Abstract
Techniques for the extraction and use of nucleic acids from formalin-fixed and paraffin-embedded (FFPE) tissues, preserved over long time periods in libraries, have been developed. However, DNA extracted from FFPE tissues is generally damaged, and long-term storage may affect DNA quality. Therefore, it is important to elucidate the effect of long-term storage on FFPE tissues and evaluate the techniques used to extract DNA from them. In the present study, the yield, purity, and integrity of DNA in FFPE tissue samples was evaluated. Two DNA extraction techniques were used: A silica-binding DNA collection method using QIAamp DNA FFPE Tissue kit (QIA) and a total tissue DNA collection method using a WaxFree DNA extraction kit (WAX). A total of 25 FFPE tissues from lung adenocarcinomas were studied, which had been surgically resected and fixed at Okayama University Hospital prior to examination and subsequent storage at room temperature for 0.5, 3, 6, 9 and 12 years. Extracted DNA was quantified using ultraviolet absorbance, fluorescent dye, and quantitative polymerase chain reaction (qPCR). The quality of the DNA was defined by the absorbance ratio of 260 to 280 nm (A260/280) and Q-score, which is the quantitative value of qPCR product size ratio. The results demonstrated that the yield of total DNA extracted using WAX was significantly greater than when QIA was used (P<0.01); however, DNA extracted using WAX included more contaminants and was significantly more fragmented compared with DNA extracted using QIA (P<0.01). Aging had no significant effect on absolute DNA yield or DNA purity, although it did significantly contribute to increased DNA degradation for both QIA and WAX extraction (QIA P=0.02, WAX P=0.03; 0.5 years vs. 3 years, QIA P<0.01, WAX P=0.03; 9 years vs. 12 years). Both extraction methods are viable depending on whether high yield or high quality of extracted DNA is required. However, due to the increased degradation with age, storage time limits the available DNA in FFPE tissues regardless of the extraction method.
Keywords: formalin-fixed and paraffin-embedded, DNA extraction, quality check, QIAamp kit, WaxFree DNA extraction kit
Introduction
Clinical tissue samples including biopsied and resected specimens by surgical operation are generally fixed with formaldehyde and embedded with paraffin for subsequent histological and pathological examinations. Novel research techniques, such as next generation sequencing, have been developed (1,2) and the usage of DNA from stored formalin-fixed and paraffin-embedded (FFPE) tissues libraries has increased. For example, comprehensive analysis of DNA alteration with next generation sequencing enables us to identify the novel genomic alterations in patients with cancer recurrence, which may provide more therapeutic targets. Indeed, various types of mutation, including epidermal growth factor receptor (EGFR) mutations and Human EGFR2 (HER2) mutations is the target of their inhibitors gefitinib, erlotinib, and afatinib (3,4).
The majority of hospitals have a store of the resected or biopsied specimens in FFPE tissue and do not routinely preserve frozen tissues. However, prolonged formalin fixation causes the crosslinking of proteins and nucleic acids (5), and random breaks in nucleotide sequences (6), resulting in fragmentation of the nucleotides. Regarding the method of DNA extraction, while digestion of tissue samples by proteinase K followed by two extractions with phenol and chloroform is a classical standard method (7), various, more convenient and simple techniques to extract DNA from stored-FFPE tissues have been developed (8,9). Considering this, it is important to evaluate DNA from FFPE tissues carefully and select DNA extraction methods that are most suitable for research purposes.
In the present study, DNA extracted from FFPE tissue was quantitatively evaluated in terms of yield, purity and age-related fragmentation, using two different, commercially available DNA extraction techniques, silica membrane methods and total tissue DNA collection methods.
Materials and methods
Clinical samples
A total of 25 FFPE tissues and frozen tissues from same lung adenocarcinomas patients were used in the present study (total of 50 samples), whose tumors were >10 mm in diameter, and which were resected surgically at Okayama University Hospital in 2002, 2005, 2008, 2011, and 2014. A set of 5 samples was randomly collected from each year. Frozen tissues were frozen immediately following pulmonary resection and stored at −80°C, and FFPE tissues were fixed in 10% formalin overnight or for up to 2 days following surgery, dependent on when the surgery was completed, at room temperature and embedded in paraffin. The 5 sets of samples were stored for ~0.5, 3, 6, 9 and 12 years at room temperature. Ethical approval was granted by the Institutional Review Board of Okayama University Hospital and informed consent was obtained in writing from all patients. The handling of these tissues was carried out according to provisions and rules of Okayama University Hospital.
DNA extraction
For each FFPE sample, two 5-µm thick sections were cut and placed on a glass slide. Subsequently, corresponding tumor portions were scraped from the slides, in order to exclude normal lung tissue, and placed into microtubes. DNA was extracted from each sample using either the QIAamp DNA FFPE Tissue kit (QIA; Qiagen AB, Sollentuna, Sweden), which utilizes a method that binds DNA to a silica membrane, or the WaxFree DNA extraction kit (WAX; Trimgen, Sparks Glencoe, MD, USA), which utilizes a method that collects total tissue DNA. For both methods, DNA was extracted following the manufacturer's protocol. Characteristics of QIA and WAX are summarized in Table I. Frozen tissue DNA was extracted using proteinase K (Qiagen AB) treatment followed by phenol-chloroform extraction, as previously described (10).
Table I.
Characteristics of each DNA extraction kit.
| Characteristic | QIAamp DNA FFPE tissue kit (Qiagen AB, Sollentuna, Sweden) | WaxFree DNA extraction kit (Trimgen, Sparks Glencoe, MD, USA) |
|---|---|---|
| Catalog number | 56404 | WF-100 |
| Crosslink removal | Incubation at 95°C for 10 min | Incubation at 90°C for 60 min |
| Purification | Wash out using silica membrane | PCR inhibitor removal using resin-enzyme mix |
| Elution volume (µl) | 30 | 120 |
| Collected DNA | Only DNA binding to silica membrane | Total tissue DNA |
| Contaminant | Limited | Protein |
FFPE, formalin-fixed, paraffin-embedded; PCR, polymerase chain reaction.
DNA quantification
DNA quantification was performed using three methods: A method based on ultraviolet (UV) absorbance at 260 nm (UV method); a method using PicoGreen double-strand DNA (dsDNA)-specific fluorescent dye (FL method); and a method based on quantitative-polymerase chain reaction (qPCR). The UV method was performed using a LabChip DS spectrophotometer (PerkinElmer, Inc., Waltham, MA, USA). The FL method was performed using a Qubit 2.0 Fluorometer and Qubit dsDNA HS Assay kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA). qPCR was performed in triplicate using three primer pairs (provided in the kit; QC kit; Kapa Biosystems, Wilmington, MA, USA) with human genomic DNA (hgDNA) and attached DNA standard QC kit (Kapa Biosystems) and producing PCR products of 41, 129, and 305 bp. The qPCR conditions were as follows: Initial denaturation step at 95°C for 3 min, followed by 40 cycles of 95°C for 10 sec and 62°C for 30 sec. The total qPCR reaction volume was 20 µl and it included: 10X KAPA SYBER FAST qPCR Master mix, premixed with 10X Primer Premix 12 µl, 50X ROX 0.4 µl, PCR-grade water 3.6 µl and 1 ng/µl hgDNA 4 µl. A total of 50 samples underwent PCR using qPCR instrument software, StepOne Software version 2.2 (Thermo Fisher Scientific, Inc.), the average concentration of each triplicated hgDNA sample was calculated from the standard curve. The quantitative value of DNA in each PCR product size (Q41, Q129, and Q305) was determined using a standard curve. Q41, Q129, and Q305 reflect the amount of DNA quantified using 41, 129 and 305 bp primer pairs, respectively.
DNA quality check
The quality of DNA extracted by QIA and WAX was evaluated using an absorbance ratio of 260 to 280 nm (A260/280) and Q-score, which is the quantitative value ratio of each PCR product size, including Q129/Q41 and Q305/Q41. Lower DNA integrity gives a lower Q-score.
EGFR L858R mutation detection
EGFR L858R mutations of DNA derived from both of frozen tissue and FFPE, described in the DNA extraction section, were examined using PCR-based direct sequencing. The final volume of the PCR mixture was 25 µl, comprised of 2 µl DNA, 200 nmol deoxynucleotide triphosphate, 2 pmol each primer, and 1 unit HotStarTaq DNA Polymerase Plus (Qiagen AB). The PCR conditions were as follows: Initial denaturation step at 95°C for 5 min, followed by 40 cycles at 94°C for 20 sec, 60°C for 30 sec, and 72°C for 20 sec. The final extension step was performed at 94°C for 20 sec. The forward primer sequence was 5′-GCAGCCAGGAACGTACTG-3′ and the reverse primer sequence was 5′-GCCTCCTTCTGCATGGTATT-3′. The size of PCR product was 108 bp. Sequencing of PCR products was performed by FASMAC Co., Ltd. (Kanagawa, Japan).
Statistical analysis
Wilcoxon signed-rank test was used to compare paired data (comparison of DNA extracted from same FFPE blocks), and the Mann-Whitney U-test was used to compare unpaired data between two groups. Data are presented as the mean ± standard deviations. All data were analyzed using GraphPad Prism, version 6.0.3, J (GraphPad Software, Inc., La Jolla, CA, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
WAX method has a higher DNA yield than the QIA method
The mean DNA yield per sample extracted by QIA and by WAX, which were determined by the UV FL and qPCR methods are displayed in Fig. 1 and Table II. The DNA yield by WAX was significantly increased compared with QIA in UV method (Fig. 1A; P<0.01). In the FL method, DNA yield by WAX was also significantly more than that of QIA (Fig. 1B; P<0.01). Notably, the yield of DNA extracted with WAX was ~10-fold greater than that extracted with QIA with the UV method, whereas it was ~2.5-fold greater with the FL method. In the qPCR method, the Q41 yield by QIA was significantly lower than that by WAX (P<0.01), whereas there was no significant difference between WAX and QIA in yield according to Q129 and Q305 analysis (Fig. 1C).
Figure 1.

The mean DNA yield per sample extracted from QIA and WAX. (A) A method based on UV absorbance at 260 nm. (B) Quantified using PicoGreen double-strand DNA-specific fluorescent dye. (C) Quantified using the quantitative polymerase chain reaction method. **P<0.01 vs. QIA. Data are presented as the mean ± standard deviations. QIA, QIAamp; WAX, WAXFree; UV, ultraviolet; Q41, the amount of DNA quantified using 41 bp primer pairs; Q129, the amount of DNA quantified using 129 bp primer pairs; Q305, the amount of DNA quantified using 305 bp primer pairs.
Table II.
Mean DNA yields on each quantification method.
| Quantification method | QIAamp DNA FFPE tissue kit (µg/5 µm thick section) | WaxFree DNA extraction kit (µg/5 µm thick section) | P-value |
|---|---|---|---|
| Ultraviolet absorbance | 1.34±0.87 | 17.34±8.03 | P<0.01 |
| Fluorescent dye | 0.51±0.47 | 1.85±1.42 | P<0.01 |
| qPCR | |||
| −Q41 | 0.33±0.29 | 0.69±0.54 | P<0.01 |
| −Q129 | 0.13±0.15 | 0.17±0.20 | P=0.06 |
| −Q305 | 0.03±0.06 | 0.03±0.06 | P=0.44 |
All data are presented as the mean ± standard deviation. FFPE, formalin-fixed, paraffin-embedded; qPCR, quantitative polymerase chain reaction; Q41, the amount of DNA quantified using 41 bp primer pairs; Q129 the amount of DNA quantified using 129 bp primer pairs; Q305, the amount of DNA quantified using 305 bp primer pairs.
DNA extracted using QIA is of better quality than that extracted by WAX
The mean A260/280 values of DNA extracted with QIA were significantly higher than when WAX was used (P<0.01; Table III and Fig. 2A), indicating that the DNA extracted with WAX included more contaminants than that extracted with QIA. In the mean Q-scores, Q129/Q41 of DNA extracted with QIA were significantly higher than that extracted with WAX (P<0.01), although Q305/Q41 of DNA by QIA were similar to that by WAX (Table III and Fig. 2B and C). This indicates that a large quantity of DNA was fragmented to lengths <305 bp when extracted with either QIA or WAX, and the amount of DNA >129 bp but <305 bp extracted by QIA was greater than that extracted by WAX.
Table III.
Mean DNA purities and integrities.
| Quantification method | QIAamp DNA FFPE tissue kit | WaxFree DNA extraction kit | P-value |
|---|---|---|---|
| A260/280 | 2.41±0.74 | 1.13±0.07 | P<0.01 |
| Q-score | |||
| −Q129/Q41 | 0.27±0.17 | 0.20±0.16 | P<0.01 |
| −Q305/Q41 | 0.05±0.07 | 0.04±0.06 | P=0.13 |
All data are presented as the mean ± standard deviation. FFPE, formalin-fixed paraffin-embedded; Q41, the amount of DNA quantified using 41 bp primer pairs; Q129 the amount of DNA quantified using 129 bp primer pairs; Q305, the amount of DNA quantified using 305 bp primer pairs.
Figure 2.
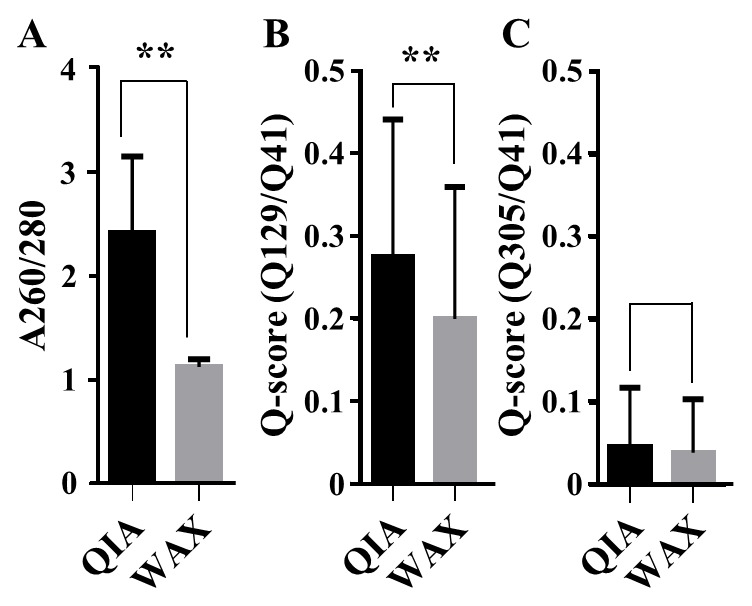
Quality of extracted DNA. (A) An absorbance ratio of 260 to 280 nm. (B and C) Quantitative value ratio of each polymerase chain reaction product size, including Q129/Q41 and Q305/Q41. **P<0.01 vs. QIA. Data are presented as the mean ± standard deviations. QIA, QIAamp; WAX, WAXFree; Q41, the amount of DNA quantified using 41 bp primer pairs; Q129, the amount of DNA quantified using 129 bp primer pairs; Q305, the amount of DNA quantified using 305 bp primer pairs, A, absorbance.
Age-related degradation of DNA
The age-related change in yield was assessed using the UV method and mean A260/280 values of DNA extracted by WAX and QIA, and no significant differences were observed (comparing DNA extracted using the same method for samples stored for different lengths of time; Fig. 3A and B).
Figure 3.

Age-related degradation of DNA. (A) The mean DNA yield per sample quantified by the UV method. (B) Quality of extracted DNA derived from each sample, quantified by an absorbance ratio of 260 to 280 nm. Quality of extracted DNA derived from each samples, quantified by qPCR with (C) Q129/Q41 and (D) Q305 and Q41. *P<0.05 vs. 0 years. #P<0.05 vs. 9 years. Data are presented as the mean ± standard deviations. UV, ultraviolet; qPCR, quantitative polymerase chain reaction; QIA, QIAamp; WAX, WAXFree; Q41, the amount of DNA quantified using 41 bp primer pairs; Q129, the amount of DNA quantified using 129 bp primer pairs; Q305, the amount of DNA quantified using 305 bp primer pairs, A, absorbance.
The Q-scores of DNAs that were extracted from FFPE tissues in each storage time are displayed in Fig. 3C and D. The Q129/Q41 and Q305/Q41 Q-scores of DNA from FFPE tissues stored for 0.5 years were significantly greater than that of tissues stored for 3 years in samples extracted using both WAX and QIA (Fig. 3C and D; QIA P=0.02, WAX P=0.03). Significant differences were also observed in the Q129/Q41 and Q305/Q41 Q-scores of DNA from FFPE tissues stored for 9 years and from those stored for 12 years (Fig. 3C and D; QIA P<0.01, WAX P=0.03). These results indicate that the fragmentation of DNA extracted using both QIA and WAX was age-related.
EGFR L858R mutation in degraded FFPE DNA was detectable
Using frozen tissue DNA, 5 samples harboring EGFR L858R mutation were detected out of 25 samples. All of these 5 EGFR L858R mutations were also detectable in corresponding FFPE-DNA, including 12 year-stored FFPE and frozen samples.
Discussion
In the present study, the age-related change in integrity of DNA extracted from FFPE tissue by WAX and QIA was evaluated. Several studies have previously evaluated the integrity of DNA from FFPE tissues or compared methods for DNA extraction from FFPE tissues (8,11–15); however, the majority of samples used in previous studies were subject to strictly controlled fixation methods and storage conditions. In the present study, the age-related degradation of DNA was assessed using various methods (UV, FL and qPCR), for FFPE tissues stored in a normal clinical hospital. The duration of formalin fixation was varied (up to 2 days) and FFPE blocks had previously been loaned to academics, thus the qualities of DNA in these FFPE tissues was generally low and unstable compared with controlled samples. The results of the present study may therefore reflect the actual status of FFPE tissues in the libraries of general clinical hospitals.
The conventional method for quantifying DNA concentration is measuring UV absorbance at 260 nm, although this is affected by many factors, such as the presence of RNA, protein, and salt (16,17). DNA purity is known to be indicated by A260/280 because the UV absorbance of protein is 280 nm (18). The mean A260/280 of DNA extracted by WAX was ~1.1 in the present study. It is reasonable to assume that the DNA extracted by QIA was purified by binding to the silica membrane, whereas the DNA extracted by WAX included more contaminants, such as proteins, due to total tissue extraction. PicoGreen dye is not affected by contaminants, however it is affected by DNA integrity (19–21). In the present study, it was demonstrated that the DNA extracted by WAX was more fragmented than that extracted by QIA. The DNA yield of WAX may be overestimated in the UV method due to contaminants and underestimated in the FL method due to fragmentation. With both the UV and FL methods, the DNA yield for samples extracted using WAX was higher than that of samples extracted using QIA; thus, the absolute dsDNA yield of samples extracted by WAX may be higher than that by QIA.
The main techniques applied following DNA extraction are PCR-based assays, and so the results of the qPCR method may provide a more meaningful indication of the amount of available DNA. In the present study, the DNA yield for samples extracted by WAX was higher than that extracted by QIA at short DNA lengths, and the amount of fragmented DNA tended to be underestimated, although the amplicon was shorter than the template DNA (18) and the DNA extracted by WAX included some PCR inhibitors (7). Developments in the last decade have allowed fragmented DNA to be used (9,22–24); it is therefore important to obtain as much DNA as possible from limited FFPE samples (25,26). WAX may therefore have some advantages over QIA with regards to DNA yield. Conversely, DNA purity was higher in samples extracted by QIA than in those extracted by WAX, and the DNA extracted by QIA was less fragmented than that extracted by WAX. Therefore, in terms of DNA quality, QIA is superior to WAX.
Several previous studies have reported that long-term storage of FFPE tissue may have no negative effects (8,27), although Adema et al (28) reported that the decrease of amplifiable DNA in paraffin blocks was age-related. The results of the present study quantitatively demonstrated that long-term storage of FFPE tissue had no effect on the amount or purity of DNA; however, DNA fragmentation was found to be age-dependent. Combined with Adema's report, this suggests that compensating for the age-related degradation of DNA using different extraction methods may be difficult, at least in general hospitals. The manufacturer of QC kit for qPCR (Kapa Biosystems) suggested that the minimum acceptable quality of sample libraries for next generation sequencing is 0.4, although the majority of DNAs extracted in the present study had a lower Q-score even with short storage times, However, the EGFR L858R mutation detected using frozen tissue DNA of 5 samples was detected in all 5 FFPE samples using direct sequencing, including in the 12 year-stored sample. This suggests that, although normal next generation sequencing using clinical FFPE tissue libraries may be difficult, it may be useful for other PCR based analyses, such as direct sequencing.
In conclusion, when comparing methods of DNA extraction from FFPE tissue samples, the total tissue extraction method provides an advantage in terms of yield, whereas the silica membrane-binding method provides an advantage in terms of quality. DNA fragmentation in FFPE tissues is age-dependent, suggesting that storage time may limit the amount of available DNA in FFPE tissue.
Acknowledgements
The authors would like to thank Ms. Yuko Hanafusa and Ms. Yayoi Kubota (Okayama University Hospital Biobank, Okayama, Japan) for their technical support.
Glossary
Abbreviations
- FFPE
formalin-fixed and paraffin-embedded
- QIA
QIAamp FFPE Tissue DNA extraction kit
- WAX
WaxFree DNA extraction kit
- dsDNA
double-strand DNA
- UV
ultraviolet
- qPCR
quantitative polymerase chain reaction
References
- 1.Hagemann IS, Devarakonda S, Lockwood CM, Spencer DH, Guebert K, Bredemeyer AJ, Al-Kateb H, Nguyen TT, Duncavage EJ, Cottrell CE, et al. Clinical next-generation sequencing in patients with non-small cell lung cancer. Cancer. 2015;121:631–639. doi: 10.1002/cncr.29089. [DOI] [PubMed] [Google Scholar]
- 2.Voelkerding KV, Dames SA, Durtschi JD. Next-generation sequencing: From basic research to diagnostics. Clin Chem. 2009;55:641–658. doi: 10.1373/clinchem.2008.112789. [DOI] [PubMed] [Google Scholar]
- 3.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 4.Suzawa K, Toyooka S, Sakaguchi M, Morita M, Yamamoto H, Tomida S, Ohtsuka T, Watanabe M, Hashida S, Maki Y, et al. Antitumor effect of afatinib, as a human epidermal growth factor receptor 2-targeted therapy, in lung cancers harboring HER2 oncogene alterations. Cancer Sci. 2016;107:45–52. doi: 10.1111/cas.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feldman M. Reactions of nucleic acids and nucleoproteins with formaldehyde. Prog Nucleic Acid Res Mol Biol. 1973;13:1–49. doi: 10.1016/S0079-6603(08)60099-9. [DOI] [PubMed] [Google Scholar]
- 6.Kibriya MG, Jasmine F, Roy S, Paul-Brutus RM, Argos M, Ahsan H. Analyses and interpretation of whole-genome gene expression from formalin-fixed paraffin-embedded tissue: An illustration with breast cancer tissues. BMC Genomics. 2010;11:622. doi: 10.1186/1471-2164-11-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hermann BG, Frischauf AM. Isolation of genomic DNA. Methods Enzymol. 1987;152:180–183. doi: 10.1016/0076-6879(87)52018-3. [DOI] [PubMed] [Google Scholar]
- 8.Okello JB, Zurek J, Devault AM, Kuch M, Okwi AL, Sewankambo NK, Bimenya GS, Poinar D, Poinar HN. Comparison of methods in the recovery of nucleic acids from archival formalin-fixed paraffin-embedded autopsy tissues. Anal Biochem. 2010;400:110–117. doi: 10.1016/j.ab.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 9.Frankel A. Formalin fixation in the ‘-omics’ era: A primer for the surgeon-scientist. ANZ J Surg. 2012;82:395–402. doi: 10.1111/j.1445-2197.2012.06092.x. [DOI] [PubMed] [Google Scholar]
- 10.Fan H, Gulley ML. DNA extraction from fresh or flozen tissues. Methods Mol Med. 2001;49:5–10. doi: 10.1385/1-59259-081-0:5. [DOI] [PubMed] [Google Scholar]
- 11.Turashvili G, Yang W, McKinney S, Kalloger S, Gale N, Ng Y, Chow K, Bell L, Lorette J, Carrier M, et al. Nucleic acid quantity and quality from paraffin blocks: Defining optimal fixation, processing and DNA/RNA extraction techniques. Exp Mol Pathol. 2012;92:33–43. doi: 10.1016/j.yexmp.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 12.Ludyga N, Grünwald B, Azimzadeh O, Englert S, Höfler H, Tapio S, Aubele M. Nucleic acids from long-term preserved FFPE tissues are suitable for downstream analyses. Virchows Arch. 2012;460:131–140. doi: 10.1007/s00428-011-1184-9. [DOI] [PubMed] [Google Scholar]
- 13.Paireder S, Werner B, Bailer J, Werther W, Schmid E, Patzak B, Cichna-Markl M. Comparison of protocols for DNA extraction from long-term preserved formalin fixed tissues. Anal Biochem. 2013;439:152–160. doi: 10.1016/j.ab.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 14.Senguven B, Baris E, Oygur T, Berktas M. Comparison of methods for the extraction of DNA from formalin-fixed, paraffin-embedded archival tissues. Int J Med Sci. 2014;11:494–499. doi: 10.7150/ijms.8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilbert MT, Haselkorn T, Bunce M, Sanchez JJ, Lucas SB, Jewell LD, Van Marck E, Worobey M. The isolation of nucleic acids from fixed, paraffin-embedded tissues-which methods are useful when? PLoS One. 2007;2:e537. doi: 10.1371/journal.pone.0000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beaven GH, Holiday ER, Johnson EA. Optical properties of nucleic acids and their components. In: The nucleic acids. In: Chargaff E, Davidson JN, editors. Chemistry and Biology. Vol. 1. Academic Press; New York, NY: 1955. pp. 493–553. [Google Scholar]
- 17.McGown E. UV absorbance measurements of DNA in microplates. Biotechniques. 2000;28(60):63–64. doi: 10.2144/00281bm11. [DOI] [PubMed] [Google Scholar]
- 18.Layne E. Spectrophotometric and tubidimetric methods for measuring proteins. Methods Enzymol. 1957;3:447–455. doi: 10.1016/S0076-6879(57)03413-8. [DOI] [Google Scholar]
- 19.Sedlackova T, Repiska G, Celec P, Szemes T, Minarik G. Fragmentation of DNA affects the accuracy of the DNA quantitation by the commonly used methods. Biol Proced Online. 2013;15:5. doi: 10.1186/1480-9222-15-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Georgiou CD, Papapostolou I. Assay for the quantification of intact/fragmented genomic DNA. Anal Biochem. 2006;358:247–256. doi: 10.1016/j.ab.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 21.Singer VL, Jones LJ, Yue ST, Haugland RP. Characterization of PicoGreen reagent and development of a fluorescence-based solution assay for double-stranded DNA quantitation. Anal Biochem. 1997;249:228–238. doi: 10.1006/abio.1997.2177. [DOI] [PubMed] [Google Scholar]
- 22.Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, Parkinson C, Chin SF, Kingsbury Z, Wong AS, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 23.Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 24.Hedegaard J, Thorsen K, Lund MK, Hein AM, Hamilton-Dutoit SJ, Vang S, Nordentoft I, Birkenkamp-Demtröder K, Kruhøffer M, Hager H, et al. Next-generation sequencing of RNA and DNA isolated from paired fresh-frozen and formalin-fixed paraffin-embedded samples of human cancer and normal tissue. PLoS One. 2014;9:e98187. doi: 10.1371/journal.pone.0098187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burgemeister R. Nucleic acids extraction from laser microdissected FFPE tissue sections. Methods Mol Biol. 2011;724:117–129. doi: 10.1007/978-1-61779-055-3_8. [DOI] [PubMed] [Google Scholar]
- 26.Cai J, Li T, Huang B, Cheng H, Ding H, Dong W, Xiao M, Liu L, Wang Z. The use of laser microdissection in the identification of suitable reference genes for normalization of quantitative real-time PCR in human FFPE epithelial ovarian tissue samples. PLoS One. 2014;9:e95974. doi: 10.1371/journal.pone.0095974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kokkat TJ, Patel MS, McGarvey D, LiVolsi VA, Baloch ZW. Archived formalin-fixed paraffin-embedded (FFPE) blocks: A valuable underexploited resource for extraction of DNA, RNA and protein. Biopreserv Biobank. 2013;11:101–106. doi: 10.1089/bio.2012.0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adema V, Torres E, Solé F, Serrano S, Bellosillo B. Paraffin treasures: Do they last forever? Biopreserv Biobank. 2014;12:281–283. doi: 10.1089/bio.2014.0010. [DOI] [PubMed] [Google Scholar]


