Abstract
The apolipoprotein B mRNA editing enzyme catalytic polypeptide-like APOBEC3A and 3B have emerged as key mutation drivers in cancer. Here we show that APOBEC3A and 3B activities impose a unique type of replication stress by inducing abasic sites at replication forks. In contrast to cells under other types of replication stress, APOBEC3A-expressing cells were selectively sensitive to ATR inhibitors (ATRi), but not to a variety of DNA replication inhibitors and DNA-damaging drugs. In proliferating cells, APOBEC3A modestly elicited ATR but not ATM. ATR inhibition in APOBEC3A-expressing cells resulted in a surge of abasic sites at replication forks, revealing an ATR-mediated negative feedback loop during replication. The surge of abasic sites upon ATR inhibition associated with increased accumulation of single-stranded DNA, a substrate of APOBEC3A, triggering an APOBEC3A-driven feedforward loop that ultimately drove cells into replication catastrophe. In a panel of cancer cell lines, ATRi selectively induced replication catastrophe in those harboring high APOBEC3A and/or 3B activities, showing that APOBEC3A and 3B activities conferred susceptibility to ATRi. Our results define an APOBEC-driven replication stress in cancer cells that may offer an opportunity for ATR-targeted therapy.
Keywords: APOBEC3A, APOBEC3B, ATR, replication stress, targeted therapy
Introduction
Genomic instability is one of the hallmarks of cancers (1). While genomic instability fuels tumorigenesis, it also offers a vulnerability of cancer cells that can be exploited therapeutically. DNA replication stress is a major source of genomic instability (2, 3). Activation of a number of oncogenes, such as MYC, RAS, and Cyclin E, induces replication stress (4–6). Inactivation of certain tumor suppressors involved in DNA repair, such as BRCA1 and BRCA2, also elevates replication stress in cancer cells (7–11). Recent cancer genomics studies by others and us revealed that cancer-associated mutations in the genes encoding mismatch DNA repair proteins and DNA polymerase epsilon (POLE), as well as cancer-associated expression of APOBEC3A and APOBEC3B, are key drivers of mutation in cancers (12, 13). Notably, the mutations inflicted by these “mutators” display a clear association with either leading or lagging strand of DNA replication forks, suggesting that they act in a replication-coupled manner (12, 14, 15). These new findings raise an important question as to whether these mutators in cancer cells induce replication stress and, if so, whether the stress induced by them can be targeted in cancer therapy.
APOBEC3A and APOBEC3B (A3A and A3B) are members of the APOBEC family of cytosine deaminases, which convert C to U in single-stranded DNA (ssDNA) (16–18). While APOBEC proteins are important for the cellular defense against foreign DNA/RNA during viral infection (19), they are not normally expressed in unstressed proliferating cells. When A3A and A3B are abnormally expressed in cancer cells, they become potent mutators of the genome. Expression of APOBEC3B is prevalent in several cancer types (17, 20–23). In-depth analysis of mutation signatures in cancers has implicated both A3A and A3B in APOBEC-mediated mutagenesis (24). An A3AB fusion gene resulting from a deletion in the APOBEC3A-APOBEC3B locus encodes A3A and associates with increased risk for breast and ovarian cancers, and with the APOBEC mutation signature in tumors (25–29). Furthermore, A3A is up regulated in a subset of leukemia (M. Weitzman, personal communications). When expressed at high levels, both A3A and A3B induce DNA double-stranded breaks (DSBs), and A3A also triggers cell cycle arrest (17, 18, 30). Together, these findings show that A3A and A3B are important drivers of mutation and genomic instability in a large subset of cancers, raising the question of how cancer cells cope with these mutators during proliferation, and whether these mutators offer an opportunity for targeted therapy.
Here, we show that A3A and A3B impose a unique type of replication stress by cytosine deamination at DNA replication forks. During DNA replication, A3A induces abasic sites in an UNG2-dependent manner, leading to modest ATR activation. Inhibition of ATR in A3A-expressing cells results in a surge of abasic sites at replication forks, revealing a previously unknown ATR-mediated feedback loop that counters A3A. The accumulation of abasic sites at replication forks upon ATR inhibition increases stalling of DNA polymerases and exposure of ssDNA, a substrate of A3A. In the absence of ATR activity, ssDNA triggers an A3A-driven feed-forward loop propelling a further buildup of abasic sites and ssDNA at replication forks, which ultimately drives cells into replication catastrophe. Interestingly, the replication stress induced by A3A renders cells sensitive to ATR inhibitors (ATRi), but not to a variety of replication inhibitors and genotoxic drugs, highlighting the unique nature of A3A-induced replication stress and the distinctive role of ATR against this stress. In a panel of cancer cell lines, ATRi rapidly induces replication catastrophe in those harboring high A3A and/or A3B activities, suggesting that the replication stress imposed by A3A and A3B may offer a promising opportunity for ATR-targeted therapy in a variety of cancers.
Materials and Methods
Cell lines
All cell lines were obtained from the Center for Molecular Therapeutics (CMT) at the MGH Cancer Center from 2015 to 2016. The CMT has obtained all cell lines described here from commercial repositories (ATTC, DSMZ, ECACC or JHSF/JCRB). Upon receipt at CMT, the cell lines were expanded and frozen stocks created. Stocks were further authenticated as follows: To identify cross-contaminated or synonymous lines, a panel of SNPs was profiled for each cell line (Sequenom, San Diego, CA) and a pair-wise comparison score calculated. In addition, we performed short tandem repeat (STR) analysis (AmpFlSTR Identifiler, Applied Biosystems, Carlsbad, CA) and matched this to an existing STR profile generated by the providing repository. From authenticated frozen stocks cells were not continuously kept in culture for more than 3 months. For the experiments described in this paper, cell lines were not continuously kept in culture for more than 3 months. All cell lines used in this study were tested for mycoplasma.
Cell culture
U2OS-derived and SKOV3-derived cell lines expressing APOBEC3A were generated by infecting U2OS cells with lentivirus expressing APOBEC3A under a Doxycycline-inducible promoter (pInducer20) and selected with G418 (400 µg/mL). U2OS derivative cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10 % Fetal Bovine Serum (FBS) and 1 % penicillin/streptomycin. For APOBEC3A expression, cells were incubated with Doxycycline (200 ng/mL) 20 h before additional treatment. For BrdU labeling, cells were incubated with 10 µM BrdU for 48 h. OVCAR5, SKOV3, NCI-H2347 and HCC78 were maintained in Roswell Park Memorial Institute 1640 Medium (RPMI 1640) GlutaMAX™-I supplemented with 10 % FBS, 1 % penicillin/streptomycin, 1 % Glucose and 1 % Sodium Pyruvate. TOV21G, OV17R, BICR6, BHY, HSC4 and BICR31 were maintained in DMEM / F12 GlutaMAX™-I supplemented with 10 % FBS and 1 % penicillin/streptomycin. SKBR3 was maintained in McCoy's 5A supplemented with 10 % FBS and 1 % penicillin/streptomycin. The cell lines above were purchased from either ATCC or Sigma-Aldrich.
Inhibitors
The kinase inhibitors used in this study were: ATRi (10 µM VE-821, Selleckchem), ATRi#2 (1 µM AZ-20, custom-made), Roscovitine (12 µM Selleckchem), DNA-PKi (2 µM NU7441, Selleckchem), ATMi (10 µM KU-55933, Selleckchem), PARP inhibitor (AZD2281, Selleckchem) and Wee1i (MK-1775, Selleckchem). When various inhibitors were used in combination with ATRi, they were added to cell cultures at the same time as ATRi unless indicated otherwise.
Antibodies
The antibodies used in this study were: γH2AX monoclonal antibody (Cell Signaling and EMD Millipore), APOBEC3B polyclonal antibody (Santa Cruz), Flag-M2 monoclonal antibody (Sigma-Aldrich), CDC7 monoclonal antibody (Abcam), PCNA monoclonal antibody (Santa Cruz), Chk1 monoclonal antibody (Santa Cruz), BrdU monoclonal antibody (BD Biosciences), PCNA polyclonal antibody (Abcam), Chk2 pT68 polyclonal antibody (Cell Signaling), Chk1 pS317 polyclonal antibody (Cell Signaling), GAPDH polyclonal antibody (EMD Millipore), UNG polyclonal antibody (Novus), RPA32 monoclonal antibody (Thermo Fisher Scientific), RPA70 polyclonal antibody (Bethyl), APE1 monoclonal antibody (Santa Cruz), Biotin polyclonal antibody (Abcam) and ATR polyclonal antibody (Bethyl).
Results
APOBEC3A and APOBEC3B activate ATR but not ATM in proliferating cancer cells
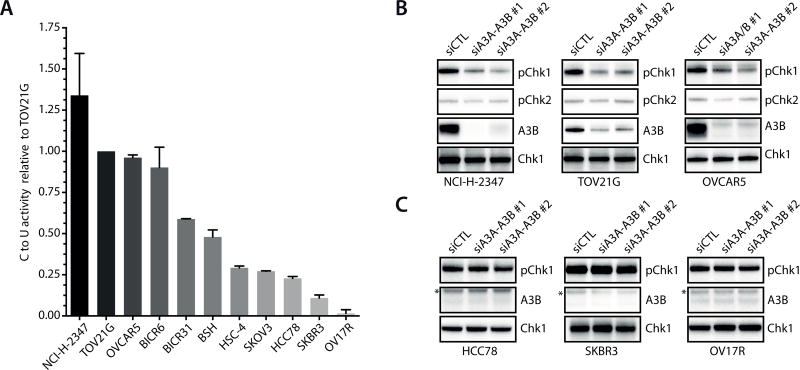
Recent studies by others and us revealed that the mutation signatures of A3A and A3B are associated with the lagging strand of DNA replication forks, suggesting that A3A and A3B act on ssDNA during DNA replication (12, 14, 15). To investigate whether A3A and A3B induce replication stress, we analyzed the expression data of A3A and A3B in a large panel of cancer cell lines (Supplementary Fig. S1A). Consistent with previous studies, A3B mRNA was detected in a large fraction of cancer cell lines (17, 21). A3A mRNA was also detected in a subset of the cell lines. We selected a group of cell lines that express A3A and A3B mRNAs at various levels according to the microarray data, and then determined the relative levels of A3A and A3B mRNAs using RT-qPCR (Supplementary Fig. S1B). Furthermore, we tested these cell lines for A3A and A3B activities using a previously described in vitro assay (Supplementary Fig. S1C–E) (17). We measured the overall A3A–A3B activity because the enzymatic activities of A3A and A3B are indistinguishable in this in vitro assay. A range of A3A–A3B activities were detected in this group of cell lines (Fig. 1A). Notably, although cell lines with high overall A3A–A3B activity tend to express high levels of A3A and A3B mRNAs, neither A3A nor A3B mRNA alone correlated with overall A3A–A3B activity precisely (Fig. 1A, Supplementary Fig. S1B), suggesting that both enzymes may contribute to the activity in cancer cells.
Figure 1. APOBEC3A and APOBEC3B activate ATR in cancer cells.
A. Relative A3A–A3B activity in a panel of cancer cell lines. A3A–A3B activity was determined from 20 µg cell extracts as showed in Supplementary Fig. S2. The activity of TOV12G is defined as 1. Cell lines are ranked according to their overall A3A–A3B activity. Error bar: S.D. (n = 3). B–C. The three high-A3A–A3B activity lines (B) and the three low-A3A–A3B activity lines (C) were transfected with control siRNA or A3A–A3B siRNAs. Levels of baseline p-Chk1 and p-Chk2 were analyzed by Western blot. *, a non-specific band.
Next, we further characterized the three cell lines with the highest A3A–A3B activity and the three cell lines with the lowest A3A–A3B activity in this panel (Fig. 1A). All these cell lines displayed baseline levels of phosphorylated Chk1 (p-Chk1) (Fig. 1B–C), suggesting that the ATR-Chk1 pathway is modestly elicited by intrinsic replication stress during proliferation. To determine if A3A and A3B induce replication stress and contribute to the ATR response, we used multiple siRNAs to knock down both A3A and A3B (Supplementary Fig. S2A–C). In TOV21G, one of the cell lines harboring high A3A–A3B activity, A3A–A3B siRNAs eliminated A3A–A3B activity while only slightly reduced S-phase cells (Supplementary Figs. S2B, 2D). The baseline p-Chk1 in the three cell lines with high A3A–A3B activity, but not that in the three lines with low A3A–A3B activity, was reduced by knockdown of A3A and A3B (Fig. 1B–C). In contrast to Chk1, Chk2, a substrate of ATM, was not phosphorylated in an A3A–A3B-dependent manner (Fig. 1B). Thus, unlike in cells arrested by high levels of A3A (31), only ATR but not ATM is modestly elicited by the endogenous A3A–A3B activity in proliferating cancer cells. These results imply that at levels tolerable in proliferating cancer cells, A3A and A3B primarily induce replication stress but not DSBs.
ATR protects cells from APOBEC-induced replication stress
To investigate the mechanism by which A3A and A3B induce replication stress, we sought to conditionally express A3A or A3B in proliferating cells. Because A3A shares similar activity with A3B and is more potent than A3B in inducing genomic instability (17), we chose to express A3A in U2OS, a cell line widely used in the studies of replication stress responses. As a negative control for wild-type A3A (A3AWT), the catalytically inactive A3A mutant (A3AE72A) was also conditionally expressed (17). At the levels expressed in these cell lines, neither A3AWT nor A3AE72A significantly affected cell proliferation (Supplementary Fig. S3A). γH2AX, a DNA damage marker, was not detectable in these cell lines (Supplementary Fig. S3B). However, a low level of p-Chk1 was detected in cells expressing A3AWT but not A3AE72A (Fig. 2A). In contrast, Chk2 was not phosphorylated in A3AWT-expressing cells (Fig. 2A). These results suggest that conditional expression of low levels of A3AWT in cycling cells induces replication stress but not DSBs, recapitulating the impact of endogenous A3A–A3B in cancer cells.
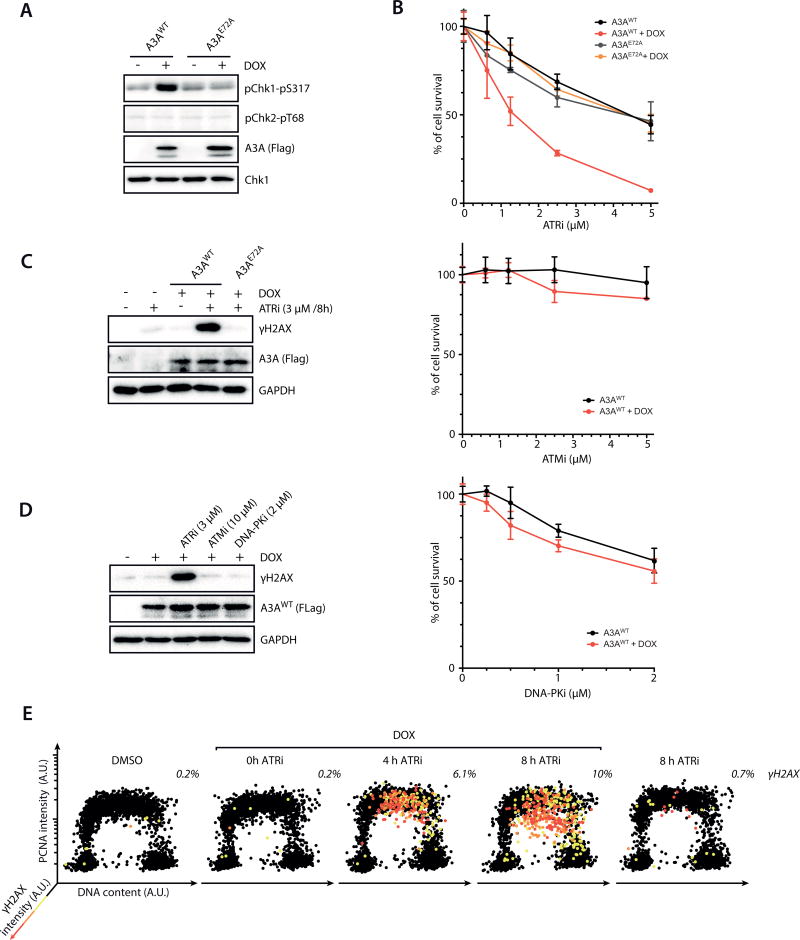
Figure 2. ATR counters APOBEC3A-induced replication stress.
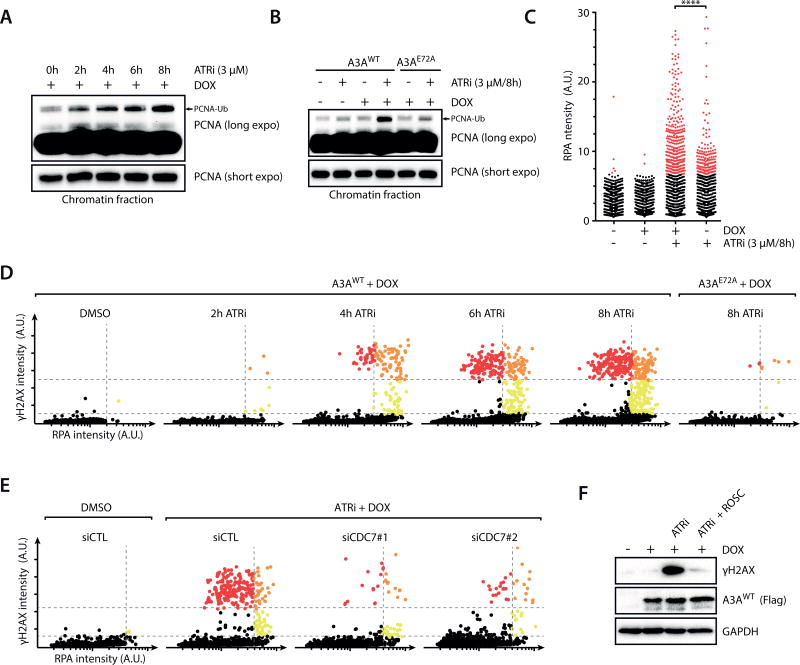
A. U2OS-derived inducible cell lines of A3AWT and A3AE72A were induced with doxycycline (DOX) or left uninduced. Levels of p-Chk1 and p-Chk2 were analyzed by Western blot. B. Cells expressing A3AWT or A3AE72A were treated with ATRi, ATMi, or DNA-PKi as indicated. Cell survival was analyzed after 5 days of continuous treatments. Error bar: S.D. (n=3). C–D. Levels of γH2AX in cells expressing A3AWT or A3AE72A after 8 h of treatments with ATRi, ATMi, or DNA-PKi. E. Cells were induced to express A3AWT or left uninduced, and treated with DMSO or ATRi (3 µM) for the indicated amounts of time. Levels of chromatin-bound PCNA, γH2AX, and DNA contents of 4,000 cells were quantified, and plotted along the 3 indicated axes. Cells were colored according to the intensity of γH2AX staining.
ATR plays a critical role in protecting cells against DNA replication stress. Indeed, the ATR inhibitor (ATRi) VE-821 (32) killed cells more effectively when A3AWT was induced (Fig. 2B). When A3AWT was expressed in an ovarian cancer cell line (SK-OV3) harboring low endogenous A3A–A3B activity, it also increased the sensitivity to VE-821 (Supplementary Fig. S3C). A second ATRi, AZ20 (33), also killed A3AWT-expressing cells preferentially (Supplementary Fig. S3D). VE-821 preferentially killed cells expressing A3AWT but not A3AE72A (Fig. 2B), suggesting that A3A activity and ATR inhibition are incompatible. Interestingly, ATRi did not affect the overall A3A–A3B deaminase activity in cells (Supplementary Fig. S3E), suggesting that A3A is not activated but acts differently upon ATR inhibition. In contrast to ATRi, inhibitors of ATM and DNA-PK (ATMi and DNA-PKi), two other PI3K-like kinases involved in the DNA damage response (DDR), did not kill A3AWT-expressing cells preferentially (Fig. 2B). These results provide further evidence that A3A induces replication stress, and that A3A activity creates a specific reliance on ATR.
To understand how ATR protects cells against A3A-induced replication stress, we analyzed the effects of ATR loss in A3A-expressing cells. Treatment of A3AWT-expressing cells with a low concentration of ATRi that only partially inhibits ATR, or with ATR siRNA, drastically increased γH2AX levels (Fig. 2C, Supplementary Fig. S4A–C) (34). The induction of γH2AX by ATRi was not observed in A3AE72A-expressing cells (Fig. 2C). The ATRi-treated A3AWT-expressing cells displaying intense γH2AX signals were also TUNEL-positive, showing that they were undergoing severe chromosome fragmentation (Supplementary Fig. S4D). In contrast to ATRi, neither ATMi nor DNA-PKi induced γH2AX in A3AWT-expressing cells (Fig. 2D). To determine when A3AWT and ATRi induce DSBs during the cell cycle, we sorted cells according to DNA content, chromatin-bound PCNA, and γH2AX staining (Fig. 2E). The cells displaying γH2AX signals were also positive for PCNA, suggesting that DSBs arise in S phase. Together, these results suggest that the action of A3A modestly elicits ATR, which in turn suppresses A3A-induced DSBs during DNA replication via a feedback loop.
APOBEC3A-induced replication stress is unique
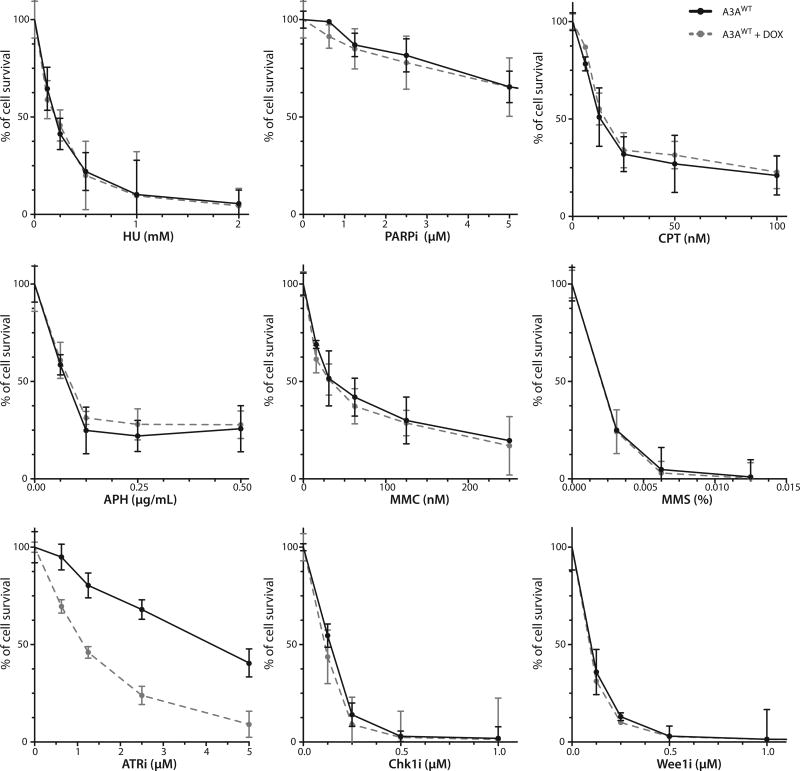
In addition to ATRi, inhibitors of several other checkpoint or DDR proteins, such as PARP, Chk1 and Wee1, are known to preferentially kill cancer cells under replication stress (35). Furthermore, several replication inhibitors and DNA-damaging drugs, such as HU (hydroxyurea), APH (aphidicolin), MMC (mitomycin C), MMS (methyl methanesulfonate), and CPT (camptothecin), also kill cells by interfering with replication. To further understand the nature of A3A-induced replication stress, we performed a “mini screen” using cells with or without A3AWT against a panel of replication inhibitors and DNA-damaging drugs. Surprisingly, none of HU, APH, MMC, MMS, and CPT preferentially killed A3AWT-expressing cells as ATRi did (Fig. 3). Moreover, none of the inhibitors of PARP, Chk1 and Wee1 showed a clear selectivity toward A3AWT-expressing cells as ATRi did (Fig. 3).
Figure 3. APOBEC3A-expressing cells are specifically sensitive to ATR inhibitor.
U2OS-derived inducible cell lines of A3AWT and A3AE72A were induced with DOX or left uninduced, and treated with increasing concentrations of the indicated inhibitors or DNA-damaging agents. Cell survival was analyzed after 5 days of continuous treatments. Error bar: S.D. (n=3).
Given that Chk1 is a downstream effector of ATR in the replication stress response, it is surprising that ATRi is more selective than Chk1i toward A3AWT-expressing cells. Even when Chk1i was used at low concentrations, continuous Chk1i treatments were unable to distinguish cells with or without A3AWT (Supplementary Fig. S4E). However, compared with uninduced cells, cells induced to express A3AWT were more sensitive to transient Chk1i treatments (Supplementary Fig. S4F). These results suggest that Chk1i also has some selectivity toward A3AWT-expressing cells, although it is not as strong as that of ATRi. We recently showed that ATRi is more selective than Chk1i toward cells under high replication stress due to activation of the DNA-PK–Chk1 pathway upon ATR inhibition (34). It is possible that the basal replication stress in U2OS cells is too high to reveal the selectivity of Chk1i toward A3AWT-expressing cells.
The selective sensitivity of A3AWT-expresssing cells to ATRi is surprising because cells defective in DNA replication, DNA repair, or the checkpoint are generally sensitive to multiple drugs/inhibitors in the panel above. If A3AWT induces replication stress in the same way as some of the drugs/inhibitors, the effects of A3AWT and these drugs/inhibitors should be additive and A3AWT expression should render cells more sensitive to these drugs/inhibitors. The inability of these drugs/inhibitors to distinguish cells with or without A3AWT suggests that the replication stress induced by A3A is unique, and that ATR has a specific role in countering this stress (see discussion).
APOBEC3A induces replication stress through abasic sites
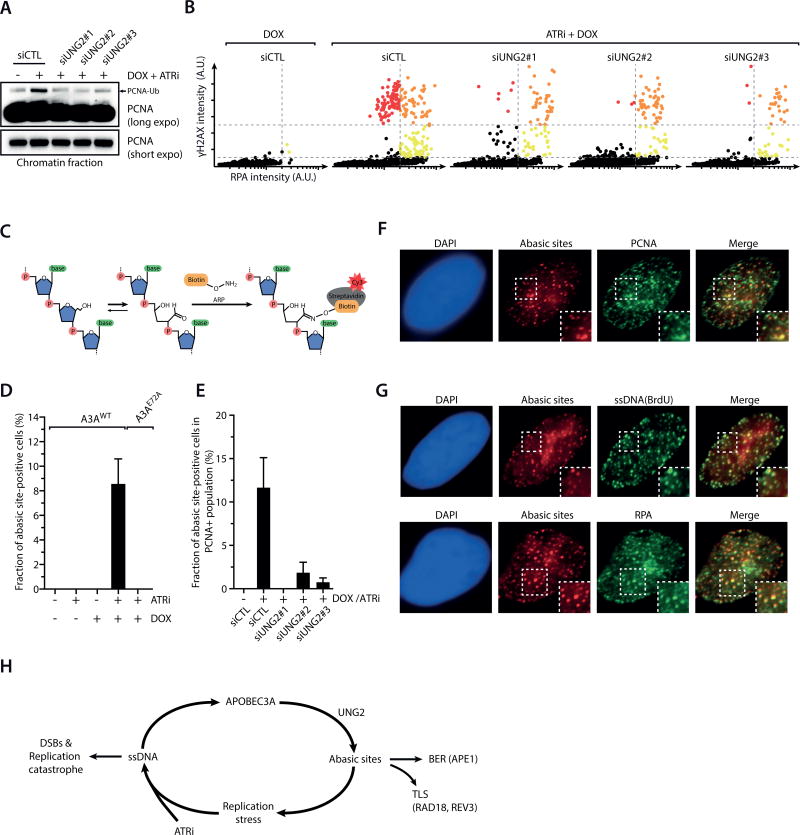
To understand why A3A-induced replication stress is unique and how ATR counters it, we next interrogated the induction of A3A-dependent DSBs by ATRi. APOBEC proteins catalyze C-to-U changes in ssDNA, which are subsequently converted to abasic (AP) sites by the uracil-DNA glycosylase UNG2 (30, 36). In S-phase cells expressing A3AWT, the induction of γH2AX by ATRi were significantly reduced by knockdown of UNG2 (Fig. 4A, Supplementary Fig. S5A). UNG2 knockdown also partially suppressed the killing of A3AWT-expressing cells by ATRi (Fig. 4B–C). These results suggest that UNG2-generated AP sites are necessary for the induction of DSBs by ATRi. Notably, UNG2 knockdown also reduced the baseline p-Chk1 in A3AWT-expressing cells (Supplementary Fig. S5B), suggesting that AP sites are required for the activation of ATR by A3AWT during replication. Thus, A3A-induced and UNG2-generated AP sites trigger modest ATR activation, and promote DSB formation when ATR is inhibited.
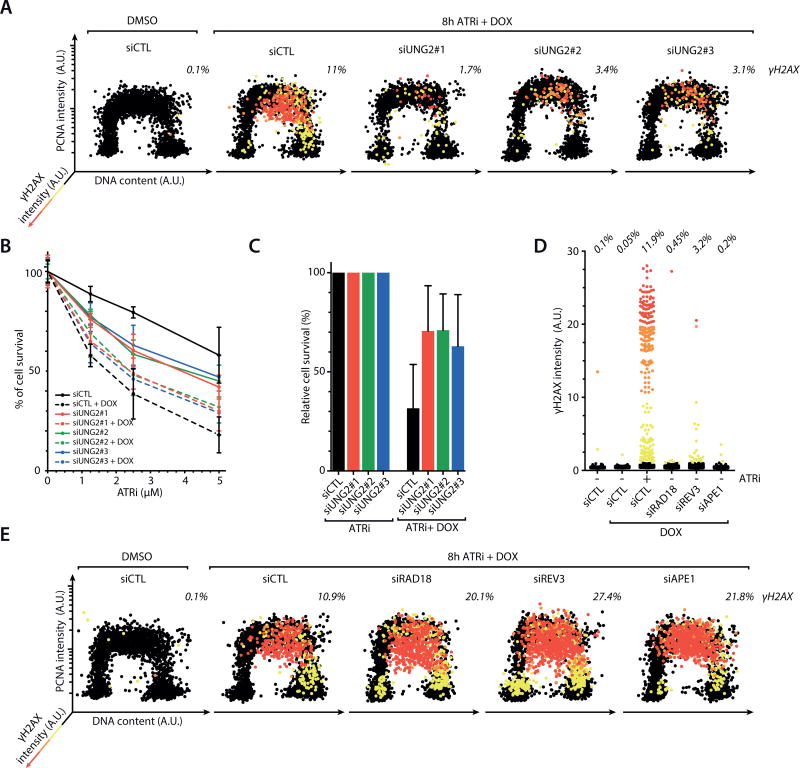
Figure 4. APOBEC3A induces DSBs through abasic sites upon ATR inhibition.
A. Inducible A3AWT cells were transfected with control siRNA or 3 independent UNG2 siRNAs, and treated with DMSO or ATRi (3 µM) + DOX. Levels of chromatin-bound PCNA, γH2AX, and DNA contents of 4,000 cells were quantified and plotted along the 3 indicated axes. Cells were colored according to the intensity of γH2AX staining. B–C. Inducible A3AWT cells were transfected with UNG2 siRNAs, and treated with increasing concentrations of ATRi (B) or with 5 µM of ATRi (C) in the presence or absence of DOX. Cell survival was analyzed after 5 days of continuous treatments. Error bar: S.D. (n=3). D. Quantification of γH2AX intensity in 2,000 inducible A3AWT cells treated with various siRNAs, ATRi, and DOX as indicated. E. Inducible A3AWT cells were treated and analyzed as in A except that RAD18, REV3, and APE1 siRNAs were used.
Several mechanisms are involved in the response to AP sites in the genome. During base excision repair (BER), AP sites are processed by the AP endonuclease APE1 and subsequently removed (37). In S phase, translesion DNA synthesis (TLS) is important for DNA replication through abasic template, allowing cells to tolerate AP sites (38, 39). To determine how ATR suppresses AP site-induced DSBs, we tested if ATR regulates APE1 or TLS. In contrast to ATR inhibition, knockdown of APE1 or key TLS factors RAD18 and REV3 only modestly increased γH2AX levels in A3AWT-expresssing cells (Fig. 4D, Supplementary Fig. S5C–D). In addition, knockdown of APE1 or the TLS factors further increased γH2AX levels in ATRi-treated A3AWT-expressing cells (Fig. 4E). These results suggest that ATR suppresses AP site-induced DSBs independently of BER and TLS.
To further investigate how ATR suppresses A3A-induced DSBs during DNA replication, we analyzed PCNA mono-ubiquitination (PCNA-Ub), which is triggered by stalling of DNA polymerases (40). While A3AWT alone induced a low level of p-Chk1 (Fig. 2A), it did not detectably induce PCNA-Ub (Fig. 5A), suggesting that polymerase stalling is modest in the presence of functional ATR. However, ATRi treatment of A3AWT-expressing cells, but not cells expressing A3AE72A, induced PCNA-Ub in a time-dependent manner (Fig. 5A–B). Consistently, A3AWT alone did not induce chromatin-bound RPA, which reflects the ssDNA resulting from polymerase stalling (Fig. 5C). The chromatin-bound RPA in A3AWT-expressing cells was significantly increased by ATRi, suggesting that A3AWT induces substantially more polymerase stalling upon ATR inhibition. The cells with high levels of chromatin-bound RPA gradually became strongly γH2AX-positive (Fig. 5D), indicating replication catastrophe driven by excessive ssDNA accumulation (41). Partial knockdown of RPA, which increases ssDNA exposure (41), significantly enhanced the induction of γH2AX even in absence of ATRi (Supplementary Fig. S5E–F). If ATRi induces replication catastrophe in A3AWT-expressing cells, this process should be suppressed by loss of CDC7 and CDKs (34), which reduces replication initiation. Indeed, the induction of PCNA-Ub, ssDNA, and γH2AX by ATRi in A3AWT-expressing cells was significantly reduced by CDC7 knockdown (Fig. 5E, Supplementary Fig. S5G–I). Roscovitine, a CDK inhibitor, also decreased ssDNA and γH2AX (Fig. 5F, Supplementary Fig. S5J). Thus, when cells undergo robust replication in the presence of A3A, ATR inhibition triggers a cascade of polymerase stalling, ssDNA accumulation, and replication catastrophe.
Figure 5. ATR inhibition in APOBEC3A-expressing cells leads to replication catastrophe.
A–B. Levels of PCNA and mono-ubiquitinated PCNA in the chromatin fractions of ATRi-treated A3AWT or A3AE72A-expressing cells were analyzed by Western blot. C. Levels of chromatin-bound RPA were analyzed in 1,000 inducible A3AWT cells after induction with DOX and treatment with ATRi as indicated. Significance was determined by t-test. ****, P<0.0001. D. Cells expressing A3AWT or A3AE72A were treated with DMSO or ATRi (3 µM) as indicated. Levels of chromatin-bound RPA and γH2AX in 5,000 cells were quantified. Cells with medium or high levels of RPA and γH2AX are divided by dotted lines and colored differently. The reduction of RPA in red cells compared to orange cells is likely due to severe chromosome fragmentation. E. Inducible A3AWT cells were transfected with control or CDC7 siRNAs, induced with DOX, and treated with ATRi (3 µM) for 8h. Levels of chromatin-bound RPA and γH2AX in 3,000 cells were quantified. F. Cells were induced to express A3AWT or left uninduced, and treated with ATRi (3 µM) and Roscovitine (12 µM) as indicated for 8 h. Levels of γH2AX were analyzed by Western blot.
ATR suppresses accumulation of abasic sites at replication forks
Given that A3A activates ATR and ATR suppresses A3A-induced DSBs, we next asked if the action of A3A is regulated by ATR. Knockdown of UNG2 reduced PCNA-Ub and γH2AX in ATRi-treated A3AWT-expressing cells (Fig. 6A–B), suggesting that A3A-induced replication stress arises from AP sites. To test if A3A induces AP sites at replication forks, we used a biotin-labeled aldehyde-reactive probe (ARP) to visualize AP sites in cells (Fig. 6C) (42). Neither A3AWT nor ATRi alone induced detectable ARP signals (Fig. 6D). However, significant levels of ARP foci were detected when both A3AWT and ATRi were present. The ARP foci were dependent on A3A activity and UNG2 (Fig. 6D–E), confirming the specificity of this assay for AP sites. Foci of AP sites were only detected in S-phase cells, and they colocalized with PCNA (Fig. 6F, Supplementary Fig. S6A–B), showing that AP sites accumulate at replication forks. Consistently, AP foci were reduced by CDC7 knockdown and roscovitine (Supplementary Fig. S6C–D), indicating the association of AP site formation with ongoing replication. APOBECs are known to act on ssDNA (19). Indeed, AP sites colocalized with RPA and the ssDNA detected by native BrdU staining (Fig. 6G). These results suggest that ATR suppresses A3A-induced DSBs by limiting the action of A3A on ssDNA at replication forks (Fig. 6H).
Figure 6. ATR inhibition increases abasic site accumulation at replication forks.
A–B. Cells were induced to express A3AWT or left uninduced, transfected with control siRNA or 3 independent UNG2 siRNAs, and treated with DMSO or ATRi (3 µM) for 8 h. Levels of PCNA and mono-ubiquitinated PCNA in the chromatin fractions were analyzed by Western blot in A, and levels of chromatin-bound RPA and γH2AX in 5,000 cells were quantified in B. C. A schematic diagram of the chemical reaction in which an Aldehyde Reactive Probe (ARP) binds to an abasic site in DNA. ARP probes were detected with Cy3-conjugated Streptavidin. D–E. Cells were transfected with control or UNG2 siRNAs (E), induced to express A3AWT, A3AE72A, or left uninduced, and treated with ATRi (3 µM) for 8h. Fractions of abasic site-positive cells in total cell populations (D) or PCNA-positive cell populations (E) were quantified. Error bar: S.D. (n = 3). F. The abasic sites induced by A3AWT and ATRi (3 µM for 8 h) colocalize with PCNA, a marker of replication forks. G. The abasic sites induced by A3AWT and ATRi (3 µM for 8h) colocalize with ssDNA (native BrdU staining) and RPA. H. A model that explains how A3A and ATRi drive a feed-forward loop that generates abasic sites, elevates DNA replication stress, and promotes replication catastrophe.
Collectively, the results above suggest that ATRi promotes formation of AP sites at replication forks by providing more ssDNA for A3A to act upon (Fig. 6H). The elevation of AP sites at forks impedes DNA Polymerases (43), propelling a further buildup of ssDNA and AP sites at replication forks, which ultimately drives cells into replication catastrophe. In this model, both A3A and ATRi are necessary drivers of a unique feed-forward loop leading to replication catastrophe, explaining the synthetic lethality between A3A activity and ATR inhibition.
The overall activity of endogenous APOBEC3A and APOBEC3B renders cancer cells susceptible to ATRi
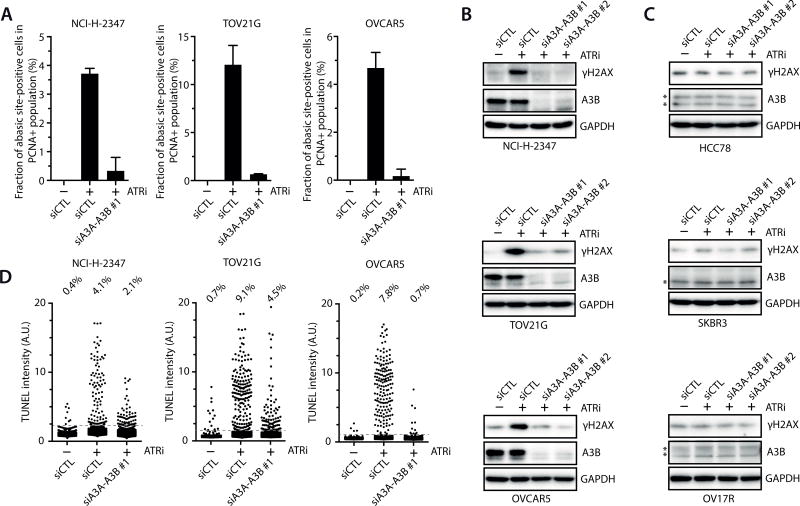
Since ATRi promotes formation of ssDNA, AP sites, and DSBs in cells induced to express A3AWT, we asked if ATRi exerts similar effects in cancer cells harboring high endogenous A3A–A3B activities. The three cancer cell lines with high overall A3A–A3B activity and the three cell lines with low A3A–A3B activity had similar fractions of S-phase cells (Fig. 1A–C, Supplementary Fig. S7A). In the three cell lines with high A3A–A3B activity, ATRi efficiently induced AP sites and γH2AX, both of which were suppressed by knockdown of A3A and A3B (Fig. 7A–B). In TOV12G, ATRi induced replication catastrophe in cells with high levels of ssDNA in a manner dependent on A3A (Supplementary Fig. S7B). In contrast, ATRi did not induce γH2AX in the three cell lines with low A3A–A3B activity (Fig. 7C). Furthermore, ATRi induced strong TUNEL signals in the three cell lines harboring high A3A–A3B activity, which were also suppressed by knockdown of A3A and A3B (Fig. 7D). In contrast, ATRi did not increase TUNEL signals in the three lines with low A3A–A3B activity in an A3A–A3B-dependent manner (Supplementary Fig. S7C). Together, these results suggest that the overall activity of endogenous A3A and A3B in cancer cells is sufficient to render them susceptible to ATR inhibition.
Figure 7. APOBEC3A and APOBEC3B render cancer cells susceptible to ATRi.
A. The three cell lines with high overall A3A–A3B activity were transfected with control siRNA or A3A–A3B siRNA, and treated with ATRi (6 µM) for 8 h. Levels of abasic sites were analyzed using biotinylated ARP. Error bar: S.D. (n = 3). B–C. The three cell lines with high overall A3A–A3B activity (B) and the three lines with low A3A–A3B activity (C) were transfected with control siRNA or A3A–A3B siRNAs, and treated with ATRi (6 µM) for 8 h. Levels of γH2AX were analyzed by Western blot. *, a non-specific band. D. The three cell lines with high overall A3A–A3B activity were transfected with control siRNA or A3A–A3B siRNA, and treated with ATRi (6 µM) for 8 h. TUNEL signals of 2,000 cells were quantified for each condition.
Discussion
APOBEC3A and APOBEC3B impose a unique type of replication stress
While DNA replication stress is known to be a major source of genomic instability, how replication stress arises in cancer cells and whether there are different types of replication stress is still incompletely understood (3). Previous studies have shown that alterations of origin firing and/or replication fork progression associate with replication stress in cancer cells (44–47). In addition, defects in the protection and/or restart of stalled replication forks, such as those caused by loss of BRCA1, BRCA2, and Fanconi Anemia proteins, could also give rise to replication stress (7–11). Recent studies by others and us have implicated A3A and/or A3B activities in the mutagenesis in cancer cells during DNA replication (12, 14, 15). However, whether this process of mutagenesis imposes replication stress is not known. In this study, we show that A3A indeed induces replication stress in a catalytic activity-dependent manner. Importantly, the A3A-induced replication stress renders cells specifically sensitive to ATRi, but not to a variety of replication inhibitors and DNA-damaging agents, suggesting that A3A imposes a unique type of replication stress distinct from that arising from other sources.
How A3A and A3B contribute to the replication stress in cancer cells still requires further investigation. While A3B is a nuclear protein, A3A is primarily induced in cytoplasm in response to foreign DNA or interferon (36, 48). A recent study suggested that APOBEC3H haplotype I, rather than A3A, was responsible for the APOBEC mutations in the absence of A3B (49). Nonetheless, the mutation signature of A3A is prevalent in several cancer types (24). The A3A-B fusion gene encoding A3A is associated with increased cancer risk and the APOBEC mutation signature in tumors (25–29). Furthermore, A3A is up regulated in a subset of leukemia (M. Weitzman, personal communications). All these findings suggest that A3A indeed affects the genome in cancer cells. Our studies suggest that the overall activity of endogenous A3A and A3B in cancer cells renders them susceptible to ATRi. A3B was recently shown to be transcriptionally induced by certain oncogenic events and DNA-damaging drugs that interfere with replication (50). Our results show that in cancer cell lines with high overall A3A–A3B activity, this activity is responsible for the baseline p-Chk1, suggesting that A3A–A3B activity is the cause of replication stress. It is possible that A3A and/or A3B are both inducers and effectors of replication stress in cancer cells, generating a feed-forward loop to promote genomic instability and drive tumorigenesis.
Why and how is the A3A–A3B-induced replication stress unique? We and others recently showed that APOBEC-induced mutations are associated with the lagging strand of replication forks, suggesting that these mutators preferentially target ssDNA on the lagging strand (12, 14, 15). The replication stress induced by A3A is dependent on UNG2, which generates abasic sites following the cytosine deamination by A3A. The abasic sites on the lagging strand interfere with DNA polymerases α and δ (43), leading to increased ssDNA exposure and thus providing more substrate to A3A. Unlike other types of replication impediments, preferential stalling of polymerases on the lagging strand may result in accumulation of ssDNA gaps without halting fork progression (51). The self-perpetuating nature of APOBEC-induced replication stress, and/or its bias toward the lagging strand, may distinguish it from other types of replication stress.
ATR counteracts a ssDNA and APOBEC-driven feed-forward loop
Our findings also suggest that ATR plays a crucial role in countering the A3A–A3B-induced replication stress. While ATR is generally important for the replication stress response, its specific role in suppressing APOBEC-induced replication stress is unique in several ways. In APOBEC-expressing cells, ATR counters intrinsic mutators but not defects in DNA replication or repair machineries. Furthermore, ATR not only suppresses the induction of abasic sites by APOBECs, but also responds to the replication stress induced by abasic sites. Since ssDNA is a substrate of APOBECs, ATR apparently acts as a brake to the feed-forward loop driven by APOBEC activity. When ATR is inhibited in APOBEC-expressing cells, the elevation of origin firing results in an increased number of replication forks and increased amounts of ssDNA at the forks. Such a surge of ssDNA in the genome would allow APOBECs and UNG2 to generate more abasic sites at replication forks, leading to additional replication stress, more ssDNA, and a further buildup of abasic sites. This ssDNA and APOBEC-dependent feed-forward loop would ultimately drive cells into replication catastrophe (Fig. 6H).
It is worth noting that the effects of ATRi on APOBEC-expressing cells are distinct from those of replication inhibitors. Although HU and APH also induce ssDNA as ATRi does, they do not block the ATR-mediated replication stress response. In the presence of functional ATR, HU or APH treatment of APOBEC-expressing cells would trigger the ATR-mediated feedback loop to restrict ssDNA accumulation, which prevents cells from going into replication catastrophe. The unique abilities of ATRi to induce ssDNA and block the ATR-mediated replication stress response may underlie its distinctive effects on APOBEC-expressing cells. While replication inhibitors do not preferentially kill APOBEC-expressing cells, they may promote APOBEC-mediated mutagenesis in cancer cells by inducing ssDNA. It would be interesting to test whether an increase of APOBEC-mediated mutagenesis renders cancer cells more responsive to immunotherapy.
Targeting cancers harboring high APOBEC3A and APOBEC3B activities with ATRi
Although expression of high levels of A3A leads to DSBs and cell cycle arrest, cancer cells expressing A3A and/or A3B are able to proliferate despite the accumulation of APOBEC mutations in their genomes. We show that the endogenous A3A and/or A3B in cancer cells, even at levels insufficient to arrest the cell cycle, induce modest levels of replication stress and confer ATRi susceptibility. While A3A expression alone does not induce significant levels of ssDNA, it primes cells for the ssDNA-forming feed-forward loop that can be activated by ATR inhibition. The endogenous A3A and/or A3B in cancer cells likely confer ATRi susceptibility in a similar manner. While our results show that ATRi is more selective than Chk1i toward U2OS cells expressing A3A, we do not exclude the possibility that Chk1i can selectively kill APOBEC-expressing cancer cells in other contexts. In cancer cells with low baseline levels of replication stress, A3A and/or A3B activity may confer susceptibility to both ATRi and Chk1i.
It is important to note that overall A3A–A3B activity in cancer cells is only one of the several factors affecting ATRi sensitivity. For example, it has been shown that loss of ATM, activation or amplification of RAS, MYC or Cyclin E, reliance on the alternative telomere-lengthening (ALT) pathway, and specific defects in DNA repair can confer ATRi sensitivity (32, 34, 52–55). Whether the A3A–A3B activity in cancer cells, either alone or in combinations with other oncogenic events, is sufficient to create a vulnerability that can be exploited therapeutically remains a critical question to be addressed in clinical settings. It is conceivable that assays for the A3A–A3B activity in cancer cells may broaden the clinical use of ATR inhibitors and improve their efficacies in cancer therapy.
Supplementary Material
Acknowledgments
We thank members of the Zou and Dyson labs for helpful discussions.
Financial Support: R. Buisson is supported by a National Institutes of Health (NIH) Pathway to Independence Award (1K99CA212154), and was supported by a Susan G. Komen Fellowship (PDF16380839) and a Marsha Rivkin Scholar Award. L. Zou is the James & Patricia Poitras Endowed Chair in Cancer Research and the Jim & Ann Orr Massachusetts General Hospital (MGH) Research Scholar. This work is supported by grants from the Department of Defense (W81XWH-14-1-0082 to L. Zou), NIH (GM076388 and CA197779 to L. Zou), Breast Cancer Alliance (to L. Zou), and the Wellcome Trust (102696 to C.H. Benes).
Abbreviations list
- APOBEC
apolipoprotein B mRNA editing enzyme catalytic polypeptide-like
- ATR
ataxia telangiectasia and Rad3 related.
Footnotes
The conflict of interest statement: The authors do not have conflicts of interest to disclose.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macheret M, Halazonetis TD. DNA replication stress as a hallmark of cancer. Annu Rev Pathol. 2015;10:425–448. doi: 10.1146/annurev-pathol-012414-040424. [DOI] [PubMed] [Google Scholar]
- 4.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 5.Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 6.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 7.Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pathania S, Bade S, Le Guillou M, Burke K, Reed R, Bowman-Colin C, et al. BRCA1 haploinsufficiency for replication stress suppression in primary cells. Nat Commun. 2014;5:5496. doi: 10.1038/ncomms6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tibbetts RS, Cortez D, Brumbaugh KM, Scully R, Livingston D, Elledge SJ, et al. Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress. Genes Dev. 2000;14:2989–3002. doi: 10.1101/gad.851000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lomonosov M, Anand S, Sangrithi M, Davies R, Venkitaraman AR. Stabilization of stalled DNA replication forks by the BRCA2 breast cancer susceptibility protein. Genes Dev. 2003;17:3017–3022. doi: 10.1101/gad.279003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haradhvala NJ, Polak P, Stojanov P, Covington KR, Shinbrot E, Hess JM, et al. Mutational Strand Asymmetries in Cancer Genomes Reveal Mechanisms of DNA Damage and Repair. Cell. 2016;164:538–549. doi: 10.1016/j.cell.2015.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shinbrot E, Henninger EE, Weinhold N, Covington KR, Goksenin AY, Schultz N, et al. Exonuclease mutations in DNA polymerase epsilon reveal replication strand specific mutation patterns and human origins of replication. Genome Res. 2014;24:1740–1750. doi: 10.1101/gr.174789.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoopes JI, Cortez LM, Mertz TM, Malc EP, Mieczkowski PA, Roberts SA. APOBEC3A and APOBEC3B Preferentially Deaminate the Lagging Strand Template during DNA Replication. Cell Rep. 2016;14:1273–1282. doi: 10.1016/j.celrep.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seplyarskiy VB, Soldatov RA, Popadin KY, Antonarakis SE, Bazykin GA, Nikolaev SI. APOBEC-induced mutations in human cancers are strongly enriched on the lagging DNA strand during replication. Genome Res. 2016;26:174–182. doi: 10.1101/gr.197046.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor BJ, Nik-Zainal S, Wu YL, Stebbings LA, Raine K, Campbell PJ, et al. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. Elife. 2013;2:e00534. doi: 10.7554/eLife.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knisbacher BA, Gerber D, Levanon EY. DNA Editing by APOBECs: A Genomic Preserver and Transformer. Trends Genet. 2016;32:16–28. doi: 10.1016/j.tig.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 20.Sasaki H, Suzuki A, Tatematsu T, Shitara M, Hikosaka Y, Okuda K, et al. gene overexpression in non-small-cell lung cancer. Biomed Rep. 2014;2:392–395. doi: 10.3892/br.2014.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leonard B, Hart SN, Burns MB, Carpenter MA, Temiz NA, Rathore A, et al. APOBEC3B upregulation and genomic mutation patterns in serous ovarian carcinoma. Cancer Res. 2013;73:7222–7231. doi: 10.1158/0008-5472.CAN-13-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet. 2013;45:977–983. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swanton C, McGranahan N, Starrett GJ, Harris RS. APOBEC Enzymes: Mutagenic Fuel for Cancer Evolution and Heterogeneity. Cancer Discov. 2015;5:704–712. doi: 10.1158/2159-8290.CD-15-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan K, Roberts SA, Klimczak LJ, Sterling JF, Saini N, Malc EP, et al. An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat Genet. 2015;47:1067–1072. doi: 10.1038/ng.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kidd JM, Newman TL, Tuzun E, Kaul R, Eichler EE. Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet. 2007;3:e63. doi: 10.1371/journal.pgen.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caval V, Suspene R, Shapira M, Vartanian JP, Wain-Hobson S. A prevalent cancer susceptibility APOBEC3A hybrid allele bearing APOBEC3B 3'UTR enhances chromosomal DNA damage. Nat Commun. 2014;5:5129. doi: 10.1038/ncomms6129. [DOI] [PubMed] [Google Scholar]
- 27.Qi G, Xiong H, Zhou C. APOBEC3 deletion polymorphism is associated with epithelial ovarian cancer risk among Chinese women. Tumour Biol. 2014;35:5723–5726. doi: 10.1007/s13277-014-1758-7. [DOI] [PubMed] [Google Scholar]
- 28.Xuan D, Li G, Cai Q, Deming-Halverson S, Shrubsole MJ, Shu XO, et al. APOBEC3 deletion polymorphism is associated with breast cancer risk among women of European ancestry. Carcinogenesis. 2013;34:2240–2243. doi: 10.1093/carcin/bgt185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nik-Zainal S, Wedge DC, Alexandrov LB, Petljak M, Butler AP, Bolli N, et al. Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nat Genet. 2014;46:487–491. doi: 10.1038/ng.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landry S, Narvaiza I, Linfesty DC, Weitzman MD. APOBEC3A can activate the DNA damage response and cause cell-cycle arrest. EMBO Rep. 2011;12:444–450. doi: 10.1038/embor.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Green AM, Landry S, Budagyan K, Avgousti D, Shalhout S, Bhagwat AS, et al. APOBEC3A damages the cellular genome during DNA replication. Cell Cycle. 2016:0. doi: 10.1080/15384101.2016.1152426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reaper PM, Griffiths MR, Long JM, Charrier JD, Maccormick S, Charlton PA, et al. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat Chem Biol. 2011;7:428–430. doi: 10.1038/nchembio.573. [DOI] [PubMed] [Google Scholar]
- 33.Foote KM, Blades K, Cronin A, Fillery S, Guichard SS, Hassall L, et al. Discovery of 4-{4-[(3R)-3-Methylmorpholin-4-yl]-6-[1-(methylsulfonyl)cyclopropyl]pyrimidin-2-y l}-1H-indole (AZ20): a potent and selective inhibitor of ATR protein kinase with monotherapy in vivo antitumor activity. J Med Chem. 2013;56:2125–2138. doi: 10.1021/jm301859s. [DOI] [PubMed] [Google Scholar]
- 34.Buisson R, Boisvert JL, Benes CH, Zou L. Distinct but Concerted Roles of ATR, DNA-PK, and Chk1 in Countering Replication Stress during S Phase. Mol Cell. 2015;59:1011–1024. doi: 10.1016/j.molcel.2015.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Connor MJ. Targeting the DNA Damage Response in Cancer. Mol Cell. 2015;60:547–560. doi: 10.1016/j.molcel.2015.10.040. [DOI] [PubMed] [Google Scholar]
- 36.Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat Struct Mol Biol. 2010;17:222–229. doi: 10.1038/nsmb.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krokan HE, Bjoras M. Base excision repair. Cold Spring Harb Perspect Biol. 2013;5:a012583. doi: 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Otterlei M, Kavli B, Standal R, Skjelbred C, Bharati S, Krokan HE. Repair of chromosomal abasic sites in vivo involves at least three different repair pathways. EMBO J. 2000;19:5542–5551. doi: 10.1093/emboj/19.20.5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao B, Xie Z, Shen H, Wang Z. Role of DNA polymerase eta in the bypass of abasic sites in yeast cells. Nucleic Acids Res. 2004;32:3984–3994. doi: 10.1093/nar/gkh710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 41.Toledo LI, Altmeyer M, Rask MB, Lukas C, Larsen DH, Povlsen LK, et al. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013;155:1088–1103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- 42.Atamna H, Cheung I, Ames BN. A method for detecting abasic sites in living cells: age-dependent changes in base excision repair. Proc Natl Acad Sci U S A. 2000;97:686–691. doi: 10.1073/pnas.97.2.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan F, Zhang Y, Rajpal DK, Wu X, Guo D, Wang M, et al. Specificity of DNA lesion bypass by the yeast DNA polymerase eta. J Biol Chem. 2000;275:8233–8239. doi: 10.1074/jbc.275.11.8233. [DOI] [PubMed] [Google Scholar]
- 44.Neelsen KJ, Zanini IM, Herrador R, Lopes M. Oncogenes induce genotoxic stress by mitotic processing of unusual replication intermediates. J Cell Biol. 2013;200:699–708. doi: 10.1083/jcb.201212058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, et al. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448:445–451. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 46.Srinivasan SV, Dominguez-Sola D, Wang LC, Hyrien O, Gautier J. Cdc45 is a critical effector of myc-dependent DNA replication stress. Cell Rep. 2013;3:1629–1639. doi: 10.1016/j.celrep.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsantoulis PK, Kotsinas A, Sfikakis PP, Evangelou K, Sideridou M, Levy B, et al. Oncogene-induced replication stress preferentially targets common fragile sites in preneoplastic lesions. A genome-wide study. Oncogene. 2008;27:3256–3264. doi: 10.1038/sj.onc.1210989. [DOI] [PubMed] [Google Scholar]
- 48.Land AM, Law EK, Carpenter MA, Lackey L, Brown WL, Harris RS. Endogenous APOBEC3A DNA cytosine deaminase is cytoplasmic and nongenotoxic. J Biol Chem. 2013;288:17253–17260. doi: 10.1074/jbc.M113.458661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Starrett GJ, Luengas EM, McCann JL, Ebrahimi D, Temiz NA, Love RP, et al. The DNA cytosine deaminase APOBEC3H haplotype I likely contributes to breast and lung cancer mutagenesis. Nat Commun. 2016;7:12918. doi: 10.1038/ncomms12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanu N, Cerone MA, Goh G, Zalmas LP, Bartkova J, Dietzen M, et al. DNA replication stress mediates APOBEC3 family mutagenesis in breast cancer. Genome Biol. 2016;17:185. doi: 10.1186/s13059-016-1042-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fu YV, Yardimci H, Long DT, Ho TV, Guainazzi A, Bermudez VP, et al. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell. 2011;146:931–941. doi: 10.1016/j.cell.2011.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toledo LI, Murga M, Zur R, Soria R, Rodriguez A, Martinez S, et al. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat Struct Mol Biol. 2011;18:721–727. doi: 10.1038/nsmb.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mohni KN, Thompson PS, Luzwick JW, Glick GG, Pendleton CS, Lehmann BD, et al. A Synthetic Lethal Screen Identifies DNA Repair Pathways that Sensitize Cancer Cells to Combined ATR Inhibition and Cisplatin Treatments. PLoS One. 2015;10:e0125482. doi: 10.1371/journal.pone.0125482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Flynn RL, Cox KE, Jeitany M, Wakimoto H, Bryll AR, Ganem NJ, et al. Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science. 2015;347:273–277. doi: 10.1126/science.1257216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sultana R, Abdel-Fatah T, Perry C, Moseley P, Albarakti N, Mohan V, et al. Ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase inhibition is synthetically lethal in XRCC1 deficient ovarian cancer cells. PLoS One. 2013;8:e57098. doi: 10.1371/journal.pone.0057098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.