatic mice compared with the controls (P=0.024), whereas the expression levels of Runx3 were significantly augmented in the lung tissues of mice pre-treated with genistein (P=0.031; Fig. 3C). These results suggested that in asthmatic mice, an increased proportion of the Runx3 protein is exported from the nucleus into the cytoplasm, where it may be inactivated and that genistein may decrease cytoplasmic Runx3 protein levels. These data indicated that reduction in nuclear Runx3 expression may be an important factor in the development of OVA-induced-asthma and that genistein may reduce airway inflammation by inhibiting the translocation of Runx3 to the cytoplasm.
ITK inhibition augments the nuclear expression of Runx3 in splenic mononuclear cells in vitro
To further test the validity of the above results, splenic mononuclear cells from OVA-immunized mice were harvested and cultured. Following exposure to OVA in vitro, total mRNA and protein were extracted from splenic mononuclear cells for PCR and western blot analysis, respectively. As shown in Fig. 4A and B, Runx3 mRNA and protein levels were consistent with those observed in vivo. No significant differences were observed between the control, OVA and ITK inhibition groups.
Figure 4.
Inhibition of ITK affects the expression of Runx3 mRNA and protein. (A) Runx3 mRNA in splenic mononuclear cells in the control, OVA and ITK inhibition groups (n=8/group). (B) Total Runx3 protein expression was detected in the splenic mononuclear cells of the control, OVA, and ITK-inhibition groups (n=6/group). (C) Nuclear expression of Runx3 was detected in the splenic mononuclear cells of the control, OVA and ITK-inhibition groups (n=6/group). *P<0.05 vs. Control group; **P<0.05 vs. OVA group. ITK, inducible T-cell kinase; Runx3, runt-related transcription factor 3; OVA, ovalbumin-immunized group.
To further evaluate the results observed in vivo, the Runx3 protein levels in OVA-stimulated splenic mononuclear cells were measured (Fig. 4C). The OVA group exhibited a significant reduction of nuclear Runx3 protein compared with the control group (P=0.012). In cells cultured with 5-(aminomethyl)benzimidazole, a specific ITK inhibitor (18), the nuclear expression of Runx3 protein was significantly higher than that in the OVA group (P=0.036). These results indicated that 5-(aminomethyl)benzimidazole may augment the nuclear expression of Runx3 in vitro by suppressing the function of ITK.
ITK inhibition may activate IFN-γ and silence IL-4 expression in vitro
IFN-γ and IL-4 levels were measured in supernatant of the cultured cells. IL-4 levels in cell supernatants in the OVA group were significantly increased compared with those in the controls (P=0.0054; Fig. 5A). Conversely, IFN-γ levels were significantly decreased in the OVA group compared with those in the controls (P=0.0072; Fig. 5B). These results suggested that OVA stimulation induces an increase in Th2 activity. However, administration of 5-(aminomethyl)benzimidazole significantly suppressed the IL-4 levels and increased IFN-γ levels, indicating a reduction in the Th2 response (P=0.0038 and 0.0028, respectively).
Figure 5.

5-Aminomethyl(benzimidazole) is able to activate IFN-γ and silence IL-4 expression in vitro. (A) An increase was observed in supernatant fluid IL-4 in the OVA group compared with the control group and a decrease was observed in the ITK inhibition group compared with the OVA group. (B) A decrease was observed in supernatant fluid IFN-γ in the OVA group compared with the control group and an increase was observed in the ITK inhibition group compared with the OVA group (n=5/group). *P<0.01 vs. Control group; **P<0.05 vs. OVA group. IL, interleukin; IFN, interferon; OVA, ovalbumin-immunized group.
Discussion
The present study investigated the role of genistein in the Runx3-regulated development of airway inflammation and airway hyper-responsiveness. In vivo, genistein attenuated OVA-induced airway inflammation and decreased airway hyper-responsiveness. The results also revealed that the locus of Runx3 is important in asthma and genistein may attenuate airway inflammation by inhibiting the re-localization of Runx3 from the nucleus to the cytoplasm. Furthermore, 5-(aminomethyl)benzimidazole activated IFN-γ and silenced IL-4 expression by inhibiting the re-localization of Runx3 in vitro.
Runx3 is a member of the runt domain family of transcription factors, which regulate lineage-specific gene expression (19). Runx3 is the smallest of the Runx genes and contains the fewest number of exons, all of which are highly conserved among the three Runx genes (20) and, when mutated, are associated with human diseases (19). Djuretic et al (2) previously demonstrated that Runx3 expression is upregulated during Th1 differentiation and functions in a positive feed forward manner. T cell-specific Runx3-deficient mice have been shown to spontaneously develop asthma-associated symptoms, including elevated serum IgE, which is a hallmark of Th2 bias (6,7). Kohu et al (21) previously demonstrated that Runx3-transgenic mice exhibit a Th1-biased phenotype, including elevated titers of serum IgG2a and IgG2b following immunization. These results support the hypothesis that Runx3 is a critical regulator of Th1 and Th2 responses in vivo.
The findings of the present study demonstrated that the loci of Runx3 may have a role in the development of asthma. A body of evidence suggests that Runx3 may function as a tumor suppressor in various cancer types (22–24). Some studies have indicated that inactivation of Runx3 protein by various chemical modifications may contribute to tumor pathogenesis, initiation and progression in specific microenvironmental contexts. Cytoplasmic localization of Runx3 has been reported in various types of cancer tissue, including colorectal (25), gastric (26), oral squamous (27), and breast cancer (28). In the present study, total Runx3 protein levels were almost equal between the asthmatic and control mice, whereas nuclear Runx3 protein levels were significantly reduced in asthmatic mice. As nuclear expression of Runx3 is necessary for the expression of target genes, it can be inferred that the cytoplasmic localization will lead to inactivation of the Runx3 protein and attenuate or augment the expression of its down-stream target genes, such as IFN-γ and IL-4, respectively. In accordance with studies by the authors of the present study (29) and others (2,30), the present findings suggest that the decrease in IFN-γ levels and increase in IL-4 levels may be related to the inactivation of Runx3 protein.
IL-2-ITK, which is predominantly expressed in T cells, is a major kinase belonging to the TEC family of non-receptor tyrosine kinases. Mueller and August (31) have previously demonstrated that ITK has a role in regulating the development of airway inflammation in a murine model of allergic asthma. Lin et al (32) reported that a selective ITK inhibitor is able to alleviate lung inflammation in a mouse model of OVA-induced asthma and other studies have indicated that phosphorylation of the Runx3 protein is a major cause of cytoplasmic sequestration (33–35). These findings suggested that Runx3 may be phosphorylated by ITK, resulting in the exclusion of Runx3 from the nucleus. Therefore, in the present study ITK was inhibited in vitro by using a selective ITK inhibitor, 5-(aminomethyl)benzimidazole (14), which consequently reactivated the Runx3 protein by increasing the nuclear expression of Runx3. Following the administration of 5-(aminomethyl) benzimidazole, IL-4 and IFN-γ release in response to OVA was restored to that in the control group, exhibiting a balance of Th1/Th2 phenotypes.
Genistein, a non-selective ITK inhibitor, is one of the soy isoflavones (IF). IFs act as phytoestrogens and prevent tumorigenesis in rodent models via various bioactivities. IFs have been previously demonstrated to target all major epigenetic mechanisms regulating gene expression in in vitro and in vivo studies (36–43). Greathouse et al (44) have recently reported that genistein-mediated phosphorylation of zeste homolog 2 was able to inhibit its activity and promote benign uterine cancers in Eker rats. These results suggest that epigenetic gene regulation may be an important novel mechanism of action of genistein, which may therefore be a potential therapeutic agent for the treatment of asthma. In the present study, mice treated with genistein exhibited an increase in nuclear Runx3 protein and, of note, reduced lung allergic inflammation and airway hyper-responsiveness. These findings suggest that genistein may diminish allergic inflammation and airway hyper-responsiveness via changing the subcellular localization of the Runx3 protein.
One limitation of the present study is that, due to the lack of commercially available anti-phospho-Runx3 antibodies, it could not be elucidated whether Runx3 protein phosphorylation was modulated in the present murine model of asthma. Furthermore, it could not be determined whether Runx3 protein is a substrate for ITK. Future studies will address these issues.
In conclusion, the findings of the present study suggested that the mislocalization of Runx3 transcription factor induces airway inflammation and hyper-responsiveness in a murine asthma model. Furthermore, genistein is able to diminish the allergic inflammation and airway hyper-responsiveness by altering the subcellular localization of Runx3 protein.
Acknowledgements
This work was supported by the Science Foundation of Wuxi Health-bureau (Wuxi, China; grant no. MS201411). The authors would like to thank Rongli Pharmaceutical Research Institute for providing the genistein and Boehringer Ingelheim International GmbH for providing 5-(aminomethyl)benzimidazole.
References
- 1.Agrawal DK, Shao Z. Pathogenesis of allergic airway inflammation. Curr Allergy Asthma Rep. 2010;10:39–48. doi: 10.1007/s11882-009-0081-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Djuretic IM, Levanon D, Negreanu V, Groner Y, Rao A, Ansel KM. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat Immunol. 2007;8:145–153. doi: 10.1038/ni1424. [DOI] [PubMed] [Google Scholar]
- 3.Collins A, Littman DR, Taniuchi I. RUNX protein in transcription factor networks that regulate T-cell lineage choice. Nat Rev Immunol. 2009;9:106–115. doi: 10.1038/nri2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yagi R, Junttila IS, Wei G, Urban JF, Jr, Zhao K, Paul WE, Zhu J. The transcription factor GATA3 actively represses Runx3 protein-regulated production of interferon-gamma. Immunity. 2010;32:507–517. doi: 10.1016/j.immuni.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pham D, Kaplan MH. Antisocial networking in T helper cells. Immunity. 2010;32:500–501. doi: 10.1016/j.immuni.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 6.Naoe Y, Setoguchi R, Akiyama K, Muroi S, Kuroda M, Hatam F, Littman DR, Taniuchi I. Repression of interleukin-4 in T helper type 1 cells by Runx/Cbf beta binding to the Il4 silencer. J Exp Med. 2007;204:1749–1755. doi: 10.1084/jem.20062456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fainaru O, Woolf E, Lotem J, Yarmus M, Brenner O, Goldenberg D, Negreanu V, Bernstein Y, Levanon D, Jung S, Groner Y. Runx3 regulates mouse TGF-beta-mediated dendritic cell function and its absence results in airway inflammation. EMBO J. 2004;23:969–979. doi: 10.1038/sj.emboj.7600085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fainaru O, Shseyov D, Hantisteanu S, Groner Y. Accelerated chemokine receptor 7-mediated dendritic cell migration in Runx3 knockout mice and the spontaneous development of asthma-like disease. Proc Natl Acad Sci USA. 2005;102:10598–10603. doi: 10.1073/pnas.0504787102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levanon D, Negreanu V, Bernstein Y, Bar-Am I, Avivi L, Groner Y. AML1, AML2, and AML3, the human members of the runt domain gene-family: cDNA structure, expression, and chromosomal localization. Genomics. 1994;23:425–432. doi: 10.1006/geno.1994.1519. [DOI] [PubMed] [Google Scholar]
- 10.Haagerup A, Bjerke T, Schiøtz PO, Binderup HG, Dahl R, Kruse TA. Asthma and atopy-a total genome scan for susceptibility genes. Allergy. 2002;57:680–686. doi: 10.1034/j.1398-9995.2002.23523.x. [DOI] [PubMed] [Google Scholar]
- 11.Yokouchi Y, Shibasaki M, Noguchi E, Nakayama J, Ohtsuki T, Kamioka M, Yamakawa-Kobayashi K, Ito S, Takeda K, Ichikawa K, et al. A genome-wide linkage analysis of orchard grass-sensitive childhood seasonal allergic rhinitis in Japanese families. Genes Immun. 2002;3:9–13. doi: 10.1038/sj.gene.6363815. [DOI] [PubMed] [Google Scholar]
- 12.Andreotli AH, Schwartzberg PL, Joseph RE, Berg LJ. T-cell signaling regulated by the Tec family kinase, Itk. Cold Spring Harb Perspect Biol. 2010;2:a002287. doi: 10.1101/cshperspect.a002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwartzberg PL, Finkelstein LD, Readinger JA. TEC-family kinases: Regulators of T-helper-cell differentiation. Nat Rev Immunol. 2005;5:284–295. doi: 10.1038/nri1591. [DOI] [PubMed] [Google Scholar]
- 14.Riether D, Zindell R, Kowalski JA, Cook BN, Bentzien J, Lombaert SD, Thomson D, Kugler SZ, Jr, Skow D, Martin LS, et al. 5-Aminomethylbenzimidazoles as potent ITK antagonists. Bioorg Med Chem Lett. 2009;19:1588–1591. doi: 10.1016/j.bmcl.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Gao F, Wei D, Bian T, Xie P, Zou J, Mu H, Zhang B, Zhou X. Genistein attenuated allergic airway inflammation by modulating the transcription factors T-bet, GATA-3 and STAT-6 in a murine model of asthma. Pharmacology. 2012;89:229–236. doi: 10.1159/000337180. [DOI] [PubMed] [Google Scholar]
- 17.Wong WF, Kohu K, Chiba T, Sato T, Satake M. Interplay of transcription factors in T-cell differentiation and function: The role of Runx. Immunology. 2011;132:157–164. doi: 10.1111/j.1365-2567.2010.03381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang S, Wang Q, Crute BE, Melnikova IN, Keller SR, Speck NA. Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Mol Cell Biol. 1993;13:3324–3339. doi: 10.1128/MCB.13.6.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levanon D, Groner Y. Structure and regulated expression of mammalian RUNX gene. Oncogene. 2004;23:4211–4219. doi: 10.1038/sj.onc.1207670. [DOI] [PubMed] [Google Scholar]
- 20.Bangsow C, Rubins N, Glusman G, Bernstein Y, Negreanu V, Goldenberg D, Lotem J, Ben-Asher E, Lancet D, Levanon D, Groner Y. The RUNX3 gene-sequence, structure and regulated expression. Gene. 2001;279:221–232. doi: 10.1016/S0378-1119(01)00760-0. [DOI] [PubMed] [Google Scholar]
- 21.Kohu K, Ohmori H, Wong WF, Onda D, Wakoh T, Kon S, Yamashita M, Nakayama T, Kubo M, Satake M. The Runx3 transcription factor augments Th1 and down-modulates Th2 phenotypes by interacting with and attenuating GATA3. J Immunol. 2009;183:7817–7824. doi: 10.4049/jimmunol.0802527. [DOI] [PubMed] [Google Scholar]
- 22.Kang HF, Dai ZJ, Bai HP, Lu WF, Ma XB, Bao X, Lin S, Wang XJ. RUNX3 gene promoter demethylation by 5-Aza-CdR induces apoptosis in breast cancer MCF-7 cell line. Onco Targets Ther. 2013;6:411–417. doi: 10.2147/OTT.S43744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang YW, Eom SY, Yim DH, Song YJ, Yun HY, Park JS, Youn SJ, Kim BS, Kim YD, Kim H. Evaluation of the relationship between dietary factors, CagA-positive Helicobacter pylori infection, and RUNX3 promoter hypermethylation in gastric cancer tissue. World J Gastroenterol. 2013;19:1778–1787. doi: 10.3748/wjg.v19.i11.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li M, Tan SY, Zhang J, You HX. Effects of paeonol on intracellular calcium concentration and expression of RUNX3 in LoVo human colon cancer cells. Mol Med Rep. 2013;7:1425–1430. doi: 10.3892/mmr.2013.1372. [DOI] [PubMed] [Google Scholar]
- 25.Ito K, Lim AC, Salto-Tellez M, Motoda L, Osato M, Chuang LS, Lee CW, Voon DC, Koo JK, Wang H, et al. RUNX3 attenuates beta-catenin/T cell factors in intestinal tumorigenesis. Cancer Cell. 2008;14:226–237. doi: 10.1016/j.ccr.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 26.Ito K, Liu Q, Salto-Tellez M, Yano T, Tada K, Ida H, Huang C, Shah N, Inoue M, Rajnakova A, et al. RUNX3, anovel tumor suppressor, is frequently inactivated in gastric cancer by proteinmislocalization. Cancer Res. 2005;65:7743–7750. doi: 10.1158/0008-5472.CAN-05-0743. [DOI] [PubMed] [Google Scholar]
- 27.Gao F, Huang C, Lin M, Wang Z, Shen J, Zhang H, Jiang L, Chen Q. Frequent inactivation of RUNX3 by promoter hypermethylation and protein mislocalization in oral squamous cell carcinomas. J Cancer Res Clin Oncol. 2009;135:739–747. doi: 10.1007/s00432-008-0508-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lau QC, Raja E, Salto-Tellez M, Liu Q, Ito K, Inoue M, Putti TC, Loh M, Ko TK, Huang C, et al. RUNX3 is frequently inactivated by dual mechanisms of protein mislocalization and promoter hypermethylation in breast cancer. Cancer Res. 2006;66:6512–6520. doi: 10.1158/0008-5472.CAN-06-0369. [DOI] [PubMed] [Google Scholar]
- 29.Duan W, Kuo IC, Selvarajan S, Chua KY, Bay BH, Wong WS. Antiinflammatory effects of genistein, a tyrosine kinase inhibitor, on a guinea pig model of asthma. Am J Respir Crit Care Med. 2003;167:185–192. doi: 10.1164/rccm.200205-420OC. [DOI] [PubMed] [Google Scholar]
- 30.Lee SH, Jeong HM, Choi JM, Cho YC, Kim TS, Lee KY, Kang BY. Runx3 inhibits IL-4 production in T cells via physical interaction with NFAT. Biochem Biophys Res Commun. 2009;381:214–217. doi: 10.1016/j.bbrc.2009.02.026. [DOI] [PubMed] [Google Scholar]
- 31.Mueller C, August A. Attenuation of immunological symptoms of allergic asthma in mice lacking the tyrosine kinase ITK. J Immunol. 2003;170:5056–5063. doi: 10.4049/jimmunol.170.10.5056. [DOI] [PubMed] [Google Scholar]
- 32.Lin TA, McIntyre KW, Das J, Liu C, O'Day KD, Penhallow B, Hung CY, Whitney GS, Shuster DJ, Yang X, et al. Selective Itk inhibitors block T-cell activation and murine lung inflammation. Biochemistry. 2004;31:11056–11062. doi: 10.1021/bi049428r. [DOI] [PubMed] [Google Scholar]
- 33.Kim HR, Oh BC, Choi JK, Bae SC. Pim-1 kinase phosphorylates and stabilizes RUNX3 and alters its subcellular localization. J Cell Biochem. 2008;105:1048–1058. doi: 10.1002/jcb.21906. [DOI] [PubMed] [Google Scholar]
- 34.Kim JH, Choi JK, Cinghu S, Jang JW, Lee YS, Li YH, Goh YM, Chi XZ, Lee KS, Wee H, Bae SC. Jab1/CSN5 induces the cytoplasmic localization and degradation of RUNX3. J Cell Biochem. 2009;107:557–565. doi: 10.1002/jcb.22157. [DOI] [PubMed] [Google Scholar]
- 35.Goh YM, Cinghu S, Hong ET, Lee YS, Kim JH, Jang JW, Li YH, Chi XZ, Lee KS, Wee H, et al. Src kinase phosphorylates RUNX3 at tyrosine residues and localizes the protein in the cytoplasm. J Biol Chem. 2010;285:10122–10129. doi: 10.1074/jbc.M109.071381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Messina M. A brief historical overview of the past two decades of soy and isoflavone research. J Nutr. 2010;140:1350S–1354S. doi: 10.3945/jn.109.118315. [DOI] [PubMed] [Google Scholar]
- 37.Li Y, Meeran SM, Patel SN, Chen H, Hardy TM, Tollefsbol TO. Epigenetic reactivation of estrogen receptor-α (ERα) by genistein enhances hormonal therapy sensitivity in ERα-negative breast cancer. Mol Cancer. 2013;12:9. doi: 10.1186/1476-4598-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, Chen H, Hardy TM, Tollefsbol TO. Epigenetic regulation of multiple tumor-related genes leads to suppression of breast tumorigenesis by dietary genistein. PLoS One. 2013;8:e54369. doi: 10.1371/journal.pone.0054369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie Q, Bai Q, Zou LY, Zhang QY, Zhou Y, Chang H, Yi L, Zhu JD, Mi MT. Genistein inhibits DNA methylation and increases expression of tumor suppressor genes in human breast cancer cells. Genes Chromosomes Cancer. 2014;53:422–431. doi: 10.1002/gcc.22154. [DOI] [PubMed] [Google Scholar]
- 40.Hirata H, Hinoda Y, Shahryari V, Deng G, Tanaka Y, Tabatabai ZL, Dahiya R. Genistein downregulates onco-miR-1260b and upregulates sFRP1 and Smad4 via demethylation and histone modification in prostate cancer cells. Br J Cancer. 2014;110:1645–1654. doi: 10.1038/bjc.2014.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Li Q, Chen H. DNA methylation and histone modifications of Wnt genes by genistein during colon cancer development. Carcinogenesis. 2013;34:1756–1763. doi: 10.1093/carcin/bgt129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xia J, Cheng L, Mei C, Ma J, Shi Y, Zeng F, Wang Z, Wang Z. Genistein inhibits cell growth and invasion through regulation of miR-27a in pancreatic cancer cells. Curr Pharm Des. 2014;20:5348–5353. doi: 10.2174/1381612820666140128215756. [DOI] [PubMed] [Google Scholar]
- 43.Howard TD, Ho SM, Zhang L, Chen J, Cui W, Slager R, Gray S, Hawkins GA, Medvedovic M, Wagner JD. Epigenetic changes with dietary soy in cynomolgus monkeys. PLoS One. 2011;6:e26791. doi: 10.1371/journal.pone.0026791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greathouse KL, Bredfeldt T, Everitt JI, Lin K, Berry T, Kannan K, Mittelstadt ML, Ho SM, Walker CL. Environmental estrogens differentially engage the histone methyltransferase EZH2 to increase risk of uterine tumorigenesis. Mol Cancer Res. 2012;10:546–557. doi: 10.1158/1541-7786.MCR-11-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]